Abstract
Background
Kabuki syndrome (KS) is a complex multi-system developmental disorder associated with mutation of genes encoding histone-modifying proteins. In addition to craniofacial, intellectual, and cardiac defects, KS is also characterized by humoral immune deficiency and autoimmune disease, yet no detailed molecular characterization of the KS-associated immune phenotype has previously been reported.
Objective
To characterize the humoral immune defects found in KS patients with KMT2D mutations.
Methods
We comprehensively characterize B cell function in a cohort (N = 13) of patients with KS (ages 4 months to 27 years).
Results
Three-quarters (77%) of the cohort had a detectable heterozygous KMT2D mutations (50% nonsense, 20% splice site, 30% missense), and 70% of the reported mutations are novel. Amongst the patients with KMT2D mutations (KMT2DMut/+), hypogammaglobulinemia was detected in all but one individual, with IgA deficiency affecting 90% of patients and a deficiency in at least one other isoform seen in 40% of patients. Total memory (CD27+) and class-switched memory B cells (IgM−) were significantly reduced in KMT2DMut/+ patients compared to controls (p-values < 0.001). KMT2DMut/+ patients also had significantly reduced rates of somatic hypermutation in IgG (p value = 0.003), but not IgA or IgM heavy chain sequences. Impaired terminal differentiation was noted in KMT2DMut/+ primary B cells. Autoimmune pathology was observed in patients with missense mutations affecting the SET domain and its adjacent domains.
Conclusions
In patients with KS, autosomal dominant KMT2D mutations are associated with the dysregulation of terminal B cell differentiation leading to humoral immune deficiency and in some cases autoimmunity. All patients with KS should undergo serial clinical immune evaluations.
Clinical Implications
KMT2DMut/+ Kabuki syndrome causes IgA deficiency in nearly all patients, although additional humoral defects (memory B cell deficiency, IgG hypogammaglobulinemia) have variable expressivity. Missense mutations in terminal domains may increase autoimmunity risk.
Keywords: Kabuki syndrome, KMT2D, KDM6A, hypogammaglobulinemia, memory B cells, CD21lo B cells, class-switch recombination, somatic hypermutation, PTIP, AICDA, AID, polymerase eta
Introduction
Kabuki syndrome (KS) or Niikawa-Kuroki syndrome is a congenital syndrome characterized by a stereotypical set of facial features, skeletal anomalies, dermatoglyphic abnormalities, intellectual disability, and post-natal growth deficiency (1, 2). Classic KS-associated facies include arched eyebrows with central notching; elongated palpebral fissures with eversion of the lateral lower eyelid; and large, prominent, cupped ears. Though consensus clinical diagnostic criteria have not been established for this rare disease, other frequently noted structural defects include congenital heart disease, cleft palate, gastrointestinal malformations, and ophthalmologic defects (3–5). Functional defects are also observed including immune dysfunction (recurrent infections and autoimmune disorders), seizures, hearing loss, and endocrinopathies (6–9).
Autosomal-dominant mutations in two epigenetic regulatory genes, lysine methyltransferase 2D (KMT2D/MLL2/ALR/MLL4; KS-1, OMIM 147920) and lysine demethylase 6A (KDM6A/UTX; KS-2, OMIM 300867), were recently found in large cohorts of patients with KS, suggesting that defects in histone-mediated regulation drive the diverse pathology of this syndrome (10, 11). KMT2D modifies gene expression via methylation of lysine 4 on histone 3 (H3K4), and mutations of this gene are the most common cause of KS (~60–70% of cases) (12, 13). KMT2D encodes a large, multi-domain protein that forms the core of one of the six mammalian complex of proteins associated with SET1 (COMPASS) protein complexes (14). KDM6A is a cofactor physically associated with the KMT2D COMPASS complex and exhibits complementary demethylase activity at lysine 27 on histone 3 (H3K27) (15). Together, the components of the KMT2D COMPASS complex remove inhibitory epigenetic marks and add activating marks, specifically H3K4 mono-, di-, or trimethylation (H3K4me1,2,or 3). KMT2D has been implicated in the regulation of hundreds of genes via regulation of enhancer and promoter elements, providing a rationale for how defects in this single gene and its binding partners can lead to a complex and heterogeneous congenital phenotype (12, 16, 17).
KMT2D binds multiple co-factors, including the paired box (PAX) transactivation domain– interacting protein (PTIP), which plays a key role in B cell terminal differentiation. PTIP has dual activities, binding regions of DNA double-strand breaks, such as those that occur during class-switch recombination (CSR), and interacting with PAX transcription factors. PTIP directly binds the B cell master transcription factor PAX5, targeting KMT2D-mediated H3K4me3 methylation to switch regions within the immunoglobulin (Ig) heavy chain locus (IGH) during CSR (18–20). B cell–specific knockout of Ptip in primary murine B cells dramatically impairs CSR (20). Studies of PTIP have demonstrated the important role of the KMT2D COMPASS complex in regulating the IGH locus during B cell differentiation, thus linking the molecular functions of the two KS-associated genes with a central mechanism of humoral immunity.
Immune dysfunction is a common feature of KS. Described as having a common variable immune deficiency-like (CVID-like) immune profile in early studies (7), this initial observation was later correlated with clinical findings in three larger cohorts of patients with heterozygous KMT2D mutations (KMT2DMut/+). Approximately half of the patients in these studies had clinical features of immune deficiency, including recurrent otitis media [47% (21)] and frequent/repeat infections [41.9% (22)/64% (23)]. A more recent study found recurrent otitis media to be a nearly universal clinical finding (85%) in KMT2DMut/+ KS patients (24). In addition, autoimmune diseases, especially immune thrombocytopenia (ITP), hemolytic anemia, and vitiligo, have also been reported in patients with KS; however, all of these early cases were in clinically diagnosed patients without a defined genetic etiology. Two new case reports of ITP in genetically defined KMT2DMut/+ patients, however, have strengthen the link between KS and autoimmunity (25, 26). Together, these foundational studies provide a general outline of the KS immune phenotype, but reveal little about the underlying pathogenesis of the observed immune dysfunction.
Despite a growing clinical interest in KS, the cellular characteristics and molecular mechanisms of KS-associated immune dysfunction remain poorly understood. A variety of recent studies, however, have now shown that histone-modifications regulate key events in terminal B cell differentiation. Specifically, CSR and the related phenomenon, somatic hypermutation (SHM), are two critical B cell processes subject to epigenetic regulation (27–29), but despite their importance to humoral immunity, these processes have never previously been evaluated in patients with KS. To address these knowledge gaps, we characterize herein the specific cellular and molecular B cell defects found in a cohort of 13 patients with KS (10 pediatric, 4 adult cases). Our findings provide mechanistic insight into the humoral immune deficiency and autoimmunity found in this understudied patient population.
Methods
Patient cohort
Patients with a clinical diagnosis of KS by a board-certified clinical geneticist were recruited to participate. All participants and/or their parents/guardians provided written consent/assent for enrollment in this study (including explicit authorization to publish images of patients). The study protocol (#2012–4636) was approved by the Institutional Review Board at Cincinnati Children’s Hospital Medical Center (CCHMC). Phenotypic data were collected from medical record review and interviews with patients, guardians and physicians.
KMT2D/KDM6A sequencing analysis and multiplex ligation-dependent probe amplification (MLPA)
All 13 patients underwent KMT2D sequencing (reference: NM_003482.3). KMT2D domains were mapped according to UniProt Database (O14686). Per the step-wise approach to genetic diagnosis of KS in clinical practice, the 3 patients (A, B, C) in whom no KMT2D mutation was identified underwent multiplex ligation-dependent probe amplification (MLPA) analysis of 23 KMT2D exons, sequencing of the KDM6A coding region, and MLPA of 26 KDM6A exons. (See Online Methods for MLPA exon numbers).
