Abstract
We identified 8 candidate thinness predisposition variants from the Illumina HumanExome chip genotyped on members of pedigrees selected for either healthy thinness or severe obesity. For validation, we tested the candidates for association with healthy thinness in additional pedigree members while accounting for effects of obesity-associated genes: NPFFR2, NPY2R, FTO, and MC4R. Significance was obtained for the interaction of FTO rs9939609 with APOH missense variant rs52797880 (minor allele frequency = 0.054). The thinness odds ratio was estimated as 2.15 (p<0.05) for the combination of APOH heterozygote with the homozygote for the non-obesity FTO allele. Significance was not obtained for any other combination of a candidate variant with an obesity gene or for any of the 8 candidates tested independently.
Keywords: Body mass index, Likelihood, Exome chip, Pedigrees
Introduction
For some individuals, thinness is a life-long healthy state rather than the result of malnutrition or disease. Nevertheless, most genetic studies of low body mass index (BMI) have focused not on healthy thinness, but on eating disorders (Helder and Collier 2011) or sarcopenia (Tan et al. 2012). This neglect is unfortunate since healthy thinness may more readily yield its underlying genes than will obesity, for which environmental factors obscure the effects of genes. Furthermore, knowledge of thinness predisposition genes promises to provide insight not only into thinness, but also into obesity. As a model for disease treatments inspired by genetic variants associated with a healthy state, drugs that inhibit PCSK9 mimic the effect of genetic variants that lower cholesterol levels (Gouni-Berthold and Berthold 2014). Similarly, thinness predisposition variants, redefined as obesity resistance variants, might be the inspiration for drugs to combat obesity. A number of BMI- and obesity-associated single nucleotide polymorphisms (SNPs) have been identified by genome-wide association studies; however, even in combination, these SNPs fall short of accounting for the heritability of BMI (Sandholt et al. 2012). The small effect sizes that characterize most associated SNPs are consistent with polygenic inheritance: the accumulation of small effects from many risk variants. Nevertheless, the ability of polygenic inheritance to roughly predict obesity risk (Domingue et al. 2014) does not preclude the existence of variants with large effects on BMI, especially variants that affect the less studied lower end of the range.
Even if unaffected by environmental factors, genetic interactions may obscure the genotype-phenotype relationship for thinness. If a causal variant promotes thinness only when co-inherited with an interacting variant, association between the causal variant and thinness may appear weak because many non-thin subjects carry the causal variant but not the interacting variant. Therefore, accounting for genetic interactions can facilitate the discovery of thinness predisposition variants. Herein, we identified candidate thinness predisposition variants from the Illumina HumanExome chip on members of pedigrees selected either for severe obesity or for healthy thinness, thereby excluding individuals with malnutrition or eating disorders, the focus of most previous genetic studies of the lower end of the BMI range (Helder and Collins 2011). We then validated the candidates in additional pedigree members while accounting as well for the effects of four obesity-associated genes, and their interactions with the candidates.
Methods
Subjects
We ascertained obesity pedigrees using the Health Family Tree, a Utah high school-based family history program designed to teach genetics and disease prevention as part of a mandatory health class (Williams et al. 1988). Students, with parental input, reported disease information and risk factors for the parents’ first-degree relatives. We identified 435 severely obese sib pairs from the Trees, chose 107 sibships for expansion, and examined 3,333 relatives of the sibs. Similarly, we identified 40 thin probands who reported multiple thin relatives and examined 400 family members including 265 thin individuals. All subjects were Utah residents with European ancestry. The obesity and thinness projects have been approved by the Institutional Review Board of the University of Utah.
For each subject, height was measured to the nearest centimeter using a Harpenden anthropometer (Holtain, Ltd). Weight was measured in a hospital gown using a Scaletronix scale (model 5100) (Scaletronix Corporation, Wheaton, IL, USA), which has an 800-pound capacity and weighing accuracy of 0.1 kg. BMI was computed as weight divided by height squared.
