Abstract
Throughout the biological sciences, the past fifteen years have seen a push towards the analysis and engineering of biological systems at the organism level. Given the complexity of even the simplest organisms, though, to elicit a phenotype of interest often requires genotypic manipulation of several loci. By traditional means, sequential editing of genomic targets requires a significant investment of time and labor, as the desired editing event typically occurs at a very low frequency against an overwhelming unedited background. In recent years, the development of a suite of new techniques has greatly increased editing efficiency, opening up the possibility for multiple editing events to occur in parallel. Termed as multiplexed genome engineering, this approach to genome editing has greatly expanded the scope of possible genome manipulations in diverse hosts, ranging from bacteria to human cells. The enabling technologies for multiplexed genome engineering include oligonucleotide-based and nuclease-based methodologies, and their application has led to the great breadth of successful examples described in this review. While many technical challenges remain, there also exists a multiplicity of opportunities in this rapidly expanding field.
Keywords: genome engineering, multiplexing, MAGE, ZFN, TALEN, CRISPR/Cas9
INTRODUCTION
Living organisms from bacteria to humans represent the most complex systems on earth, which despite centuries of research effort, still elude a complete understanding by scientists. Synthetic biologists harness the principles of the central dogma to engineer biological systems for improved and novel functions.1-4 Specifically, a desired phenotype can be generated by deleting or incorporating the corresponding genetic elements. Techniques enabling the precise alteration of genomic DNA sequences in vivo, termed as “genome engineering”, have been substantially developed in recent years and pushed the frontiers of cell biology, gene therapy, and industrial and agricultural biotechnology. Nonetheless, genome engineering in the early days was challenging, largely due to the inefficiency of introducing single-site mutations and inability to rapidly alter targeting sequences at will, which ultimately led to a high cost. As a result, conventional techniques usually target one locus on a genome in a single round, which severely limits the turnover of the design-build-test cycle.5 While this does not typically pose a problem for generating single modifications, such as correcting a point mutation associated with a monogenic disease,6 it does make combinatorial testing of multiple genome modifications unfeasible, which is usually required in strain and cell line development and complex disease modeling.7, 8
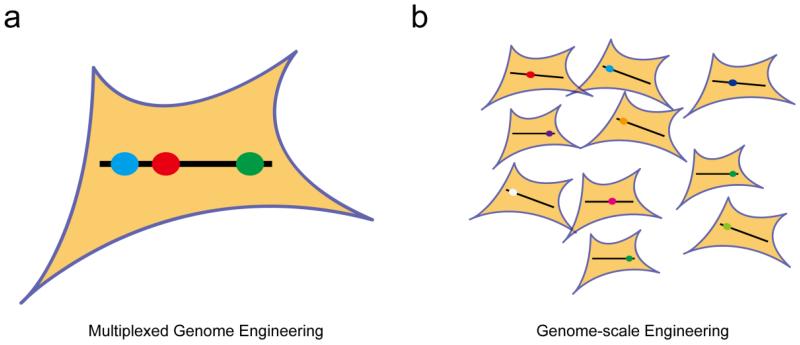
Within this context, there exists an urgent need for tools enabling multiplexed and high-throughput genome engineering. To avoid possible confusion, we define “multiplex” as generating altered DNA sequences in two or more loci within a single genome in a single round of mutagenesis (Fig. 1a). As a result, population-based genome engineering with one modification per cell (Fig. 1b) will not be considered in this review. Besides, epigenome and transcriptome engineering (including RNA editing and regulation of gene expression levels) that do not introduce a change in DNA sequences will not be discussed. Readers who are interested in these topics are referred to other recent reviews.9-12
Figure 1.
Definition of multiplexed genome engineering in this review. (a) In multiplexed genome engineering, multiple mutations (colored ovals) are simultaneously introduced into a single genome (black line). (b) In genome-scale engineering, each genome receives one specific but different mutation.
In this review, we will focus our discussion on recent technological advances in multiplexed genome engineering in various organisms, mostly from the past four years. We will first introduce oligonucleotide-mediated multiplexed genome engineering via homologous recombination in microbes. We will then move on to nuclease-mediated multiplexed genome engineering, with or without homologous recombination, in microbes as well as higher order organisms. Specifically, zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered, regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) will be discussed. Finally, we will conclude by offering our perspectives on current challenges and future developments in the field of multiplexed genome engineering.
OLIGONUCLEOTIDE-MEDIATED MULTIPLEXED GENOME ENGINEERING
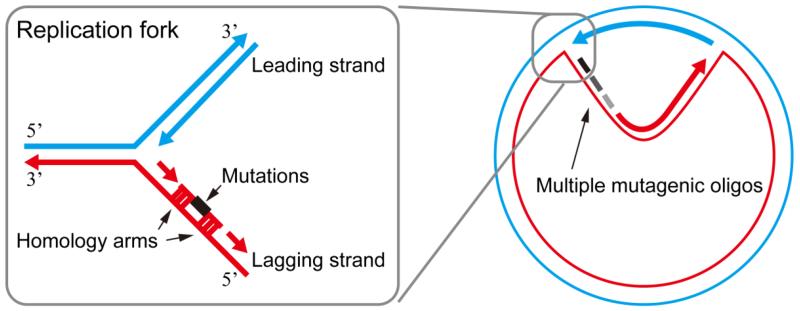
In bacteria, homologous recombination has been utilized as a robust mechanism for genome engineering.13 It was first demonstrated at the end of 20th century that phage-derived proteins can be used to replace DNA fragments in Escherichia coli, through the homologous recombination mechanism.14-16 Mechanistically, with the help from heterologous-expressed RecE/RecT or λ-red system, single-stranded DNA oligonucleotides harboring deletions, insertions or mismatches anneal to the lagging strand during DNA replication, resulting in the incorporation of customized sequence modifications in the newly synthesized genome (Fig. 2). This technique is termed as recombination-mediated genome engineering, or recombineering. Experimentally, multiple oligonucleotides can be transformed simultaneously into a single cell to allow editing of multiple loci at the same time.
Figure 2.
Recombineering based multiplexed genome engineering. Mutagenic oligonucleotides recombine with the host genome to cause mutations during DNA replication. Multiple mutagenic oligonucleotides (black and grey lines) can be simultaneously introduced into the genome.
The burst of multiplexed recombineering came in 2009 when the Church laboratory developed a high throughput multiplexed engineering methodology in Escherichia coli, termed as Multiplex Automated Genome Engineering (MAGE).17 The relatively high efficiency of recombineering in MAGE makes it possible to omit selections for modified mutants, resulting in a highly diversified mutant pool where single cells can have multiple edits across the genome. The first example using MAGE successfully increased lycopene production by more than five-fold in E. coli, through optimizing the 1-deoxy-d-xylulose-5-phosphate (DXP) biosynthesis pathway.17 MAGE was also used to genetically recode the E. coli genome, replacing all the TAG stop codons with TAA codons.18, 19 The released TAG codon was further repurposed as a dedicated sense codon for synthetic amino acids and was introduced into 22 essential genes in the E. coli genome by MAGE, generating recoded organisms that are safe for use in the natural environment.20 Of note, to enable multiple edits (typically more than 10) within a single cell, conjugative assembly genome engineering (CAGE) was used together with MAGE to merge different sets of edits through horizontal transfer. Besides random mutations and codon replacement, MAGE has also been used to introduce more focused functionalities such as promoter sequences. In one study, a MAGE variant called ‘coselection’ MAGE (CoS-MAGE) was used to combinatorially insert multiple T7 promoters simultaneously into 12 genomic operons, enabling the study of gain-of-function modifications and epistatic interactions within gene networks.21 Due to its popularity, other studies focusing on improving the efficiency22-24, in silico design25 and reducing the cost26 of MAGE have also been described.
Although groundbreaking, applications of MAGE are largely restricted to the model microorganism E. coli. Its success is not easily transferred to other microbial systems because of the inefficiency of homologous recombination using short oligonucleotides in other species. For instance, Yeast Oligo-Mediated Genome Engineering (YOGE) method used a similar strategy as MAGE, but in Saccharomyces cerevisiae.27 After strain engineering, including knocking out mismatch repair proteins, overexpressing DNA recombinases, and optimization of transformation and oligonucleotide design, frequencies of recombination in yeast was significantly improved. YOGE was then applied to simultaneously modify two loci in the yeast genome in a cycled manner. Although successful, the efficiency of double modifications falls below 0.01% after three cycles, despite the use of the best performing oligonucleotides. Phenotypic screening or site-specific nuclease-mediated negative selection may be coupled with YOGE to further increase the efficiency of multiplexed yeast genome engineering.28
Another study explored the possibility of multiplexed genome engineering in naturally competent microorganisms. Natural competence refers to the ability of diverse microbial species to take up DNA from the environment and integrate it into the genome by homologous recombination.29 Taking advantage of natural competence, a multiplexed genome engineering method called Multiplex Genome Editing by Natural Transformation (MuGENT) was developed.30 The effectiveness of this method was demonstrated by combinatorially editing the Vibrio cholerae genome via gene deletions, promoter replacements and tuning translation initiation of five genes to optimize the natural transformation trait. MuGENT was further used to simultaneously edit four redundant genes in another species, Streptococcus pneumoniae, lacking mismatch repair, demonstrating its broad utility as a multiplexed genome engineering tool.
