Abstract
The prolactin (PRL) and growth hormone (GH) gene families represent species-specific expansions of pregnancy-associated hormones/cytokines. In this review we examine the structure, expression patterns, and biological actions of the pregnancy-specific PRL and GH families.
Review
Prolactin (PRL) and growth hormone (GH) are hormones/cytokines responsible for the coordination of a wide range of biological processes in vertebrates. They can act as classic endocrine modulators (hormones) via entry into the circulation or locally (cytokines) through juxtacrine, paracrine and autocrine modes of action. In the mouse, rat, and cow and likely other mammalian species there are large families of paralogous genes closely related to PRL [1-4]. The human and rhesus monkey genomes and likely other primate genomes possess a single PRL family gene, but unlike the rat and mouse contain an expanded GH gene family [2,3]. Sheep and goat genomes each contain duplicated GH loci [5,6]. Members of the PRL family of genes have been given a variety of names, including placental lactogens (PLs), PRL-like proteins (PLPs), PRL-related proteins (PRPs), proliferin (PLF), and PLF-related protein (PLF-RP), whereas, members of the GH family are referred to as PLs, GH variant (GH-V), and chorionic somatomammotropins [1-3]. Unfortunately, in some instances the literature contains nomenclature that is confusing. PRL and GH family ligands are expressed in cell- and temporal-specific patterns in the anterior pituitary and uteroplacental compartment [1-3,7-9]. An overriding theme characteristic of the PRL and GH families is their association with pregnancy and regulatory mechanisms controlling viviparity. In this minireview we examine the structure, expression patterns, and biological actions of the PRL and GH families from rodents (primarily rat and mouse), ruminants (primarily ovine and bovine), and primates (primarily human).
Species-specific gene family expansion
The annotation of genes from genome sequencing projects has provided considerable insight into the existence of species-specific gene expansion. Although the biology of most of these expanded gene families is not well understood, the majority appear to be associated with reproduction and host defense [10]. Gene families arise by gene duplication and natural selection [11]. PRL gene families of rodents and ruminants and the GH gene family of primates represent excellent examples of species-specific gene expansion. While the mouse and rat PRL families are largely orthologous, the expanded bovine PRL family is not orthologous with rodent PRL families, which suggests the independent utilization of the ancestral PRL template [12]. Primates have utilized an evolutionarily related template (ancestral GH) [2], while genomes for other mammalian species may not possess either an expanded PRL or GH family [13]. The overall breadth of our understanding of the diversity of PRL and GH families is both modest and biased because of the few species investigated.
Structural features
PRL and GH genes are structurally similar and evolved from a common ancestral precursor. Lists of PRL and GH family members from the mouse and rat, bovine and ovine, and human and rhesus monkey are provided in Tables 1,2 and 3. The PRL family genes in the mouse and rat are clustered on chromosomes 13 [4] and 17 [14], respectively. To date at least 26 separate mouse genes encoding members of the PRL family have been localized within a one-megabase segment [4]. The alignment of genes along the mouse PRL family locus primarily reflects sequence conservation. Commonalities in cell-specific and/or temporal-specific aspects of gene expression do not appear to be main factors governing alignment within the locus. The solitary GH genes are situated on chromosomes 11 and 10 of the mouse and rat genomes, respectively [15,16]. Eight identified PRL family members from the cow are located on chromosome 23 [17]. An expanded five-member GH gene cluster spans approximately 48 kb on chromosome 17 of the human genome, whereas the sole human PRL gene is present on chromosome 6 [2]. Members of the human GH family are closely related structural variants, whereas members of the rat, mouse, and bovine PRL families exhibit considerable sequence diversity.
Table 1.
Members of the mouse and rat PRL and GH gene families
| Member | Abbreviation | Mouse GenBank Accession No. | Rat GenBank Accession No. |
| Prolactin family members* | |||
| Prolactin | PRL | NM_011164 | NM_012629 |
| Placental lactogen-Iα | PL-Iα | AF525162 | D21103 |
| Placental lactogen-Iβ | PL-Iβ | NM_172155 | --- |
| Placental lactogen-Iγ | PL-Iγ | NM_172156 | --- |
| Placental lactogen-I variant | PL-Iv | --- | NM_033233 |
| Placental lactogen-II | PL-II | M14647 | M13749 |
| Proliferin-1 | PLF-1 | NM_031191 | --- |
| Proliferin-2 | PLF-2 | K03235 | --- |
| Proliferin-3 | PLF-3 | NM_011954 | --- |
| Proliferin-4 | PLF-4 | AF128884 | --- |
| Proliferin-related protein | PLF-RP | NM_011120 | NM_053364 |
| Prolactin-like protein-A | PLP-A | NM_011165 | NM_017036 |
| Prolactin-like protein-B | PLP-B | NM_011166 | M31155 |
| Prolactin-like protein-C | PLP-C | --- | NM_173110 |
| Prolactin-like protein-C variant | PLP-Cv | --- | NM_020079 |
| Prolactin-like protein-Cα | PLP-Cα | NM_011167 | --- |
| Prolactin-like protein-Cβ | PLP-Cβ | NM_023332 | NM_134385 |
| Prolactin-like protein-Cγ | PLP-Cγ | NM_023741 | --- |
| Prolactin-like protein-Cδ | PLP-Cδ | NM_028477 | --- |
| Prolactin-like protein-D | PLP-D | --- | NM_022537 |
| Prolactin-like protein-H | PLP-H | --- | NM_021580 |
| Decidual prolactin-related protein | DPRP | NM_010088 | NM_022846 |
| Prolactin-like protein-E | PLP-E | NM_008930 | --- |
| Prolactin-like protein-F | PLP-F | NM_011168 | NM_022530 |
| Prolactin-like protein-I | PLP-I | AF525154 | NM_153736 |
| Prolactin-like protein-J | PLP-J | NM_013766 | NM_031316 |
| Prolactin-like protein-K | PLP-K | NM_025532 | NM_138861 |
| Prolactin-like protein-L | PLP-L | NM_023746 | NM_138527 |
| Prolactin-like protein-M | PLP-M | NM_019991 | NM_053791 |
| Prolactin-like protein-N | PLP-N | AF525156 | NM_153738 |
| Prolactin-like protein-O | PLP-O | NM_026206 | --- |
| Growth hormone family member | |||
| Growth hormone | GH | NM_008117 | V01237 |
*Please note that additional candidate rat PRL family genes have been identified from the rat genome project, which have not yet been investigated.
Table 2.
Members of the bovine and ovine PRL and GH gene families
| Member | Abbreviation | Bovine GenBank Accession No. | Ovine GenBank Accession No. |
| Prolactin family members* | |||
| Prolactin | PRL | V00112 | M27057 |
| Placental lactogen | PL | NM_181007 | M31660 |
| Prolactin related protein-I | PRP-I | NM_174159 | --- |
| Prolactin related protein-II | PRP-II | M27239 | --- |
| Prolactin related protein-III | PRP-III | NM_174160 | --- |
| Prolactin related protein-IV | PRP-IV | M33269 | --- |
| Prolactin related protein-V | PRP-V | X15975 | --- |
| Prolactin related protein-VI | PRP-VI | X59504 | --- |
| Growth hormone family members | |||
| Growth hormone | GH | NM_180996 | AH005460 |
| Growth hormone2-Z | GH2-Z | --- | AH005492 |
*Please note that the list of bovine and ovine PRL family members is probably not complete. Additional bovine PRL family members and possibly ovine PRL members may be discovered as bovine and ovine genome projects move forward.
Table 3.
Members of the primate PRL and GH gene families
| Member | Abbreviation | Human GenBank Accession No. | Rhesus Monkey GenBank Accession No. |
| Prolactin family member | |||
| Prolactin | PRL | NM_000948 | U09018 |
| Growth hormone family members* | |||
| Growth hormone-N | GH-N | NM_000515 | L16556 |
| Chorionic somatomammotropin-L | CS-L, rhCS3 | NM_001318 | L16554 |
| Chorionic somatomammotropin-A | CS-A, rhCS1 | NM_001317 | L16552 |
| Growth hormone variant | GH-V | NM_002059 | U02293 |
| Chorionic somatomammotropin-B | CS-B, rhCS2 | NM_020991 | L16553 |
*Please note that it is difficult to precisely assign orthologs for the human and rhesus monkey CS genes and that multiple transcripts have been identified for each human GH family member. GenBank accession numbers are only presented for transcript variant 1.
Two primary exon-intron arrangements are evident: i) a prototypical five exon-four intron structure notable for both PRL and GH and many other members of the PRL and GH families and ii) a six exon-five intron structure present in members of the rodent PLP-C subfamily [2,4,7]. The latter exon/intron arrangement has been identified in both the rat and mouse and represents the addition of a short exon between exons II and III of the prototypical PRL or GH exon/intron organization. The mouse PLP-E gene is related to the latter subgroup but is unusual in that it has a seven exon-six intron structure, with two short exons situated between the prototypical exons II and III [4]. Each of the human GH family transcripts can undergo alternative processing [2]. The human PRL gene has two physically separated promoters, which are responsible for tissue-specific expression [18]. The extent of alternative promoter usage or post-transcriptional processing of PRL/GH family transcripts in other species is not well appreciated.
