Abstract
Current treatment options for human African trypanosomiasis (HAT) are ineffective, and they have several well-known clinical limitations. In our continued efforts to identify chemotypes that can be developed into clinically useful drugs, we screened a targeted compound library against the major cathepsin L (rhodesain) in T. brucei. We report the antirhodesain activity and antitrypanosomal activity of the compounds in this letter. The identified compounds can serve as starting points for structure- and/or phenotype-based lead optimization strategy against Trypanosoma brucei.
Keywords: Rhodesain, Antitrypanosomal, Malaria Box, Dianiline, Trypanosomes
According to the World Health Organization (WHO), the human African trypanosomiasis (HAT) caused by the kinetoplastid protozoan Trypanosoma brucei rhodeseinse and Trypanosoma brucei gambiense is one of the neglected tropical diseases (NTDs) that lack medicines that are both effective and safe for patients. This has necessitated a push, over the past decade, for the discovery and development of new chemical entities into effective medicines for HAT and other neglected diseases. While the incidence of HAT has decreased in the last decade according to the WHO, the need to develop new and effective drugs remains a principal goal in the fight against the disease [1]. In this letter, we report the antitrypanosomal activity of compound 1 and 7, (4,4′-[methylenebis(1H-benzimidazole-6,2-diyl)]-dianiline) and (N-[4-(5-benzoyl-1H-benzimidazol-2-yl)phenyl]-2-chlorobenzamide), respectively. The discovery of the antitrypanosomal activity of these compounds was initiated by an in vitro screening of the Malaria Box provided by the Medicines for Malaria Venture (MMV) against the major cathepsin L of T. brucei rhodesiense (rhodesain). Rhodesain, a cysteine protease, is druggable and it is a well-validated drug target in T. brucei. It is essential for the survival and infectivity of the parasite. Its important role in the ability of the parasite to proliferate has been investigated by various groups [2].
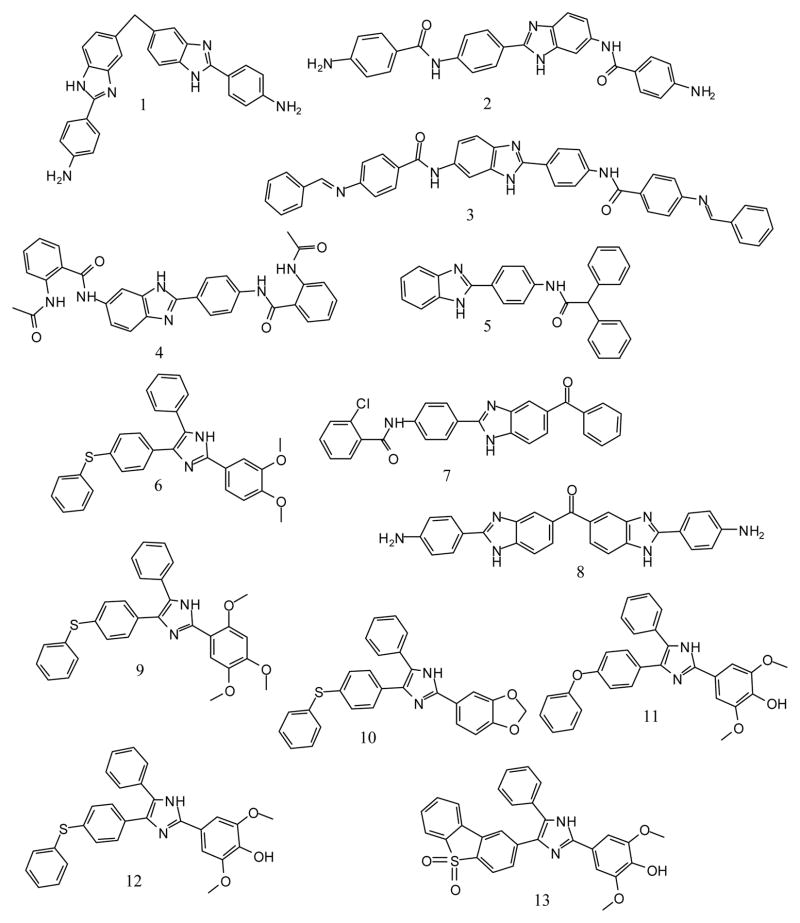
Steverding and co-workers have also shown that pharmacological inhibition of rhodesain is lethal to the parasites [3]. The initial screen of the MMV Malaria box led to the identification of compounds 14 and 22 as non-covalent inhibitors of rhodesain. We then carried out a sub-structure similarity search using compound 14 and 22 to obtain a series of compounds that can be evaluated for possible inhibitory activity on rhodesain and more importantly, to explore if the compounds in the series could have superior antitrypanosomal activity than compounds 14 and 22 (Figure 1). These structural analogues were subsequently tested for their inhibitory activity against rhodesain. Their antitrypanosomal activity as well as their cytotoxicity were also evaluated. In addition, the plausible interaction(s) between rhodesain and compound 1 was modelled via molecular docking.
