Abstract
Decreased hydroxymethylated cytosine (5-hydroxymethycytosine, 5-hmC) is reported to correlate with melanocyte dysplasia. The purpose of this study was to assess the diagnostic utility of this observation. 5-hmC immunohistochemistry was performed on tissue microarrays containing 171-melanocytic lesions from two different institutions. An immunohistochemical staining score representing the percentage and intensity of nuclear staining was assigned. The performance characteristics of 5-hmC immunohistochemistry for discriminating between a nevus and melanoma were determined. Additional cases of melanoma arising in a nevus (n = 8), nodal nevi (n = 5) and melanoma micrometastases to a lymph node (n = 6) were also assessed. Pronounced 5-hmC loss was observed in melanomas when compared with nevi (mean ± standard deviation = 6.71 ± 11.78 and 55.19 ± 23.66, respectively, p < 0.0001). While the mean immunohistochemical staining score values for melanocytic nevi and melanoma were distinct, there was considerable variability in immunohistochemical staining score within a single diagnostic category. The sensitivity and specificity of this assay for nevus vs. melanoma is 92.74 and 97.78%, respectively. Distinct biphasic staining patterns were observed in cases of melanoma arising in association with a nevus. Relative changes of 5-hmC expression within a single lesion may be more informative than absolute values when using 5-hmC as a diagnostic adjunct.
Keywords: 5-hydroxymethylcytosine, epigenetics, melanoma, nevus, dermatopathology
The classic model of melanomagenesis is that mutations initiate and promote tumor formation and progression. Indeed, the melanoma genome contains one of the highest somatically acquired nucleotide mutation frequencies of any human cancer.1 These mutations are dominated by C to T transitions, which are generated by ultraviolet A and B electromagnetic radiation.1 The mutations reproducibly occur in genes belonging to one of the three, near mutually exclusive, major oncogenic pathways that sustains proliferative signaling. In decreasing order of frequency, the most common mutations involve BRAF, NRAS and C-Kit genes.2–5 In addition to point mutations, chromosomal gains and losses are also prevalent in melanocytic tumors.6,7 It is now known that in addition to changes in the sequence of genomic DNA, epigenetic aberrations also play a role in melanoma formation.
Gene expression is tightly and dynamically regulated by a multi-layer epigenetic system that includes DNA methylation, histone modification and non-coding RNAs. Pathologic alterations in the epigenome are thought to contribute to tumor development and virulence (reviewed in8). For example, hypermethylation of the promoters of tumor suppressor genes may lead to gene silencing and condensation or opening of the chromatin structure via histone modification may alternately prevent or facilitate gene expression, depending on the type and position of the modification. In cutaneous melanoma, genes impacting melanoma progression and metastases such as those involving DNA repair, cell cycle regulation and invasion, have all been shown to be silenced by DNA hypermethylation.9–11 Demethylation, on the other hand, has been associated with the aberrant re-expression of certain genes, which are typically silenced early in development.12 His-tone modifications in melanoma are not as well understood, though there is evidence to suggest that the histone variant macroH2A is lost in melanoma relative to nevi.13 MacroH2A is associated with chromatin condensation and its loss results in an increased proliferative and migratory phenotype.
DNA methylation associated with gene regulation occurs at the 5-carbon position of the cytosine base, resulting in 5-methylcytosine (5-mC). It is a single step catalyzed by DNA methyltransferases. The demethylation process is more complicated and requires multiple steps, the first of which is performed by the Ten Eleven Translocase family enzymes and results in the formation of 5-hydroxymethycytosine (5-hmC).14,15 5-hmC is the most abundant intermediate product of the demethylation pathway, and recently Lian et al. described 5-hmC loss as an ‘epigenetic hallmark’ of melanoma.16 In their initial description, they showed that loss of 5-hmC was associated with higher stage lesions and worse prognosis in patients with melanoma.16 In a follow-up report, the authors went on to further define 5-hmc loss in specific-histopathologic subtypes of melanocytic lesions, whereby they demonstrated progressive loss of 5-mhC with increasing nuclear diameter in a spectrum of melanocytic lesions that included benign nevi, low-grade dysplastic nevi, high-grade dysplastic nevi and invasive melanoma.17 While the mean/median differences between 5-hmC expression in different diagnostic categories were presented in these earlier reports, the diagnostic sensitivity and sensitivity of the 5-hmC immunohistochemical assay were not determined. The purpose of this study was to characterize further the immunohistochemical assay for 5-hmC expression to determine its potential as a diagnostic adjunct in melanocytic lesions.