Translation products generated from the PRL and GH family transcripts exhibit sequence conservation, similarities in the positioning of cysteines, and likely conformation. All proteins are synthesized with a signal peptide of approximately 30 amino acids and are targeted for secretion. Many of the mature proteins receive post-translational addition of carbohydrates. Glycosylation patterns are cell type- and protein-dependent and may influence the bioavailability and/or biological activities of the proteins [19]. The three-dimensional structure for members of the PRL and GH families is predicted to be composed of a four-helix bundle connected in a unique "up-up-down-down" arrangement [20]. This structural arrangement facilitates interactions of PRL and GH with their receptors and their activation of intracellular signaling pathways. It is assumed that the natural variants of PRL and GH possess similar structures; however, it is not known how these structural features impact their biological activities.
Expression patterns
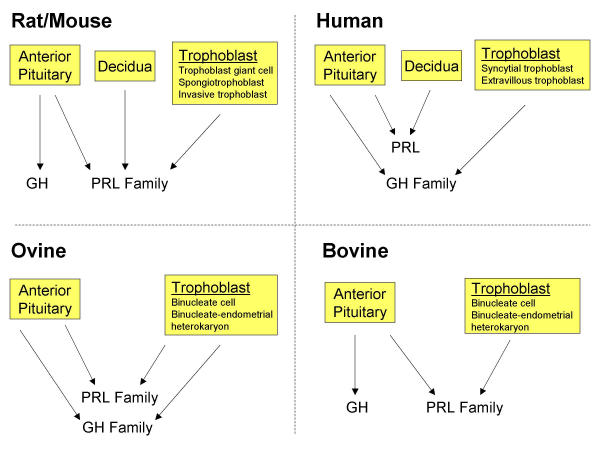
Members of the PRL and GH families of cytokines are produced in the anterior pituitary and at the maternal-fetal interface (Fig. 1). In mammalian species the anterior pituitary produces both PRL and GH. Somatotrophs, lactotrophs and somatolactotrophs of the anterior pituitary are important sources of both PRL and GH [21]. Anterior pituitary synthesis and secretion of PRL and GH are responsive to the integration of neural and endocrine signals. These two hormones, their sites of synthesis in the anterior pituitary, and their regulation are highly conserved across mammalian species. This degree of conservation is not as evident for members of the PRL and GH families expressed at the maternal-fetal interface. Decidual cells of primates and rodents produce members of the PRL family, as do rodent and ruminant trophoblast cells, while, primate and ovine trophoblast cells express members of the GH gene family (Fig. 1).
Figure 1.
Sources of PRL and GH family cytokines in rat and mouse, human, ovine, and bovine species. Abbreviations: PRL, prolactin; GH, growth hormone.
Uterus
During pregnancy, differentiated uterine stromal cells in primates and rodents are referred to as decidual cells and are prominent sources of PRL family gene expression [22,23]. Cellular events during decidual cell formation, a process referred to as decidualization; include proliferation and differentiation of uterine stromal cells. Decidual tissue effectively provides a barrier between the developing embryo and the remainder of the uterus. The control of decidualization differs in primates and rodents. Formation of decidual cells in primates is progesterone-dependent and occurs during the luteal phase of each menstrual cycle. Decidualization in the mouse and rat is similarly progesterone-dependent but also requires activatory signals provided by the implanting blastocyst [24]. In rodents, these events begin at the site of implantation within the antimesometrial endometrium (farthest from the incoming blood supply). Mesometrial and antimesometrial decidua are morphologically and functionally distinct. In primates, PRL, the sole member of the primate PRL family, is expressed in the uterine decidua [23,25]. As introduced above, lactotroph- and decidual cell-specific PRL expression are under the control of two physically separated promoters situated within the 5' flanking region of the prolactin gene and are responsive to distinct regulatory signals [18,23]. PRL of decidual origin is also glycosylated differently than is PRL from the anterior pituitary, which may contribute to distinct biological actions [26]. In rodents (mouse and rat), decidual cells localized within the antimesometrial uterine compartment express at least four members of the PRL family [decidual PRP, PLP-B, PLP-J, and PRL; [27-30]]. The regional expression pattern of the rodent decidual PRL family ligands is likely a key to ultimately understanding their physiology during the establishment of pregnancy.
Trophoblast cells
Trophoblast cells arise through the differentiation of a multilineage pathway and show elements of species specificity in their development and organization into placental structures. Rodents and primates possess a hemochorial placenta, an invasive form of placentation characterized by maternal blood directly bathing trophoblast cells; whereas ruminants possess a synepithelialchorial placenta, with minimal invasion and maximal cellular separation between maternal and fetal compartments [31,32]. Nutrient delivery is histiotrophic, via maternal uterine gland secretions, and/or hemotrophic, via the maternal blood [31,33-35]. Histiotrophic nutrition occurs throughout pregnancy in ruminants and during early phases of gestation in rodents and primates. Hemotrophic nutrition predominates following establishment of the hemochorial placenta.
Rodents
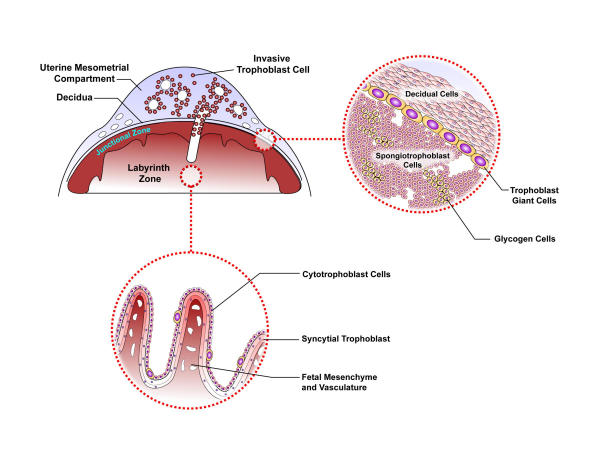
Three regions within the mature mouse and rat uteroplacental site possess trophoblast cells: the labyrinth zone, the junctional zone, and the uterine mesometrial compartment [Fig. 2]. PRL family members are expressed in cell type-, location-, and temporal-specific profiles [36]. In rodents, three different trophoblast cell lineages are capable of expressing members of the PRL family: trophoblast giant cells, spongiotrophoblast cells, and invasive trophoblast cells [3,4,37,38]. Each cell type expresses a unique subset of PRL family members. Locations of hormone/cytokine production influence post-translational processing and temporal and spatial access to biological targets. Trophoblast giant cells form the early interface with the maternal environment. Once the chorioallantoic placenta is established trophoblast giant cells localize to the junctional zone and during the last week of gestation this same cell type arises within the labyrinth zone. The labyrinth zone of the chorioallantoic placenta is principally involved in bidirectional nutrient/waste transport between maternal and fetal compartments, functions assumed by syncytial trophoblast cells [39]. The transient development of trophoblast giant cells within the labyrinth zone may facilitate hormone/cytokine delivery into the fetal compartment. Transcriptional control of trophoblast giant cell-specific gene expression has been investigated using the Rcho-1 trophoblast cell line [41,42]. Spongiotrophoblast cells arise just prior to midgestation, are prominent cellular constituents of the junctional zone, and a major source of most PRL family members throughout the latter half of gestation. Secretory products of spongiotrophoblast cells are particularly abundant in maternal circulation. An in vitro cell culture system for studying spongiotrophoblast cell expression of PRL family members has been established [42]. During the last week of gestation, a population of trophoblast cells exits the chorioallantoic placenta. These invasive trophoblast cells are more prominent in the rat where they invade deep into the uterine mesometrial compartment forming intimate relationships with spiral arteries, producing a unique subset of PRL family members [37,38]. The invasive trophoblast cells have first access to blood-borne nutrients, toxic compounds, signaling molecules, and cells entering the uterine environment. In vitro systems for investigating invasive trophoblast cells have not been described. The two remaining lineages of rodent trophoblast cells, glycogen cells and syncytial trophoblast cells, do not express members of the rodent PRL family. However, mouse syncytial trophoblast cells, which share transport properties with human syncytial trophoblast cells, do possess the transcriptional machinery needed to activate transgenic DNA constructs containing the locus control region of the human GH gene family [43]. The conservation suggests an interesting analogous relationship between syncytial trophoblast cells of humans and rodents. Promoter regions for the mouse and rat PL-II genes have been identified that direct placental-specific expression in transgenic mice [14,44-46]. At least one member of the PRL family has been shown to exhibit allele-specific expression patterns. These experiments were performed in deer mice, Peromyscus [47]. Peromyscus PL-Iv expression is restricted to the paternal allele and to spongiotrophoblast cells of the chorioallantoic placenta. Interestingly, spongiotrophoblast cell development is regulated by a maternally imprinted transcription factor, Mash2 [48], and is abnormally affected by disruptions in genomic imprinting, as seen in placentas from cloned mice [49] and following interspecies breeding [50,51]. At present, it is not known whether genomic imprinting is a feature shared among other members of the Peromyscus PRL family or between PRL and GH family loci from other species; nor is the biological significance of the allele-specific expression pattern appreciated.
Figure 2.
A schematic representation of a late gestation rat uteroplacental compartment. Please note that trophoblast cells located in the uterine mesometrial compartment, junctional zone, and labyrinth zone all contribute to the production of PRL family ligands.