Splice site analysis
To confirm the presence of mis-spliced mutant variant transcripts, putative splice site variants were analyzed using NNSPLICE software (30). cDNA from patients 3 and 7, who have mutations in KMT2D splice sites, was generated from peripheral blood mononuclear cells (PBMCs). Primers flanking the predicted splice site mutation amplified regions of interest. Bands were purified, cloned, and sequenced.
Clinical laboratory evaluations
Immunoglobulin (Ig) levels, post-vaccination serology, lymphocyte subpopulation data, and mitogen studies were derived from physician review of medical records. All clinical evaluations were performed at Clinical Laboratory Improvement Amendments (CLIA)–certified clinical laboratories and reported with the age-matched normal ranges. B cell flow cytometry was performed in the Diagnostic Immune Laboratory, CCHMC.
Somatic hypermutation analysis
V(D)J libraries were generated from PBMC-derived cDNA, cloned, and sequenced. Briefly, we combined the BIOMED-2 FR1 pool of variable gene primers (31) with individual antisense, constant region–specific (μ, γ, α) primers to amplify pools of isotype-specific Ig-derived clones (32, 33). After gel purification, these libraries were cloned (TOPO TA cloning, Life Technologies), screened for inserts, and 20–30 clones per library were sequenced. Ig sequences were aligned with the human IGH locus using the IgBLAST software and the IMTG database (34). Non-productive clones were excluded from analysis. Mutation analysis performed with SHMTool software (35).
In vitro terminal B cell differentiation
B cell differentiation was induced using a modified version of the protocol described by Ettinger et al. (36). (See Online Methods for culture and staining details). Cells were analyzed after 3 days or 7 days of culture by flow cytometry (LSR-II flow cytometer, BD Biosciences).
Statistical Analysis
Patient-derived B cell data was compared to age-matched controls using a linear regression model in which the p-value was calculated on the basis of the F-test (GLM procedure, SAS ver9.4 software). All other statistical analyses were performed using an unpaired Student t test. p < 0.05 was considered statistically significant.
Results
Cohort characteristics
Each patient’s KS phenotype was evaluated for the presence of 4 of the 5 cardinal manifestations of KS (Table 1) (dermatoglyphic analysis not done) (37) Images of each patient’s face and right ear are provided (Supplemental Figure E1). Additional KS-associated structural and functional anomalies were also evaluated. The frequency of selected traits in our cohort were calculated and compared with the meta-analysis of Bogershausen and Willnik (4 studies, total n = 287 patients) (9). All but two categories of KS-associated traits showed similar frequency rates between our cohort and the meta-analysis; congenital heart disease and genitourinary anomalies were more common in our cohort than in the meta-analysis cohorts. Adult KS patients also exhibit a variety of CVID-associated comorbidities, especially splenomegly and pulmonary disease (Supplemental Table E2, A).
TABLE 1.
KS Phenotype
| SUBJECT : | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | A | B | C | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age at Enrollment | 0 yrs, 4 m |
0 yrs, 5 m |
0 yrs, 11 m |
3 yrs, 8 m |
6 yrs, 10 m |
15 yrs, 5 m |
15 yrs, 6 m |
23 yrs, 0 m |
24 yrs, 10 m |
27 yrs, 2 m |
2 yrs, 11 m |
12 yrs, 0 m |
13 yrs, 6 m |
Present in this Cohort |
Meta- analysis (Ref. 9) |
|
| CARDINAL FEATURES | Typical Facial Features | + | + | + | + | + | + | + | + | + | + | + | + | + | 100% | 87% |
| Skeletal Anomalies | − | + | + | + | + | − | + | + | − | + | + | + | + | 77% | ND | |
| Mild-Moderate Intellectual disability | NA | NA | NA | NA | + | + | + | + | + | + | + | + | + | 100%* | ND | |
| Post-natal growth deficiency | + | − | + | + | − | + | + | + | + | + | − | + | + | 77% | 70% | |
| ADDITIONAL FEATURES | Congenital Heart Defects | + | + | + | + | + | + | + | − | − | + | − | − | + | 69% | 46% |
| Genitourinary anomalies | − | + | − | + | − | + | + | + | + | + | − | + | + | 69% | 44% | |
| Cleft lip and/or palate | + | − | + | + | + | − | + | − | + | − | − | − | − | 46% | 37% | |
| Gastrointestinal anomalies | − | − | − | − | − | − | + | − | + | − | − | − | − | 15% | ND | |
| Ophthalmologic anomalies | + | + | − | − | − | + | + | + | − | − | − | + | + | 54% | ND | |
| Autoimmune Complications | − | − | − | − | + | + | − | + | − | − | − | − | − | 23% | ND | |
| Recurrent Infections OM/PNA/UTI | −/−/− | −/−/− | −/−/− | +/+/+ | +/−/− | +/+/+ | +/+/− | +/+/+ | +/+/− | −/−/− | +/−/− | +/−/− | +/+/− | 77% | 61% |
LEGEND: yrs, years; m, months; NA, not available; ND not done. OM, otitis media; PNA, pneumonia; UTI, urinary tract infection
indicates percentage of subjects over 5 years of age.
Molecular genetic evaluation
All of the patients underwent KMT2D gene sequencing, and 10 out of 13 (77%) were found to have mutations predicted to be pathogenic (Supplemental Figure E2), a rate consistent with prior reports (9, 10). Seven of these KMT2D mutations were novel. Half of the 10 identified KMT2DMut/+ patients harbored nonsense/frameshift mutations predicted to generate a truncated protein or induce nonsense-mediated decay (NMD) of the mutated transcript (38).
Two patients, 3 and 7, had splice site mutations identified (Supplemental Figure E2). Bioinformatic analysis of patient 3’s mutation predicted a deleterious effect (loss of intron 26 acceptor site). This prediction was confirmed by PCR amplification of a mutant transcript fragment (Mut) from patient-derived PBMC cDNA (Supplemental Figure E3,A). Cloning and sequencing of this aberrant band revealed retained intronic sequence encoding a premature stop codon. Patient 7 was also found to have a splice site mutation (adjacent to the donor site of intron 29), and bioinformatic analysis predicted reduced confidence in the functionality of the site. Similar PCR analysis of PBMC-derived cDNA from patient 7 failed to detect a mutant band (Supplemental Figure E3, B).
Three patients, 5, 6, and 8, had missense mutations in the 3’ end of the KMT2D locus (exons 48, 51) within highly conserved functional domains (Supplemental Figure E2). The previously reported missense mutation identified in patient 8 (c.16294C>T) was recently shown to disrupt the Kabuki interaction surface (KIS), which forms a secondary active site in the COMPASS complex (39).
The three patients (A, B, C) who were clinically diagnosed with KS but in whom no KMT2D mutation were detected (clinical KS, cKS) also underwent KMT2D exon copy number variant testing (MLPA) and KDM6A sequencing and copy number testing (MLPA). In all three cases, these additional studies failed to detect a pathogenic mutation, insertion, or deletion in these two previously described KS-associated genes.
Autoimmune disease clinical histories
Autoimmune disease is commonly associated with humoral immune deficiency and is observed in ~20% of patients with CVID (40). Three of our patients have a history of marked autoimmune disease (patients 5, 6, and 8; 23% of total cohort; detailed clinical histories provided in Supplemental Figure E4). All 3 patients experienced ITP, and in 2 cases, ITP developed within several months of contracting Epstein-Barr virus (EBV) (patient 5 and 8). Patient 6 also developed vitiligo, which required phototherapy. Patient 8 has had recurrent ITP, autoimmune neutropenia, autoimmune hepatitis, and insulin-dependent diabetes mellitus. Interestingly, these patients all harbor similar missense mutations in the 3’-end of the KMT2D gene near the SET domain region (Supplemental Figure E2).