Genotypes
We selected a discovery sample comprised of 504 members of the thin and obese pedigrees, chosen for distant relatedness within pedigrees in order to increase statistical power at reduced cost, and a validation sample comprised of 3,569 additional pedigree members. We used the HumanExome chip to genotype the discovery sample and 351 members of the validation sample. We used the LightScanner (BioFire Diagnostics, SLC, UT) to genotype the remaining members of the validation sample for 8 candidate thinness-associated variants selected based on analysis of the discovery sample. Genotypes of 13 obesity-associated SNPs were available on 3,661 members of the pedigrees (Hunt et al. 2011), henceforth designated the OBgene sample. The Illumina HumanExome chip contains >240,000 variants selected from exome sequencing of >10,000 individuals, including subjects in studies of obesity-related conditions (e.g., NHLBI Exome Sequencing Project, T2D diabetes project, Lipid Extremes, BMI Extremes). Any variant observed ≥2 times in these samples and passing quality control was included on the chip (see: genome.sph.umich.edu/wiki/Exome_Chip_Design for additional information about this chip platform). Duplicate blind repeated controls within the sample showed over 99.8% genotype consistency. Standard quality control was applied.
We merged all sources of genotypes for a combined sample of 5,517 subjects each with genotypes for one or more of the 21 variants (8 thinness-candidate and 13 obesity-associated) using PLINK (Purcell et al. 2007). For each variant, we computed each individual's genotype probability in jPAP allowing an error rate of 2% for genotypes uncorrected for Mendelian errors. Similarly, we haplotyped SNP pairs within each obesity-associated gene and computed each individual's diplotype probabilities. Each probability accounted for allele or haplotype frequencies and genotypes of relatives. A dosage effect of a variant or haplotype for a subject was computed as 0/1/2, corresponding to the number of copies of the allele or haplotype, weighted by the corresponding genotype/diplotype probability to account for genotyping errors and to impute missing genotypes. For interaction effects, dosages of two variants or haplotypes were multiplied, reversing the dosage for any that decreased thinness predisposition.
Discovery Statistical Analysis
To identify thinness candidates among the autosomal exome chip variants, we applied 3 statistical tests to the complete discovery sample. First, the transmission disequilibrium test (TDT) in jPAP (Hasstedt 2005; Hasstedt and Thomas 2011) detected over-transmission of the minor allele from a heterozygous parent to a thin (BMI ≤23 kg/m2) offspring. Second, a test designated ASSOC in jPAP tested each autosomal exome chip variant for an additive (genotypes coded as 0/1/2) effect on inverse-transformed BMI while accounting for gender, age, and heritability. Finally, famSKAT (Chen et al. 2013), the family-based implementation of the sequence kernel association test SKAT (Wu et al. 2011) evaluated sets of variants within a gene for association with BMI, accounting for gender, age, and heritability. Correction for multiple testing was unnecessary in the discovery phase of the study.
The TDT statistic equaled twice the natural logarithm of the ratio of the likelihood maximized over the transmission probability (τ) to the likelihood with τ = 0.5; the ASSOC test statistic equaled twice the natural logarithm of the ratio of the likelihood maximized over a negative effect on BMI to the likelihood with effect size of zero. Since the null hypothesis constrained τ to the boundary for TDT and constrained the effect size to the boundary for ASSOC, we obtained P-values assuming the statistic approximated a 50:50 mixture of chi square distributions with 0 and 1 degree of freedom (Self and Liang 1987). P-values for famSKAT results were computed using Kuonen's saddlepoint method (Kuonen 1999).
For the TDT we restricted the test to rarer (minor allele frequency (MAF) ≤5%) variants to increase the likelihood that the allele was present in ungenotyped parents only when present in an offspring. jPAP probabilistically inferred missing parental genotypes using information from measured genotypes and the allele frequency. Because genotype inference was not independent of τ, we simulated all missing genotypes fixing τ = 0.5 and allele frequency = 0.001, estimated τ, computed the TDT statistic for the simulated genotypes, then averaged τ and the TDT statistic over 1000 replicates.
Validation Statistical Analysis
For the validation analyses, BMI and age restrictions were applied to the discovery, OBgene, and validation samples; “complete” designates samples without these restrictions. Thinness was defined as below the 15th percentile of gender- and age-specific BMI from US reference data for 1999-2002 (McDowell et al. 2005), the approximate years of data collection. Controls were restricted to BMI between the 15th and 85th percentiles, thereby excluding severely obese subjects to detect variants specific for thinness rather than more generally for non-obesity. Age was restricted to 20 to 79 years to eliminate thinness attributable to youth or advanced age.