Ruminants
Three trophoblast cell types are associated with placentation in ruminants: i) uninucleate trophoblast cells, ii) binucleate trophoblast cells, and iii) heterokaryons formed by the fusion of binucleate trophoblast cells with endometrial epithelial cells. Uninucleate trophoblast cells serve as precursors for binucleate cell formation. Binucleate trophoblast cells and trophoblast-endometrial heterokaryons of sheep and cattle produce PLs [1,52]. PRP-I shows a similar expression pattern in the bovine placenta [53]. Expression of PLs and PRP-I begins around the time of the initial embryo-uterine interactions [1,7,53]. The cellular localization and ontogeny of placental expression for other members of the bovine PRL family have not been reported. Ovine placental GH is expressed in each of the above cell populations during a restricted period of gestation (days 35 to 70), and is controlled by mechanisms distinct from the regulation of GH production by the anterior pituitary [6]. A new bovine trophoblast cell differentiation culture system capable of binucleate cell formation and PL synthesis offers great promise for future in vitro analyses of PRL and GH family gene expression [54,55].
Primates
Transcripts for members of the GH family (GH-V, CS-A, CS-B, and CS-L) have each been detected in the human placenta [2,23]. CS-A and CS-B genes encode for identical mature secreted proteins, collectively referred to as CS, and are regulated similarly [2,23]. Although, CS-L transcripts have been identified, they likely contribute to low levels of translated products [2]. Two principal trophoblastic structures exist within the human placenta: villous and extravillous trophoblast cells. Syncytial trophoblast cells of the villous portion of the chorioallantoic placenta are a major source of GH family members, as are extravillous invasive trophoblast [2,23,56]. The latter cell population is analogous with the invasive trophoblast of the rodent placenta and merits additional investigation regarding their expression of GH family cytokines. Transcription of GH family genes is regulated by a locus control region located 23 kb upstream of the cluster [43,57]. Four members of the rhesus monkey GH family (CS-1, CS-2, CS-3, and GH-V), possessing extensive homology with human GH family members, are expressed in the monkey chorioallantoic placenta [58].
Other sites
There is some evidence for the activation or reactivation of PRL and GH family members in a variety of tissues, especially those exhibiting pathology. Local PRL production has been detected in the reproductive tract, mammary tissue, immune cells, brain, and skin [21]; GH and GH-V biosynthesis have been demonstrated in hematopoietic lineages [2,6]; and CS transcripts detected in the testis [59]. In the mouse, PLF can be expressed in tissues undergoing angiogenesis, such as wounds and tumors [60,61], and thrombocytopenia activates PLP-E expression in hematopoietic tissues (bone marrow and spleen; [62]. Such findings are not surprising since the targets that these cytokines act on during the establishment of pregnancy (see below) may also be relevant in diseased tissues.
Hormone/cytokine delivery and tissue distribution
A number of members of the PRL and GH family are present in maternal circulation and a few members have been detected in the fetus, presumably acting as classic hormones [6,8,23,52,63,64]. Ligand access and availability to target cells can be influenced by post-translational modifications, interactions with circulating binding proteins, and/or association with extracellular matrices. Addition of carbohydrate influences clearance rates of rodent PLs [63,65]. Circulating proteins such as the extracellular domain of the GH receptor (GH-R) and α2-macroglobulin bind members of the PRL and GH cytokine families [66-68]. Some members of the PRL family appear to be restricted in their distribution because of interactions with the extracellular matrix. Decidual PRP and human PRL bind to heparin containing molecules and accumulate in the decidual extracellular matrix [69-71]. Such a location is ideal for gaining access to cells that traverse the decidua (e.g. inflammatory and trophoblast cells) and in limiting cytokine exposure to extrauterine sites. A few members of the PRL and GH families have been reported to enter the fetal circulation, whereas others are selectively excluded from the fetus [63,64,72-76]. Human CS gains entry into the fetal circulation, while GH-V is excluded [23]. Although GH-V is secreted into the maternal circulation of the human, ovine placental GH has a more restricted sphere of influence within the uteroplacental compartment and fetus [6]. The decidual source of ligands may also facilitate their entry into amniotic fluid and ultimately the fetus [23]. Temporal profiles of secretion into maternal and fetal compartments are not equivalent and can vary during the course of pregnancy within a species and among species. For example, in sheep, concentrations of PL in maternal circulation attain relatively high levels and progressively increase as gestation proceeds, whereas fetal concentrations are considerably lower and peak at midgestation [73]. PL concentrations in the cow show a similar profile in the fetus but, in contrast to sheep, are low in maternal circulation throughout gestation [72]. Our current information about hormone delivery and transport within maternal and fetal compartments is limited to only a few examples and thus provides us with only glimpse of what to expect.
Biological actions
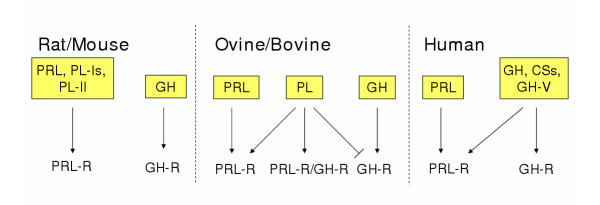
Biology of the PRL and GH families of cytokines can be subdivided into two broad categories: i) classical and ii) nonclassical. Classical actions correspond to biological effects mediated through interactions with PRL and/or GH receptors (Fig. 3). Nonclassical actions reflect all other mechanisms of ligand-mediated biological activities.
Figure 3.
Classical ligand-receptor relationships in rat and mouse, ovine and bovine, and human species. Arrows denote stimulatory actions; lines without arrows reflect inhibitory actions. Abbreviations: PRL, prolactin; PL-Is, placental lactogen-Iα, β, γ, and PL-I variant; PL-II, placental lactogen-II; GH, growth hormone; PL, placental lactogen; CSs, chorionic somatomammotropins; GH-V, growth hormone variant; PRL-R, PRL receptor; GH-R, GH receptor; PRL-R/GH-R, PRL-R and GH-R heterodimer.
Classical actions
Ligand-receptor systems contributing to classical actions exhibit species differences. In some species, the PRL receptor (PRL-R) is the target of ligands produced at the maternal-fetal interface, while in other species, hormones/cytokines are produced that activate both PRL and GH receptors.
Rodents
The mammary gland and corpus luteum represent two classic biological targets of a subset of the PRL family [77-83]. The physiological responses of these targets to PRL family members are mediated by PRL-Rs. Ligands activating PRL-Rs during pregnancy arise from maternal (lactotroph and decidual: PRL), trophoblast (PL-Is, PL-Iv, PL-II), and fetal (lactotroph: PRL) tissues (Fig. 3). Production of these ligands is orchestrated in a precise temporal pattern that ensures the presence of activators of the PRL-R signaling system throughout gestation [3,63,84-86] (Fig. 4). The PRL-R agonists stimulate mammary epithelial cell growth and differentiation and help maintain corpus luteum integrity and progesterone and relaxin biosynthesis. Phenotypes of PRL-/- and PRL-R-/- null mutant mouse models support the importance of the PRL-R signaling pathway in mammary gland and luteal biology [87]. Investigations using gene-targeting approaches are inherently more complex for studying the biology of the multiple PL genes expressed in trophoblast cells. In addition to the mammary gland and corpus luteum, evidence has arisen for PRL and the PLs modulating pregnancy-dependent changes in brain function, pancreas biology, and the immune system [88-92]. Local production of PRL by decidual cells may also contribute to decidual cell survival [93,94]. The existence of multiple ligands activating the PRL-R signaling pathway provides for complementarity and specialization in ligand regulation, availability, and possibly action [87].
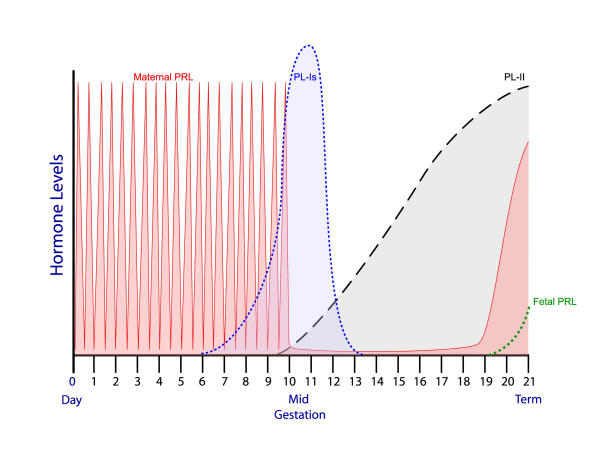
Figure 4.
A schematic representation of lactogenic hormone profiles during pregnancy in the mouse or rat. Early pregnancy is dominated by the twice-daily surges of PRL produced by the maternal anterior pituitary; midgestation by the products of three PL-I genes; and the second half of gestation by PL-II. The fetal anterior pituitary begins producing PRL at the end of pregnancy. Abbreviations: PRL, prolactin, PL-Is, placental lactogen-Iα, β, γ; PL-II, placental lactogen-II.
Insights into intracellular events subsequent to PRL-R activation by members of the placental PRL family are minimal. It is assumed that PLs activate signal transduction pathways downstream of the PRL-R similar to those activated by PRL. Beyond the abilities of members of the PL-I subfamily to stimulate the Jak-Signal Transducer and Activator of Transcription (STAT) pathway in Nb2 lymphoma cells [95,96] and cultured luteinized granulosa cells [80], information on intracellular signaling pathways triggered by classical members of the placental PRL family is lacking. It seems likely that at least some aspects of PRL signaling are mimicked by PLs; however, whether these uteroplacental ligands reproduce the full spectrum of PRL action or elicit unique responses is unknown. The autocrine/paracrine actions of PRL within the decidua appear to be mediated by activation of classic PRL ligand-PRL-R signal transduction, including stimulation of Jak2-STAT5 and phosphatidylinositol 3-kinase/Akt signaling pathways [93,94].