Materials and methods
Specimen procurement and tissue microarray format
One hundred seventy one benign and malignant melanocytic lesions were gathered from two separate institutions for analysis. All tissue specimens were originally procured per routine surgical pathology protocol and arranged for analysis using tissue microarray technology. This study was approved by the review board at both institutions. The tissue microarray from our institution (cohort I) consisted of 43 melanocytic lesions that were acquired from 1974 to 2008, Table 1. Each specimen was tiled in technical triplicate or quadruplicate cores. Selected melanoma cases were tiled as biological replicates (i.e. primary vs. metastatic melanoma specimen from the same patient). The tissue microarray from the outside institution (cohort II) contained 128 melanocytic lesions. Each case was cored once and did not have associated clinical information. In addition to the tissue microarrays, whole mount sections of melanoma arising in association with a nevus (n = 8), micrometastatic melanoma to lymph nodes (n = 6) and nodal melanocytic nevi (n = 5) were stained for 5-hmC expression.
Table 1.
Melanocytic proliferations included in the tissue microarrays from two different institutions
| Cohort I (n = 43) | Cohort II (n = 128) | |
|---|---|---|
| Nevi | 13 | 32 |
| Primary melanoma | 16 | 32 |
| Metastatic melanoma | 14 | 64 |
Pathologic assessment
The immunohistochemical protocol used in this report was performed according to the previously optimized and validated method by Haffner et al.18 For each staining run, a positive control slide of representative tissue sections from primary prostatic carcinoma was included. The nuclei of the basal cell layer within benign prostatic glands are immunoreactive with 5-hmC antibody, whereas the nuclei of malignant glands are non-immunoreactive with 5-hmC (data not shown). In each experimental tissue microarray core, at least 40 individual nuclei were counted to ensure uniform results. Forty nuclei per core cutoff were chosen because we determined that the relative coefficient of variation for 5-hmC immunolabeling within any given core is <20% if 20 lesional nuclei are counted. Each case was scored by a trained pathologist (NR and JT). The 5-hmC staining intensity was scored as none (0), weak (1), moderate (2) or marked (3), according to Lian et al.16 More specifically, 0 = no immunolabeling; 1 = less intense than immunolabeling of in adjacent benign cells, e.g. normal keratinocyte or lymphocyte nuclei in the core; 2 = comparable with normal nuclei; and 3 = more intense than normal nuclei. An immunohistochemical staining score was developed, ranging from 0 to 100. This score represents the percentage of cells expressing 5-hmC multiplied by the intensity of staining/maximum possible staining intensity (i.e. 3). For example, if 100% of melanocyte nuclei demonstrate marked (score 3) staining intensity, the immunohistochemical staining score = (100 × 3)/3 or 100. Sequential whole mount sections from 10 cases from the cohort I tissue microarray were also stained for 5-hmC and Ki-67, the latter of which was performed according to standard automated methods. The immunohistochemical staining score for 5-hmC was compared with the percentage of nuclei within a given lesion that stained positive for Ki-67.
Statistical analysis
Five-hmC scores were analyzed as interval data sets using two-sided Student’s t-test. A p-value ≤0.05 was considered statistically significant.