TOOLS FOR NUCLEASE-MEDIATED MULTIPLEXED GENOME ENGINEERING
It is a well-known phenomenon that the introduction of a double-strand break (DSB) to the chromosomal DNA of a living cell significantly stimulates DNA repair functions,31 primarily through either non-homologous end-joining (NHEJ) or homologous recombination (HR). This phenomenon is unsurprising from an evolutionary perspective, as DSBs can be deleterious or lethal to the organism if unaddressed. From an engineering perspective, the stimulated repair response can be exploited as a means to prime specific loci of interest for genome editing events. In other words, by targeting a DSB to a precise chromosomal location, that location can become a “hotspot” for DNA sequence revision. Modification of the target site can then be achieved either via error-prone NHEJ or through HR by supplementing an editing template in trans that dictates the desired remodeled sequence.
While agents such as radiation or chemical mutagens can be used to introduce chromosomal DSBs in vivo, practical application of DSB-mediated genome editing necessitates precise control over the specific breakage point. This level of control can be afforded by endonucleases, enzymes that recognize specific DNA sequences and cleave within or near their recognition site. However, the most commonly used and well-studied restriction endonucleases (of the Type II class) recognize sequences that are too short to provide uniqueness in the total genomic context. As a result, additional cleavage events would occur at undesired loci bearing the same recognition sequence as the desired target, rendering these endonucleases impractical for all but the smallest genomes. Homing endonucleases (HEs), in contrast, recognize much longer DNA sequences, typically 14 – 40 nucleotides in length.32 This length typically satisfies the requirement for uniqueness, even in large metazoan and plant genomes. The key limitation, though, is that HEs have proven to be recalcitrant to extensive reprogramming of their DNA recognition sequences. HEs can be envisioned as bespoke enzymes, made to measure over evolutionary time for a particular recognition sequence. Consequently, DNA recognition and cleavage activity are difficult to decouple, as both are intricately interwoven in the overall protein fold. HE-mediated genome engineering is thus limited to cases where a wild type HE scaffold can be identified with sequence specificity that already closely matches the desired target32, 33
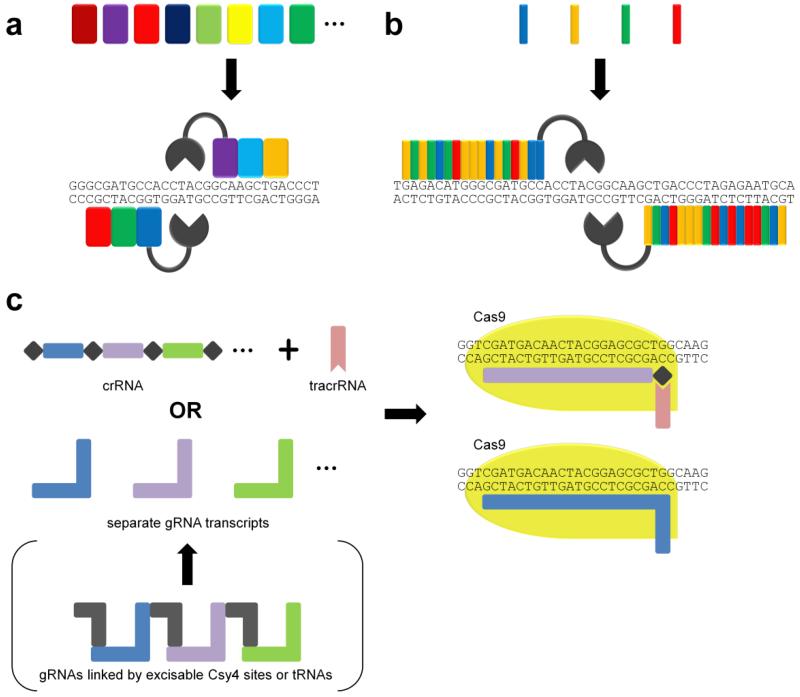
For practical application of endonucleases in multiplexed genome engineering, what is needed is a set of “off-the-shelf” DNA recognition modules that can readily be assembled to generate custom endonucleases with designer specificity (Fig. 3). At the protein level, this modularity was first discovered in zinc finger (ZF) structural motifs, each of which can recognize a specific 3-nucleotide triplet (Fig. 3a).34 Later on, the specificity code of the transcription activator-like effector (TALE) repeat domains, each of which recognizes a specific nucleotide on a 1:1 basis (Fig. 3b), was deciphered, further expanding the DNA recognition toolbox.35, 36 By fusing a set of DNA recognition domains with a nonspecific nuclease domain (such as that of the FokI endonuclease), these DNA binding proteins can be repurposed as site-specific endonucleases – namely ZFNs and TALENs. Most recently, the CRISPR-associated protein Cas9 has been demonstrated as perhaps the most versatile reprogrammable endonuclease. Its DNA sequence specificity is conferred by a short RNA sequence with complementarity to its recognition site. Multiple short RNA sequences can be transcribed either separately or as a nuclease-cleavable polycistronic array (Fig. 3c).37-39 Thus, targeting Cas9 to multiple loci in vivo requires only the transcription of the corresponding guide RNA sequences, rather than construction of entirely new multi-domain proteins. As a result, though ZFNs and TALENs continue to see applications for multiplex genome engineering, it is Cas9 that has largely become the tool of choice for multiplexed genome editing.
Figure 3.
Nuclease-mediated multiplexed genome engineering can be achieved using (a) ZFNs, (b) TALENs, and (c) CRISPR/Cas9 (note that elements are not drawn to scale).
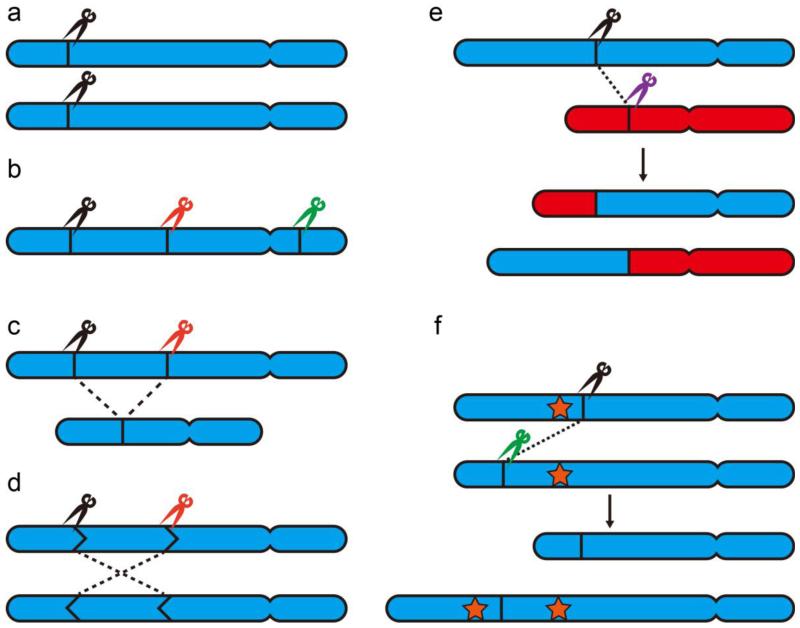
By targeting multiple genomic loci simultaneously, three general classes of genome modifications are possible that would be difficult, tedious, or impossible to generate via single-locus targeting (Fig. 4). The first can be termed multi-allelic modification (Fig. 4a), in which a single engineered nuclease is employed to target multiple alleles in a multiploid organism. Though conceptually synonymous with single-locus editing (one nuclease/one target), the demands placed on editing efficiency make this a non-trivial extension of the single-locus platform. Further, the simultaneous generation of homozygous mutations provides clear advantages over alternative methods that would require screening of a population for the desired heterozygous mutation, followed by breeding and screening of progenies for the desired homozygous genotype. The second class involves deployment of multiple nucleases to enact simultaneous independent editing events (Fig. 4b). Again, this approach greatly facilitates the genome engineering process compared to single-locus methods that would require sequential selection or screening for each mutation of interest, often paired with intermediate counter-selection to remove selectable markers. Finally, the third class involves the concerted action of multiple nucleases to achieve outcomes inaccessible to single-locus targeting (Fig. 4c-4f). These outcomes include the manipulation of large fragments of chromosomal DNA, including excision, inversion, and translocation.