Figure 1.
Structures of compounds studied in this work.
The malaria Box, a generous gift from MMV, was supplied by SCYNEXIS, Inc. (North Carolina, USA) [4]. Compounds 1-13 were obtained from ChemBridge Corporation (San Diego, CA) while compounds 15–21 were obtained from Vitas-M Laboratory (Hong Kong). 1H NMR spectra of the compounds provided by the vendors are provided in the supporting document. Rhodesain was expressed in Pichia pastoris and purified as previously described [5]. The inhibition assays were carried out in 50 mM sodium acetate buffer pH 5.5, containing 2 mM DTT and 0.001% Triton X-100. The enzyme test compound mixtures were pre-incubated for 5 min, followed by the addition of 100 μL of the substrate, Z-Phe-Arg-AMC (prepared in the same buffer system). Fluorescence (RFU/sec) resulting from proteolytic cleavage of the substrate was monitored at 25 °C (λex 355 nm and λem 460 nm) on a PolarStar Omega plate reader (BMG LABTECH, Germany). IC50 values were obtained by non-linear regression (Sigmoidal dose–response variable slope model) using GraphPad Prism 6. All inhibition data were carried out in triplicate [6].
Antitrypanosomal activity of the compounds was assessed using the Alamar blue assay [7]. T. brucei brucei 427 strain was used. T. brucei growing in HMI-9 medium were dispensed into 96 well plates at about 50,000 cells per well and treated with the compounds for 24 hours. After 24 hours, the cells are treated with Alamar blue and incubated for 4 hours. After the 4 hours of incubation, fluorescence signals (λex 530 nm; λem 590 nm) in the plates were read. All compounds were initially tested at 20 μM followed by a dose dependent assay for those compounds that showed significant inhibitory activity at 20 μM. Suramin was used as positive control.
The effect of the compounds on the viability of human liver carcinoma Hep G2 cells was assessed using the MTT assay. Hep G2 cells (CRL-11997™) were grown in DMEM:F12 medium containing 10 % FBS, 1 % penicillin/streptomycin and maintained in a humidified 5 % CO2 incubator at 37° C until reaching 80–90 % confluency. Cells were treated with 0.25 % trypsin-EDTA, counted and suspended in medium. About 50,000 cells/well were dispensed into 96-well plates and treated with compounds for 72 hours. Thereafter, DMEM:F12 media containing MTT (5 mg/mL in PBS) was added to the cells and incubated for 1 hr. The MTT-containing medium was then gently removed and replaced with DMSO (200 μL per well), the plate was then mix gently to allow the formazan crystals to dissolve. Absorbance was measured on a POLARstar Omega plate reader at 550 nm. All compounds were tested in triplicates at 20 μM. Compounds 1, 7 and 13 were also tested in an 8 points dose response assay (0.75 to 50 μM). 10 % SDS solution was used as positive control. For molecular docking, the structures of the compounds were built using SPARTAN ’10 for Windows. The ligand geometries were optimized using the MMFF 94 force field [8]. The docking simulations were carried out using the MolDock docking algorithm of the Molegro Virtual Docker as previously described [9, 10]. The x-ray crystal structure of rhoesain was used for the docking calculation (PDB ID: 2P7U).