Results
Immunohistochemistry for 5-hmC using tissue microarrays
The tissue microarray cohorts are summarized in Table 1. Cohort I consisted of 13 nevi, 16 primary melanomas and 14 metastatic melanomas. Cohort II consisted of 32 nevi, 32 primary malignant melanomas and 64 metastatic melanomas. Immunohistochemistry for 5-hmC was performed on the tissue microarrays and an immunohistochemical staining score was assigned. Representative examples of staining in a nevus and a primary melanoma are shown in Fig. 1. One hundred percent (45/45) of nevi demonstrated immunolabeling for 5-hmC, where only 55% (69/126 cases) of melanoma cases were immunoreactive. Across multiple samples from an individual case, the proportion of 5-hmC immunolabeling in lesional nuclei was relatively uniform, and cases demonstrated a constant staining intensity.
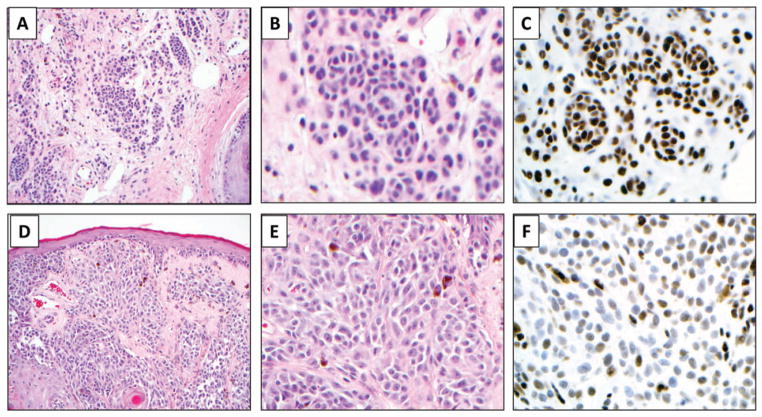
Fig. 1.
Representative photomicrographs showing 5-hydroxymethycytosine (5-hmC) immunolabeling in a melanocytic nevus (A–C), markedly decreased 5-hmC expression in a primary melanoma (D–F). 5-hmC nuclear labeling is also observed in background normal cells including keratinocytes and lymphocytes (not shown). Panels A and D at ×100; B, C, E, and F at 400×, original magnification.
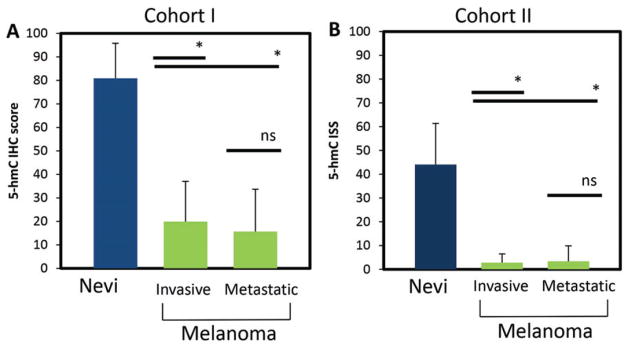
Using the tissue microarray from cohort I, we detected a marked depression of 5-hmC immunolabeling in malignant melanomas relative to the 5-hmC immunolabeling in nevi (mean immunohistochemical staining score ± SD = 17.97 ± 17.83 and 80.90 ± 14.98, respectively, p < 0.001) (Fig. 2A). To extend these findings, we also evaluated 5-hmC expression using an additional 128 cases from an outside institution (cohort II). Similar to the cohort I, we observed a reduction in 5-hmC expression between nevi and malignant melanoma (p < 0.0001) (Fig. 2B). When the results of both tissue microarrays were combined, nevi and melanoms showed a mean immunohistochemical staining score ± standard deviation of 55.19 ± 23.66 and 6.71 ± 11.78, respectively, p < 0.0001. We observed no statistically significant difference between the 5-hmC immunohistochemical staining score when primary and metastatic melanoma cases were compared in either cohort independently or both cohorts together.
Fig. 2.