Figure 4.
Different types of nuclease-mediated multiplexing. (a) Multi-allelic modification. One nuclease or nuclease pair cleaves multiple copies of the same site. (b) Multi-gene disruption. Multiple nucleases/nuclease pairs target different loci across the genome. (c) Chromosomal excision. Two distant cleavages and splicing of the two break points cause the loss of a large chromosome fragment. (d) Chromosomal inversion. Two distant cleavages cause the inversion of a large chromosome fragment. (e) Chromosomal translocation. Two cleavages on two different chromosomes cause the translocation of chromosome arms. (f) Chromosomal duplication (a special case of chromosomal translocation). Two cleavages on two sites of two identical chromosomes result in the duplication of chromosome fragment in between the two cleavage sites. The brown star represents a duplicated gene.
APPLICATIONS OF NUCLEASE-MEDIATED MULTIPLEXED GENOME ENGINEERING
Microbes
In bacterial systems, nuclease-mediated multiplexed genome engineering was only recently brought into play with the rapid development of the Cas9-based platform. This is perhaps due to the relative ease with which the most widely studied bacterial hosts can be sequentially manipulated by recombineering, diminishing the time- and effort-saving benefits of multiplexing. Nevertheless, Cas9-mediated bacterial genome editing has been demonstrated, starting with a landmark study in Streptococcus pneumoniae.40 Here, the authors leveraged the native CRISPR/Cas system of the host by introducing an engineered CRISPR array containing two spacers and a selection marker, along with two editing templates dictating the desired mutations. Following co-transformation of all three elements, genotyping confirmed that 6/8 transformants were correctly editing at both loci, while the remaining two were edited at only one locus.
More recently, heterologous Cas9-mediated multiplexed genome editing has been demonstrated in Streptomyces spp.41 and E. coli.42 In Streptomyces lividans, two guide RNA (gRNA) cassettes and two editing templates were assembled into a shuttle plasmid containing a codon-optimized cas9 gene.41 Following conjugative transfer of the plasmid from an E. coli donor, four exconjugants were genotyped, and all were found to contain both desired short deletions. This approach was also demonstrated for the deletion of full gene clusters in S. lividans and Streptomyces albus by supplying gRNAs targeting both ends of the cluster along with a single editing template to bridge the gap between the two DSBs. In E. coli, a two-plasmid system was employed to simultaneously introduce double and triple deletions with efficiencies of 97% and 47%, respectively.37 Additionally, the authors performed simultaneous deletion and insertion at separate loci with efficiency of 78%. Notably, their plasmid system included the λ-red exo, beta, and gam genes to provide the recombination rates necessary for efficient DSB repair, and longer homology arms (≥250 bp) were found to significantly enhance efficiency relative to short arms (40 bp).
In the model eukaryotic microbe S. cerevisiae, multiple groups have recently explored the multiplexing potential of Cas9. As an intrinsic benefit of this host, naturally high HR proficiency enables precision genome editing with relatively short editing templates (≤100 bp). For example, in the Homology-Integrated CRISPR/Cas (HI-CRISPR) system, editing templates are directly embedded with the spacer sequences in a CRISPR array, enabling an all-in-one approach in which Cas9, multiple spacers, and multiple editing templates are encoded on a single plasmid.43 As a demonstration of this platform, two sets of three genes each were simultaneously disrupted with 8 bp deletions, with efficiencies ranging from 27% to 100%. In an alternative approach, gRNAs were fused to a self-cleaving ribozyme sequence to protect their 5’ ends from exonuclease activity.44, 45 Using this so-called Multiplex CRISPR (CRISPRm) approach, highly efficient barcoding was achieved for up to three alleles in haploid and diploid laboratory strains, as well as for a single allele in a polyploid industrial strain. Most recently, up to five genes were simultaneously disrupted via co-transformation of a plasmid containing five gRNA cassettes with five separate 90 bp double-stranded oligonucleotide editing templates.46 Disruption efficiency of 100% was confirmed for the quintuple knockout by genotyping a small subset of transformants. Besides merely gene inactivation, integration of metabolic pathway genes into up to three different loci of the yeast genome has been realized by different groups,47-49 which broadened the application scope of CRISPR/Cas and facilitated metabolic engineering in this important host.
Although TALENs have primarily only been expressed in S. cerevisiae as a prototyping platform for eventual application in other organisms,50 a recent study demonstrated their application for multiplexed yeast genome editing as well.51 Here, four plasmids (each containing one TALEN of a heterodimer pair) were simultaneously introduced to the cell in order to disrupt two acyl-CoA synthetases, with the goal of improving intracellular free fatty acid accumulation. Of note, while separation of the TALENs on multiple plasmids aids plasmid stability both in construction and in vivo, it limits the number of TALENs that can be simultaneously introduced based on the number of available orthogonal selection markers.
Plants
As TALEs were originally identified in plant pathogens, it is logical that TALEN-mediated multiplexed genome editing has been attempted in plants. In Oryza sativa, TALENs were used to generate NHEJ-induced disruptions in the binding elements for pathogenic TALEs located in the promoter region of a blight susceptibility gene.52 Following introduction of TALEN pairs via Agrobacterium-mediated transformation, the progeny of primary transgenic plants were screened. In total, 18/53 genotyped plants contained biallelic mutations, of which 10 were homozygous. Expanding to greater numbers of targets has also led to success, but with predictably lower efficiency. For example, three pairs of TALENs were introduced via bombardment to O. sativa to disrupt three separate genes with the goal of improving rice fragrance and grain yield.53 Screening of 207 plants revealed that only four were disrupted in all three positions, and none of them were homozygous at all three loci. In an allohexaploid wheat, three nearly identical copies of a mildew resistance locus were targeted with a single pair of TALENs.54 Although only 27 plants were subsequently screened, one was identified to be heterozygous at all three loci. After rounds of self-pollination, triple homozygous progeny could be obtained, revealing the desired disease-resistant phenotype.
Cas9-mediated multiplexing has been evaluated in a variety of plants, including not only the model system Arabidopsis thaliana but also cereal crops such as corn and rice. In A. thaliana, when a single gRNA was employed to target two synonymous loci, a low mutation rate was observed at both sites, and presence of the desired double mutant was not confirmed.55 Similarly, when two gRNAs were deployed to target the same gene, deletion of the region between the two loci was only observed in 5/91 genotyped transformants. However, in a later study, a significantly higher efficiency was observed when two gRNAs were used to target genes CHLI1 and CHLI2, eliciting the anticipated albino phenotype of a double mutant in 24/36 plants.56 Further, targeting three nearly synonymous loci with two gRNAs elicited the phenotype of a double or triple mutant in 26/33 members of the T1 generation; notably, though, the diversity of mutations found in a given transgenic plant suggested that editing had occurred after division of the fertilized egg. In Zea mays, chromosomal deletion was attempted by deploying two adjacent gRNAs, resulting in the expected deletion in 12/20 screened plants.
In O. sativa, multiplexed Cas9-mediated editing has proven to be very efficient. In one study, biallelic mutations were detected in 3/9 plants following bombardment of calli with plasmids containing a codon-optimized cas9 and a gRNA.57 In another study, biallelic editing with a codon-optimized cas9 was observed with efficiencies ≥87%.58 Notably, the efficiency dropped significantly when the gRNA was replaced with a tracrRNA/crRNA configuration. Introduction of two distant gRNAs was employed as a means to delete chromosomal fragments ranging from 115 – 245 kb in rice protoplasts. While the desired large deletions could be confirmed by PCR, sequencing of plants generated from transformed calli revealed deletion of the large fragment from one chromosome only, with mutation of the gRNA target sites on the homologous chromosome. Elsewhere, deployment of one gRNA targeting up to four homologous genes enabled six triple mutants to be identified from 13 regenerated rice plants, including five with triple mono-allelic mutations, and one with two mono-allelic mutations and one bi-allelic mutation.59 Note that the fourth potential target for this gRNA was unmodified in all plants screened, likely due to a single nucleotide difference close to the 3’ end of the spacer that abrogated targeting. Most recently, an ingenious method to introduce many gRNAs in a single transcript was demonstrated in rice.39 In this example, up to eight gRNAs were fused together with interstitial tRNA coding sequences as so-called polycistronic tRNA-gRNA (PTG) genes. This setup enables the plant’s native tRNA processing machinery to splice out the individual gRNA coding sequences, which can then be used for multiplexed targeting. In one example, deployment of 8 gRNAs (four pairs targeting adjacent sites in four genes) in protoplasts enabled 4 – 20 % deletion efficiency between the two targeted sites in any single gene; individual clones, however, were not screened for the possible quadruple deletion. In intact plants, two-target PTGs enabled biallelic editing in up to 76% of screened plants, and an eight-target PTG created biallelic mutations in at least 5 loci in 7/14 plants, with mutations confirmed at all eight sites.
