Abstract
Leiomyosarcoma (LMS) belongs to the class of genetically complex sarcomas and shows numerous, often non-recurrent chromosomal imbalances and aberrations. We investigated a group of LMS using NGS platform to identify recurrent genetic abnormalities and possible therapeutic targets. Targeted exome sequencing of 230 cancer-associated genes was performed on 35 primary soft tissue and visceral (extra-uterine) LMS. Sequence data were analyzed to identify single nucleotide variants, small insertions/deletions (indels), and copy number alterations. Key alterations were further investigated using FISH assay. The study group included patients with median age of 64 years and median tumor size of 7 cm. The primary sites included retroperitoneal/intra-abdominal, extremity, truncal and visceral. Thirty one tumors were high grade LMS, while 4 were low grade. Losses of chromosomal regions involving key tumor suppressor genes PTEN (10q), RB1 (13q), CDH1 (16q) and TP53 (17p) were the most frequent genetic events. Gains mainly involved chromosome regions 17p11.2 (MYOCD) and 15q25-26 (IGF1R). The most frequent mutations were identified in the TP53 gene in 13 of 35 (37%) cases. FISH analysis showed amplification of the myocardin (MYOCD) gene in 5 of 25 (20%) cases analyzed. None of the four low grade LMS showed losses or mutations of PTEN or TP53 genes. Genetic complexity is the hallmark of LMS with losses of important tumor suppressor genes being a common feature. MYOCD, a key gene associated with smooth muscle differentiation, is amplified in a subset of both retroperitoneal and extremity LMS. Further studies are necessary to investigate the significance of gains/amplifications in the development of these tumors.
Keywords: Leiomyosarcoma, TP53, Myocardin, PTEN, RB1
INTRODUCTION
Leiomyosarcomas (LMS) are one of the most common sarcomas, comprising about 15 - 25% of all histologic subtypes. These malignant neoplasms may involve any body part, including skin, soft tissue, bone, and visceral organs such as uterus, urinary bladder, etc. In the soft tissue, extra-uterine LMS comprise about 10-12% of the soft tissue sarcomas seen (Toro et al., 2006). LMS are more common in the middle aged and elderly population and are infrequent in children. Soft tissue LMS present as large lesions usually seen in the extremities or the retroperitoneal location (Weiss, 2002; Miettinen and Fetsch, 2006). Retroperitoneal LMS usually present with a larger size than the LMS of the extremities. The morphologic criteria that have been used to diagnose and grade LMS include cellular atypia, increased mitotic activity and presence of necrosis (Weiss, 2002). Clinically, they are aggressive tumors with a metastatic rate of 40-45% and, with current modes of therapy, a 5-year survival rate of 65 to 80 % dependent on tumor location, size and grade (Svarvar et al., 2007; Gladdy et al., 2013). In a recent study by Gladdy et al., tumor size and grade were the only independent predictors of disease specific survival. This study also showed that a large fraction of the patients with tumors in the abdominal / retroperitoneal location show late local and distant recurrences after 5 years (Gladdy et al., 2013).
LMS belong to the class of sarcomas with complex genomic alterations characterized by non- recurrent structural and copy number alterations (Guillou and Aurias, 2010). Cytogenetic studies have shown that the loss of 1p12, 2p, 13q, 10q and 16q are most frequent and gains are seen in chromosomes 17p, 8q, and 5p. The 5p region is frequently amplified in LMS. The 10q and 13q region losses most likely affect the tumor suppressor genes PTEN and Rb (Yang et al., 2009; Guillou and Aurias, 2010).
Chromosome arm 17p amplification is one of the most frequent amplifications seen in LMS (Larramendy et al., 2006). The target of this amplification is a gene encoding myocardin (MYOCD), a transcriptional cofactor of serum response factor (SRF) regulating smooth muscle development and differentiation. Perot et al. reported that the MYOCD gene is highly amplified and overexpressed in retroperitoneal LMS (Perot et al., 2009). This study showed that MYOCD induces smooth muscle differentiation and promotes cell migration. In another study, MYOCD overexpression induced cell phenotypic switch in a human uterine LMS cell line from a dedifferentiated to a differentiated phenotype (Kimura et al., 2010).