5-hydroxymethycytosine (5-hmC) immunolabeling is markedly attenuated in both primary and metastatic melanoma when compared with melanocytic nevi. The mean immunohistochemical staining score ± standard deviation for nevi, primary invasive melanoma and metastatic melanoma are shown for both cohorts. *p < 0.001 (two-sided Student’s t-test). n.s., not statistically significant.
Diagnostic utility of 5-hmC immunohistochemistry in the evaluation of melanocytic proliferations
Using the results of both tissue microarrays combined, a receiver operating characteristic curve for 5-hmC immunohistochemical staining score was developed (Fig. 3). The balance between sensitivity and specificity was maximized at an immunohistochemical staining scoreof 17. At this immunohistochemical staining score, indicating a relative reduction in 5-hmC expression, this assay has a sensitivity of 92.74% and specificity of 97.78% for the identification of melanoma.
Fig. 3.
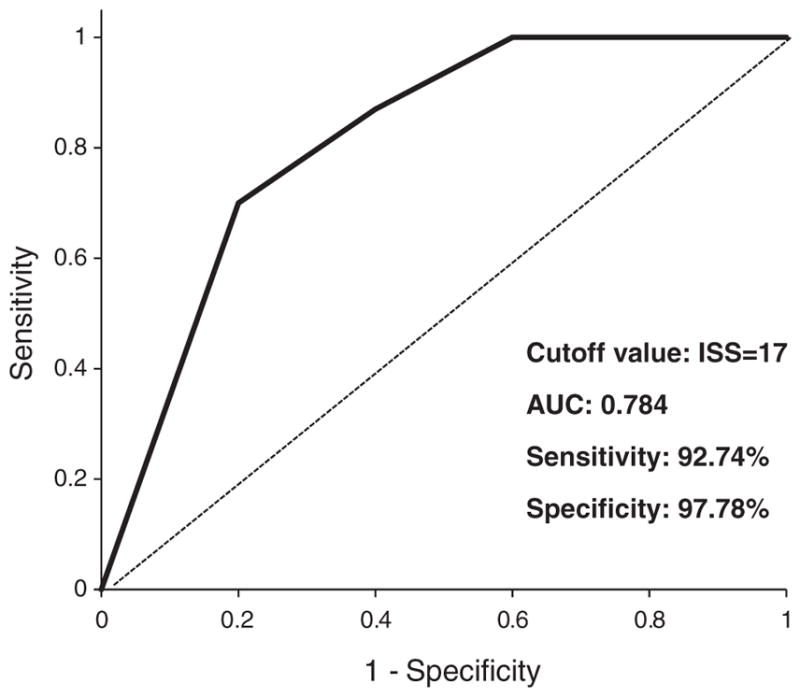
Performance characteristics of the 5-hydroxymethycytosine (5-hmC) immunohistochemical assay. A receiver operating characteristic (ROC) curve was generated for 5-hmC immunohistochemical staining score for distinguishing nevi from malignant melanoma, using the combined cohorts. The cutoff was set at an immunohistochemical staining score = 17 to achieve optimal sensitivity and specificity. AUC, area under the curve.