Ruminants
PLs have been postulated to affect maternal (corpus luteum function, uterine gland development, intermediary metabolism, and mammary gland development) and fetal physiological processes [7]. The general implication is that PLs contribute to normal physiological adjustments occurring during pregnancy; promoting corpus luteum function, uterine and mammary gland development, and facilitating nutrient delivery and utilization by the fetus. These predictions have not always been supported by empirical observation. The absence of supportive evidence may at times be attributed to experimental design or may instead reflect a more subtle and potentially unique role of PLs in the biology of pregnancy (see below). Some of the most impressive physiological data relates to the local actions of PLs on uterine histiotrophic nutrient delivery, which is of paramount importance in ruminants [34]. PL binds to endometrial gland epithelia and is capable of increasing uterine gland density and uterine milk protein expression [97,98]. These in vivo actions of PL are dependent upon the hormonal milieu and require exposure to progesterone and interferon tau. Ovine placental GH likely contributes to the pregnancy-dependent functional changes in uterine gland function [97,98] and may also influence fetal tissues prior to the ontogeny of fetal GH production. A point of consideration is warranted regarding comparisons of the biology of PLs in sheep and cattle. Ovine and bovine PLs have undergone significant sequence divergence, especially when compared to the minimal divergence for their respective PRLs and GHs [99]. These structural changes have contributed to some differences in regulation and bioavailability and potentially in activities of the PLs from each species.
Ruminant PLs offer a unique mode of action. It has been long known that PLs of ruminants interact with both PRL and GH receptors [7,9,52]. Early evidence suggested that ruminant PLs were dual-acting agonists for both PRL-R and GH-R signaling pathways and there was some suggestion of a unique receptor system for ruminant PLs. Many of these early experiments were performed with receptors isolated from non-ruminant species. It is now known that ruminant PLs effectively signal through PRL-R homodimers, PRL-R/GH-R heterodimers, and in the absence of PRL-R may act as a GH-R antagonist [[9]; Fig. 3]. Some of the GH-like effects of PLs may be mediated by their interaction with PRL-R/GH-R heterodimers. Ruminant PLs activate Jak/STAT and mitogen activated protein kinase (MAPK) signaling pathways [7,9,100]. The nature of PL signal transduction appears to differ depending upon whether PRL-R homodimers or PRL-R/GH-R heterodimers are activated. Ligand-receptor heterodimer interaction results in prolonged STAT3 activation, which may culminate in distinct cellular responses [100]. Existence of a separate ruminant PL receptor remains elusive. At this juncture, ovine placental GH is thought to act via mechanisms identical to GH produced by the anterior pituitary [6,7].
Primates
Classical PRL-R and GH-R mediated actions in primates include those stimulated by decidual PRL and placental members of the GH family. Human decidual PRL may accumulate in the decidual extracellular matrix due to its association with heparin containing molecules [69] and its targets include, intrauterine (e.g. modulatory effects on uterine gland development and function, angiogenesis, trophoblast cell development and function, and immune regulation), amniotic, and possibly fetal tissues [23,25]. There is an intriguing parallel between the intrauterine effects of decidual PRL and those of ruminant PLs on histiotrophic nutrient delivery. Similarly, CSs and GH-V have been implicated in the modulation of maternal physiology and directly or indirectly as regulators of fetal growth and development, as suggested for ruminant PLs [7,23].
Genetic deletions impacting CS and GH-V have been described and offer insight into the biology of members of the placental GH family [2,23,101-103]. In several instances the deficiencies of CS and GH-V had no demonstrable impact on the progression of pregnancy [101,102]; however, a patient with CS and GH-V deficiencies exhibiting pregnancy-associated pathologies (including preeclampsia and intrauterine growth restriction) has been described [103]. Pathologies in the latter clinical presentation may or may not be related to the absence of CS and GH-V proteins. Although, the anecdotal nature of the clinical findings makes it difficult to make absolute conclusions about the biology of the placental GH family, they suggest that the CS and GH-V genes may be dispensable in some pregnancies.
The basis for much of the speculation concerning CS and GH-V biology relates to their close structural similarity with pituitary-derived GH (GH-N) and the nature of their interactions with PRL and GH receptors [2,7,23]. Unlike rodent and ruminant GHs, primate GH-Ns function as potent agonists for both PRL-R and GH-R signaling. In comparison to GH-N, CSs are equipotent activators of PRL-R signaling and weak GH-R agonists, whereas GH-V is a more potent GH-R agonist, and a less potent PRL-R agonist. Introduction of these subtle functional variants into maternal and fetal circulation during pregnancy would seem to be an effective means of modulating PRL-R and GH-R activation and thus maternal and fetal physiology. Based on PRL-R and GH-R expression profiles, PRL- or CS-activated PRL-R signaling is probably more significant in the fetus than is GH-R signaling. Some evidence exists for a unique CS receptor present in human fetal tissues; however, the molecular structure of the receptor has not been reported [23]. Signaling via PRL-R/GH-R heterodimers has not been described for members of the primate GH family but would seem to be a means of expanding the diversity of signals emanating from the ligands. PRL-R and GH-R signal transduction cascades activated by PRL and GH family ligands arising at the maternal-fetal interface have not been extensively investigated but are likely similar to those activated by anterior pituitary-derived PRL and GH and involve the Jak/STAT pathway among other signaling pathways.
Nonclassical actions
All members of the PRL family do not utilize the PRL receptor-signaling pathway. Distinct targets and biological actions are evident and are collectively referred to as nonclassical. The nonclassical ligands are generated as the primary products of individual genes and as post-translational products of PRL.
Much of our knowledge of nonclassical ligands arising as primary products of individual genes has been garnered from experimentation with the mouse and rat. Within the mouse PRL family locus five genes encode for PRL-R agonists. These include the PRL gene, three PL-I genes, and a PL-II gene. Evidence for PRL-R usage by protein products of the remaining 21 genes is not available. Some have not been examined but others do not utilize the PRL receptor. Cellular targets for nonclassical members of the PRL family are intriguing in that each is known to undergo profound adjustments during the course of pregnancy. They include endothelial cells, erythrocyte and megakaryocyte precursors, natural killer cells, eosinophils and hepatocytes (Table 4). PLF and PLF-RP target endothelial cells and have agonist and antagonist effects on angiogenesis, respectively [3,104]. PLF may also be involved in coordinating responses to tissue injury [60]. Increased maternal demands for erythrocytes and platelets are stimulated through the actions of PLP-E and PLP-F [105-107], while PLP-A modulates the behavior of uterine natural killer cells, a specialized cell population contributing to maternal immune responses and uteroplacental vascular remodeling [108,109]. Specific interactions have been identified for decidual PRP with eosinophils [71] and for PLP-C and PLP-N with hepatocytes [S. Ohboshi, J. Bustamante, M.J. Soares, unpublished results]. The biological responses of these targets to the ligands have not been reported. Other PRL family ligands (rat PLP-B, PLP-D, and PLP-H; bovine PRP-I) do not activate the PRL-R signaling pathway; however, their physiological targets have not been identified [68,110,111]. As is true, for classical members, the overall physiological significance of nonclassical members of the PRL family is yet to be determined.
Table 4.
Nonclassical targets for members of the PRL family
| Biological Targets | Ligands (function) | References |
| Endothelial cells | PLF (angiogenic) PLF-RP (anti-angiogenic) 16kDa-PRL (anti-angiogenic) |
104, 112–115 |
| Hematopoietic precursors | PLP-E and PLP-F (promote erythrocyte and megakaryocyte development) | 105–107 |
| Natural killer cells | PLP-A (modulator of the uterine natural killer cell phenotype) | 108, 109 |
| Eosinophils | Decidual PRP (unknown) | 71 |
| Hepatocytes | PLP-C, PLP-N (unknown) | S. Ohboshi, J. Bustamante, M.J. Soares, unpublished results |
Insights into the mechanism of action of nonclassical PRL family members are fragmentary. PLF effects on endothelial cells appear to be mediated by the insulin-like growth factor II/mannose 6-phosphate receptor and involve G proteins regulating MAPK activity [112,113]. These actions appear to require a mannose 6-phosphate modification to PLF. In contrast, even though receptors for PLF-RP and PLP-E have not been identified, some insights into the signaling pathways affected by the ligands have been determined. PLF-RP inhibits angiogenesis by inhibiting arachidonic acid release [114], while PLP-E stimulates hematopoiesis through signal transduction cascades involving the gp130 co-receptor and Jak/STAT pathways [105,106]. The heparin-binding capacity of human decidual PRL and rat decidual PRP may modulate their ligand-receptor interactions, as well as those for other heparin-binding ligands.
PRL can be proteolytically processed into an N-terminal 16 kDa species possessing novel nonclassical actions [87,115]. 16 kDa PRL targets endothelial cells and is a potent angiostatic factor. PRL cleavage can occur at its site of synthesis or at its target tissues and may be mediated by cathepsin D. The 16 kDA PRL proteolytic cleavage product does not interact with the PRL-R. The receptor transducing the angiostatic actions of 16 kDa PRL has not been identified but its association with 16 kDa PRL leads to an inhibition of the MAPK signaling pathway [87,115]. As stated above, PLF-RP is also angiostatic but it is unclear whether 16 kDA PRL and PLF-RP share a common receptor and mechanism of action. Generation of the 16 kDa PRL at the maternal-fetal interface and its involvement in controlling uteroplacental vascular remodeling has not been determined. Nonetheless, some have hypothesized that excessive decidual production of 16 kDa PRL may lead to preeclampsia, a pregnancy disorder associated with defective uteroplacental vascular remodeling [116]. It is logical that other members of the PRL and GH families might be susceptible to similar proteolytic processing. At this time, naturally occurring truncations of other PRL and GH family ligands have not been described.