In this study, we investigate a cohort of primary soft tissue LMS, using next generation sequencing methodology, to determine recurrent genetic events and potential site-specific alterations.
MATERIAL AND METHODS
The soft tissue sarcoma database at MSKCC was searched for primary soft tissue and visceral (extra-uterine) leiomyosarcoma. The H&E and immunostained slides were retrieved from the pathology files and were reviewed to confirm the diagnosis. Diagnostic criteria included tumors that showed immunohistochemical staining for smooth muscle actin and desmin. Tumors that had adequate frozen tissue and matched normal tissue were selected for the study. The study was approved by the Institutional Review Board 02-060.
Targeted Exome Sequencing
We profiled genomic alterations in key cancer-associated genes using the IMPACT assay (Integrated Mutation Profiling of Actionable Cancer Targets), which utilizes solution phase hybridization-based exon capture and deep-coverage massively parallel DNA sequencing.(Won et al., 2013; Cheng et al., 2015) Custom oligonucleotides were designed to capture all protein-coding exons and select introns of commonly implicated oncogenes, tumor suppressor genes, and members of pathways deemed actionable by targeted therapies. Tumors and patient-matched normal tissue were run in parallel for every case. Samples in this project were analyzed on two different versions of IMPACT. 30 cases (60 tumor/normal matched samples) were captured using probes corresponding to 230 cancer genes (Agilent Technologies, SureSelect custom panel). 5 cases (10 tumor/normal matched samples) were captured using probes corresponding to an expanded set of 275 cancer genes (Nimblegen SeqCap custom panel), including the 230 genes used in the first version. For both capture platforms, we first prepared barcoded sequence libraries according to the manufacturers’ protocols (New England Biolabs, Kapa Biosystems) using 500 ng of genomic DNA as input. Libraries were pooled at equimolar concentrations (100 ng per library) and input to a single exon capture reaction as previously described (Wagle et al., 2012; Won et al., 2013). To prevent off-target hybridization, we spiked in a pool of blocker oligonucleotides complementary to the full sequences of all barcoded adaptors to a final total concentration of 10 micromolar. DNA was subsequently sequenced on an Illumina HiSeq 2000 to generate paired-end 75-bp reads. Sequence data were demultiplexed using CASAVA, and reads were aligned to the reference human genome (hg19) using the Burrows-Wheeler Alignment tool (Li and Durbin, 2009). Local realignment and quality score recalibration were performed using the Genome Analysis Toolkit (GATK) according to GATK best practices (DePristo et al., 2011). We achieved mean unique sequence coverage of 377X per sample. Sequence data were analyzed to identify three classes of somatic alterations: single-nucleotide variants, small insertions/deletions (indels), and copy number alterations. Single-nucleotide variants and indels were called using muTect and SomaticIndelDetector, respectively (DePristo et al., 2011; Cibulskis et al., 2013). The mean sequence coverage was calculated using the DepthOfCoverage tool in GATK and was used to compute copy number as described previously (Wagle et al., 2012; Cheng et al., 2015). Increases and decreases in the coverage ratios (tumor: normal) were used to infer amplifications and deletions, respectively.