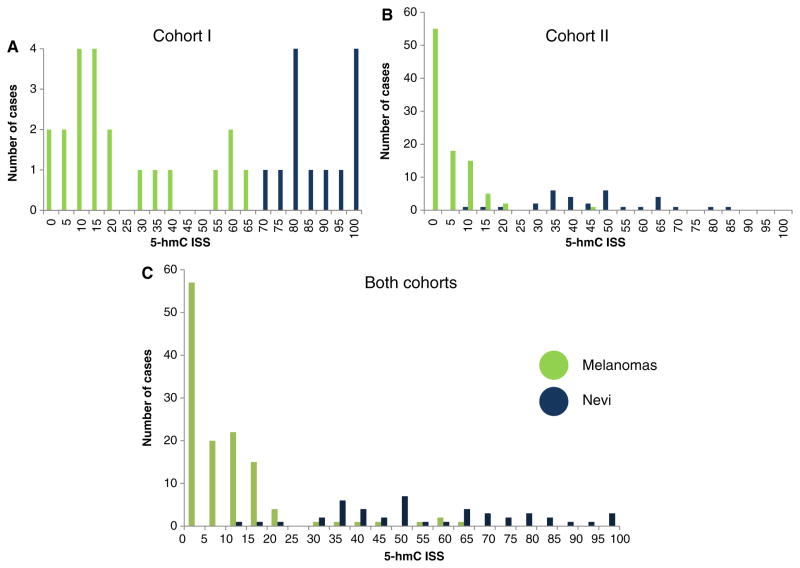
While the mean 5-hmC levels between nevi and melanoma are significantly different, the variance in immunohistochemical staining score is of great interest if the stain is to be used as a diagnostic adjunct. The variance of the immunohistochemical staining scores for both cohorts is shown in Fig. 4A,B, respectively. The overlap between categories was much less when each tissue microarray was analyzed individually. When the two tissue microarrays were analyzed together, we noted considerable variability between cases within the same diagnostic category, with melanoma demonstrating more variability than nevi [relative coefficient of variation for nevi and melanoma: 42.8 and 175.0, respectively] (Fig. 4C), for both combined. The difference in immunohistochemical staining scores between the two different tissue microarrays suggests that pre-analytical variables such as specimen handling and preparation may impact on assay performance.
Fig. 4.
Histogram showing the distribution of immunohistochemical staining scores for 5-hydroxymethycytosine (5-hmC) expression for melanocytic nevi and melanoma. An immunohistochemical staining score of zero (0) indicates no 5-hmC immunolabeling in any of the evaluable lesional nuclei, where an immunohistochemical staining score of 100 indicates that all lesional nuclei were markedly/strongly immunoreactive (see Methods for detailed description for calculating immunohistochemical staining score). When the two cohorts were evaluated separately, (A) and (B) respectively, the potential discriminatory ability of 5-hmC immunohistochemistry between nevi and melanoma is more evident than when both cohorts are combined (C).
Immunohistochemistry for 5-hmC on whole mount sections
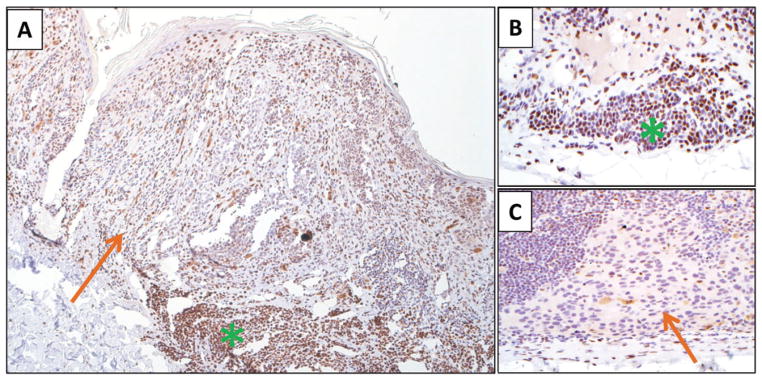
Because 5-hmC expression is an early event in melanomagenesis, we hypothesized that 5-hmC loss may have utility in evaluating melanomas arising in association with a nevus. We examined whole mount sections of such cases (n = 8), and we noted marked attenuation of 5-hmC in the malignant component, with retention of 5-hmC expression in the adjoining benign nevomelanocytes (mean 5-hmC immunohistochemical staining score 12.5 vs. 84.16, respectively p < 0.001). While the means are significantly different, the real strength in this observation is related to the fact that each of the eight cases demonstrated a clear biphasic pattern of expression. The staining quality made it possible to delineate nevomelanocytes and malignant melanocytes within a single lesion (Fig. 5A). In areas with a well-nested in situ component, loss of 5-hmC expression was also readily identified. Loss of expression was more challenging to assess when cells were present as single cells, either demonstrating pagetoid scatter or arranged in a lentiginous array along the dermal–epidermal junction, because of the difficulty in assessing partial loss on a single-cell basis in a background of keratinocyte staining. Micrometastatic melanoma to lymph nodes (n = 6) and nodal nevi (n = 5) were also assessed for 5-hmC expression (Fig. 5B,C). Although interpretation was challenging in micrometastases composed of singular cells or very small deposits, distinctions between nodal nevi and melanoma were often apparent. We also compared 5-hmC expression on n = 10 whole mount sections from nevi and melanoma to Ki-67 expression and did not observe a correlation between these two factors (R2 = 0.003, p = 0.99). This finding suggests that epigenetic markers such as 5-hmC have the potential to reflect status changes in melanocytes beyond proliferative activity, and thus could potentially serve a complimentary role to stains that are already in routine use.