Immune evaluations
Lymphocyte Evaluations
Peripheral lymphocyte subpopulations were evaluated for 12 out of the 13 patients, and the majority of patients had normal or minor deviations from age-matched control values(41) (Supplemental Table E1). Similarly, mitogen studies were performed in 11 out of 13 subjects, with >80% having essentially normal results. Two patients (6,8) had diminished, but not absent, mitogen responses (Supplemental Table E1). With regard to lymphocyte subpopulations, four out of 12 patients (33%), were found to have a reduced percentage of CD19+ B cells (vs. age-matched controls). Four patients (2, 5, 6, 8) had lymphopenia and abnormal lymphocyte subpopulations at the time of enrollment. Patient 2 has a large, retroperitoneal lymphatic malformation, resulting in significant chylous ascites and loss of peripheral lymphocytes, especially T cells; patient 2’s absolute B cell count was low (556 cells/mcl vs. age-adjusted normal range 700–2500 cells/mcl), despite an elevated B cell percentage because of preferential loss of T cells. Patient 5 underwent thymectomy at 3 days of life, with intermittent lymphopenia noted since that time. Patient 6 was noted to have lymphocyte subpopulation levels within normal, age-matched ranges at initial evaluation (5.5 years of age) but subsequently developed lymphopenia with onset of autoimmune disease (Supplemental Figure E4). Similarly, patient 8 had a normal absolute lymphocyte count at 13 years of age prior to contracting EBV, but since that time has had chronic borderline lymphopenia (Supplemental figure E4).
Immunoglobulin levels
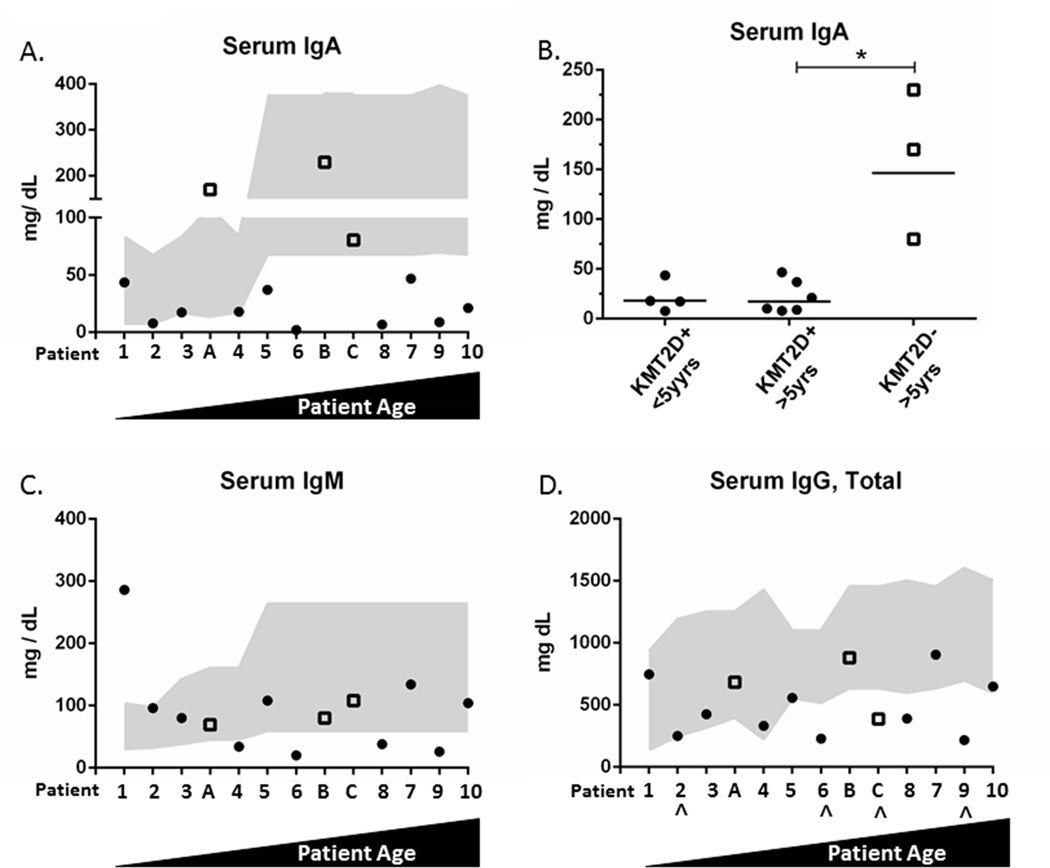
We found that 90% of our KMT2DMut/+ patients had IgA deficiency, whereas our 3 cKS patients (A–C) had normal or above-normal serum IgA levels (Figure 1A). In patients greater than 5 years of age, the IgA levels were significantly decreased in KMT2DMut/+ patients compared with cKS patients (A–C) (p value = 0.003) (Figure 1A, right panel). Low serum IgM was noted in 40% of KMT2DMut/+ patients (4, 6, 8, and 9) but in none of the cKS patients (Figure 1B). Low IgG was also noted in 40% of patients with KMT2D mutations (patients 6, 8, and 9) and in 1 cKS patient (Figure 1C). Three KMT2DMut/+ patients (6, 8, 9) had pan-hypogammaglobulinema.
Figure 1. Serum Ig levels.
Patients arranged by ascending age. Gray shading represents age-matched normal range (2.5th–97.5th percentile). (A) IgA levels in KMT2DMut/+ patients (#1–10, solid circles) and cKS patients (A–C, hollow squares). (B) IgA values (bar=mean) are significantly reduced in KMT2DMut/+ patients > 5 yrs old versus cKS patients. *p < 0.05. (C, D) IgM levels and IgG levels. IgG values for patients 2, 6, C, and 9 (^) are the last levels acquired before IVIG/SCIG treatment.
Specific antibody titers against tetanus were also evaluated for all patients greater than 2 years of age except for patients 4 and 5. All patients had protective tetanus titers except for patient 6. One month after revaccination, patient 6 mounted a 10-fold increase in her anti-tetanus titers (still below protective level), but this level returned to her previously low baseline by 1 year later. An alternative measure of vaccine responsiveness was available for patient 5, who mounted protective responses to hepatitis A and B vaccinations at 6 years of age. Of note, 5 adult KMT2DMut/+ patients (patients 6–10) underwent pre- and post-vaccination pneumococcal titer testing, and 3 out of 5 (60%) failed to mount a normal response (defined as 2-fold increase in 70% of tested titers)(Supplemental Table E2) (42).