Metazoans
The importance of metazoans in diverse research areas, such as functional genomics and disease modeling, along with the time and labor intensity of their genetic manipulation, have motivated significant exploration of multiplexing technologies in these organisms. ZFNs, for example, have been introduced to CHO cells to create biallelic knockout lines. In a proof of concept study, three sites were targeted, and all three biallelic knockout events were observed at frequencies of >1 % without the use of selection.60 The TALEN technology has been explored in a variety of metazoans, although largely as proofs of concept. In the silkworm Bombyx mori, two adjacent TALEN pairs were introduced to generate a deletion of 792 bp, and 14% efficiency was observed in the germline of the G1 brood.61 Taking this approach further, deletions, duplications, and inversions of an 8.9 Mb chromosomal fragment were observed when two distant TALEN pairs were introduced, although the specific efficiency of these events was not quantified.62 Small and large deletions have also been demonstrated by similar means in zebrafish. In one example, non-coding RNA clusters ranging from 1.2 kb to 79.8 kb were deleted with efficiencies ranging from 1.9% – 4.2%.63, 64 In another study, deletions of 200 bp – 4.2 kb could be introduced with 1 – 15% efficiency.65 Note, however, that efficiency was not always directly proportional to deletion size. Deletions of 39 kb or 69 kb could be introduced with 3.2 and 4.9% efficiency, respectively, and a 5.5 Mb deletion (introduced by a combination of one ZFN and a TALEN pair) was introduced with 0.7% efficiency. Inversion events were also observed in several deletion experiments, but with lower efficiency as compared to the corresponding deletion events (<2%).
In the frogs Xenopus laevis and Xenopus tropicalis, eight single genes were targeted with TALENs, all of which exhibited ≥61.9 % efficiency.66 Although not specifically measured, the high efficiencies observed at each site suggest that multiallelic editing has occurred in a subset of the transformed embryos. Of note, this study directly compared the TALENs to ZFNs, the latter of which exhibited greater toxicity at comparable doses and lower efficiency at lower doses. A subsequent study targeted three genes (two tyrosinase paralogs and EGFP) with two TALEN pairs or two genes (histone chaperone paralogs) with two TALEN pairs, resulting in 40 – 90 % disruption efficiency at each locus.67 Note that in this study, mutations that did not result in a frameshift (e.g., 6 bp deletion) were not counted as effective disruptions. The frequency of simultaneous editing at all targeted loci was not directly reported. In livestock, biallelic mutations have been introduced via injection of TALEN mRNA to bovine and porcine embryos.68 With optimization of mRNA dosage, 2/4 bovine embryos and 6/56 porcine embryos (from 214 total, after an initial screen for EGFP fluorescence as an injection control) displayed biallelic mutation at the targeted locus. Further, five of six TALEN pairs tested in fibroblast culture showed biallelic modification in up to 17% of selected colonies. Deletion and inversion of a 6.5 kb region could be achieved with efficiencies of 10% and 4%, respectively, and modified fibroblasts could subsequently be used as nuclear donors for cloning.
Not surprisingly, the greatest attention for multiplexing in metazoan systems has been paid to Cas9. Though primarily in the proof-of-concept phase, results obtained so far have largely justified this interest. In the fruit fly Drosophila melanogaster, an optimized Cas9 toolkit was recently developed and shown to drive efficient biallelic editing in somatic and germline cells by either NHEJ or HR.69 Supplying a donor sequence dictating insertion of gfp into the wg coding region, for example, led to GFP expression in 38% of the offspring of injected embryos. Targeting different loci in somatic cells enabled markerless biallelic editing suitable for phenotypic loss-of-function screening. Alternatively, by inserting the cas9 sequence and a gRNA into the locus of interest, inheritance of the cas9-containing allele can be driven to near 100%, as recently demonstrated in a powerful gene drive system.70 Such tools have clear applications in the engineering of entire insect populations, which could be a valuable means to prevent the spread of insect-borne diseases.
Cas9-mediated multiplexing has also been attempted in silkworms, with the goals of introducing deletions or simultaneously editing multiple loci. In one example, three gRNAs were employed to target three sites in the BmKu70 gene.71 The efficiency, however, was quite low, with only 2/101 double-locus mutants and 1/101 triple-locus mutants observed. Two mutants were also observed with deletions between two of the targeted sites. In a second example, two gRNAs were shown to enable deletion or inversion of a 3 kb region, although the efficiency was not reported.72 To further test the limits of the system, ten gRNAs were deployed to target a total of six genes. While editing was observed at each locus, simultaneous targeting of each site in a single cell was not confirmed.
In zebrafish, while TALENs were largely explored for the introduction of small and large deletions, Cas9 based applications have focused more on the simultaneous introduction of small mutations in one or more targets. For example, using a codon-optimized cas9 sequence, an efficiency of 75 – 99 % in single targeting of four endogenous loci and a reporter was observed, indicating a significant proportion of biallelic mutants.73 Simultaneous targeting of all five loci also revealed high targeting efficiency at all five sites, and sequencing of a single embryo confirmed the presence of indels in each locus. Independent studies have also confirmed high single-locus editing efficiency,74, 75 demonstrated simultaneous targeting of five loci to edit three genes,74 and shown deletion and inversion of a 7.1 kb fragment.75 In X. tropicalis, high biallelic single-targeting efficiency with Cas9 was also confirmed, as was biallelic double-targeting modification.76 Overall, the targeting efficiencies in X. tropicalis were comparable to those obtained with the TALEN technology.
In mammals and mammalian cell lines, numerous instances of Cas9-based multiplexing have been reported. In mice, a side-by-side comparison of TALENs and Cas9 showed that Cas9 was superior for the introduction of biallelic mutations.77 In an early example in mouse embryonic stem cells, five genes were simultaneously targeted with five gRNAs, resulting in 10/96 clones modified by NHEJ at all eight possible alleles.78 In mouse zygotes transformed with two gRNAs, 22/28 contained biallelic mutations. HR-mediated repair was also attempted, but with significantly lower efficiencies, suggesting NHEJ to be the dominant repair mechanism. In a separate study, three genes were targeted with three gRNAs in 60 embryos.79 Of the nine pups obtained, all had mutations at all three loci, and 7/9 were mutated at all alleles. Here, a 95-base polyadenine tail was added to the cas9 transcript to improve translation efficiency. Two-cut deletions could also be introduced using adjacent gRNAs; in mice, 3/12 co-injected embryos contained biallelic deletions between the two target sites.80 An analogous experiment in rats yield only 1/15 rats separately disrupted at both targeted loci. In other studies, however, greater targeting efficiency in rats was achieved. For example, two gRNAs were designed to target the Tet1 and Tet2 genes, resulting in 15/24 rats mutated at both sites.81 A second set of two gRNAs targeting the same two genes yielded similarly high efficiency (15/20), with a significant proportion exhibiting biallelic mutations in both genes. Deploying two gRNAs each to target four genes simultaneously yielded 6/25 rats with mutations at all four loci. Finally, in CHO cells, Cas9 has been fused to GFP via a 2A linkage to enable an initial enrichment of Cas9-expressing transfected cells.82 In this experiment, 97 clones were deep sequenced after deployment of three gRNAs, resulting in 23/97 disrupted at two loci and 34/97 disrupted at all three loci.