Fluorescence In Situ Hybridization (FISH)
FISH on interphase nuclei from paraffin-embedded 4-micron sections was performed applying custom probes using bacterial artificial chromosomes (BAC), covering and flanking genes that were identified. BAC clones were chosen according to USCS genome browser (http://genome.ucsc.edu), see Supplementary Table 1. The BAC clones were obtained from BACPAC sources of Children’s Hospital of Oakland Research Institute (CHORI) (Oakland, CA) (http://bacpac.chori.org). DNA from individual BACs was isolated according to the manufacturer’s instructions, labeled with different fluorochromes in a nick translation reaction, denatured, and hybridized to pretreated slides. Slides were then incubated, washed, and mounted with DAPI in an antifade solution, as previously described (Antonescu et al., 2010). The genomic location of each BAC set was verified by hybridizing them to normal metaphase chromosomes. Two hundred successive nuclei were examined using a Zeiss fluorescence microscope (Zeiss Axioplan, Oberkochen, Germany), controlled by Isis 5 software (Metasystems, Newton, MA).
Statistical Analysis
Disease-specific survival (DSS) and distant-recurrence-free survival (DRFS) were the clinical endpoints. Deaths confirmed to be caused by the disease (7 among the 35 patients) were treated as events for DSS; other deaths (2/35) were considered censored observations. The time to disease-specific death was defined as the time from operation until death. Distant recurrences (17/35) were treated as events for DRFS; deaths without DR (2/35) were considered censored observations. The time to distant recurrence were defined as the time from operation until date of distant recurrence. The clinicopathologic variables examined include tumor grade, size, location, and genomic abnormalities. The DSS and DRFS probabilities were estimated using the Kaplan-Meier method. Their associations with clinicopathologic variables were examined using the log-rank test for categorical variables and the Cox-regression score test for continuous variables.
RESULTS
Pathologic Features
Thirty-five patients with primary soft tissue and visceral (extra-uterine) LMS were selected for the study. (Supplementary Table 2) Tumors occurred in 15 females and 20 males with ages ranging from 37 to 84 years (mean, 61 years; median, 64 years). The primary sites of the tumors selected included: 17 from retroperitoneal/intra-abdominal location, eight from the lower extremity, four from the thorax/trunk, three from gastrointestinal viscera and two from the genitourinary viscera. Tumors ranged in size from 2.2-37 cm (mean, 9.9 cm; median, 7 cm). Histologically, 31 tumors were characterized as high grade, displaying moderate to severe cytologic atypia, increased mitotic activity and presence of necrosis. Four of the tumors were low grade LMS with mild atypia, low mitotic activity and absence of necrosis.
Mutations in the TP53 Gene are a Frequent Abnormality in High Grade LMS
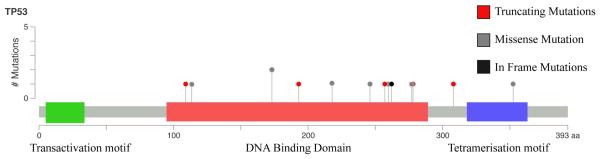
Thirteen of the 35 cases (37%) showed presence of TP53 mutations. (Fig. 1) In two of the cases, an additional TP53 alteration was identified. Overall, five frameshift mutations, nine missense mutations and one in-frame deletion were noted. (Fig. 2) Six of the cases with TP53 mutations also showed concurrent deletions of the TP53 gene. Apart from TP53, the gene with the most common abnormalities was RB1, with mutations being identified in three cases (8.5 %) (two frameshift insertions and one frameshift deletion mutation). Two of the three cases also showed deletions of the RB1 gene. All other gene mutations identified were non-recurrent and limited to one or two cases (Supplementary Table 3). No gene mutations were identified in any of the four low grade LMS.
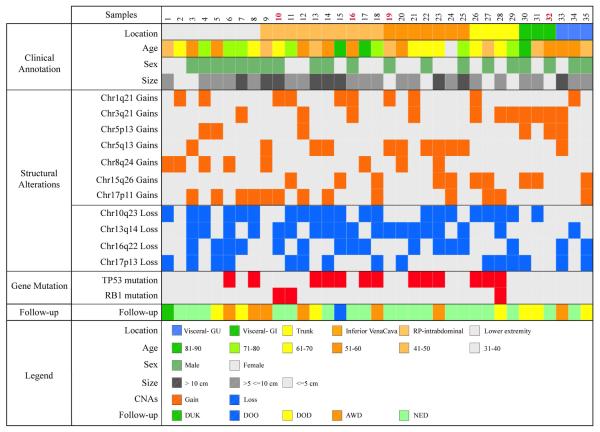
Figure 1.