B cell subpopulations
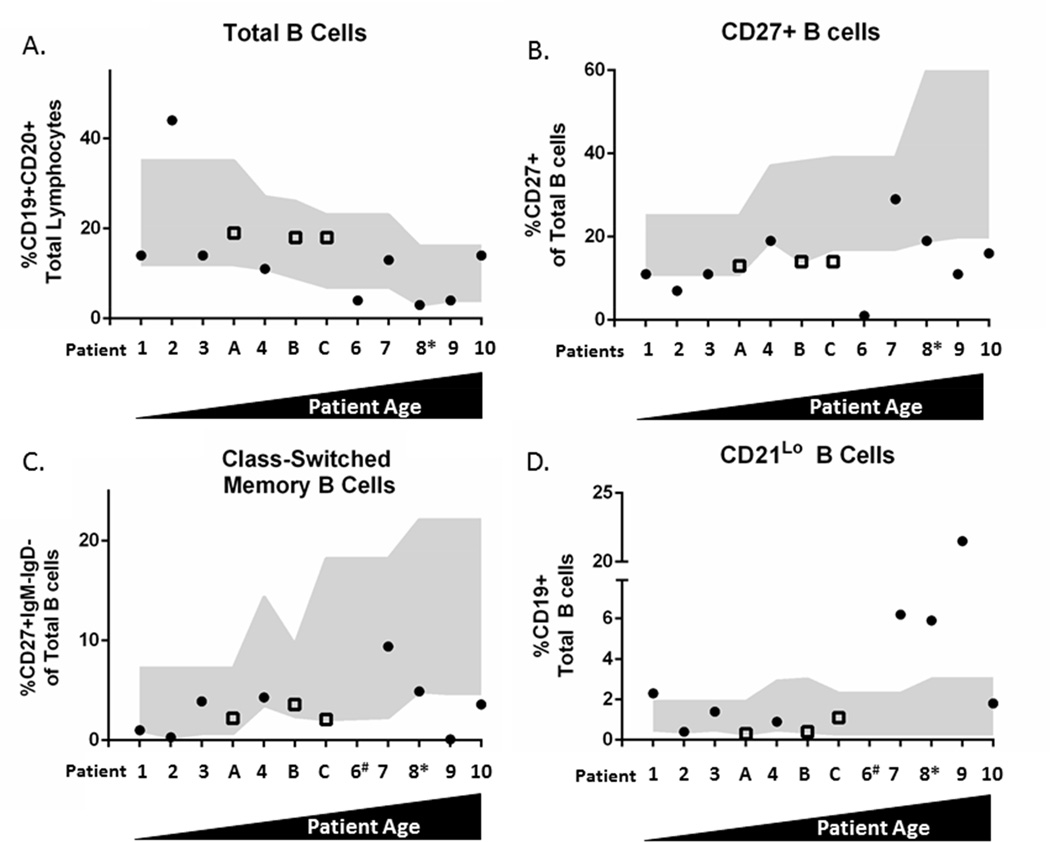
Given our finding of hypogammaglobulinemia in the majority of our KMT2DMut/+ patients, we next evaluated peripheral B cell subpopulations in 12 out of 13 patients in our cohort (patient 5- no data; patient 6- not all data available). Consistent with our lymphocyte subpopulation analysis, 4 (33%, 4 of 12) of our patients had a low percentage of total B cells (CD19+/CD20+, Figure 2A); all 4 were KMT2DMut/+ patients (44%, 4 of 9). We next evaluated total memory B cells (CD27+) and found that 89% (8 of 9) of KMT2DMut/+ patients were deficient in this cell type, a feature shared with 67% (2 of 3) of cKS patients (Figure 2B). When compared to control subjects, we found that KMT2DMut/+ patients had a significant reduction in memory B cells after adjusting for age (p-value = 0.0005). Class-switched memory B cells (IgM−, IgD−, CD27+) were also low in 5 KMT2DMut/+ patients (63%) and in 1 patient lacking a KMT2D mutation (Figure 2C). Compared to age-adjusted control subjects, we found that KMT2DMut/+ patients had a significant reduction in class-switched memory B cells (p-value = 0.0077). Finally, we evaluated CD19+CD21lo B cells, a cell population previously associated with CVID in adults (43) and found that 4 KMT2DMut/+ patients had an expanded population of these cells; no expansion of these cells was seen in cKS patients (Figure 2D). When compared to control subjects, KMT2DMut/+ patients had a significant expansion in CD19+CD21lo B cells after adjusting for age (p-value < 0.0001). Collectively, these findings indicate that many patients with KS exhibit abnormal terminal B cell differentiation.
Figure 2. B cell subset analysis.
Patients arranged by ascending age; gray shading represents age-matched normal range (5th–95th percentile). (A) Total B cells (CD19+CD20+) in KMT2DMut/+ (solid circles) and cKS patients (hollow squares). (B) Total memory B cells (CD19+, CD27+). (C) Class-switched memory B cells (CD19+CD27+IgM−IgD−). (D) CD19+ CD21lo B cells. # = data were not available for patient 6 (n = 11). *Received anti-CD20 (rituximab) antibody therapy 3 yrs prior to analysis; had normal total B cell count pre-therapy. Lo, low.
Somatic hypermutation (SHM) rates
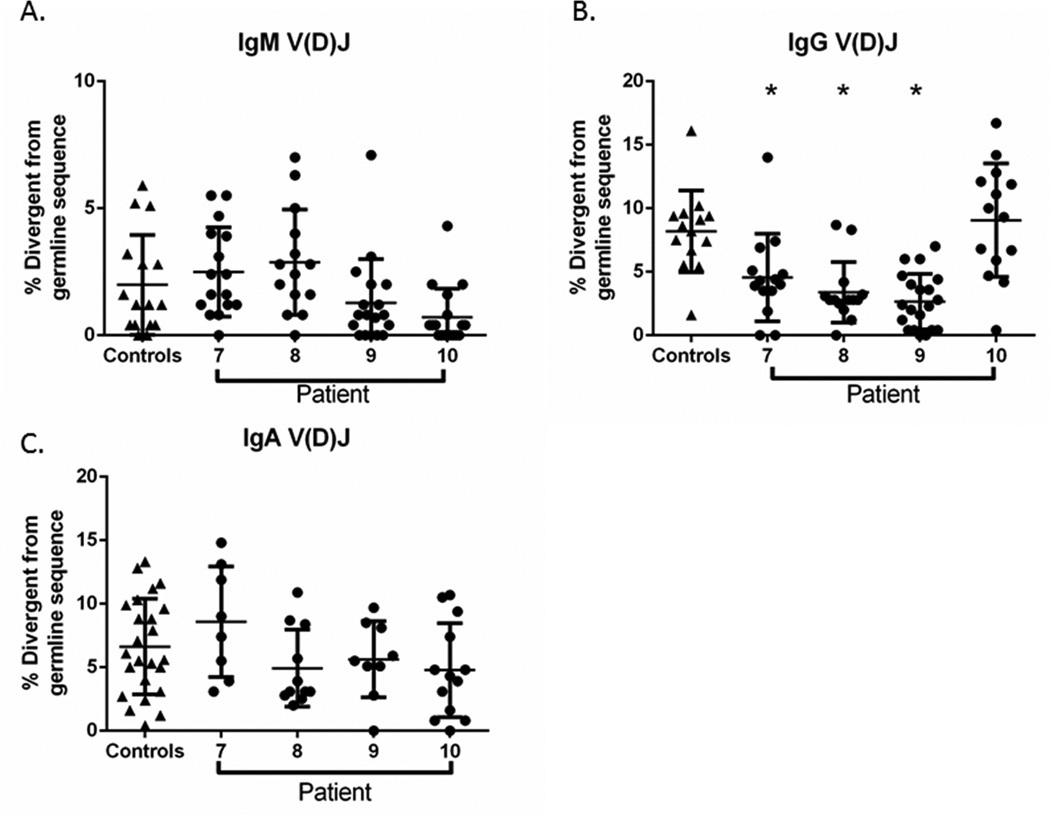
A critical step in terminal B cell differentiation is post-activation SHM of the IGH locus. We therefore quantified the rate of SHM in V(D)J transcripts cloned from KMT2DMut/+ B cells to investigate whether this process was disrupted by KMT2D genetic lesions. We focused our analysis on our 4 adult KMT2DMut/+ patients (7–10) because of the availability of age-matched controls. Using a set of diverse variable gene primers (BioMED-2 FR1) paired with a constant region primer (IgM, pan IgG, or IgA specific), we isolated and sequenced patient-specific V(D)J clones and quantified their divergence from germline sequence. SHM rates for IgM and IgA V(D)J clones from KMT2DMut/+ patients did not significantly differ from rates derived from control subject clones (Figure 3A, B) and were consistent with previously published rates of SMH in adults (44–47). Interestingly, the SHM rate for IgG V(D)J clones was significantly lower in 3 out of 4 KMT2DMut/+ patients when compared to controls clones (p-values <0.003)(Figure 3C). These findings show that a subset of adult patients with KS have diminished IgG affinity maturation in circulating memory B cells. IgG affinity maturation appears to be more susceptible to KMT2D insufficiency than IgA SHM, possibly the result of the divergent cytokine signaling mechanisms and cell trafficking that drive mucosal (IgA) vs systemic (IgG) humoral immunity(48).
Figure 3. Somatic hypermutation (SHM) rate.
Isoform-specific V(D)J clones analyzed in adult KMT2DMut/+ patients(circles) and age-matched adult controls (triangles). Presented as percent (%) divergent from germline IGH sequence. (A,B, C) IgM, IgG and IgA mutations rates in KMT2DMut/+ patients and controls. IgG mutation rates (B) were significantly lower in 3 of 4 KMT2DMut/+ patients than in controls (*p < 0.05). All data (A,B, C) are present as mean ± standard deviation of the mean.