Fig. 5.
Representative photomicrographs demonstrating 5-hmC immunolabeling in a melanoma arising in association with a precursor melanocytic nevus (A), a nodal nevus (B) and a micrometastatic deposit of melanoma in a lymph node (C). The malignant melanocytes demonstrate loss of 5-hmC expression (orange arrows), while the benign melanocyte population demonstrates strong nuclear staining in the majority of cells (green asterisk). Panel A at ×40; Panels B and C at ×400, original magnification.
Discussion
Methylation is a mechanism by which gene expression is controlled throughout the course of cellular development and differentiation. Specifically, DNA methylation occurs at the 5-position of cytosine to form 5-mC. When this modification occurs at a promoter site, it is known to silence gene expression, helping to maintain a differentiated state.19 In contrast, when the gene itself is methylated, transcription is activated, rather than silenced.20 Not surprisingly, tumor cells are known to demonstrate aberrant methylation patterns of key genes. 5-hmC is reportedly lost in a broad array of cancer types, including glioblastoma, prostate, breast, gastric and colon cancers18,21–23 making it one of the most common features of human cancers known to date.
The results of our study indicate that the loss of 5-hmC is an early change in melanoma tumor formation. Two findings support this model. First, 5-hmC loss was observed in the in situ component of lesions. Second, the vast majority of the 126 invasive and metastatic malignant melanomas examined had attenuated or absent global 5-hmC nuclear levels by immunohistochemistry. These findings confirm and extend the observations of other authors. In their sentinel study, Lian et al. showed that 5-hmC is markedly decreased in primary and metastatic melanomas when compared with nevi.16 Two smaller studies by Gambiechler et al. and Uchiyama et al. also observed comparable attenuation of 5-hmC immunolableing in malignant vs. benign melanocytic lesions.24,25
Diagnostic scenarios in the assessment of melanocytic lesions where ancillary immunohistochemical or molecular studies have the potential for therapeutic impact include: (i) characterizing the biology of borderline lesions, such as distinguishing between a severely dysplastic/Clark’s nevus with unusual features and an early malignant melanoma, (ii) defining the trailing edge of a lentigo maligna on sun-damaged skin, and (iii) improved characterization of atypical Spitz tumors, among others. Early reports on the patterns of immunohistochemical detection of 5-hmC in melanocytic lesions have raised the possibility that this assay could play a role in each of these circumstances. Specifically, 5-hmC loss has been reported to decrease with increasing malignant potential in melanocytic lesions from benign nevi to dysplastic nevi with low-grade atypia to severely dysplastic nevi, to primary melanoma and to metastatic disease.17 Our findings highlight the variance possible within such groups, indicating that it the current semi-quantitative immunohistochemical approach for assessing 5-hmC levels would not be useful in the finer distinctions of grading-melanocytic lesions, e.g. low grade vs. high grade or high grade vs. early melanoma.