Conclusions
A few final observations regarding the biology of the PRL and GH families may be helpful.
For the purposes of this presentation, discussions about classical and nonclassical biological actions were segregated; however, this distinction for a ligand need not be mutually exclusive. It is certainly possible that some PRL or GH family ligands might be capable of activating both classical and nonclassical pathways. Post-transcriptional or post-translational processing may lead to the generation of ligands with expanded signaling capabilities.
Under some circumstances ligand families and their corresponding receptor families have undergone coordinate evolution [11]. At first analysis, this does not seem to be the case for the PRL and GH families. There is no evidence for expanded PRL-R or GH-R gene families. Thus, expanded receptor systems must have been achieved by other means. These may include: i) post-transcriptional and/or post-translational processing of the PRL-R and GH-R resulting in an expansion of ligand specificities and downstream signaling events; ii) the use of the PRL or GH family ligands as backbones for other receptor activators, such as carbohydrates; iii) utilization of distinct classes of receptor signaling molecules. Evidence for the latter speculation is seen in the insulin family, where some family members use receptor tyrosine kinases and others use G protein-coupled receptors [117]. The difficulty is that we are not aware of the selective pressures that drove the expansion of PRL and GH families of ligands. These evolutionary forces presumably led to meaningful advantages for the survival of each species. This discussion also makes the assumption that all members of the PRL and GH families act as ligands, which still remains to be proven.
Research on the biology of the pregnancy-associated PRL and GH families is laden with experiments questioning their physiological significance. This is best illustrated by data, cited above, that pregnancies can proceed in the absence of CSs and GH-V in humans [2,23,101-103]. Preliminary findings in the mouse suggest that null mutations of individual uteroplacental PRL family genes may also be compatible with uneventful pregnancies (R. Ain, G. Dai, A. Godwin, M.J. Soares, unpublished results). Such observations may seem to be in conflict with evolutionary dogma. Why would the PRL and GH family gene expansions be retained if they were not advantageous to the organism? Some insight may be obtained from ideas presented by Kaplan and Grumbach over two decades ago [118]. They proposed that CSs serve as regulatory molecules ensuring adaptation to physiological stressors, especially those creating increased metabolic demands. Dorshkind and Horseman [92] have similarly advanced the proposal that PRL is a key regulatory molecule involved in immune system adaptations to stress. The selective pressures driving the PRL and GH family expansions did not occur in the laboratory or with the availability of a reliable food supply and modern medical care. Consequently, the true biology of the PRL and GH families may only become apparent when the organisms are presented with environmental challenges. The nature of the challenges may differ depending upon the niche the species occupies. Maybe such logic also helps explain why there is considerable diversity among rodent, ruminant, and primate PRL and GH families.
Finally, the limited species conservation violates a popular tenet of modern biomedical research and has driven talented scientists from this important field. Understanding the physiology of the PRL and GH families will provide access to important regulatory processes of pregnancy. In some instances, cross species similarities may prevail, while in other cases the differences may be most compelling. Nonetheless, our appreciation for the biology of pregnancy increases.
Acknowledgments
Acknowledgments
Reviews have been cited, where possible, instead of an exhaustive list of primary research reports. Consequently, I apologize to scientists who have contributed to research on the biology of the PRL and GH families and whose work has not been cited. Research from the author's laboratory is supported by grants from the National Institutes of Health (HD20676, HD39878) and the Hall Family Foundation. I would like to acknowledge members of my laboratory (past and present) for their valuable contributions and discussions. I thank Mr. Stanton Fernald for his assistance with some of the figures.
References
- Schuler LA, Kessler MA. Bovine placental prolactin-related hormones. Trends Endocrinol Metab. 1992;3:334–338. doi: 10.1016/1043-2760(92)90112-E. [DOI] [PubMed] [Google Scholar]
- Cooke NE, Liebhaber SA. Molecular biology of the growth hormone-prolactin gene system. Vitamins Horm. 1995;50:385–459. doi: 10.1016/s0083-6729(08)60659-7. [DOI] [PubMed] [Google Scholar]
- Soares MJ, Linzer DIH. The rodent prolactin family and pregnancy. In: Horseman ND, editor. Prolactin. Norwell, MA, Kluwer Academic Publishers; 2001. pp. 139–167. [Google Scholar]
- Wiemers DO, Shao L-J, Ain R, Dai G, Soares MJ. The mouse prolactin gene family locus. Endocrinology. 2003;144:313–325. doi: 10.1210/en.2002-220724. [DOI] [PubMed] [Google Scholar]
- Wallis M, Lioupis A, Wallis OC. Duplicate growth hormone genes in sheep and goat. J Mol Endocrinol. 1998;21:1–5. doi: 10.1677/jme.0.0210001. [DOI] [PubMed] [Google Scholar]
- Lacroix M-C, Guibourdenche J, Frendo J-L, Pidoux G, Evain-Brion D. Placental growth hormones. Endocrine. 2002;19:73–79. doi: 10.1385/ENDO:19:1:73. [DOI] [PubMed] [Google Scholar]
- Anthony RV, Limesand SW, Fanning MD, Liang R. Placental lactogens and growth hormone. In: Bazer FW, editor. In The Endocrinology of Pregnancy. Totowa, NJ Humana Press Inc; 1998. pp. 461–490. [Google Scholar]
- Soares MJ, Dai G, Orwig KE, Peters TJ, Müller HM. The uteroplacental prolactin family and pregnancy. Biol Reprod. 1998;58:273–284. doi: 10.1095/biolreprod58.2.273. [DOI] [PubMed] [Google Scholar]
- Gertler A, Djiane J. Mechanism of ruminant placental lactogen action: molecular and in vivo studies. Mol Genet Metab. 2002;75:189–201. doi: 10.1006/mgme.2002.3303. [DOI] [PubMed] [Google Scholar]
- Waterston RH, Lindblad-Toh K, Birney E, Rogers J, Abril JF, Agarwal P, Agarwala R, Ainscough R, Alexandersson M, An P, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Fryxell KJ. The coevolution of gene family trees. Trends Genet. 1996;12:364–369. doi: 10.1016/S0168-9525(96)80020-5. [DOI] [PubMed] [Google Scholar]
- Wallis M. The expanding growth hormone/prolactin family. J Mol Endocrinol. 1992;9:185–188. doi: 10.1677/jme.0.0090185. [DOI] [PubMed] [Google Scholar]
- Talamantes F, Ogren L, Markoff E, Woodard S, Madrid J. Phylogenetic distribution, regulation of secretion, and prolactin-like effects of placental lactogens. Fed Proc. 1980;39:2582–2587. [PubMed] [Google Scholar]
- Öztürk A, Fresnoza A, Savoie A, Duckworth HW, Duckworth ML. Defining regulatory regions in the rat prolactin gene family locus usinga large P1 genomic clone. Endocrinology. 2003;144:4742–4754. doi: 10.1210/en.2003-0591. [DOI] [PubMed] [Google Scholar]
- Jackson-Grusby LL, Pravtcheva D, Ruddle FH, Linzer DIH. Chromosomal mapping of the prolactin/growth hormone gene family in the mouse. Endocrinology. 1988;122:2462–2466. doi: 10.1210/endo-122-6-2462. [DOI] [PubMed] [Google Scholar]
- Cooke NE, Szpirer C, Levan G. The related genes encoding growth hormone and prolactin have been dispersed to chromosomes 10 and 17 in the rat. Endocrinology. 1986;119:2451–2454. doi: 10.1210/endo-119-6-2451. [DOI] [PubMed] [Google Scholar]
- Dietz A, Georges M, Threadgill DW, Womack JE, Schuler LA. Somatic cell mapping, polymorphism, and linkage analysis of bovine prolactin related proteins and placental lactogens. Genomics. pp. 137–143. [DOI] [PubMed]
- Telgmann R, Gellersen B. Marker genes of decidualization: activation of the decidual prolactin gene. Human Reprod Update. 1998;4:472–479. doi: 10.1093/humupd/4.5.472. [DOI] [PubMed] [Google Scholar]