Human cells
Compared to animal models, genome engineering in human cells provides a direct way to study human genetics and offers critical insights into the genesis of diseases. Abnormal genome rearrangements are hallmarks of complex diseases such as cancer. Thus, it is not surprising that the major application of multiplexed genome engineering in human cells is to treat or model complex diseases. Using nucleases for genomic deletion is a common proof-of-concept demonstration of multiplexed genome engineering. For example, ZFNs were used to generate deletions from several-hundred base pairs to 15 mega base pairs in human cancer cell lines, at frequencies from 10−3 to 10−1.83 ZFNs have also been engineered into nickases that only cleave one DNA strand in a DNA duplex, which is preferably repaired by the error-free homologous recombination pathway. Using two pairs of zinc finger nickases, which generated 4 chromosomal single strand breaks in homologous CCR2 and CCR5 loci, a 15 kb genomic deletion with precise junctions was created, though at low frequencies (0.01 % and 0.04 %).84 Other than genomic deletion, ZFNs were also employed for generating chromosomal translocations, which are signatures of various cancers and lead to expression of oncogenic fusion genes. In one study, DSBs were induced at two loci on two chromosomes in both human embryonic stem cells (hES cells) and hES cell-derived mesenchymal precursor cells, leading to translocations at frequencies less than 10−6.85 Another study used ZFNs to model chromosomal translocations associated with Ewing sarcoma in hES cell-derived mesenchymal precursor cells.86 Using an analogous strategy, TALENs were also used to model chromosomal translocations in anaplastic large cell lymphoma (ALCL) as well as to revert the ALCL translocations in a patient derived cell line. The overall reported efficiencies for chromosomal translocations using either ZFNs or TALENs in this particular study were between 10−4 and 10−2. In castration-resistant prostate cancer cells, two TALEN pairs were introduced to generate intragenic androgen receptor gene rearrangements, including both deletion and inversion, with efficiencies not reported.87
It is not surprising that CRISPR/Cas quickly became much more popular than ZFNs and TALENs in human cells, once again due to its ease in manipulation and avoidance of introducing multiple bulky proteins. The innate multiplexing nature of the crRNA expressing array was utilized for simultaneously editing EMX1 and PVALB loci in human cells, by expressing two targeting spacers in a single crRNA expression cassette.88 A 118 bp genomic deletion was also verified in 3 out of 182 amplicons after expressing two crRNAs targeting the EMX1 locus. In another study, a 19 bp deletion was realized by expressing two gRNAs simultaneously, with efficiencies not quantified.89 Due to its great promise in multiplexed genome engineering with more than just two edits, vector systems that allow simultaneous expression of multiple gRNAs on a single plasmid were also reported. For example, an all-in-one plasmid bearing two to seven gRNA expression cassettes can be sequentially assembled.90 Editing efficiencies for an individual locus by a 7-gRNA plasmid varied from 4.3 % to 37.8 %, indicating the possibility of multi-locus editing. This vector system was also used to generate deletions, although the efficiencies were not reported. A second study assembled four gRNA expression cassettes through Golden Gate assembly into a single lentiviral vector, with each gRNA expressed under a different promoter.91 Using this vector, two loci in IL1RN and one locus each in HBG1 and AAVS1 were simultaneously edited. The individual editing efficiency of the four loci varied from 17.9 % to 33.3 % in HEK293T cells, and 4.8 % to 18.4 % in fibroblasts. Note that these efficiencies are comparable to those generated with single gRNA bearing vectors, again indicating the possibility of multi-locus editing. In dealing with diseases, CRISPR/Cas was utilized to generate a single large genomic deletion in Duchenne muscular dystrophy (DMD) patient myoblasts, which can correct up to 62% of DMD mutations.92 Although the efficiency of deletion was not quantified, this study showcased the potential of CRISPR/Cas in treating genetic diseases as well as cancers.
Factors affecting multiplexing capability
Thanks to the development of the aforementioned techniques, genome engineering has been greatly accelerated through multiplexing. Nonetheless, the level of multiplexing is still restricted, typically at only a few edits a time (Table 1). Increased multiplexing is highly desirable, yet technically challenging. What lies at the center of multiplexed genome engineering is a cost-effective way to generate mutagenizing reagents, be they oligonucleotides or nucleases. Ideally, it is the cheapest to use chemically synthesized short DNA oligos to directly introduce mutations into the genome, such as in the case of MAGE and YOGE. These methods are also cloning free, which further reduces the cost. However, these methods can only be applied to microbial systems. ZFNs and TALENs, although have been demonstrated for multiplexed genome engineering, suffer from high-cost, sophisticated assembly workflows. In contrast, Cas9 specificity solely relies on the gRNA sequence, which is very easy and cheap to synthesize. Thus, Cas9 seems a better choice over ZFNs and TALENs for multiplexed applications.
Table 1.
Reported efficiency values for nuclease-mediated multiplexed genome engineering
| Organism | Endonuclease | Type of Modification | Targets | Efficiency (%) | Reference |
|---|---|---|---|---|---|
| Streptococcus pneumoniae | Cas9 | multi-locus deletion | 2 | 75 | 40 |
| Streptomyces sp. | Cas9 | multi-locus deletion | 2 | 100 | 41 |
| Cas9 | large deletion | 2 | 100 | 41 | |
| Escherichia coli | Cas9 | multi-locus deletion | 2 | 97 | 42 |
| Cas9 | multi-locus deletion/insertion | 2 | 78 | 42 | |
| Cas9 | multi-locus deletion | 3 | 47 | 42 | |
| Saccharomyces cerevisiae | TALEN | multi-locus deletion | 2 | ~5 (phenotype) | 51 |
| Cas9 | multi-allelic insertion | 1 | 70 - 100 | 44, 45 | |
| Cas9 | multi-locus deletion | 3 | 27 - 100 | 43 | |
| Cas9 | multi-locus deletion | 5 | 100 | 46 | |
| Cas9 | multi-locus insertion | 2 | 43 | 45 | |
| Cas9 | multi-locus insertion | 3 | 19 | 45 | |
| Rice (Oryza sativa) | TALEN | multi-allelic deletion | 1 | 34 | 52 |
| TALEN | multi-locus deletion | 3 | 1.9 | 53 | |
| Cas9 | multi-allelic deletion | 1 | 33 - 100 | 39, 57 - 59 | |
| Cas9 | large deletion | 2 | 4 - 20 | 39 | |
| Wheat (Triticum aestivum L.) | TALEN | multi-allelic deletion | 1 | 3.7 | 54 |
| Arabidopsis thaliana | Cas9 | large deletion | 2 | 5.5 | 55 |
| Cas9 | multi-locus deletion | 2 | 67 - 79 | 56 | |
| Corn (Zea mays) | Cas9 | large deletion | 2 | 60 | 56 |
| Silkworn (Bombyx mori) | TALEN | large deletion | 2 | 14 | 61 |
| Cas9 | multi-locus deletion | 3 | 1 | 71 | |
| Zebrafish (Danio rerio) | TALEN | large deletion | 2 | 0.7 - 15 | 63 - 65 |
| TALEN | inversion | 2 | < 2 | 64 | |
| Fruitfly (Drosophila melanogaster) |
Cas9 | multi-allelic deletion | 1 | > 99 | 70 |
| Livestock (cow and swine) | TALEN | multi-allelic deletion | 1 | 0 - 17 | 68 |
| TALEN | large deletion | 2 | 10 | 68 | |
| TALEN | inversion | 2 | 4 | 68 | |
| Mouse | Cas9 | multi-locus deletion | 2 | 79 | 78 |
| Cas9 | multi-locus deletion | 3 | 78 | 79 | |
| Cas9 | multi-locus deletion | 5 | 10 | 78 | |
| Cas9 | large deletion | 2 | 25 | 80 | |
| Rat | Cas9 | multi-locus deletion | 2 | 6.7, 63 - 75 | 80, 81 |
| Cas9 | multi-locus deletion | 4 | 24 | 81 | |
| Chinese hamter ovary | ZFN | multi-locus deletion | 3 | > 1 | 60 |
| Cas9 | multi-locus deletion | 3 | 35 | 82 | |
| Human cell lines | ZFN | large deletion | 2 | 0.01 - 10 | 83, 84 |
| ZFN | translocation | 2 | < 0.0001 - 0.01 | 85, 86 | |
| TALEN | translocation | 2 | 0.1 - 1 | 86 | |
| Cas9 | large deletion | 2 | 1.6 | 88 |
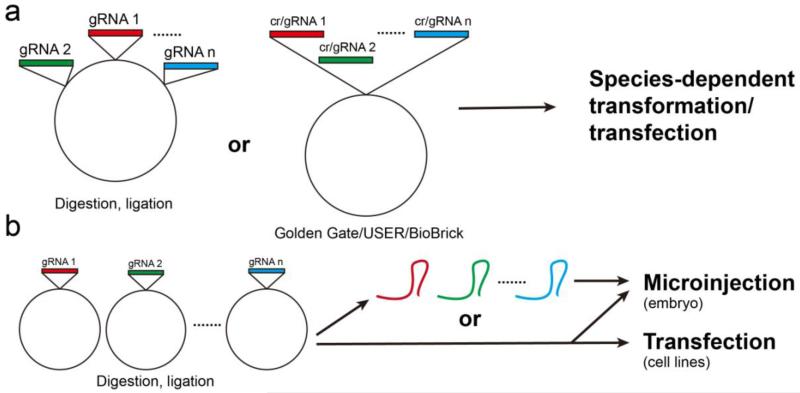
Another factor affecting multiplexing capability is the delivery of mutagenizing reagents. It is important to ensure that multiple oligonucleotides or nuclease-encoding plasmids/mRNAs can be efficiently introduced into a single cell. Taking CRISPR/Cas9 as an example, there are two general strategies to introduce multiple cr/gRNAs (Fig. 5). One strategy is to construct all the cr/gRNAs into one vector, by either sequential restriction digestion and ligation45 or one-step assembly using customized protocols, such as Golden Gate39, 41, 43, 56, USER46 and BioBrick42 (Fig. 5a). Multiplexing capability in this strategy is restricted by the number of cr/gRNAs that can be cloned into one vector and functionally expressed. Another strategy is to construct each gRNA into one separate vector and introduce mixed plasmids or in vitro transcribed RNAs simultaneously into a cell (Fig. 5b). Multiplexing capability in this strategy is thus restricted by the number of constructs a cell can take at a time. Again, CRISPR/Cas9 has the advantage over ZFNs and TALENs, since multiple short cr/gRNA sequences can be readily made and easily delivered, as compared to the long gene fragments encoding multiple pairs of ZFNs and TALENs.