A comprehensive profile of the genomic alterations in LMS and associated clinical information with samples sorted by location of tumor. Low grade LMS cases are identified in red font. Copy number alterations were identified in all of the cases. Chromosomal losses were more frequently identified than gains and were most frequently seen in regions of known tumor suppressor genes, such as PTEN (10q23), RB1 (13q14), and TP53 (17p13). An additional region with frequent losses identified was in the gene CDH1 (16q22.1). Gains were most frequently seen in chromosome regions 17p, 15q, 8q, 1q and 3q. The most frequent gains were seen in the 17p11.2 region.
Figure 2.
Schematic representation of the TP53 gene showing the sites of mutations identified. Thirteen of the 35 cases (37%) showed presence of TP53 mutations. Overall, five frameshift mutations, 9 missense mutations and 1 in-frame deletion were noted.
Copy Number Alterations are Frequently Identified in LMS
Copy number alterations were identified in all of the cases. (Fig. 1) Chromosomal losses were more frequently identified than gains and were most frequently seen in regions of known tumor suppressor genes, such as PTEN (21 of 35, 60%), RB1 (19 of 35, 54%), and TP53 (15 of 35, 43%). An additional region with frequent losses identified was in the gene CDH1 (16 of 35, 46%). Losses of multiple tumor suppressor genes was a frequent event with 27 of the 35 (77%) cases showing losses in more than one of the above four genes. Among the low grade LMS, three of the four cases showed hemizygous losses in the RB1 gene, of which two had concurrent losses of the CDH1 gene. None of the four cases showed losses of the PTEN and / or TP53 genes.
Gains were most frequently seen in chromosome bands 17p11, 15q26, 8q24, 1q21 and 3q21. The 17p11.2 region showed the most frequent gains, identified in 12 of the 35 cases (34%). The genes in this region that were part of the gene panel included FLCN (folliculin) and MAP2K4 (mitogen-activated protein kinase 4). (Fig. 3) In four of these cases gains were also seen in ALOX12B (arachidonate 12-lipoxygenase, 12R type), a gene located in 17p13.1 indicating that in some cases the gains are seen in the amplicon extending from 17p11.2 to 17p13.1.
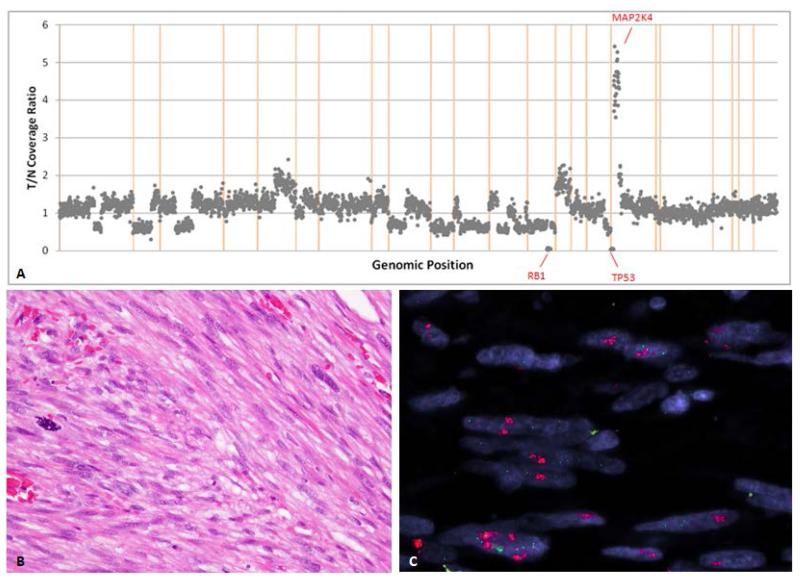
Figure 3.