- Manzella SM, Dharmesh SM, Cohick CB, Soares MJ, Baenziger JU. Developmental regulation of a pregnancy-specific oligosaccharide structure, NeuAcα2, 6GalNAcβ1, 4GlcNAc on select members of the rat placental prolactin family. J Biol Chem. 1997;272:4775–4782. doi: 10.1074/jbc.272.8.4775. [DOI] [PubMed] [Google Scholar]
- Goffin V, Shiverick KT, Kelly PA, Martial JA. Sequence-function relationships within the expanding family of prolactin, growth hormone, placental lactogens, and related proteins in mammals. Endocr Rev. 1996;17:385–410. doi: 10.1210/er.17.4.385. [DOI] [PubMed] [Google Scholar]
- Ben-Jonathan N, Mershon JL, Allen DL, Steinmetz RW. Extrapituitary prolactin: distribution, regulation, functions, and clinical aspects. Endocr Rev. 1996;17:639–669. doi: 10.1210/er.17.6.639. [DOI] [PubMed] [Google Scholar]
- Orwig KE, Rasmussen CA, Soares MJ. Decidual signals in the establishment of pregnancy: the prolactin family. Trophoblast Research. 1997;10:329–343. [Google Scholar]
- Handwerger S, Brar A. Human uteroplacental lactogens: physiology and molecular biology. In: Horseman ND, editor. Prolactin. Norwell, MA, Kluwer Academic Publishers; 2001. pp. 169–187. [Google Scholar]
- DeFeo VJ. Decidualization. In: Wynn RM, editor. In Cellular Biology of the Uterus. New York Appleton-Century-Crofts; 1967. pp. 191–290. [Google Scholar]
- Jabbour HN, Critchley HOD. Potential roles of decidual prolactin in early pregnancy. Reproduction. 2001;123:197–205. doi: 10.1530/rep.0.1210197. [DOI] [PubMed] [Google Scholar]
- Lee DW, Markoff E. Synthesis and release of glycosylated prolactin by human decidua in vitro. J Clin Endocrinol Metab. 1986;62:990–993. doi: 10.1210/jcem-62-5-990. [DOI] [PubMed] [Google Scholar]
- Roby KF, Deb S, Gibori G, Szpirer C, Levan G, Kwok SCM, Soares MJ. Decidual prolactin related protein: identification, molecular cloning, and characterization. J Biol Chem. 1993;268:3136–3142. [PubMed] [Google Scholar]
- Prigent-Tessier A, Tessier C, Hirosawa-Takamori M, Boyer C, Ferguson-Gottschall S, Gibori G. Rat decidual prolactin. Identification, molecular cloning, and characterization. J Biol Chem. 1999;274:37982–37989. doi: 10.1074/jbc.274.53.37982. [DOI] [PubMed] [Google Scholar]
- Toft DJ, Linzer DIH. Prolactin (PRL)-like protein J, a novel member of the PRL/growth hormone family, is exclusively expressed in maternal decidua. Endocrinology. 1999;140:5095–5101. doi: 10.1210/en.140.11.5095. [DOI] [PubMed] [Google Scholar]
- Dai G, Wang D, Liu B, Kasik JW, Müller H, White RA, Hummel GS, Soares MJ. Three novel paralogs of the rodent prolactin gene family. J Endocrinol. 2000;166:63–75. doi: 10.1677/joe.0.1660063. [DOI] [PubMed] [Google Scholar]
- Rossant J, Cross JC. Extraembryonic lineages. In: Rossant J, Tam PPL, editor. In Mouse Development. New York, Academic Press; 2002. pp. 155–180. [Google Scholar]
- Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Hempstock J, Jaunizux E. Nutrition of the human fetus during the first trimester. Placenta. 2001;22:S70–S77. doi: 10.1053/plac.2001.0639. [DOI] [PubMed] [Google Scholar]
- Bazer FW. Uterine protein secretions: relationship to development of the conceptus. J Anim Sci. 1975;41:1376–1382. doi: 10.2527/jas1975.4151376x. [DOI] [PubMed] [Google Scholar]
- Enders AC, Welsh AO. Structural interactions of trophoblast and uterus during hemochorial placenta formation. J Exp Zool. 1993;266:578–587. doi: 10.1002/jez.1402660608. [DOI] [PubMed] [Google Scholar]
- Dai G, Lu L, Tang S, Peal MJ, Soares MJ. Prolactin family miniarray: a tool for evaluating uteroplacental-trophoblast endocrine cell phenotypes. Reproduction. 2002;124:755–765. doi: 10.1530/rep.0.1240755. [DOI] [PubMed] [Google Scholar]
- Ain R, Canham LN, Soares MJ. Gestational stage-dependent intrauterine trophoblast cell invasion in the rat and mouse: novel endocrine phenotype and regulation. Dev Biol. 2003;260:176–190. doi: 10.1016/S0012-1606(03)00210-0. [DOI] [PubMed] [Google Scholar]
- Wiemers DO, Ain R, Ohboshi S, Soares MJ. Migratory trophoblast cells express a newly identified member of the prolactin gene family. J Endocrinol. 2003;179:335–346. doi: 10.1677/joe.0.1790335. [DOI] [PubMed] [Google Scholar]
- Knipp GT, Audus KL, Soares MJ. Nutrient transport across the placenta. Adv Drug Delivery Rev. 1999;38:41–58. doi: 10.1016/S0169-409X(99)00005-8. [DOI] [PubMed] [Google Scholar]
- Orwig KE, Wolfe MW, Cohick CB, Dai G, Peters TJ, Soares MJ. Trophoblast-specific regulation of endocrine-related genes. Trophoblast Research. 1998;11:65–85. [Google Scholar]
- Peters TJ, Chapman BM, Soares MJ. Trophoblast differentiation: an in vitro model for trophoblast giant cell development. In: Tuan RS, Lo CW, editor. Developmental Protocols. Humana Press, Inc, Totowa, NJ; 2000. pp. 301–311. [DOI] [PubMed] [Google Scholar]
- Lu X-J, Deb S, Soares MJ. Spontaneous differentiation of trophoblast cells along the spongiotrophoblast pathway: expression of the placental prolactin gene family and modulation by retinoic acid. Dev Biol. 1994;163:86–97. doi: 10.1006/dbio.1994.1125. [DOI] [PubMed] [Google Scholar]
- Su Y, Liebhaber SA, Cooke NE. The human growth hormone gene cluster locus control region supports position-independent pituitary- and placenta-specific expression in transgenic mice. J Biol Chem. 2000;275:7902–7909. doi: 10.1074/jbc.275.11.7902. [DOI] [PubMed] [Google Scholar]
- Shida MM, Ross SR, Linzer DIH. Placental-specific expression in transgenic mice from the mouse placental lactogen-II gene promoter. Proc Natl Acad Sci, USA. 1992;89:3864–3868. doi: 10.1073/pnas.89.9.3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah P, Sun Y, Szpirer C, Duckworth ML. Rat placental lactogen II gene: characterization of gene structure and placental-specific expression. Endocrinology. 1998;139:967–973. doi: 10.1210/en.139.3.967. [DOI] [PubMed] [Google Scholar]
- Lin J, Linzer DIH. Identification of trophoblast-specific regulatory elements in the mouse placental lactogen II gene. Mol Endocrinol. 1998;12:418–427. doi: 10.1210/me.12.3.418. [DOI] [PubMed] [Google Scholar]
- Vrana PB, Matteson PG, Schmidt JV, Ingram RS, Joyce A, Prince KL, Dewey MJ, Tilghman SM. Genomic imprinting of a placental lactogens gene in Peromyscus. Dev Genes Evol. 2001;211:523–532. doi: 10.1007/s00427-001-0188-x. [DOI] [PubMed] [Google Scholar]
- Guillemot F, Caspary T, Tilghman SM, Copeland NG, Gilbert DJ, Jenkins NA, Anderson DJ, Joyner AL, Rossant J, Nagy A. Genomic imprinting of Mash2, a mouse gene required for trophoblast development. Nature Genetics. 1995;9:235–241. doi: 10.1038/ng0395-235. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Yoshida N, Hattori N, Ohgane J, Yanagimachi R, Shiota K. Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod. 2001;65:1813–1821. doi: 10.1095/biolreprod65.6.1813. [DOI] [PubMed] [Google Scholar]
- Rogers JF, Dawson WD. Foetal and placental size in a Peromyscus species cross. J Reprod Fertil. 1970;21:255–262. doi: 10.1530/jrf.0.0210255. [DOI] [PubMed] [Google Scholar]
- Zechner U, Reule M, Burgoyne PS, Schubert A, Orth A, Hameister H, Fundele R. Paternal transmission of X-linked placental dysplasia in mouse interspecific hybrids. Genetics. 1997;146:1399–1405. doi: 10.1093/genetics/146.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony RV, Liang R, Kayl EP, Pratt SL. The growth hormone/prolactin gene family in ruminant placentae. J Reprod Fertil. 1995;Suppl 49:83–95. [PubMed] [Google Scholar]
- Yamada O, Todoroki J, Kizaki K, Takahashi T, Imai K, Patel OV, Schuler LA, Hashizume K. Expression of prolactin-related protein I at the fetomaternal interface during the implantation period in cows. Reproduction. 2002;124:427–437. doi: 10.1530/rep.0.1240427. [DOI] [PubMed] [Google Scholar]
- Nakano H, Takahashi T, Imai K, Hashizume K. Expression of placental lactogen and cytokeratins in bovine placental binucleate cells in culture. Cell Tissue Res. 2001;303:263–270. doi: 10.1007/s004410000316. [DOI] [PubMed] [Google Scholar]