To test for simultaneous effects of alleles and haplotypes on thinness, we applied logistic regression (LR) in jPAP (Hasstedt and Thomas 2011). We estimated prevalence, heritability, and odds ratios for genotypic dosage effects. Significance was determined from 95% 1-tailed or 2-tailed confidence intervals (CI), depending on presence or absence of a prior knowledge of directionality of the effect. Because all effects were tested simultaneously, no multiple testing correction was required.
Results
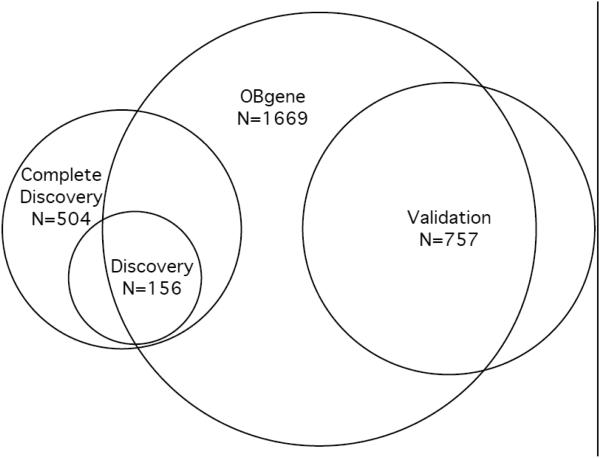
The discovery and validation samples comprised non-overlapping subsets of subjects genotyped for 8 candidate thinness variants either separately or from the exome chip; the OBgene sample comprised individuals genotyped for 13 obesity-associated SNPs; the overlap sample comprised members of the validation sample who were also either in the OBgene sample or were genotyped using the exome chip (Figure 1). Every sample subset included more controls than thin subjects and more women than men; controls were on average older than thin subjects (Table 1).
Fig. 1.
Samples used in the tests for association with thinness. Except for the Complete discovery Sample, all samples were restricted to BMI < 85th percentile and 20 ≤ age < 80 years. The Overlap Sample (Table 1) comprised the overlap of the OBgene Sample with the validation Sample with the inclusion of an additional 28 subjects from the validation Sample who were genotyped using the exome chip
Table 1.
Counts and mean age (restricted to 20 yrs ≤age < 80 yrs) by gender, phenotype, and sample.
| Sample | Discovery | Validation | OBgene | Overlap | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | Mean Age | N | Mean Age | N | Mean Age | N | Mean Age | |||||||||
| Phenotype | M | F | M | F | M | F | M | F | M | F | M | F | M | F | M | F |
| Thina | 34 | 54 | 28.15 | 29.33 | 62 | 87 | 42.47 | 43.24 | 137 | 208 | 42.93 | 42.36 | 57 | 79 | 42.23 | 42.89 |
| Controlsb | 17 | 51 | 40.82 | 42.63 | 233 | 375 | 49.01 | 48.14 | 558 | 766 | 50.54 | 49.30 | 179 | 290 | 48.87 | 48.78 |
| Total | 51 | 105 | 32.37 | 35.79 | 295 | 462 | 47.63 | 47.21 | 695 | 974 | 49.04 | 47.82 | 236 | 369 | 47.26 | 47.52 |
Gender-, age-specific for BMI < 15th percentile
Gender-, age-specific for BMI 15th to 85th percentile
To investigate the role of obesity-associated genes on thinness, we tested for simultaneous effects of the FTO SNP and of all common (frequency > 1%) 2-SNP haplotypes of 13 SNPs within NPFFR2, NPY2R, and MC4R (Table 2) in the OBgene sample (1669 members of 909 pedigrees). Starting with all effects, we successively eliminated the least significant until only significant effects remained. One NPFFR2 haplotype increased thinness predisposition; FTO allele T, 2 haplotypes of NPY2R and 1 haplotype of MC4R decreased thinness predisposition (Table 3).
Table 2.