To explore the mechanism driving this reduction in the SHM rate, we next performed mutation analysis on IgG clones derived from KMT2DMut/+ patients (Supplemental Table E3). SHM-associated mutations are targeted to the V(D)J and occur in non-random patterns with certain nucleotide motifs being preferentially targeted (“hotspot”) or ignored (“coldspot”). We observed no differences in the fraction of transition or transversion mutations detected in V(D)J sequences derived from KMT2DMut/+ patients versus from normal controls. Likewise, the percentage of total V(D)J mutations occurring at activation-induced cytidine deaminase (AICDA, also known as AID) “hotspot” and “coldspot” motifs also did not differ between KMT2DMut/+ patients and controls. In contrast, we did observe a trend toward reduced utilization of the preferred polymerase η (eta) motif in KMT2DMut/+ patients (KS average 9.01% vs. Control 15.96%, p = 0.058). This ~40% reduction was seen in all KMT2DMut/+ patients examined, including patient 10 who had a normal overall SHM rate.
In vitro terminal B cell differentiation
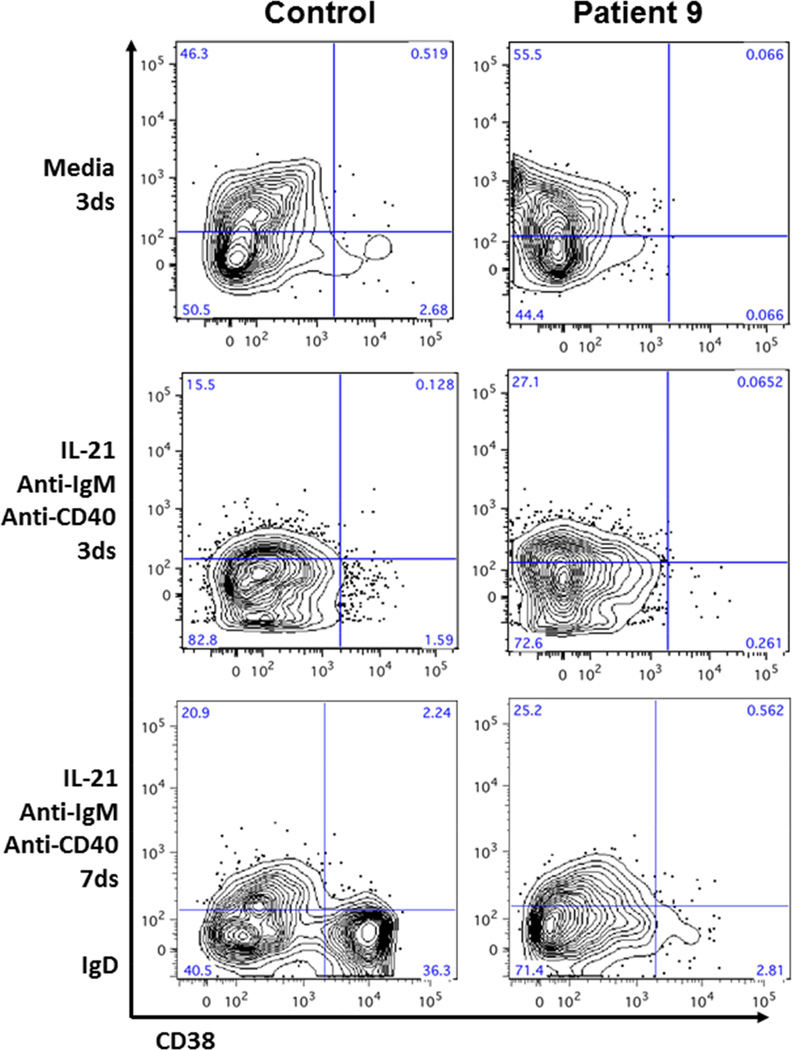
In order to clarify whether the humoral defects seen in many of our patients with KS were the result of intrinsic B cell defects (cell autonomous or nonautonomous), we induced in vitro class-switch recombination in B cells isolated from an adult KMT2DMut/+ patient with hypogammaglobulinemia, but no prior exposure to rituximab or other immune suppressive medication (patient 9). Total B cells were isolated from patient 9 and two age-matched control patients and incubated in media alone or media plus germinal center differentiation factors/stimuli (IL-21, anti-CD40 antibody, anti-IgM antibody). Cells were analyzed after 3 days and 7 days of incubation by flow cytometry. Control cells exposed to the differentiation factors efficiently downregulated IgD and induced CD38 (Figure 4), a marker of memory and plasma cell differentiation (36). B cells derived from patient 9 appeared to initiate normal differentiation at day 3 of culture by downregulating surface IgD but failed to upregulate CD38 on IgD− cells by day 7 of culture (2.8% versus 36.3% IgD−CD38+ in control cells). CD27 was also examined and followed the same pattern as CD38 (data not shown). These findings suggest a specific role for KMT2D signaling during germinal center–mediated terminal differentiation.
Figure 4. In vitro B cell differentiation.
Isolated total B cells cultured in media alone or media plus differentiation factors. Flow cytometry performed on day 3 and day 7 of culture. Performed in duplicate with representative flow plots shown.
Discussion
Based on our results and accumulating evidence from multiple model systems (27, 49, 50), we believe the hypogammaglobulinemia, reduced memory B cells, and impaired SHM observed in our KMT2DMut/+ patients are likely the result of disrupted epigenetic regulation of the IGH locus. During B cell terminal differentiation, the IGH locus undergoes a variety of dynamic, stepwise epigenetic modifications. In activated naive mature B cells, H3K4me3 modifications accumulate throughout the V(D)J region and IGH switch regions. These targeted H3K4me3 marks are associated with an open chromatin state, and initiate a cascade of secondary H3 histone modifications that drive AICDA/AID recruitment to the switch regions, and ultimately induce recombination of the IGH locus (51). The V(D)J sequence also is subject to similar epigenetic regulation during SHM, with initial H3K4me3 accumulation at the V(D)J coding sequence leading to secondary histone modifications, recruitment of AICDA/AID and the error-prone DNA polymerase-η (pol-η), and subsequent mutation-driven affinity maturation of the antibody (49). Impaired H3K4 trimethylation of the IGH locus reduces both CSR and SHM efficiency (27, 50), linking these independent, but simultaneous processes to epigenetic regulatory mechanisms. These findings are highly relevant to KS-associated immune dysfunction since KMT2D catalyzes H3K4 methylation at the IGH locus by binding it’s COMPASS cofactor PTIP (20), therefore supporting a direct link between KMT2D function and CSR/SHM (Supplemental Figure E5).
The CVID-like phenotype observed in our KMT2DMut/+ patients implicates altered epigenetic signaling as an etiology for human B cell dysfunction. Multiple knockout murine models have shown that a variety of chromatin modifier genes (Ezh2, Suv39h1, Spt6) have important roles in terminal B cell differentiation, CSR, and SHM (27–29). Future investigations into the etiology of CVID may benefit from closer inspection of B cell epigenetic regulation. Other important aspects of B cell differentiation may also be affected in some KMT2DMut/+ patients, such as impaired deletion of autoreactive B cell clones. One intriguing observation is that all three of our KMT2DMut/+ patients with autoimmune disease were found to have missense point mutations in the terminal carboxyl end of KMT2D gene. Of the two other KMT2DMut/+ patients reported in the literature to have developed ITP, one patient exhibited a highly similar missense mutation in the same gene region (exon 52, c.16384G>C, p.D5462H) (26). A recent enzymatic study of the KMT2D pre-SET domain region modeled five previously reported KS missense mutations and found that all clustered onto a solvent-exposed surface termed the KIS (39). These mutations interfered with KMT2D’s formation of a second active site with critical protein cofactors rendering the protein only capable of performing histone monomethylation. Additional studies, perhaps using animal models that recapitulate human genetic lesions, are needed to clarify the in vivo effects of specific KMT2D point mutations on the development of autoreactive B cells.