- Nakano H, Shimada A, Imai K, Takezawa T, Takahashi T, Hashizume K. Bovine trophoblastic cell differentiation on collagen substrata: formation of binucleate cells expressing placental lactogen. Cell Tissue Res. 2002;307:225–235. doi: 10.1007/s00441-001-0491-x. [DOI] [PubMed] [Google Scholar]
- Tarrade A, Kuen RL, Malassine A, Tricottet V, Blain P, Vidaud M, Evain-Brion D. Characterization of human villous and extravillous trophoblasts isolated from first trimester placenta. Lab Invest. pp. 1199–2001. [DOI] [PubMed]
- Jones BK, Monks BR, Liebhaber SA, Cooke NE. The human growth hormone gene is regulated by a multicomponent locus control region. Mol Cell Biol. 1995;15:7010–7021. doi: 10.1128/mcb.15.12.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golos TG, Durning M, Fisher JM, Fowler PD. Cloning of four GH/chorionic somatomammotropins-related cDNAs differentially expressed during pregnancy in the rhesus monkey placenta. Endocrinology. 1993;133:1744–1752. doi: 10.1210/en.133.4.1744. [DOI] [PubMed] [Google Scholar]
- Untergasser G, Kranewitter W, Schwarzler P, Madersbacher S, Dirnhofer S, Berger P. Organ-specific expression pattern of the human growth hormone/placental lactogen gene-cluster in the testis. Mol Cell Endocrinol. 1997;130:53–60. doi: 10.1016/S0303-7207(97)00073-7. [DOI] [PubMed] [Google Scholar]
- Fassett JT, Nilsen-Hamilton M. Mrp3, a mitogen-regulated protein/proliferin gene expressed in wound healing and in hair follicles. Endocrinology. 2001;142:2129–2137. doi: 10.1210/en.142.5.2129. [DOI] [PubMed] [Google Scholar]
- Toft DJ, Rosenberg SB, Bergers G, Volpert O, Linzer DIH. Reactivation of proliferin gene expression is associated with increased angiogenesis in a cell culture model of fibrosarcoma tumor progression. Proc Natl Acad Sci USA. 2001;98:3055–13059. doi: 10.1073/pnas.231364798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S, Lin J, Linzer DIH. Reactivation of a hematopoietic endocrine program of pregnancy contributes to recovery from thrombocytopenia. Mol Endocrinol. 2002;16:1386–1393. doi: 10.1210/me.16.6.1386. [DOI] [PubMed] [Google Scholar]
- Ogren L, Talamantes F. Prolactins of pregnancy and their cellular source. Int Rev Cytol. 1988;112:1–65. doi: 10.1016/s0074-7696(08)62005-7. [DOI] [PubMed] [Google Scholar]
- Handwerger S. The physiology of placental lactogens in human pregnancy. Endocr Rev. 1991;12:329–336. doi: 10.1210/edrv-12-4-329. [DOI] [PubMed] [Google Scholar]
- Kelly PA, Shiu RPC, Robertson MC, Friesen HG. Characterization of rat chorionic mammotropin. Endocrinology. 1975;96:1187–1195. doi: 10.1210/endo-96-5-1187. [DOI] [PubMed] [Google Scholar]
- Leung DW, Spencer DA, Cachianes G, Hammonds RG, Collins C, Henzel WJ, Barnard R, Waters MJ, Wood WI. Growth hormone receptor and serum binding protein: purification, cloning, and expression. Nature. 1987;330:537–543. doi: 10.1038/330537a0. [DOI] [PubMed] [Google Scholar]
- Southard JN, Talamantes F. High molecular weight forms of placental lactogens: evidence for lactogens-α2 macroglobulin complexes in rodents and humans. Endocrinology. 1989;125:791–800. doi: 10.1210/endo-125-2-791. [DOI] [PubMed] [Google Scholar]
- Kessler MA, Schuler LA. Purification and properties of placental prolactin-related protein-I. Placenta. 1997;18:29–36. doi: 10.1016/S0143-4004(97)90068-0. [DOI] [PubMed] [Google Scholar]
- Rasmussen CA, Hashizume K, Orwig KE, Xu L, Soares MJ. Decidual prolactin-related protein: heterologous expression and characterization. Endocrinology. 1996;137:5558–5566. doi: 10.1210/en.137.12.5558. [DOI] [PubMed] [Google Scholar]
- Khurana S, Kuns R, Ben-Jonathan N. Heparin-binding property of human prolactin: a novel aspect of prolactin biology. Endocrinology. 1999;140:1026–1029. doi: 10.1210/en.140.2.1026. [DOI] [PubMed] [Google Scholar]
- Wang D, Ishimura R, Walia DS, Müller H, Dai G, Hunt JS, Lee NA, Lee JJ, Soares MJ. Eosinophils are cellular targets of the novel uteroplacental heparin-binding cytokine, decidual/trophoblast prolactin-related protein. J Endocrinol. 2000;167:15–29. doi: 10.1677/joe.0.1670015. [DOI] [PubMed] [Google Scholar]
- Beckers JF, deCoster R, Wouters-Ballman P, Fromont-Lienard C, van der Zwalman P, Ectors F. Dosage radioimmunologique de l'hormone placentaire somatotrope et mammotrope bovine. Annales Med ecine Veterinaire. 1982;126:9–21. [Google Scholar]
- Kappes SM, Warren WC, Pratt SL, Liang R, Anthony RV. Quantification and cellular localization of ovine placental lactogen messenger ribonucleic acid expression during mid- and late gestation. Endocrinology. 1992;131:2829–2838. doi: 10.1210/en.131.6.2829. [DOI] [PubMed] [Google Scholar]
- Frankenne F, Closset J, Gomez F, Scippo ML, Smal J, Hennen G. The physiology of growth hormones (GHs) in pregnant women and partial characterization of the placental GH variant. J Clin Endocrinol Metab. pp. 1171–1180. [DOI] [PubMed]
- Freemark M, Kirk K, Pihoker C, Robertson MC, Shiu RPC, Driscoll P. Pregnancy lactogens in the rat conceptus and fetus: circulating levels, distribution of binding, and expression of receptor messenger ribonucleic acid. Endocrinology. 1993;133:1830–1842. doi: 10.1210/en.133.4.1830. [DOI] [PubMed] [Google Scholar]
- Jackson D, Linzer DIH. Proliferin transport and binding in the mouse fetus. Endocrinology. 1997;138:149–155. doi: 10.1210/en.138.1.149. [DOI] [PubMed] [Google Scholar]
- Thordarson G, Villalobos R, Colosi P, Southard J, Ogren L, Talamantes F. Lactogenic response of cultured mouse mammary epithelial cells to mouse placental lactogen. J Endocrinol. 1986;109:263–274. doi: 10.1677/joe.0.1090263. [DOI] [PubMed] [Google Scholar]
- Dai G, Imagawa W, Liu B, Levan G, Szpirer C, Kwok SCM, Soares MJ. Rcho-1 trophoblast cell placental lactogens: complementary DNAs, heterologous expression, and biological activities. Endocrinology. 1996;137:5020–5027. doi: 10.1210/en.137.11.5020. [DOI] [PubMed] [Google Scholar]
- Galosy SS, Talamantes F. Luteotropic actions of placental lactogens at midpregnancy in the mouse. Endocrinology. 1995;136:3993–4003. doi: 10.1210/en.136.9.3993. [DOI] [PubMed] [Google Scholar]
- Peters CA, Maizels ET, Robertson MC, Shiu RP, Soloff MS, Hunzicker-Dunn M. Induction of relaxin messenger RNA expression in response to prolactin receptor activation requires protein kinase C delta signaling. Mol Endocrinol. 2000;14:576–590. doi: 10.1210/me.14.4.576. [DOI] [PubMed] [Google Scholar]
- Sugino N, Hirosawa-Takemori M, Zhong L, Telleria CM, Shiota K, Gibori G. Hormonal regulation of copper-zine superoxide dismutase messenger ribonucleic acid in the rat corpus luteum: induction by prolactin and placental lactogens. Biol Reprod. 1998;59:599–605. doi: 10.1095/biolreprod59.3.599. [DOI] [PubMed] [Google Scholar]
- Telleria CM, Zhong L, Deb S, Srivastava RK, Park KS, Sugino N, Park-Sarge OK, Gibori G. Differential expression of the estrogen receptors alpha and beta in the rat corpus luteum of pregnancy: regulation by prolactin and placental lactogens. Endocrinology. 1998;139:2432–2442. doi: 10.1210/en.139.5.2432. [DOI] [PubMed] [Google Scholar]
- Zhong L, Parmer TG, Robertson MC, Gibori G. Prolactin-mediated inhibition of 20α-hydroxysteroid dehydrogenase gene expression and the tyrosine kinase system. Biochem Biophys Res Commun. 1997;235:587–592. doi: 10.1006/bbrc.1997.6833. [DOI] [PubMed] [Google Scholar]
- Robertson MC, Gillespie B, Friesen HG. Characterization of the two forms of rat placental lactogen (rPL): rPL-I and rPL-II. Endocrinology. 1982;111:1862–1866. doi: 10.1210/endo-111-6-1862. [DOI] [PubMed] [Google Scholar]
- Soares MJ, Faria TN, Roby KF, Deb S. Pregnancy and the prolactin family of hormones: coordination of anterior pituitary, uterine, and placental expression. Endocr Rev. 1991;12:402–423. doi: 10.1210/edrv-12-4-402. [DOI] [PubMed] [Google Scholar]
- Gunnet JW, Freeman ME. The mating-induced release of prolactin: a unique neuroendocrine response. Endocr Rev. 1983;4:44–61. doi: 10.1210/edrv-4-1-44. [DOI] [PubMed] [Google Scholar]