Obesity-associated SNPs included in the analysis.
| SNP | Gene | Chromosome | Position (bp)a | N | MAF |
|---|---|---|---|---|---|
| rs12510838 | NPFFR2 | 4 | 72961538 | 3661 | 0.1776 |
| rs4129733 | NPFFR2 | 4 | 72963020 | 3661 | 0.3063 |
| rs9291171 | NPFFR2 | 4 | 72981626 | 3661 | 0.2882 |
| rs11940196 | NPFFR2 | 4 | 73003569 | 3661 | 0.3614 |
| rs12649641 | NPY2R | 4 | 156125333 | 3875 | 0.3880 |
| rs12507396 | NPY2R | 4 | 156129044 | 3661 | 0.1114 |
| rs17376826 | NPY2R | 4 | 156130948 | 3910 | 0.0359 |
| rs10461238 | NPY2R | 4 | 156132216 | 3661 | 0.4447 |
| rs10461239 | NPY2R | 4 | 156132447 | 3661 | 0.0460 |
| rs2880415 | NPY2R | 4 | 156136027 | 3661 | 0.4527 |
| rs9939609 | FTO | 16 | 53820527 | 4110 | 0.4356 |
| rs17782313 | MC4R | 18 | 57851097 | 4131 | 0.2602 |
| rs477181 | MC4R | 18 | 57896038 | 3898 | 0.3570 |
Build GRCh37
Table 3.
Thinness OR and 95% 2-tailed CI for obesity-associated haplotypes on the OBgene sample.
| Gene | Haplotypea | Frequency | ORb | CI | |
|---|---|---|---|---|---|
| Lower | Upper | ||||
| NPFFR2 | GTXX | 0.1774 | NS | ||
| NPFFR2 | AXGX | 0.1116 | NS | ||
| NPFFR2 | XGXG | 0.3140 | NS | ||
| NPFFR2 | XGXA | 0.0130 | NS | ||
| NPFFR2 | XTXG | 0.0414 | 1.90 | 1.50 | 2.42 |
| NPFFR2 | XXAA | 0.6308 | NS | ||
| NPY2R | AXXXXT | 0.1792 | 0.79 | 0.65 | 0.96 |
| NPY2R | AXXXXA | 0.2168 | NS | ||
| NPY2R | CXXXXT | 0.2859 | 0.88 | 0.79 | 0.99 |
| NPY2R | CXXXXA | 0.3181 | NS | ||
| FTO | A | 0.4356 | 0.78 | 0.73 | 0.85 |
| MC4R | TG | 0.2098 | NS | ||
| MC4R | TT | 0.0594 | 0.20 | 0.07 | 0.58 |
| MC4R | GG | 0.1612 | NS | ||
| MC4R | GT | 0.5696 | NS | ||
Haplotype designated by base, with order corresponding to Table 2. X indicates the SNP was not included.
NS indicates restriction of the OR to 1 due to non-significance at an earlier analysis stage.
To identify candidates for thinness predisposition from the exome chip variants, we required high significance on one or more of 3 statistical tests applied to the complete discovery sample (Figure 1) comprising 504 members of 46 pedigrees (Table 4). Two of the selected candidates were common; for the remaining 6 candidates, MAF ≤ 6%. TDT and ASSOC analyses evaluated variants individually; famSKAT analysis evaluated sets of variants within a gene from which we selected for follow-up the variant with the largest minor allele frequency. ASSOC and famSKAT treated BMI as a continuous variable; TDT included only sibships with thin (BMI ≤ 23 kg/m2) members. ASSOC and famSKAT tested all autosomal exome chip variants; TDT tested only variants with MAF ≤ 5%. TDT and ASSOC tested for thinness predisposition of minor alleles; famSKAT did not require assumptions of or imply directionality of the effect. We tested whether the 8 candidate variants, each identified individually, maintained significance when tested simultaneously, accounting for the significant alleles/haplotypes of NPFFR2, NPY2R, FTO, and MC4R (Table 3), and following restriction of BMI and age in the discovery sample (156 members of 41 pedigrees ranging from 1 to 13 genotyped members, Figure 1, Table 1). Three of the 8 variants (in FSIP2, HCN1, and APOH) attained significance and A2M occurred only in thin subjects (Table 5). Failed significance for the SNTB1, SPATS2, and SCN4A variants may have been due to the reduced numbers of heterozygotes, 8, 15, and 3 respectively, following age and BMI restrictions. Failed significance for the GATM variant may have reflected a non-ideal choice from among the variants showing gene-based significance selected as the variant with the largest minor allele frequency.