Schematic representation of copy number alterations in Case 3 (A) showing gains of 17p11-13 and losses of 10q, 13q, 16q and 17p. (B) H&E image showing typical morphology of the tumor composed of spindle cells in fascicles with cytologic atypia and mitotic activity. (C) FISH for myocardin (MYOCD) gene showing amplification (red signal).
Gains in the 15q24-26 region involving the IGF1R and the NTRK3 genes were identified in 10 of the 35 cases. FISH for IGF1R, performed on these 10 cases showed amplification in only two cases.
MYOCD Amplification is Seen in a Small Subset of LMS
MYOCD, a gene which has been previously shown to be amplified in LMS (Perot et al., 2009), is located approximately 600kb from the 17p11.2 region of gain, that spans MAP2K4. MYOCD was not included among the genes that were sequenced on the NGS platform. Thus, we have investigated the presence of MYOCD gene amplifications using gene-specific BAC probes by FISH in 25 of the 35 LMS cases of the study, including all 12 LMS with 17p11.2 gains by IMPACT. MYOCD amplification was identified in five cases including two retroperitoneal/ intraabdominal cases and three cases from the lower extremity. (Fig. 3) In addition, we have tested 15 other high grade LMS from various anatomic sites that are not part of the study and none of the cases showed MYOCD amplification. Thus, in our experience, the incidence of MYOCD gene amplification is 13%.
Follow-up and Correlation of Genetic Alterations with Clinical Parameters
Clinical follow-up was available on all 35 cases ranging from 2-144 months (median, 62 months). At last follow-up, 15 had no evidence of disease (NED), five were alive with disease (AWD), 10 had died of disease (DOD), two died of other causes (DOO) and two patients died of unknown causes (DUK).
The most frequent genetic alterations identified (mutations of TP53, gains of 17p, losses of 10q, 13q, 16q and 17p) did not show any correlation with clinical parameters, distant recurrences or survival by statistical analysis. No site-specific genetic alterations were noted.
DISCUSSION
DNA copy number alterations are frequently identified in LMS, which belong to the group of sarcomas with complex genomics. Prior studies, using comparative genomic hybridization, have catalogued the chromosomal abnormalities commonly observed in LMS. The most frequent regions of chromosomal losses reported are 1p12, 2p, 13q, 10q and 16q and the most frequent gains are in chromosome arms 17p, 15q, 8q, and 5p (Hu et al., 2001; Larramendy et al., 2006; Yang et al., 2009; Barretina et al., 2010). Our study, using paired-end deep sequencing methodology confirms this finding, with the most common chromosomal losses being seen in 10q23 (PTEN), 13q14 (RB1), 16q22 (CDH1) and 17p13 (TP53).
PTEN, located in 10q23, is one of the most frequently involved genes in LMS. Hu et al., in their study of extra-uterine LMS, suggest that loss of 10q is associated with aggressive behavior (Hu et al., 2005). Cases of intra-abdominal LMS with PTEN mutations have been reported (Saito et al., 2003). PTEN was the most commonly affected gene in our study, with 22 of the 35 cases (63%) showing losses. Hernando et al, recently showed that mice with homozygous deletion of PTEN alleles developed widespread smooth muscle cell hyperplasia and abdominal LMS, with a rapid onset and increased incidence (Hernando et al., 2007). Their study also highlighted the critical role of the AKT-mTOR pathway in smooth muscle transformation and LMS development.
Loss of 13q14 harboring the RB1 gene, along with loss of 10q23, is one of the most frequent losses reported in LMS. In our study, 19 of the 35 cases (54%) showed losses of genes in the 13q14 region. Loss of 16q22 was identified in 16 of the 35 cases (46%) in our study. Although this region harbors the CDH1 gene, the critical tumor suppressor gene at this locus remains uncertain.