Figure 5.
Common vector construction and delivery methods of multiplexed CRISPR/Cas systems. (a) Multiplexing at vector construction stage. Multiple gRNA or crRNA sequences can be constructed into one vector by either sequential digestion and ligation or one step assembly using customized methods. (b) Multiplexing at gRNA delivery stage. Each gRNA was constructed into one separate vector. Multiple vectors or in vitro transcribed gRNAs can be mixed and simultaneously delivered by microinjection or transfection.
It is worth noting that each multiplexed genome engineering technique comes with advantages and limitations (Table 2). Thus, careful considerations must be taken when choosing from different tools. In microbial systems, though conceptually similar, oligonucleotide-mediated multiplexed genome engineering methods are customized for each species and are not easily transferable between different organisms. Among nuclease-mediated methods, ZFNs and TALENs have smaller protein sizes than Cas9 (from Streptococcus pyogenes), which is an advantage in single-locus genome engineering applications. However, in multiplexed genome engineering applications, introduction of multiple pairs of ZFNs or TALENs requires efficient delivery methods. In addition, mismatched ZFN or TALEN dimers could form, which aggravates off-target effects93. In contrast, while off-target effects can still be a concern with Cas9, mismatching is not an issue, as Cas9 functions as a monomer and its targeting specificity resides solely in the cr/gRNA.
Table 2.
Advantages and limitations of various multiplexed genome engineering methods
| Methods | Advantages | Limitations |
|---|---|---|
| Oligonucleotide-mediated | ||
| MAGE |
|
|
| YOGE |
|
|
| MuGENT |
|
|
| Nuclease-mediated | ||
| ZFN |
|
|
| TALEN |
|
|
| CRISPR/Cas9 |
|
|
CONCLUSIONS AND FUTURE PROSPECTS
While true multiplexing has only recently been established as a viable genome engineering paradigm, a great number of very promising examples have already emerged. In diverse organisms ranging from bacteria to human cells, tools are being developed to enact complex genomic manipulations that would otherwise be tedious, difficult, or impossible to achieve by traditional means. Such tools have numerous applications in the fundamental and applied sciences, including multi-locus functional genomics studies in model organisms, the development of efficient industrial production strains, and the creation of models to study and treat complex diseases.
Moving forward, the development of these multiplexing tools beyond proof-of-concept studies will likely depend on two factors. First, continued optimization is necessary to increase the efficiency with which desired mutations can be introduced while reducing instances of off-target modification. This might include competency improvement to enhance the uptake of mutagenic oligonucleotides, promoter engineering to create optimal expression profiles for key elements (such as TALENs, Cas9, or gRNAs) in each organism of interest, or transcript optimization to improve the expression or stability of such elements. Protein engineering will also aid in the development of more efficient genome editing tools, as has been demonstrated with TALENs31, 94 and Cas9,43 as will the continued development of tools and protocols to aid the selection of specific target sequences95-97 and increase the likelihood of desired editing events.98, 99
Second, ongoing advances in high-throughput processing and laboratory automation will greatly influence future multiplexed genome engineering efforts. With increasing efficiency and a desire to target more and more loci simultaneously, the sizes of genome-wide libraries will increase exponentially. As a result, tools to rapidly generate targeting elements and screen such large libraries100 will become a necessity. For the assembly of TALENs, generalized and scalable assembly schemes have already been developed.101, 102 Genome-wide application of Cas9 as well has benefitted from high-throughput generation of gRNAs103 and the development of tools to screen the resulting libraries. Looking forward, automated platforms will likely become a staple to facilitate many aspects of multiplexed genome engineering, including DNA cloning, transformation, and cell culture.104 Further, one can envision a future in which even decision-making processes are hardwired into laboratory robots, such that automated data analysis can close the loop of the design-build-test cycle.105, 106 Thus, with advances in both scale and efficiency, multiplexed genome engineering is poised to build upon its early successes and achieve further growth and applicability.
Acknowledgements
We thank the National Institutes of Health (GM077596), Roy J. Carver Charitable Trust (13-4257), and Carl R. Woese Institute for Genomic Biology at the University of Illinois at Urbana-Champaign for financial support in our development and application of genome engineering technologies. We thank all Zhao group members for giving critical comments during the preparation of this review.
Footnotes
In addition, we state that there is no conflict of interest.
References
- 1.Liang J, Luo Y, Zhao H. Synthetic biology: putting synthesis into biology. Wiley Interdiscip Rev Syst Biol Med. 2011;3:7–20. doi: 10.1002/wsbm.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo Y, Lee JK, Zhao H. Challenges and opportunities in synthetic biology for chemical engineers. Chem Eng Sci. 2013;103 doi: 10.1016/j.ces.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abil Z, Xiong X, Zhao H. Synthetic biology for therapeutic applications. Mol Pharm. 2015;12:322–331. doi: 10.1021/mp500392q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Synthetic Biology: Tools and Applications. Academic Press-Elsevier; Amsterdam: 2013. [Google Scholar]
- 5.Carr PA, Church GM. Genome engineering. Nat Biotechnol. 2009;27:1151–1162. doi: 10.1038/nbt.1590. [DOI] [PubMed] [Google Scholar]
- 6.Sun N, Liang J, Abil Z, Zhao H. Optimized TAL effector nucleases (TALENs) for use in treatment of sickle cell disease. Mol Biosyst. 2012;8:1255–1263. doi: 10.1039/c2mb05461b. [DOI] [PubMed] [Google Scholar]
- 7.Alper HS, Wittmann C. Editorial: how multiplexed tools and approaches speed up the progress of metabolic engineering. Biotechnol J. 2013;8:506–507. doi: 10.1002/biot.201300167. [DOI] [PubMed] [Google Scholar]
- 8.Wang HH, Church GM. Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods Enzymol. 2011;498:409–426. doi: 10.1016/B978-0-12-385120-8.00018-8. [DOI] [PubMed] [Google Scholar]
- 9.Si T, HamediRad M, Zhao H. Regulatory RNA-assisted genome engineering in microorganisms. Curr Opin Biotechnol. 2015 doi: 10.1016/j.copbio.2015.08.003. in press. [DOI] [PubMed] [Google Scholar]
- 10.Si T, Xiao H, Zhao H. Rapid prototyping of microbial cell factories via genome-scale engineering. Biotechnol Adv. 2014 doi: 10.1016/j.biotechadv.2014.11.007. DOI: 10.1016/j.biotechadv.2014.1011.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kabadi AM, Gersbach CA. Engineering synthetic TALE and CRISPR/Cas9 transcription factors for regulating gene expression. Methods. 2014;69:188–197. doi: 10.1016/j.ymeth.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jurkowski TP, Ravichandran M, Stepper P. Synthetic epigenetics-towards intelligent control of epigenetic states and cell identity. Clin Epigenetics. 2015;7:18. doi: 10.1186/s13148-015-0044-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pines G, Freed EF, Winkler JD, Gill RT. Bacterial recombineering - genome engineering via phage-based homologous recombination. ACS Synth Biol. 2015 doi: 10.1021/acssynbio.5b00009. DOI: 10.1021/acssynbio.1025b00009. [DOI] [PubMed] [Google Scholar]
- 14.Murphy KC. Use of bacteriophage lambda recombination functions to promote gene replacement in Escherichia coli. J Bacteriol. 1998;180:2063–2071. doi: 10.1128/jb.180.8.2063-2071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res. 1999;27:1555–1557. doi: 10.1093/nar/27.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet. 1998;20:123–128. doi: 10.1038/2417. [DOI] [PubMed] [Google Scholar]