We did, however, note the potential utility of this stain for distinguishing different melanocyte populations within a single lesion, i.e. malignant melanocytes and benign nevomelanocytes. This observation should be considered preliminary because of the limited number of cases assessed, but warrants further study. This approach may be useful for assessing the Breslow thickness of a melanoma arising in association with a nevus or for assessing whether a precursor nevus is present vs. pseudomaturation of a melanoma. Potential pre-analytic influences are also circumvented, as the expression values are all relative. As the accurate determination of Breslow thickness is the most important Tumor-Node-Metastasis (TNM) staging feature in early melanomas, such a tool could potentially have impact with regard to prognostication and subsequent treatment.
Recently, 5-hmC immunohistochemistry has also been proposed for the assessment of nodal nevi.26 Our sample size of nodal nevi and melanoma micrometastases in lymph nodes was limited, but our findings indicate that 5-hmC loss is seen in micrometastatic deposits, as anticipated. This finding was apparent when considering aggregated metastatic deposits, but the immunohistochemistry was more challenging to assess when the lesion was composed of single cells or very small cellular aggregates, because of the staining pattern of the background lymph node. Similarly, the reliable scoring of 5-hmC loss in an intraepidermal melanocytic proliferation when composed of single cells was challenging to resolve from the background keratinocyte staining. This may be related to the fact that a decrease in the 5-hmC immunolabeling in keratinocytes that overly invasive malignant melanoma may also occur.17 Irrespective of whether an epigenetic field effect becomes apparent, the utility of assessing partial nuclear loss of a marker by immunohistochemistry is impractical, especially when required on a single-cell level and would be made near-impossible if adjacent keratinocytes also demonstrate loss. Thus, in our estimation, it is doubtful that this marker will play a role in assessing lentigo maligna. The potential assessment of atypical Spitz tumors has also been posited,17 and tests are currently underway in our laboratories to determine whether this marker may play a role in the assessment of atypical Spitz tumors when compared with spitzoid melanoma.
In conclusion, using collections of melanocytic proliferations from two different academic institutions, we confirmed that 5-hmC reduction is a feature of the vast majority of malignant melanomas. The sensitivity and specificity of the current method for determining 5-hmC expression is insufficient to use as a single marker. Its greatest potential at this time may be to illuminate relative changes within a single lesion that may be undergoing transformation. It is possible that in the future, immunohistochemical panels for challenging melanocytic lesions may include one or more epigenetic markers to inform the assessment of biologic potential.
Acknowledgments
The authors acknowledge Alan K. Meeker, Marcella Sutherland and Ellen Tully from the Microarray Core facility. They thank Dr Corrado Spadafora for helpful comments.
Footnotes
Author contribution
All authors confirmed they have contributed meaningfully to the intellectual content of this publication, and this study was supported in part by the Resident Research Fund grant from the Department of Pathology at the Johns Hopkins Hospital (NR) and by the Fred and Janet Sanfilippo Research Fund (NR).