TP53 mutations are the most common mutations identified in cancer. The spectrum of TP53 missense mutations is very broad with more than 4,000 different alterations reported (Leroy et al., 2014). Based on our study, recurrent gene mutations are infrequent in LMS. TP53 gene mutations were the most common abnormalities identified in 37% of the cases. This is in keeping with the 39% rate of TP53 mutations reported in a study by Ito et al. (2011) The TP53 genetic alterations in our study included both frameshift alterations and missense mutations. A majority of the mutations were in the DNA-binding domain of the gene. TP53 mutations were seen concurrent with the copy number alterations of the tumor suppressor genes, such as PTEN, RB1 and TP53. The effect of these different missense mutations in LMS is unclear, given that they are often seen in tumors with co-existent genetic abnormalities. It is also uncertain if TP53 mutation, in a tumor with preexisting losses of the TP53 or other tumor suppressor genes, such as PTEN and/or RB1 leads to an aggressive behavior.
Gain of 17p11.2 involving the MAP2K4 and FLCN genes were identified in 12 of the 35 cases. This region also covers MYOCD, a gene that has been implicated in the pathogenesis of LMS. MYOCD, a serum response factor (SRF) transcriptional cofactor, is essential for cardiac and smooth muscle development and differentiation (Du et al., 2003; Wang et al., 2003; Pipes et al., 2006). MYOCD gene amplification has been reported previously in LMS, with a predilection for retroperitoneal location (Perot et al., 2009). In their study, 17p11.2 gains were observed, by array-CGH, in 10 of the 19 (52%) LMS, including eight of the 11 retroperitoneal LMS. They also noted that MYOCD is the most overexpressed gene in the amplified region. In our study, 17p11.2 gains were seen in 12 of 35 (34%) LMS cases. MYOCD amplification, as detected by FISH studies, was identified in five cases that were equally distributed among the retroperitoneal and extremity tumors. Our findings suggest that MYOCD gene amplification is seen only in a small subset of LMS and is not restricted to the retroperitoneal cases. Because MYOCD was amplified in only a subset of cases with 17p11.2 region gains in our study, it is possible that other genes in this region may be involved in smooth muscle development and / or differentiation.
Hu et al., in their study of 17 LMS, showed that copy number loss affecting chromosome arm 10q is associated with aggressive tumor behavior (Hu et al., 2005). In our study, statistical analysis was limited by the small sample size, but none of the genetic alterations correlated with clinical parameters (size, site and grade) or with aggressive behavior (distant recurrence, survival). Additional studies with a larger number of cases are required to study this correlation.
Low grade LMS of the soft tissue are relatively rare and usually show well-differentiated smooth muscle differentiation with minimal atypia and mitotic activity. Among the four cases included in our study, three showed losses of 13q14, including two cases with 16q22 losses. One case showed no aberrations. All four cases lacked losses in 10q23 or 17p13, gains in 17p11.2 or TP53 mutations. Our findings suggest that 13q14 loss might be an early genetic alteration in the progression of the disease. This finding is in keeping with results from other studies identifying loss of 13q as one of the most common abnormalities in LMS (Hu et al., 2001).
In summary, LMS show a complex genetic profile. Deep-sequencing methodology reveals recurrent mutations of TP53, seen in more than one third of the cases. The low mutation frequency in these tumors suggests that leiomyosarcoma genesis is driven primarily by copy number alterations. Chromosomal losses involving tumor suppressor genes PTEN, RB1 and TP53 are common events and emerge as the hallmark genetic abnormalities in these tumors. Our study showed no correlation between the genetic events and the clinical behavior of the disease. MYOCD gene amplification is present in a small subset of LMS and does not correlate with the anatomic site of the tumor.
Supplementary Material
Acknowledgements
The authors thank and acknowledge the efforts of Agnès Viale and the MSKCC Genomics Core Laboratory where sequencing was performed.