Table 4.
Candidate variants with P-values from 3 statistical tests performed on the complete discovery sample.
| Variant | Gene | Chr | Position(bp)a | N | MAFb | TDT | ASSOC | famSKAT |
|---|---|---|---|---|---|---|---|---|
| rs79762465 | FSIP2 | 2 | 186654867 | 502 | 0.041 | 1.5×10−8 | 1.2×10−5 | 2.0 × 10−5 |
| rs981782 | HCN1 | 5 | 45285718 | 403 | 0.462 | NA | 0.21 | 2.7 × 10−10 |
| rs61762674 | SNTB1 | 8 | 121561028 | 503 | 0.002 | 1.5×10−3 | 0.04 | 0.01 |
| rs55761427 | A2M | 12 | 9243017 | 501 | 0.011 | 3.1×10−3 | 0.13 | 0.01 |
| rs142230440 | SPATS2 | 12 | 49884473 | 456 | 0.004 | 1.2×10−4 | 1.8 × 10−5 | 1.3 × 10−4 |
| rs1288775 | GATM | 15 | 45661678 | 501 | 0.264 | NA | 0.02 | 1.8 × 10−7 |
| rs41280102 | SCN4A | 17 | 62028920 | 456 | 0.009 | 1.3×10−3 | 0.42 | 0.01 |
| rs52797880 | APOH | 17 | 64216854 | 503 | 0.054 | NA | 4.7 × 10−5 | 4.0× 10−6 |
Build GRCh37
Estimated using N>3000 with genotypes
Table 5.
Thinness OR (per allele) and 95% 1-tailed lower CI in the discovery and validation samples and for interaction with FTO in the overlap sample
| Sample | Discovery | Validation | Overlap | |||
|---|---|---|---|---|---|---|
| Interaction | None | None | FTO | |||
| Gene | OR | CI | OR | CI | OR | CI |
| FSIP2 | 2.87 | 1.39 | 1.00 | NA | 1.00 | NA |
| HCN1 a | 1.52 | 1.28 | 1.02 | 0.93 | 1.01 | 0.92 |
| SNTB1 | 5.27 | 0.56 | 1.00 | NA | 1.00 | NA |
| A2M | ∞ | NA | 1.53 | 0.46 | 1.19 | 0.55 |
| SPATS2 | 3.43 | 0.88 | 1.00 | NA | 1.00 | NA |
| GATM | 1.24 | 0.91 | 1.00 | NA | 1.00 | NA |
| SCN4A | 1.00 | NA | 1.00 | NA | 1.00 | NA |
| APOH | 2.36 | 1.08 | 1.21 | 0.73 | 1.47 | 1.01 |
Major allele.
The validation sample comprised 757 members of 187 pedigrees ranging from 1 to 83 genotyped members (Table 1); 30 pedigrees within the validation sample also contained subjects from the discovery sample. Because of the pedigree overlap, we made an ascertainment correction in the LR analysis by dividing the likelihood of the validation and discovery samples combined by the likelihood of the discovery sample. The combined discovery and validation samples included 913 members of 198 pedigrees ranging from 1 to 95 genotyped members. None of the 8 candidates attained significance (Table 5).
Next we tested for interaction between the thinness candidates and the obesity genes. For each significant obesity allele/haplotype in turn, we repeated the LR analysis, replacing each candidate dosage with its product with the obesity allele/haplotype dosage. To preclude imputation of either candidate or obesity gene variants, we replaced the validation sample with the overlap sample. The combined discovery and overlap sample included 761 members of 175 pedigrees ranging from 1 to 84 members (Table 1), with ascertainment correction again the discovery sample. The APOH variant attained significance in interaction with the FTO SNP (Table 5). None of the 8 candidates attained significance in interaction with any other obesity alleles/haplotypes (results not shown).