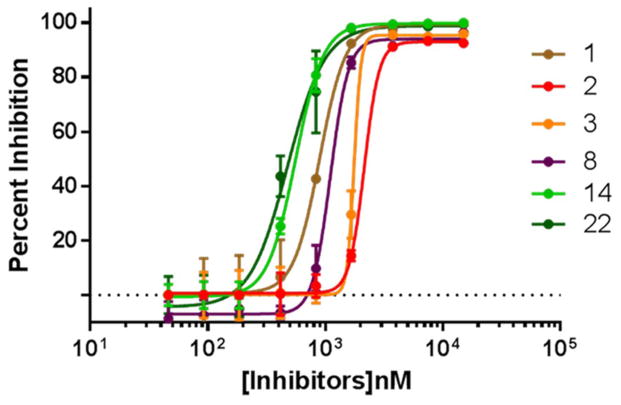
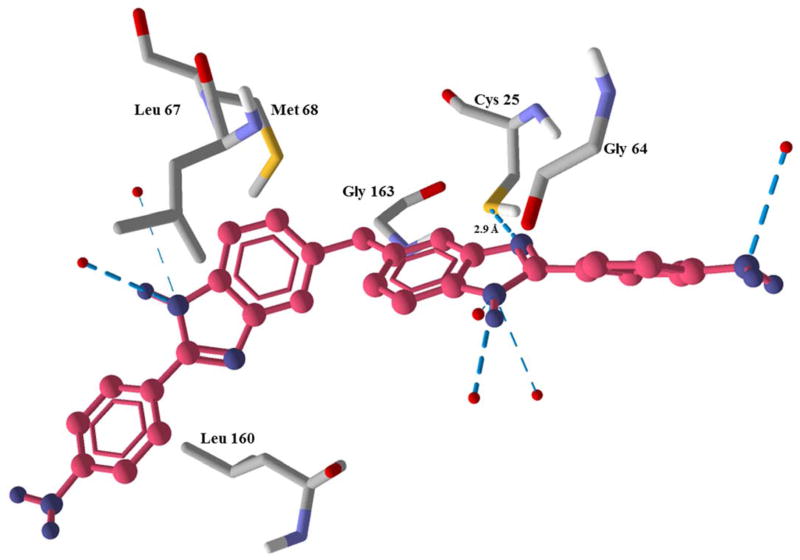
Compounds 14 and 22 were discovered to be non-covalent inhibitors of rhodesain with IC50 values of 0.56 μM and 0.47 μM, respectively (Table 1). We then assembled a series of compounds (1-13 and 15-21) based on the sub-structure features of compound 14 and 22 with the goal of identifying a promising hit that can be developed as a new drug lead. The inhibitory activities of the compounds are shown in Table 1 and Figure 2. The most active (against the cysteine protease) out of the sub-structure analogues was compound 1, its IC50 value against rhodesain was 0.89 μM, IC50 against T. brucei was 5.80 μM, and it displayed low toxicity to Hep G2 cells at 20 μM. Compound 1 is structurally interesting in the sense that it mimics the bidentate structural feature of the clinically used antitrypanosomal drug, pentamidine, and the simplicity of its structure makes it a good candidate for both structure- and phenotype-based lead discovery and optimization [13]. In addition, if the free amine groups in the compound are converted to nitro groups, its antitrypanosomal activity may be enhanced, since nitro group seems to be an important structural motif in many potent antitrypanosomal agents [14]. Molecular modeling of the possible interactions between compound 1 and rhodesain suggest that the benzimidazoyl motifs of compound 1 have H-bond and steric interactions with Cys 25 of rhodesain’s catalytic triad as well as residues Leu 67 and Met 68 while its dianiline motifs have steric interactions with Gly 64 and Leu 160. Compound 1 was also predicted to coordinate with 6 water molecules via its amine groups (Figure 3). The coordination with the water molecules may be important to the stability of the ligand-protein complex. Compounds 2, 3, 5, 8, 9 and 11 showed significant inhibitory activity against rhodesain, however, they displayed little to no antitrypanosomal activity at 20 μM (Table 1).