Supported in part by: Cancer Center Support Grant of the National Institute of Health/National Cancer Institute (P30CA008748); P50 CA 140146-01 (S.S., C.R.A.); Farmer Family Foundation. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflict of interest: none
REFERENCES
- Antonescu CR, Zhang L, Chang NE, Pawel BR, Travis W, Katabi N, Edelman M, Rosenberg AE, Nielsen GP, Dal Cin P, Fletcher CD. EWSR1-POU5F1 fusion in soft tissue myoepithelial tumors. A molecular analysis of sixty-six cases, including soft tissue, bone, and visceral lesions, showing common involvement of the EWSR1 gene. Genes Chromosomes Cancer. 2010;49:1114–1124. doi: 10.1002/gcc.20819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, Chiang DY, Reva B, Mermel CH, Getz G, Antipin Y, Beroukhim R, Major JE, Hatton C, Nicoletti R, Hanna M, Sharpe T, Fennell TJ, Cibulskis K, Onofrio RC, Saito T, Shukla N, Lau C, Nelander S, Silver SJ, Sougnez C, Viale A, Winckler W, Maki RG, Garraway LA, Lash A, Greulich H, Root DE, Sellers WR, Schwartz GK, Antonescu CR, Lander ES, Varmus HE, Ladanyi M, Sander C, Meyerson M, Singer S. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, Chandramohan R, Liu ZY, Won HH, Scott SN, Brannon AR, O’Reilly C, Sadowska J, Casanova J, Yannes A, Hechtman JF, Yao J, Song W, Ross DS, Oultache A, Dogan S, Borsu L, Hameed M, Nafa K, Arcila ME, Ladanyi M, Berger MF. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladdy RA, Qin LX, Moraco N, Agaram NP, Brennan MF, Singer S. Predictors of survival and recurrence in primary leiomyosarcoma. Ann Surg Oncol. 2013;20:1851–1857. doi: 10.1245/s10434-013-2876-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillou L, Aurias A. Soft tissue sarcomas with complex genomic profiles. Virchows Arch. 2010;456:201–217. doi: 10.1007/s00428-009-0853-4. [DOI] [PubMed] [Google Scholar]
- Hernando E, Charytonowicz E, Dudas ME, Menendez S, Matushansky I, Mills J, Socci ND, Behrendt N, Ma L, Maki RG, Pandolfi PP, Cordon-Cardo C. The AKT-mTOR pathway plays a critical role in the development of leiomyosarcomas. Nat Med. 2007;13:748–753. doi: 10.1038/nm1560. [DOI] [PubMed] [Google Scholar]
- Hu J, Khanna V, Jones M, Surti U. Genomic alterations in uterine leiomyosarcomas: potential markers for clinical diagnosis and prognosis. Genes Chromosomes Cancer. 2001;31:117–124. doi: 10.1002/gcc.1125. [DOI] [PubMed] [Google Scholar]
- Hu J, Rao UN, Jasani S, Khanna V, Yaw K, Surti U. Loss of DNA copy number of 10q is associated with aggressive behavior of leiomyosarcomas: a comparative genomic hybridization study. Cancer Genet Cytogenet. 2005;161:20–27. doi: 10.1016/j.cancergencyto.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Ito M, Barys L, O’Reilly T, Young S, Gorbatcheva B, Monahan J, Zumstein-Mecker S, Choong PF, Dickinson I, Crowe P, Hemmings C, Desai J, Thomas DM, Lisztwan J. Comprehensive mapping of p53 pathway alterations reveals an apparent role for both SNP309 and MDM2 amplification in sarcomagenesis. Clin Cancer Res. 2011;17:416–426. doi: 10.1158/1078-0432.CCR-10-2050. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Morita T, Hayashi K, Miki T, Sobue K. Myocardin functions as an effective inducer of growth arrest and differentiation in human uterine leiomyosarcoma cells. Cancer Res. 2010;70:501–511. doi: 10.1158/0008-5472.CAN-09-1469. [DOI] [PubMed] [Google Scholar]