KS is a complex, heterogeneous disease and patients are at increased risk for recurrent sinopulmonary infections and serious autoimmune sequela, as seen in our adult KMT2DMut/+ patients(Supplemental Table E2, A&B). Unfortunately, no disease-specific clinical treatments are currently approved for use in patients with KS, though this may change now that the underlying gene defects have been identified. Several lysine demethylase (KDM) inhibitors have recently been described and may prove useful in directly targeting H3K4 methylation defects (52). B cells are interesting initial targets for therapeutic epigenetic treatment given their lifelong, ongoing differentiation from stem cells, providing a broad developmental window for aberrant epigenetic signaling to be normalized by pharmacologic intervention.
In summary, we show that greater than 80% of KMT2DMut/+ patients have hypogammaglobulinemia and diminished memory B cell populations. In addition, we present evidence associating KMT2D mutations with expansion of the CVID-associated CD21lo B cell population, impaired SHM in IgG transcripts, and disrupted terminal B cell differentiation We also note a clustering of missense mutations in the terminal region of the KMT2D gene and that such mutations may increase the risk for autoimmune disease in KMT2DMut/+ patients. In summary, this report reveals that epigenetic dysregulation of the B cell compartment can lead to clinically relevant primary immune deficiency and autoimmunity in humans.
Supplementary Material
Acknowledgement
Disclosures
A.W.L. received investigator-initiated funding from CSL Behring for this study. AWL also supported by NIH K12 (HD028827) Child Health Research Career Development Award
Abbreviations used
- AICDA
activation-induced cytidine deaminase
- CCHMC
Cincinnati Children’s Hospital Medical Center
- cKS
Clinical Kabuki syndrome (no identifiable KMT2D/KDM6A mutations)
- COMPASS
complex of proteins associated with SET1
- CSR
class-switch recombination
- H3K4
histone 3, lysine 4
- Ig
immunoglobulin
- IGH
immunoglobulin heavy chain locus
- KDM6A
lysine demethylase 6A
- KIS
Kabuki interaction surface
- KMT2D
lysine methyltransferase 2D
- KMT2DMut/+
autosomal dominant mutation in KMT2D
- KS
Kabuki syndrome
- PTIP
paired box (PAX) transactivation domain–interacting protein
- SHM
somatic hypermutation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Niikawa N, Matsuura N, Fukushima Y, Ohsawa T, Kajii T. Kabuki make-up syndrome: a syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. The Journal of pediatrics. 1981;99(4):565–569. doi: 10.1016/s0022-3476(81)80255-7. PubMed PMID: 7277096. [DOI] [PubMed] [Google Scholar]
- 2.Kuroki Y, Suzuki Y, Chyo H, Hata A, Matsui I. A new malformation syndrome of long palpebral fissures, large ears, depressed nasal tip, and skeletal anomalies associated with postnatal dwarfism and mental retardation. The Journal of pediatrics. 1981;99(4):570–573. doi: 10.1016/s0022-3476(81)80256-9. PubMed PMID: 7277097. [DOI] [PubMed] [Google Scholar]
- 3.Matsumoto N, Niikawa N. Kabuki make-up syndrome: a review. American journal of medical genetics Part C, Seminars in medical genetics. 2003;117C(1):57–65. doi: 10.1002/ajmg.c.10020. PubMed PMID: 12561059. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong L, Abd El Moneim A, Aleck K, Aughton DJ, Baumann C, Braddock SR, Gillessen-Kaesbach G, Graham JM, Jr, Grebe TA, Gripp KW, Hall BD, Hennekam R, Hunter A, Keppler-Noreuil K, Lacombe D, Lin AE, Ming JE, Kokitsu-Nakata NM, Nikkel SM, Philip N, Raas-Rothschild A, Sommer A, Verloes A, Walter C, Wieczorek D, Williams MS, Zackai E, Allanson JE. Further delineation of Kabuki syndrome in 48 well-defined new individuals. American journal of medical genetics Part A. 2005;132A(3):265–272. doi: 10.1002/ajmg.a.30340. PubMed PMID: 15690370. [DOI] [PubMed] [Google Scholar]
- 5.Schrander-Stumpel CT, Spruyt L, Curfs LM, Defloor T, Schrander JJ. Kabuki syndrome: Clinical data in 20 patients, literature review, and further guidelines for preventive management. American journal of medical genetics Part A. 2005;132A(3):234–243. doi: 10.1002/ajmg.a.30331. PubMed PMID: 15690368. [DOI] [PubMed] [Google Scholar]
- 6.Kawame H, Hannibal MC, Hudgins L, Pagon RA. Phenotypic spectrum and management issues in Kabuki syndrome. The Journal of pediatrics. 1999;134(4):480–485. doi: 10.1016/s0022-3476(99)70207-6. PubMed PMID: 10190924. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman JD, Ciprero KL, Sullivan KE, Kaplan PB, McDonald-McGinn DM, Zackai EH, Ming JE. Immune abnormalities are a frequent manifestation of Kabuki syndrome. American journal of medical genetics Part A. 2005;135(3):278–281. doi: 10.1002/ajmg.a.30722. PubMed PMID: 15887282. [DOI] [PubMed] [Google Scholar]
- 8.Ming JE, Russell KL, McDonald-McGinn DM, Zackai EH. Autoimmune disorders in Kabuki syndrome. American journal of medical genetics Part A. 2005;132A(3):260–262. doi: 10.1002/ajmg.a.30332. PubMed PMID: 15523604. [DOI] [PubMed] [Google Scholar]
- 9.Bogershausen N, Wollnik B. Unmasking Kabuki syndrome. Clinical genetics. 2013;83(3):201–211. doi: 10.1111/cge.12051. PubMed PMID: 23131014. [DOI] [PubMed] [Google Scholar]
- 10.Ng SB, Bigham AW, Buckingham KJ, Hannibal MC, McMillin MJ, Gildersleeve HI, Beck AE, Tabor HK, Cooper GM, Mefford HC, Lee C, Turner EH, Smith JD, Rieder MJ, Yoshiura K, Matsumoto N, Ohta T, Niikawa N, Nickerson DA, Bamshad MJ, Shendure J. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nature genetics. 2010;42(9):790–793. doi: 10.1038/ng.646. PubMed PMID: 20711175; PubMed Central PMCID: PMC2930028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lederer D, Grisart B, Digilio MC, Benoit V, Crespin M, Ghariani SC, Maystadt I, Dallapiccola B, Verellen-Dumoulin C. Deletion of KDM6A, a histone demethylase interacting with MLL2, in three patients with Kabuki syndrome. American journal of human genetics. 2012;90(1):119–124. doi: 10.1016/j.ajhg.2011.11.021. PubMed PMID: 22197486; PubMed Central PMCID: PMC3257878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Molecular and cellular biology. 2007;27(5):1889–1903. doi: 10.1128/MCB.01506-06. PubMed PMID: 17178841; PubMed Central PMCID: PMC1820476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ansari KI, Mandal SS. Mixed lineage leukemia: roles in gene expression, hormone signaling and mRNA processing. The FEBS journal. 2010;277(8):1790–1804. doi: 10.1111/j.1742-4658.2010.07606.x. PubMed PMID: 20236313. [DOI] [PubMed] [Google Scholar]
- 14.Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes & development. 2011;25(7):661–672. doi: 10.1101/gad.2015411. PubMed PMID: 21460034; PubMed Central PMCID: PMC3070929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. PubMed PMID: 17761849. [DOI] [PubMed] [Google Scholar]