References
- 1.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubinstein JC, Sznol M, Pavlick AC, et al. Incidence of the V600K mutation among melanoma patients with BRAF mutations, and potential therapeutic response to the specific BRAF inhibitor PLX4032. J Transl Med. 2010;8:67. doi: 10.1186/1479-5876-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massi D, Pinzani P, Simi L, et al. BRAF and KIT somatic mutations are present in amelanotic melanoma. Melanoma Res. 2013;23:414. doi: 10.1097/CMR.0b013e32836477d4. [DOI] [PubMed] [Google Scholar]
- 4.Griewank KG, Westekemper H, Murali R, et al. Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin Cancer Res. 2013;19:3143. doi: 10.1158/1078-0432.CCR-13-0163. [DOI] [PubMed] [Google Scholar]
- 5.Colombino M, Lissia A, Capone M, et al. Heterogeneous distribution of BRAF/NRAS mutations among Italian patients with advanced melanoma. J Transl Med. 2013;11:202. doi: 10.1186/1479-5876-11-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Botton T, Yeh I, Nelson T, et al. Recurrent BRAF kinase fusions in melanocytic tumors offer an opportunity for targeted therapy. Pigment Cell Melanoma Res. 2013;26:845. doi: 10.1111/pcmr.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner T, He J, Yelensky R, et al. Kinase fusions are frequent in Spitz tumours and spitzoid melanomas. Nat Commun. 2014;5:3116. doi: 10.1038/ncomms4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JJ, Murphy GF, Lian CG. Melanoma epigenetics: novel mechanisms, markers, and medicines. Lab Invest. 2014;94:822. doi: 10.1038/labinvest.2014.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howell PM, Liu SH, Ren SP, et al. Epigenetics in human melanoma. Cancer Control. 2009;16:200. doi: 10.1177/107327480901600302. [DOI] [PubMed] [Google Scholar]
- 10.Sigalotti L, Covre A, Fratta E, et al. Epigenetics of human cutaneous melanoma: setting the stage for new therapeutic strategies. J Transl Med. 2010;8:56. doi: 10.1186/1479-5876-8-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schinke C, Mo YK, Yu YT, et al. Aberrant DNA methylation in malignant melanoma. Melanoma Res. 2010;20:253. doi: 10.1097/CMR.0b013e328338a35a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fratta E, Coral S, Covre A, et al. The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol Oncol. 2011;5:164. doi: 10.1016/j.molonc.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapoor A, Goldberg MS, Cumberland LK, et al. The histone variant macroH2A suppresses melanoma progression through regulation of CDK8. Nature. 2010;468:1105. doi: 10.1038/nature09590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito S, D’Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang HK, Zhang X, Clark E, et al. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 16.Lian CG, Xu Y, Ceol C, et al. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell. 2012;150:1135. doi: 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson AR, Dresser KA, Zhan Q, et al. Loss of 5-hydroxymethylcytosine correlates with increasing morphologic dysplasia in melanocytic tumors. Mod Pathol. 2014;27:936. doi: 10.1038/modpathol.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haffner MC, Chaux A, Meeker AK, et al. Global 5-hydroxymethylcytosine content is significantly reduced in tissue stem/progenitor cell compartments and in human cancers. Oncotarget. 2011;2:627. doi: 10.18632/oncotarget.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maunakea AK, Nagarajan RP, Bilenky M, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lister R, Pelizzola M, Dowen RH, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kraus TF, Globisch D, Wagner M, et al. Low values of 5-hydroxymethylcytosine (5hmC), the “sixth base,” are associated with anaplasia in human brain tumors. Int J Cancer. 2012;131:1577. doi: 10.1002/ijc.27429. [DOI] [PubMed] [Google Scholar]
- 22.Yang Q, Wu K, Ji M, et al. Decreased 5-hydroxymethylcytosine (5-hmC) is an independent poor prognostic factor in gastric cancer patients. J Biomed Nanotechnol. 2013;9:1607. doi: 10.1166/jbn.2013.1713. [DOI] [PubMed] [Google Scholar]
- 23.Kudo Y, Tateishi K, Yamamoto K, et al. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer Sci. 2012;103:670. doi: 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gambichler T, Sand M, Skrygan M. Loss of 5-hydroxymethylcytosine and ten-eleven translocation 2 protein expression in malignant melanoma. Melanoma Res. 2013;23:218. doi: 10.1097/CMR.0b013e32835f9bd4. [DOI] [PubMed] [Google Scholar]
- 25.Uchiyama R, Uhara H, Uchiyama A, et al. 5-Hydroxymethylcytosine as a useful marker to differentiate between malignant melanomas and benign melanocytic nevi. J Dermatol Sci. 2014;73:161. doi: 10.1016/j.jdermsci.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 26.Lee JJ, Granter SR, Laga AC, et al. 5-Hydroxymethylcytosine expression in metastatic melanoma versus nodal nevus in sentinel lymph node biopsies. Mod Pathol. 2014;28:218. doi: 10.1038/modpathol.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]