- Larramendy ML, Kaur S, Svarvar C, Bohling T, Knuutila S. Gene copy number profiling of soft-tissue leiomyosarcomas by array-comparative genomic hybridization. Cancer Genet Cytogenet. 2006;169:94–101. doi: 10.1016/j.cancergencyto.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Leroy B, Anderson M, Soussi T. TP53 mutations in human cancer: database reassessment and prospects for the next decade. Hum Mutat. 2014;35:672–688. doi: 10.1002/humu.22552. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen M, Fetsch JF. Evaluation of biological potential of smooth muscle tumours. Histopathology. 2006;48:97–105. doi: 10.1111/j.1365-2559.2005.02292.x. [DOI] [PubMed] [Google Scholar]
- Perot G, Derre J, Coindre JM, Tirode F, Lucchesi C, Mariani O, Gibault L, Guillou L, Terrier P, Aurias A. Strong smooth muscle differentiation is dependent on myocardin gene amplification in most human retroperitoneal leiomyosarcomas. Cancer Res. 2009;69:2269–2278. doi: 10.1158/0008-5472.CAN-08-1443. [DOI] [PubMed] [Google Scholar]
- Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- Saito T, Oda Y, Kawaguchi K, Takahira T, Yamamoto H, Tamiya S, Tanaka K, Matsuda S, Sakamoto A, Iwamoto Y, Tsuneyoshi M. PTEN/MMAC1 gene mutation is a rare event in soft tissue sarcomas without specific balanced translocations. Int J Cancer. 2003;104:175–178. doi: 10.1002/ijc.10918. [DOI] [PubMed] [Google Scholar]
- Svarvar C, Bohling T, Berlin O, Gustafson P, Folleras G, Bjerkehagen B, Domanski HA, Sundby Hall K, Tukiainen E, Blomqvist C. Clinical course of nonvisceral soft tissue leiomyosarcoma in 225 patients from the Scandinavian Sarcoma Group. Cancer. 2007;109:282–291. doi: 10.1002/cncr.22395. [DOI] [PubMed] [Google Scholar]
- Toro JR, Travis LB, Wu HJ, Zhu K, Fletcher CD, Devesa SS. Incidence patterns of soft tissue sarcomas, regardless of primary site, in the surveillance, epidemiology and end results program, 1978-2001: An analysis of 26,758 cases. Int J Cancer. 2006;119:2922–2930. doi: 10.1002/ijc.22239. [DOI] [PubMed] [Google Scholar]
- Wagle N, Berger MF, Davis MJ, Blumenstiel B, Defelice M, Pochanard P, Ducar M, Van Hummelen P, Macconaill LE, Hahn WC, Meyerson M, Gabriel SB, Garraway LA. High-throughput detection of actionable genomic alterations in clinical tumor samples by targeted, massively parallel sequencing. Cancer Discov. 2012;2:82–93. doi: 10.1158/2159-8290.CD-11-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang DZ, Pipes GC, Olson EN. Myocardin is a master regulator of smooth muscle gene expression. Proc Natl Acad Sci U S A. 2003;100:7129–7134. doi: 10.1073/pnas.1232341100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss SW. Smooth muscle tumors of soft tissue. Adv Anat Pathol. 2002;9:351–359. doi: 10.1097/00125480-200211000-00004. [DOI] [PubMed] [Google Scholar]
- Won HH, Scott SN, Brannon AR, Shah RH, Berger MF. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Expe. 2013 doi: 10.3791/50710. 50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Du X, Chen K, Ylipaa A, Lazar AJ, Trent J, Lev D, Pollock R, Hao X, Hunt K, Zhang W. Genetic aberrations in soft tissue leiomyosarcoma. Cancer Lett. 2009;275:1–8. doi: 10.1016/j.canlet.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.