- 16.Guo C, Chang CC, Wortham M, Chen LH, Kernagis DN, Qin X, Cho YW, Chi JT, Grant GA, McLendon RE, Yan H, Ge K, Papadopoulos N, Bigner DD, He Y. Global identification of MLL2-targeted loci reveals MLL2's role in diverse signaling pathways. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(43):17603–17608. doi: 10.1073/pnas.1208807109. PubMed PMID: 23045699; PubMed Central PMCID: PMC3491484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu D, Gao X, Morgan MA, Herz HM, Smith ER, Shilatifard A. The MLL3/MLL4 branches of the COMPASS family function as major histone H3K4 monomethylases at enhancers. Molecular and cellular biology. 2013;33(23):4745–4754. doi: 10.1128/MCB.01181-13. PubMed PMID: 24081332; PubMed Central PMCID: PMC3838007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, Ge K. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. The Journal of biological chemistry. 2007;282(28):20395–20406. doi: 10.1074/jbc.M701574200. PubMed PMID: 17500065; PubMed Central PMCID: PMC2729684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwab KR, Patel SR, Dressler GR. Role of PTIP in class switch recombination and long-range chromatin interactions at the immunoglobulin heavy chain locus. Molecular and cellular biology. 2011;31(7):1503–1511. doi: 10.1128/MCB.00990-10. PubMed PMID: 21282469; PubMed Central PMCID: PMC3135287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daniel JA, Santos MA, Wang Z, Zang C, Schwab KR, Jankovic M, Filsuf D, Chen HT, Gazumyan A, Yamane A, Cho YW, Sun HW, Ge K, Peng W, Nussenzweig MC, Casellas R, Dressler GR, Zhao K, Nussenzweig A. PTIP promotes chromatin changes critical for immunoglobulin class switch recombination. Science. 2010;329(5994):917–923. doi: 10.1126/science.1187942. PubMed PMID: 20671152; PubMed Central PMCID: PMC3008398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Bogershausen N, Alanay Y, Simsek Kiper PO, Plume N, Keupp K, Pohl E, Pawlik B, Rachwalski M, Milz E, Thoenes M, Albrecht B, Prott EC, Lehmkuhler M, Demuth S, Utine GE, Boduroglu K, Frankenbusch K, Borck G, Gillessen-Kaesbach G, Yigit G, Wieczorek D, Wollnik B. A mutation screen in patients with Kabuki syndrome. Human genetics. 2011;130(6):715–724. doi: 10.1007/s00439-011-1004-y. PubMed PMID: 21607748. [DOI] [PubMed] [Google Scholar]
- 22.Micale L, Augello B, Fusco C, Selicorni A, Loviglio MN, Silengo MC, Reymond A, Gumiero B, Zucchetti F, D'Addetta EV, Belligni E, Calcagni A, Digilio MC, Dallapiccola B, Faravelli F, Forzano F, Accadia M, Bonfante A, Clementi M, Daolio C, Douzgou S, Ferrari P, Fischetto R, Garavelli L, Lapi E, Mattina T, Melis D, Patricelli MG, Priolo M, Prontera P, Renieri A, Mencarelli MA, Scarano G, della Monica M, Toschi B, Turolla L, Vancini A, Zatterale A, Gabrielli O, Zelante L, Merla G. Mutation spectrum of MLL2 in a cohort of Kabuki syndrome patients. Orphanet journal of rare diseases. 2011;6:38. doi: 10.1186/1750-1172-6-38. PubMed PMID: 21658225; PubMed Central PMCID: PMC3141365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banka S, Veeramachaneni R, Reardon W, Howard E, Bunstone S, Ragge N, Parker MJ, Crow YJ, Kerr B, Kingston H, Metcalfe K, Chandler K, Magee A, Stewart F, McConnell VP, Donnelly DE, Berland S, Houge G, Morton JE, Oley C, Revencu N, Park SM, Davies SJ, Fry AE, Lynch SA, Gill H, Schweiger S, Lam WW, Tolmie J, Mohammed SN, Hobson E, Smith A, Blyth M, Bennett C, Vasudevan PC, Garcia-Minaur S, Henderson A, Goodship J, Wright MJ, Fisher R, Gibbons R, Price SM, D CdS, Temple IK, Collins AL, Lachlan K, Elmslie F, McEntagart M, Castle B, Clayton-Smith J, Black GC, Donnai D. How genetically heterogeneous is Kabuki syndrome?: MLL2 testing in 116 patients, review and analyses of mutation and phenotypic spectrum. European journal of human genetics : EJHG. 2012;20(4):381–388. doi: 10.1038/ejhg.2011.220. PubMed PMID: 22126750; PubMed Central PMCID: PMC3306863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin JL, Lee WI, Huang JL, Chen PK, Chan KC, Lo LJ, You YJ, Shih YF, Tseng TY, Wu MC. Immunologic assessment and KMT2D mutation detection in Kabuki syndrome. Clinical genetics. 2014 doi: 10.1111/cge.12484. PubMed PMID: 25142838. [DOI] [PubMed] [Google Scholar]
- 25.Brackmann F, Krumbholz M, Langer T, Rascher W, Holter W, Metzler M. Novel MLL2 mutation in Kabuki syndrome with hypogammaglobulinemia and severe chronic thrombopenia. Journal of pediatric hematology/oncology. 2013;35(7):e314–e316. doi: 10.1097/MPH.0b013e3182707fa8. PubMed PMID: 23042018. [DOI] [PubMed] [Google Scholar]
- 26.Giordano P, Lassandro G, Sangerardi M, Faienza MF, Valente F, Martire B. Autoimmune haematological disorders in two Italian children with Kabuki syndrome. Italian journal of pediatrics. 2014;40:10. doi: 10.1186/1824-7288-40-10. PubMed PMID: 24460868; PubMed Central PMCID: PMC3917534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Begum NA, Stanlie A, Nakata M, Akiyama H, Honjo T. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation. The Journal of biological chemistry. 2012;287(39):32415–32429. doi: 10.1074/jbc.M112.351569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradley SP, Kaminski DA, Peters AH, Jenuwein T, Stavnezer J. The histone methyltransferase Suv39h1 increases class switch recombination specifically to IgA. Journal of immunology. 2006;177(2):1179–1188. doi: 10.4049/jimmunol.177.2.1179. PubMed PMID: 16818776. [DOI] [PubMed] [Google Scholar]
- 29.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, Shen H, Yang SN, Wang L, Ezponda T, Martinez-Garcia E, Zhang H, Zheng Y, Verma SK, McCabe MT, Ott HM, Van Aller GS, Kruger RG, Liu Y, McHugh CF, Scott DW, Chung YR, Kelleher N, Shaknovich R, Creasy CL, Gascoyne RD, Wong KK, Cerchietti L, Levine RL, Abdel-Wahab O, Licht JD, Elemento O, Melnick AM. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer cell. 2013;23(5):677–692. doi: 10.1016/j.ccr.2013.04.011. PubMed PMID: 23680150; PubMed Central PMCID: PMC3681809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reese MG, Eeckman FH, Kulp D, Haussler D. Improved splice site detection in Genie. Journal of computational biology : a journal of computational molecular cell biology. 1997;4(3):311–323. doi: 10.1089/cmb.1997.4.311. PubMed PMID: 9278062. [DOI] [PubMed] [Google Scholar]
- 31.van Dongen JJ, Langerak AW, Bruggemann M, Evans PA, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JH, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, Smith JL, Morgan GJ, Kneba M, Macintyre EA. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17(12):2257–2317. doi: 10.1038/sj.leu.2403202. PubMed PMID: 14671650. [DOI] [PubMed] [Google Scholar]
- 32.Berkowska MA, Driessen GJ, Bikos V, Grosserichter-Wagener C, Stamatopoulos K, Cerutti A, He B, Biermann K, Lange JF, van der Burg M, van Dongen JJ, van Zelm MC. Human memory B cells originate from three distinct germinal center-dependent and -independent maturation pathways. Blood. 2011;118(8):2150–2158. doi: 10.1182/blood-2011-04-345579. PubMed PMID: 21690558; PubMed Central PMCID: PMC3342861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruminy P, Etancelin P, Couronne L, Parmentier F, Rainville V, Mareschal S, Bohers E, Burgot C, Cornic M, Bertrand P, Lenormand B, Picquenot JM, Jardin F, Tilly H, Bastard C. The isotype of the BCR as a surrogate for the GCB and ABC molecular subtypes in diffuse large B-cell lymphoma. Leukemia. 2011;25(4):681–688. doi: 10.1038/leu.2010.302. PubMed PMID: 21233831. [DOI] [PubMed] [Google Scholar]