Table 1.
IC50 values (μM) of compounds 1 - 22.
| Compound | Rhodesain | T. brucei | Hep G2a |
|---|---|---|---|
| 1 | 0.89 ± 0.10 | 5.80 ± 1.00 | 84.71 ± 5.79 |
| 2 | 2.15 ± 0.11 | > 20 | 83.42 ± 9.16 |
| 3 | 1.78 ± 0.20 | > 20 | 92.20 ± 2.07 |
| 4 | > 20 | 19.32 ± 0.06 | 92.67 ± 6.48 |
| 5 | 3.04 ± 0.11 | > 20 | 62.54 ± 1.94 |
| 6 | 2.41 ± 0.11 | 20.02 ± 0.40 | 86.31 ± 4.80 |
| 7 | > 20 | 2.83 ± 0.26 | 86.19 ± 9.67 |
| 8 | 1.10 ± 0.10 | > 20 | 97.18 ± 4.67 |
| 9 | 3.38 ± 0.12 | 18.31 ± 0.52 | 98.14 ± 6.06 |
| 10 | > 20 | 12.93 ± 0.29 | 80.78 ± 7.91 |
| 11 | 2.36 ± 0.11 | > 20 | 109.00 ± 5.33 |
| 12 | > 20 | > 20 | 86.97 ± 3.62 |
| 13 | > 20 | 9.81 ± 0.81 | 88.35 ± 6.52 |
| 14 | 0.56 ± 0.10 | 11.79 ± 1.51 | 62.09 ± 7.92 |
| 15 | > 20 | > 20 | 74.01 ± 1.24 |
| 16 | > 20 | > 20 | 40.20 ± 10.98 |
| 17 | > 20 | > 20 | 86.49 ± 9.96 |
| 18 | > 20 | > 20 | 65.53 ± 8.64 |
| 19 | > 20 | 14.75 ± 0.15 | 81.79 ± 10.71 |
| 20 | > 20 | 17.06 ± 0.29 | 88.83 ± 13.35 |
| 21 | > 20 | 17.49 ± 1.71 | 99.72 ± 5.96 |
| 22 | 0.47 ± 0.10 | 1.93b | 5.37c |
Figure 2.
IC50 plots for compounds 1-3, 8, 14 and 22.
Figure 3.
Modelled complex of rhodesain and compound 1. The blue dash depicts H-bond interaction and the red dots are water molecules.
Compound 7 and 13 on the other hand did not display any inhibitory activity against rhodesain but they were active against T. brucei. Compound 7 showed considerable selectivity for the parasite over Hep G2 cells. Since compound 7 is not an inhibitor of rhodesain, its moderate antitrypanosomal activity suggests that it is an inhibitor of another important drug target in T. brucei. Identification of its molecular target would be helpful for hypothesis-driven lead design and discovery. In general, the presence of free alcohol or primary amine groups on terminal aromatic rings appears to positively correlate with higher rhodesain inhibitory activity.
In summary, compounds from the malaria box were evaluated for their inhibitory activity against the major cathepsin L in T. brucei. Compounds 14 and 22 displayed good inhibitory activity against the cysteine protease and their structural analogues, compounds 1 and 7 are inhibitors of rhodesain and T. brucei, respectively, and they can serve as starting points for structure and/or phenotype based lead optimization and development against T. brucei.
Acknowledgments
This work was carried out in part by resources made available by the US National Institutes of Health (SC2GM109782 and G12MD007581) and the Medicines for Malaria Venture. We thank Dr. Huey-Min Hwang and Dr. Stephen Ekunwe of the Department of Biological Sciences, Jackson State University for providing access to some of their laboratory equipment.