- 17.Wang HH, Isaacs FJ, Carr PA, Sun ZZ, Xu G, Forest CR, Church GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma NJ, Moonan DW, Isaacs FJ. Precise manipulation of bacterial chromosomes by conjugative assembly genome engineering. Nat Protoc. 2014;9:2285–2300. doi: 10.1038/nprot.2014.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, et al. Recoded organisms engineered to depend on synthetic amino acids. Nature. 2015;518:89–93. doi: 10.1038/nature14095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang HH, Kim H, Cong L, Jeong J, Bang D, Church GM. Genome-scale promoter engineering by coselection MAGE. Nat Methods. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang HH, Xu G, Vonner AJ, Church GM. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–7347. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosberg JA, Gregg CJ, Lajoie MJ, Wang HH, Church GM. Improving lambda red genome engineering in Escherichia coli via rational removal of endogenous nucleases. PLoS ONE. 2012;7:e44638. doi: 10.1371/journal.pone.0044638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lajoie MJ, Gregg CJ, Mosberg JA, Washington GC, Church GM. Manipulating replisome dynamics to enhance lambda red-mediated multiplex genome engineering. Nucleic Acids Res. 2012;40:e170. doi: 10.1093/nar/gks751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonde MT, Klausen MS, Anderson MV, Wallin AI, Wang HH, Sommer MO. MODEST: a web-based design tool for oligonucleotide-mediated genome engineering and recombineering. Nucleic Acids Res. 2014;42:W408–415. doi: 10.1093/nar/gku428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonde MT, Kosuri S, Genee HJ, Sarup-Lytzen K, Church GM, Sommer MO, Wang HH. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth Biol. 2015;4:17–22. doi: 10.1021/sb5001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DiCarlo JE, Conley AJ, Penttila M, Jantti J, Wang HH, Church GM. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013;2:741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lorenz MG, Wackernagel W. Bacterial gene transfer by natural genetic transformation in the environment. Microbiol Rev. 1994;58:563–602. doi: 10.1128/mr.58.3.563-602.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dalia AB, McDonough E, Camilli A. Multiplex genome editing by natural transformation. Proc Natl Acad Sci U S A. 2014;111:8937–8942. doi: 10.1073/pnas.1406478111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun N, Bao Z, Xiong X, Zhao H. SunnyTALEN: a second-generation TALEN system for human genome editing. Biotechnol Bioeng. 2014;111:683–691. doi: 10.1002/bit.25154. [DOI] [PubMed] [Google Scholar]
- 32.Sun N, Abil Z, Zhao H. Recent advances in targeted genome engineering in mammalian systems. Biotechnol J. 2012;7:1074–1087. doi: 10.1002/biot.201200038. [DOI] [PubMed] [Google Scholar]
- 33.Takeuchi R, Lambert AR, Mak AN, Jacoby K, Dickson RJ, Gloor GB, Scharenberg AM, Edgell DR, Stoddard BL. Tapping natural reservoirs of homing endonucleases for targeted gene modification. Proc Natl Acad Sci U S A. 2011;108:13077–13082. doi: 10.1073/pnas.1107719108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 35.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 36.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 37.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and programmable regulation of gene networks with an integrated RNA and CRISPR/Cas toolkit in human cells. Mol Cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie K, Minkenberg B, Yang Y. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci U S A. 2015;112:3570–3575. doi: 10.1073/pnas.1420294112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang WY, Bikard D, Cox D, Zhang F, Marraffini LA. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cobb RE, Wang Y, Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol. 2014 doi: 10.1021/sb500351f. DOI: 10.1021/sb500351f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang Y, Chen B, Duan C, Sun B, Yang J, Yang S. Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl Environ Microbiol. 2015;81:2506–2514. doi: 10.1128/AEM.04023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bao Z, Xiao H, Liang J, Zhang L, Xiong X, Sun N, Si T, Zhao H. Homology-Integrated CRISPR-Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2014 doi: 10.1021/sb500255k. DOI: 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- 44.Ryan OW, Cate JH. Multiplex engineering of industrial yeast genomes using CRISPRm. Methods Enzymol. 2014;546:473–489. doi: 10.1016/B978-0-12-801185-0.00023-4. [DOI] [PubMed] [Google Scholar]
- 45.Ryan OW, Skerker JM, Maurer MJ, Li X, Tsai JC, Poddar S, Lee ME, DeLoache W, Dueber JE, Arkin AP, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3 doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jakociunas T, Sonde I, Herrgard M, Harrison SJ, Kristensen M, Pedersen LE, Jensen MK, Keasling JD. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–222. doi: 10.1016/j.ymben.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 47.Mans R, van Rossum HM, Wijsman M, Backx A, Kuijpers NG, van den Broek M, Daran-Lapujade P, Pronk JT, van Maris AJ, Daran JM. CRISPR/Cas9: a molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015;15 doi: 10.1093/femsyr/fov004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Horwitz Andrew A, Walter Jessica M, Schubert Max G, Kung Stephanie H, Hawkins K, Platt Darren M, Hernday Aaron D, Mahatdejkul-Meadows T, Szeto W, Chandran Sunil S, et al. Efficient multiplexed integration of synergistic alleles and metabolic pathways in yeasts via CRISPR-Cas. Cell Systems. 2015 doi: 10.1016/j.cels.2015.02.001. DOI: http://dx.doi.org/10.1016/j.cels.2015.1002.1001. [DOI] [PubMed] [Google Scholar]
- 49.Jakociunas T, Rajkumar AS, Zhang J, Arsovska D, Rodriguez A, Jendresen CB, Skjodt ML, Nielsen AT, Borodina I, Jensen MK, et al. CasEMBLR: Cas9-facilitated multiloci genomic integration of in vivo assembled DNA parts in Saccharomyces cerevisiae. ACS Synth Biol. 2015 doi: 10.1021/acssynbio.5b00007. DOI: 10.1021/acssynbio.1025b00007. [DOI] [PubMed] [Google Scholar]
- 50.Sun N, Zhao H. Transcription activator-like effector nucleases (TALENs): a highly efficient and versatile tool for genome editing. Biotechnol Bioeng. 2013;110:1811–1821. doi: 10.1002/bit.24890. [DOI] [PubMed] [Google Scholar]
- 51.Aouida M, Li L, Mahjoub A, Alshareef S, Ali Z, Piatek A, Mahfouz MM. Transcription activator-like effector nucleases mediated metabolic engineering for enhanced fatty acids production in Saccharomyces cerevisiae. J Biosci Bioeng. 2015 doi: 10.1016/j.jbiosc.2015.02.017. DOI: 10.1016/j.jbiosc.2015.1002.1017. [DOI] [PubMed] [Google Scholar]
- 52.Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- 53.Shan Q, Zhang Y, Chen K, Zhang K, Gao C. Creation of fragrant rice by targeted knockout of the OsBADH2 gene using TALEN technology. Plant Biotechnol J. 2015 doi: 10.1111/pbi.12312. DOI: 10.1111/pbi.12312. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu JL. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 55.Li JF, Norville JE, Aach J, McCormack M, Zhang D, Bush J, Church GM, Sheen J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat Biotechnol. 2013;31:688–691. doi: 10.1038/nbt.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- 58.Zhou H, Liu B, Weeks DP, Spalding MH, Yang B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014;42:10903–10914. doi: 10.1093/nar/gku806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Endo M, Mikami M, Toki S. Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol. 2015;56:41–47. doi: 10.1093/pcp/pcu154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu PQ, Chan EM, Cost GJ, Zhang L, Wang J, Miller JC, Guschin DY, Reik A, Holmes MC, Mott JE, et al. Generation of a triple-gene knockout mammalian cell line using engineered zinc-finger nucleases. Biotechnol Bioeng. 2010;106:97–105. doi: 10.1002/bit.22654. [DOI] [PubMed] [Google Scholar]