- 34.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic acids research. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. (Web Server issue) PubMed PMID: 23671333; PubMed Central PMCID: PMC3692102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maccarthy T, Roa S, Scharff MD, Bergman A. SHMTool: a webserver for comparative analysis of somatic hypermutation datasets. DNA repair. 2009;8(1):137–141. doi: 10.1016/j.dnarep.2008.09.006. PubMed PMID: 18952008; PubMed Central PMCID: PMC2659805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ettinger R, Sims GP, Fairhurst AM, Robbins R, da Silva YS, Spolski R, Leonard WJ, Lipsky PE. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. Journal of immunology. 2005;175(12):7867–7879. doi: 10.4049/jimmunol.175.12.7867. PubMed PMID: 16339522. [DOI] [PubMed] [Google Scholar]
- 37.Niikawa N, Kuroki Y, Kajii T, Matsuura N, Ishikiriyama S, Tonoki H, Ishikawa N, Yamada Y, Fujita M, Umemoto H, et al. Kabuki make-up (Niikawa-Kuroki) syndrome: a study of 62 patients. American journal of medical genetics. 1988;31(3):565–589. doi: 10.1002/ajmg.1320310312. PubMed PMID: 3067577. [DOI] [PubMed] [Google Scholar]
- 38.Micale L, Augello B, Maffeo C, Selicorni A, Zucchetti F, Fusco C, De Nittis P, Pellico MT, Mandriani B, Fischetto R, Boccone L, Silengo M, Biamino E, Perria C, Sotgiu S, Serra G, Lapi E, Neri M, Ferlini A, Cavaliere ML, Chiurazzi P, Monica MD, Scarano G, Faravelli F, Ferrari P, Mazzanti L, Pilotta A, Patricelli MG, Bedeschi MF, Benedicenti F, Prontera P, Toschi B, Salviati L, Melis D, Di Battista E, Vancini A, Garavelli L, Zelante L, Merla G. Molecular analysis, pathogenic mechanisms, and readthrough therapy on a large cohort of Kabuki syndrome patients. Human mutation. 2014;35(7):841–850. doi: 10.1002/humu.22547. PubMed PMID: 24633898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinsky SA, Hu M, Vought VE, Ng SB, Bamshad MJ, Shendure J, Cosgrove MS. A non-active-site SET domain surface crucial for the interaction of MLL1 and the RbBP5/Ash2L heterodimer within MLL family core complexes. Journal of molecular biology. 2014;426(12):2283–2299. doi: 10.1016/j.jmb.2014.03.011. PubMed PMID: 24680668; PubMed Central PMCID: PMC4066448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cunningham-Rundles C. Autoimmune manifestations in common variable immunodeficiency. Journal of clinical immunology. 2008;28(Suppl 1):S42–S45. doi: 10.1007/s10875-008-9182-7. PubMed PMID: 18322785; PubMed Central PMCID: PMC2694614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comans-Bitter WM, de Groot R, van den Beemd R, Neijens HJ, Hop WC, Groeneveld K, Hooijkaas H, van Dongen JJ. Immunophenotyping of blood lymphocytes in childhood. Reference values for lymphocyte subpopulations. The Journal of pediatrics. 1997;130(3):388–393. doi: 10.1016/s0022-3476(97)70200-2. PubMed PMID: 9063413. [DOI] [PubMed] [Google Scholar]
- 42.Orange JS, Ballow M, Stiehm ER, Ballas ZK, Chinen J, De La Morena M, Kumararatne D, Harville TO, Hesterberg P, Koleilat M, McGhee S, Perez EE, Raasch J, Scherzer R, Schroeder H, Seroogy C, Huissoon A, Sorensen RU, Katial R. Use and interpretation of diagnostic vaccination in primary immunodeficiency: a working group report of the Basic and Clinical Immunology Interest Section of the American Academy of Allergy, Asthma & Immunology. The Journal of allergy and clinical immunology. 2012;130(3 Suppl):S1–S24. doi: 10.1016/j.jaci.2012.07.002. PubMed PMID: 22935624. [DOI] [PubMed] [Google Scholar]
- 43.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, Vlkova M, Hernandez M, Detkova D, Bos PR, Poerksen G, von Bernuth H, Baumann U, Goldacker S, Gutenberger S, Schlesier M, Bergeron-van der Cruyssen F, Le Garff M, Debre P, Jacobs R, Jones J, Bateman E, Litzman J, van Hagen PM, Plebani A, Schmidt RE, Thon V, Quinti I, Espanol T, Webster AD, Chapel H, Vihinen M, Oksenhendler E, Peter HH, Warnatz K. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111(1):77–85. doi: 10.1182/blood-2007-06-091744. PubMed PMID: 17898316. [DOI] [PubMed] [Google Scholar]
- 44.Weill JC, Weller S, Reynaud CA. Human marginal zone B cells. Annual review of immunology. 2009;27:267–285. doi: 10.1146/annurev.immunol.021908.132607. PubMed PMID: 19302041. [DOI] [PubMed] [Google Scholar]
- 45.Fecteau JF, Cote G, Neron S. A new memory CD27-IgG+ B cell population in peripheral blood expressing VH genes with low frequency of somatic mutation. Journal of immunology. 2006;177(6):3728–3736. doi: 10.4049/jimmunol.177.6.3728. PubMed PMID: 16951333. [DOI] [PubMed] [Google Scholar]
- 46.Chong Y, Ikematsu H, Yamaji K, Nishimura M, Kashiwagi S, Hayashi J. Age-related accumulation of Ig V(H) gene somatic mutations in peripheral B cells from aged humans. Clinical and experimental immunology. 2003;133(1):59–66. doi: 10.1046/j.1365-2249.2003.02185.x. PubMed PMID: 12823279; PubMed Central PMCID: PMC1808746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Klein U, Kuppers R, Rajewsky K. Evidence for a large compartment of IgM-expressing memory B cells in humans. Blood. 1997;89(4):1288–1298. PubMed PMID: 9028952. [PubMed] [Google Scholar]
- 48.Bemark M, Boysen P, Lycke NY. Induction of gut IgA production through T cell-dependent and T cell-independent pathways. Annals of the New York Academy of Sciences. 2012;1247:97–116. doi: 10.1111/j.1749-6632.2011.06378.x. PubMed PMID: 22260403. [DOI] [PubMed] [Google Scholar]
- 49.Borchert GM, Holton NW, Edwards KA, Vogel LA, Larson ED. Histone H2A and H2B are monoubiquitinated at AID-targeted loci. PloS one. 2010;5(7):e11641. doi: 10.1371/journal.pone.0011641. PubMed PMID: 20661291; PubMed Central PMCID: PMC2905439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stanlie A, Aida M, Muramatsu M, Honjo T, Begum NA. Histone3 lysine4 trimethylation regulated by the facilitates chromatin transcription complex is critical for DNA cleavage in class switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(51):22190–22195. doi: 10.1073/pnas.1016923108. PubMed PMID: 21139053; PubMed Central PMCID: PMC3009800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li G, Zan H, Xu Z, Casali P. Epigenetics of the antibody response. Trends in immunology. 2013;34(9):460–470. doi: 10.1016/j.it.2013.03.006. PubMed PMID: 23643790; PubMed Central PMCID: PMC3744588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Helin K, Dhanak D. Chromatin proteins and modifications as drug targets. Nature. 2013;502(7472):480–488. doi: 10.1038/nature12751. PubMed PMID: 24153301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.