References
- 1.World Health Organization Report. [Accessed 02/26/2015];Investing to overcome the global impact of neglected tropical diseases. 2015 http://apps.who.int/iris/bitstream/10665/152781/1/9789241564861_eng.pdf?ua=1.
- 2.Ettari R, Tamborini L, Angelo IC, Micale N, Pinto A, De Micheli C, Conti P. Inhibition of rhodesain as a novel therapeutic modality for human African trypanosomiasis. J Med Chem 2013. 2013;56:5637–5658. doi: 10.1021/jm301424d. [DOI] [PubMed] [Google Scholar]
- 3.Steverding D, Sexton DW, Wang X, Gehrke SS, Wagner GK, Caffrey CR. Trypanosoma brucei: chemical evidence that cathepsin L is essential for survival and a relevant drug target. Int J Parasitol. 2012;42:481–488. doi: 10.1016/j.ijpara.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 4.Medicines for Malaria Venture. [Accessed 02/25/15];The Malaria Box. http://www.mmv.org/malariabox.
- 5.Caffrey CR, Hansell E, Lucas KD, Brinen LS, Hernandez AA, Cheng J, Gwaltney SL, Roush WR, Stierhof YD, Bogyo M, Steverding D, McKerrow JH. Active site mapping, biochemical properties and subcellular localization of rhodesain, the major cysteine protease of Trypanosoma brucei rhodesiense. Mol & Biochem Parasitol. 2001;118:61–73. doi: 10.1016/s0166-6851(01)00368-1. [DOI] [PubMed] [Google Scholar]
- 6.Ogungbe IV, Crouch RA, Haber WA, Setzer WN. Phytochemical investigation of Verbesina turbacensis Kunth: trypanosome cysteine protease inhibition by (−)-bornyl esters. Nat Prod Commun. 2010;5:1161–1166. [PubMed] [Google Scholar]
- 7.Räz B, Iten M, Grether-Bühler Y, Kaminsky R, Brun R. The Alamar Blue assay to determine drug sensitivity of African trypanosomes (T.b. rhodesiense and T.b. gambiense) in vitro. Acta Trop. 1997;68:139–147. doi: 10.1016/s0001-706x(97)00079-x. [DOI] [PubMed] [Google Scholar]
- 8.SPARTAN ’10 for Windows. Wavefunction, Inc; Irvine, California: 2010. [Google Scholar]
- 9.Thomsen R, Christensen MH. MolDock: a new technique for high-accuracy molecular docking. J Med Chem. 2006;49:3315–3321. doi: 10.1021/jm051197e. [DOI] [PubMed] [Google Scholar]
- 10.Ogungbe IV, Hill GM, Crouch RA, Vogler B, Nagarkoti M, Haber WA, Setzer WN. Prenylated isoflavonoids from Rhynchosia edulis. Nat Prod Commun. 2011;6:1637–1644. [PubMed] [Google Scholar]
- 11.Macdonald S, Willis P, Kowalczyk P, Spangenberg T, Burrows J, Wells T. [Accessed 02/25/15];Medicines for Malaria Venture (MMV) http://dx.doi.org/10.6019/CHEMBL2028040.
- 12.Gold B, Warrier T, Somersan S, Lopez-Quezada L, Roberts J, Little D, Nathan C. [Accessed 02/25/15];Malaria Box Tuberculosis Screening Data. http://dx.doi.org/10.6019/CHEMBL2094260.
- 13.Porcheddu A, Giacomelli G, De Luca L. New pentamidine analogues in medicinal chemistry. Curr Med Chem. 2012;19:5819–5836. doi: 10.2174/092986712804143268. [DOI] [PubMed] [Google Scholar]
- 14.Patterson S, Wyllie S. Nitro drugs for the treatment of trypanosomatid diseases: past, present, and future prospects. Trends parasitol. 2014;30:289–298. doi: 10.1016/j.pt.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]