- Goffin V, Binart N, Touraine P, Kelly PA. Prolactin: the new biology of an old hormone. Annu Rev Physiol. 2002;64:47–67. doi: 10.1146/annurev.physiol.64.081501.131049. [DOI] [PubMed] [Google Scholar]
- Mann PE, Bridges RS. Lactogenic hormone regulation of maternal behavior. Prog Brain Res. 2001;133:251–262. doi: 10.1016/S0079-6123(01)33019-4. [DOI] [PubMed] [Google Scholar]
- Shingo T, Gregg C, Enwere E, Fujikawa H, Hassam R, Geary C, Cross JC, Weiss S. Pregnancy-stimulated neurogenesis in the adult female forebrain mediated by prolactin. Science. 2003;299:117–120. doi: 10.1126/science.1076647. [DOI] [PubMed] [Google Scholar]
- Sorenson RL, Brelje TC. Adaptation of Islets of Langerhans to pregnancy: beta cell growth, enhanced secretion, and the role of lactogenic hormone. Horm Metab Res. 1997;29:301–307. doi: 10.1055/s-2007-979040. [DOI] [PubMed] [Google Scholar]
- Yu-Lee L-Y. Prolactin modulation of immune and inflammatory responses. Rec Prog Horm Res. 2002;57:435–455. doi: 10.1210/rp.57.1.435. [DOI] [PubMed] [Google Scholar]
- Dorshkind K, Horseman ND. Anterior pituitary hormones, stress, and immune system homeostasis. BioEssays. 2001;23:288–294. doi: 10.1002/1521-1878(200103)23:3<288::AID-BIES1039>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- Tessier C, Prigent-Tessier A, Ferguson-Gottschall S, Gu Y, Gibori G. PRL antiapoptotic effect in the rat decidua involves the PI3K/protein kinase B-mediates inhibition of caspase-3 activity. Endocrinology. 2001;142:4086–4094. doi: 10.1210/en.142.9.4086. [DOI] [PubMed] [Google Scholar]
- Prigent-Tessier A, Barkai U, Tessier C, Cohen H, Gibori G. Characterization of a rat uterine cell line, UIII cells: prolactin (PRL) expression and endogenous regulation of PRL-dependent genes; estrogen receptor β, α2-macroglobulin, and decidual PRL involving the Jak2 and Stat5 pathway. Endocrinology. 2001;142:1242–1250. doi: 10.1210/en.142.3.1242. [DOI] [PubMed] [Google Scholar]
- Cohick CB, Dai G, Xu L, Deb S, Kamei T, Levan G, Szpirer C, Szpirer J, Kwok SCM, Soares MJ. Placental lactogen-I variant utilizes the prolactin receptor signaling pathway. Mol Cell Endocrinol. 1996;116:49–58. doi: 10.1016/0303-7207(95)03695-4. [DOI] [PubMed] [Google Scholar]
- Hattori N, Nukada T, Oda M, Tanaka S, Ogawa T, Shiota K. Evaluation of the role of N-linked oligosaccharides in rat placental lactogen action by site-directed mutagenesis. Endocrine J. 1998;45:659–674. doi: 10.1507/endocrj.45.659. [DOI] [PubMed] [Google Scholar]
- Spencer TE, Stagg AG, Taylor KM, Johnson GA, Gertler A, Gootwine E, Bazer FW. Effects of recombinant ovine interferon tau, placental lactogen, and growth hormone on ovine endometrial function. Biol Reprod. 1999;61:1409–1418. doi: 10.1095/biolreprod61.6.1409. [DOI] [PubMed] [Google Scholar]
- Noel S, Herman A, Johnson GA, Gray CA, Stewart MD, Bazer FW, Gertler A, Spencer TE. Ovine placental lactogen specifically binds to endometrial glands of the ovine uterus. Biol Reprod. 2003;68:772–780. doi: 10.1095/biolreprod.102.009183. [DOI] [PubMed] [Google Scholar]
- Wallis M. Remarkably high rat of molecular evolution of ruminant placental lactogens. J Mol Evol. 1993;37:86–88. doi: 10.1007/BF00170466. [DOI] [PubMed] [Google Scholar]
- Biener E, Martin C, Daniel N, Frank SJ, Centonze VE, Herman B, Djiane J, Gertler A. Ovine placental lactogen-induced heterodimerization of ovine growth hormone and prolactin receptors in living cells is demonstrated by fluorescence resonance energy transfer microscopy and leads to prolonged phosphorylation of signal transducer and activator of transcription (STAT)1 and STAT3. Endocrinology. 2003;144:3532–3540. doi: 10.1210/en.2003-0096. [DOI] [PubMed] [Google Scholar]
- Nielsen PV, Pedersen H, Kampmann EM. Absence of human placental lactogens in an otherwise uneventful pregnancy. Am J Obstet Gynecol. 1979;135:322–326. doi: 10.1016/0002-9378(79)90698-7. [DOI] [PubMed] [Google Scholar]
- Wurzel JM, Parks JS, Herd JE, Nielsen PV. A gene deletion is responsible for absence of human chorionic somatomammotropins. DNA. 1982;1:251–257. doi: 10.1089/dna.1.1982.1.251. [DOI] [PubMed] [Google Scholar]
- Rygaard K, Revot A, Esquivel-Escobedo D, Beck BL, Barrera-Saldana HA. Absence of human placental lactogens and placental growth hormone (HGH-V) during pregnancy: PCR analysis of the deletion. Hum Genet. 1998;102:87–92. doi: 10.1007/s004390050658. [DOI] [PubMed] [Google Scholar]
- Jackson D, Volpert O, Bouck N, Linzer DIH. Stimulation and inhibition of angiogenesis by placental proliferin and proliferin-related protein. Science. 1994;266:1581–1584. doi: 10.1126/science.7527157. [DOI] [PubMed] [Google Scholar]
- Lin J, Linzer DIH. Induction of megakaryocyte differentiation by a novel pregnancy-specific hormone. J Biol Chem. 1999;274:21485–21489. doi: 10.1074/jbc.274.30.21485. [DOI] [PubMed] [Google Scholar]
- Bittorf T, Jaster R, Soares MJ, Seiler J, Brock J, Friese K, Müller H. Induction of erythroid proliferation and differentiation by a trophoblast-specific cytokine involves activation of the JAK/STAT pathway. J Mol Endocrinol. 2000;25:253–262. doi: 10.1677/jme.0.0250253. [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Lin J, Linzer DIH. Reactivation of a hematopoietic endocrine program of pregnancy contributes to recovery from thrombocytopenia. Mol Endocrinol. 2002;16:1386–1393. doi: 10.1210/me.16.6.1386. [DOI] [PubMed] [Google Scholar]
- Müller H, Liu B, Croy BA, Head JR, Hunt JS, Dai G, Soares MJ. Uterine natural killer cells are targets for a trophoblast cell-specific cytokine, prolactin-like protein-A. Endocrinology. 1999;140:2711–2720. doi: 10.1210/en.140.6.2711. [DOI] [PubMed] [Google Scholar]
- Ain R, Tash JS, Soares MJ. Prolactin-like protein-A is a functional modulator of natural killer cells at the maternal-fetal interface. Mol Cell Endocrinol. 2003;204:65–74. doi: 10.1016/S0303-7207(03)00125-4. [DOI] [PubMed] [Google Scholar]
- Cohick CB, Xu L, Soares MJ. Placental prolactin-like protein-B: heterologous expression and characterization of placental and decidual species. J Endocrinol. 1997;152:291–302. doi: 10.1677/joe.0.1520291. [DOI] [PubMed] [Google Scholar]
- Iwatsuki K, Oda M, Sun W, Tanaka S, Ogawa T, Shiota K. Molecular cloning and characterization of a new member of the rat placental prolactin (PRL) family, PRL-like protein H. Endocrinology. 1998;139:4976–4983. doi: 10.1210/en.139.12.4976. [DOI] [PubMed] [Google Scholar]
- Volpert O, Jackson D, Bouck N, Linzer DIH. The insulin-like growth factor II/mannose 6-phosphate receptor is required for proliferin-induced angiogenesis. Endocrinology. 1996;137:3871–3876. doi: 10.1210/en.137.9.3871. [DOI] [PubMed] [Google Scholar]
- Groskopf JC, Syu L-J, Saltiel AR, Linzer DIH. Proliferin induces endothelial cell chemotaxis through a G protein-coupled, MAP kinase-dependent pathway. Endocrinology. 1997;138:2835–2840. doi: 10.1210/en.138.7.2835. [DOI] [PubMed] [Google Scholar]
- Bengtson NW, Linzer DIH. Inhibition of tumor growth by the antiangiogenic placental hormone, proliferin-related protein. Mol Endocrinol. 2000;14:1934–1943. doi: 10.1210/me.14.12.1934. [DOI] [PubMed] [Google Scholar]
- Corbacho AM, de la Escalera GM, Clapp C. Roles of prolactin and related members of the prolactin/growth hormone/placental lactogens family in angiogenesis. J Endocrinol. 2002;173:219–238. doi: 10.1677/joe.0.1730219. [DOI] [PubMed] [Google Scholar]
- Parra A, Ramirez-Peredo J. The possible role of prolactin in preeclampsia: a hypothesis revisited a quarter century later. Med Hypotheses. 2001;59:378–384. doi: 10.1016/S0306-9877(02)00124-X. [DOI] [PubMed] [Google Scholar]
- Hsu SY, Nakabayashi K, Nishi SJ, Kudo M, Sherwood OD, Hsueh AJ. Activation of orphan receptors by the hormone relaxin. Science. 2002;295:671–674. doi: 10.1126/science.1065654. [DOI] [PubMed] [Google Scholar]
- Kaplan SL, Grumbach MM. Chorionic somatomammotropins in primates: secretion and physiology. In: Novy MJ, Resko JA, editor. Fetal Endocrinology. New York, Academic Press; 1981. pp. 127–139. [Google Scholar]