- 61.Ma SY, Zhang SL, Wang F, Liu Y, Liu YY, Xu HF, Liu C, Lin Y, Zhao P, Xia QY. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0045035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma S, Wang X, Liu Y, Gao J, Zhang S, Shi R, Chang J, Zhao P, Xia Q. Multiplex genomic structure variation mediated by TALEN and ssODN. BMC Genomics. 2014;15:41. doi: 10.1186/1471-2164-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Luo D, Zhao H, Zhu Z, Hu W, Cheng CH. Inheritable and precise large genomic deletions of non-coding RNA genes in zebrafish using TALENs. PLoS ONE. 2013;8:e76387. doi: 10.1371/journal.pone.0076387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu Y, Luo D, Lei Y, Hu W, Zhao H, Cheng CH. A highly effective TALEN-mediated approach for targeted gene disruption in Xenopus tropicalis and zebrafish. Methods. 2014;69:58–66. doi: 10.1016/j.ymeth.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 65.Gupta A, Hall VL, Kok FO, Shin M, McNulty JC, Lawson ND, Wolfe SA. Targeted chromosomal deletions and inversions in zebrafish. Genome Res. 2013;23:1008–1017. doi: 10.1101/gr.154070.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lei Y, Guo XG, Liu Y, Cao Y, Deng Y, Chen XF, Cheng CHK, Dawid IB, Chen YL, Zhao H. Efficient targeted gene disruption in Xenopus embryos using engineered transcription activator-like effector nucleases (TALENs) Proc Natl Acad Sci U S A. 2012;109:17484–17489. doi: 10.1073/pnas.1215421109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sakane Y, Sakuma T, Kashiwagi K, Kashiwagi A, Yamamoto T, Suzuki KT. Targeted mutagenesis of multiple and paralogous genes in Xenopus laevis using two pairs of transcription activator-like effector nucleases. Dev Growth Differ. 2014;56:108–114. doi: 10.1111/dgd.12105. [DOI] [PubMed] [Google Scholar]
- 68.Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, Voytas DF, Long CR, Whitelaw CB, Fahrenkrug SC. Efficient TALEN-mediated gene knockout in livestock. Proc Natl Acad Sci U S A. 2012;109:17382–17387. doi: 10.1073/pnas.1211446109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Port F, Chen HM, Lee T, Bullock SL. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc Natl Acad Sci U S A. 2014;111:E2967–2976. doi: 10.1073/pnas.1405500111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gantz VM, Bier E. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science. 2015;348:442–444. doi: 10.1126/science.aaa5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ma S, Chang J, Wang X, Liu Y, Zhang J, Lu W, Gao J, Shi R, Zhao P, Xia Q. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci Rep. 2014;4:4489. doi: 10.1038/srep04489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu YY, Ma SY, Wang XG, Chang JS, Gao J, Shi R, Zhang JD, Lu W, Liu Y, Zhao P, et al. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect Biochem Molec. 2014;49:35–42. doi: 10.1016/j.ibmb.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 73.Jao LE, Wente SR, Chen W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc Natl Acad Sci U S A. 2013;110:13904–13909. doi: 10.1073/pnas.1308335110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yin L, Maddison LA, Li M, Kara N, LaFave MC, Varshney GK, Burgess SM, Patton JG, Chen W. Multiplex conditional mutagenesis using transgenic expression of Cas9 and sgRNAs. Genetics. 2015 doi: 10.1534/genetics.115.176917. DOI: 10.1534/genetics.1115.176917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ota S, Hisano Y, Ikawa Y, Kawahara A. Multiple genome modifications by the CRISPR/Cas9 system in zebrafish. Genes Cells. 2014;19:555–564. doi: 10.1111/gtc.12154. [DOI] [PubMed] [Google Scholar]
- 76.Guo X, Zhang T, Hu Z, Zhang Y, Shi Z, Wang Q, Cui Y, Wang F, Zhao H, Chen Y. Efficient RNA/Cas9-mediated genome editing in Xenopus tropicalis. Development. 2014;141:707–714. doi: 10.1242/dev.099853. [DOI] [PubMed] [Google Scholar]
- 77.Yasue A, Mitsui SN, Watanabe T, Sakuma T, Oyadomari S, Yamamoto T, Noji S, Mito T, Tanaka E. Highly efficient targeted mutagenesis in one-cell mouse embryos mediated by the TALEN and CRISPR/Cas systems. Sci Rep. 2014;4:5705. doi: 10.1038/srep05705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujii W, Onuma A, Sugiura K, Naito K. One-step generation of phenotype-expressing triple-knockout mice with heritable mutated alleles by the CRISPR/Cas9 system. J Reprod Dev. 2014;60:324–327. doi: 10.1262/jrd.2013-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li DL, Qiu ZW, Shao YJ, Chen YT, Guan YT, Liu MZ, Li YM, Gao N, Wang LR, Lu XL, et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat Biotechnol. 2013;31:681–683. doi: 10.1038/nbt.2661. [DOI] [PubMed] [Google Scholar]
- 81.Li W, Teng F, Li T, Zhou Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31:684–686. doi: 10.1038/nbt.2652. [DOI] [PubMed] [Google Scholar]
- 82.Grav LM, Lee JS, Gerling S, Beuchert Kallehauge T, Holmgaard Hansen A, Kol S, Lee GM, Ebdrup Pedersen L, Faustrup Kildegaard H. One-step generation of triple knockout CHO cell lines using CRISPR Cas9 and fluorescent enrichment. Biotechnol J. 2015 doi: 10.1002/biot.201500027. DOI: 10.1002/biot.201500027. [DOI] [PubMed] [Google Scholar]
- 83.Lee HJ, Kim E, Kim JS. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim E, Kim S, Kim DH, Choi BS, Choi IY, Kim JS. Precision genome engineering with programmable DNA-nicking enzymes. Genome Res. 2012;22:1327–1333. doi: 10.1101/gr.138792.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci U S A. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah GE, Zhang L, Holmes MC, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nyquist MD, Li Y, Hwang TH, Manlove LS, Vessella RL, Silverstein KA, Voytas DF, Dehm SM. TALEN-engineered AR gene rearrangements reveal endocrine uncoupling of androgen receptor in prostate cancer. Proc Natl Acad Sci U S A. 2013;110:17492–17497. doi: 10.1073/pnas.1308587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sakuma T, Nishikawa A, Kume S, Chayama K, Yamamoto T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci Rep. 2014:4. doi: 10.1038/srep05400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ousterout DG, Kabadi AM, Thakore PI, Majoros WH, Reddy TE, Gersbach CA. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sollu C, Pars K, Cornu TI, Thibodeau-Beganny S, Maeder ML, Joung JK, Heilbronn R, Cathomen T. Autonomous zinc-finger nuclease pairs for targeted chromosomal deletion. Nucleic Acids Res. 2010;38:8269–8276. doi: 10.1093/nar/gkq720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bedell VM, Wang Y, Campbell JM, Poshusta TL, Starker CG, Krug RG, Tan WF, Penheiter SG, Ma AC, Leung AYH, et al. In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–U133. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 96.Prykhozhij SV, Rajan V, Gaston D, Berman JN. CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS ONE. 2015;10:e0119372. doi: 10.1371/journal.pone.0119372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Farboud B, Meyer BJ. Dramatic enhancement of genome editing by CRISPR/Cas9 through improved guide RNA design. Genetics. 2015;199:959–971. doi: 10.1534/genetics.115.175166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Maruyama T, Dougan SK, Truttmann MC, Bilate AM, Ingram JR, Ploegh HL. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. doi: 10.1038/nbt.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chu VT, Weber T, Wefers B, Wurst W, Sander S, Rajewsky K, Kuhn R. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33:543–548. doi: 10.1038/nbt.3198. [DOI] [PubMed] [Google Scholar]
- 100.Xiao H, Bao Z, Zhao H. High throughput screening and selection methods for directed enzyme evolution. Ind Eng Chem Res. 2014 doi: 10.1021/ie503060a. 141014124302009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, Baller JA, Somia NV, Bogdanove AJ, Voytas DF. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang J, Chao R, Abil Z, Bao Z, Zhao H. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synth Biol. 2014;3:67–73. doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- 103.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet. 2015;16:299–311. doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chao R, Yuan Y, Zhao H. Building biological foundries for next-generation synthetic biology. Sci China Life Sci. 2015 doi: 10.1007/s11427-015-4866-8. DOI: 10.1007/s11427-11015-14866-11428. [DOI] [PubMed] [Google Scholar]
- 105.King RD. Functional genomic hypothesis generation and experimentation by a robot scientist. Nature. 2004;427:244–247. doi: 10.1038/nature02236. [DOI] [PubMed] [Google Scholar]
- 106.King RD, Rowland J, Oliver SG, Young M, Aubrey W, Byrne E, Liakata M, Markham M, Pir P, Soldatova LN, et al. The automation of science. Science. 2009;324:85–89. doi: 10.1126/science.1165620. [DOI] [PubMed] [Google Scholar]