Thinness was more than twice as common for APOH rs52797880 heterozygotes paired with FTO rs9939609 T/T (71%) compared to any other genotype pair (21-30%) (Table 6). It should be noted that these estimates ignore relatedness in the sample. However, in analysis that accounted for relatedness (Table 5), the per-allele OR estimate of 1.47 corresponds to 2-allele OR estimate of 2.15; dosage effect of 2 was assigned for the heterozygous APOH/homozygous FTO pairing.
Table 6.
Percentage of the overlap sample that was thin, by APOH and FTO genotypes.
| APOH | ||||||
|---|---|---|---|---|---|---|
| FTO | T/T | T/C | C/C | |||
| N | Thin (%) | N | Thin (%) | N | Thin (%) | |
| T/T | 232 | 30 | 31 | 71 | 0 | |
| T/A | 330 | 25 | 51 | 29 | 2 | 100 |
| A/A | 98 | 28 | 19 | 21 | 0 | |
Discussion
This study identified APOH (apolipoprotein H (beta-2-glycoprotein I)) rs52797880 as a potential thinness predisposition variant in interaction with FTO rs9939609. The chance of thinness was more than double for APOH rs52797880 genotype T/C in combination with FTO rs9939609 genotype T/T compared to all other genotypes. Small numbers precluded evaluation of the association for homozygous genotype APOH C/C.
Located on chromosome 17q24.2, APOH participates in a variety of physiologic pathways including lipoprotein metabolism, coagulation, and the production of antiphospholipid autoantibodies (Athanasiadis et al. 2013). More relevant to thinness, APOH regulates diet-induced obesity in zebrafish and mammals (Oka et al. 2010) and an APOH SNP is associated with BMI in diabetics (Ruaño et al. 2009). In addition, APOH plasma levels are associated with metabolic and cardiovascular risk factors (Castro et al. 2010) and APOH transcript levels differed significantly by porcine body composition (Ponsuksili et al. 2005) and by lipids in beef cattle (Romao et al. 2014).
APOH variant rs52797880 is a missense substitution of threonine for isoleucine with no predicted effect on function and no reported disease associations. However, rs1801690, a possibly deleterious variant at 8.5 kb and with LD r2 = 0.77 in our sample, is a missense substitution of serine for triptophan that associates with serum lipid levels (Guo et al. 2015). Although less strongly associated in the complete discovery analysis, rs1801690 interacts more strongly with FTO genotypes in the sample subset genotyped using the exome chip. Among 27 rs1801690 heterozygotes, thinness occurred in 8 of 8 with FTO T/T, 0 of 4 with A/A, and 8 of 7 with T/A. In comparison, among 27 rs52797880 heterozygotes, thinness occurred in 8 of 8 with FTO T/T, 3 of 7 with A/A, and 7 of 12 with T/A.
The nature of the interaction between APOH and FTO remains to be explained, although both are associated with lipid levels (Asselbergs et al. 2012). The effect of FTO on BMI may be mediated through impaired responsiveness to satiety (Benedict et al. 2014) and BMI-increasing alleles of FTO SNPs are associated with increased protein intake (Tanaka et al. 2013). However, obesity-associated FTO intronic variants and FTO activity appear functionally connected not with FTO but with two neighboring genes: IRX3 and RPGRIP1L, suggesting that noncoding variants exert a functional impact through the alteration of gene expression (Tung et al. 2014).
An NPFFR2 haplotype also contributed to thinness predisposition, while haplotypes of NPY2R and MC4R decreased thinness predisposition. Haplotypes of each of these obesity genes were previously shown to be associated with obesity (Hunt et al. 2011). Therefore, the obesity genes affect BMI across the range, affecting risk at the lower as well as the upper end of the range. Upon restricting controls to BMI<50th percentile, only effects of FTO and MC4R remained (results not shown), possibly because of smaller sample size.
In summary, we identified APOH as an obesity-resistance gene that interacts with FTO rs9939609 to more than double the occurrence of thinness.
Acknowledgments
This project was supported by National Institutes of Health grants DK093151 and DK073550. We thank all members of the pedigrees for their participation in this study.
References
- Asselbergs FW, Guo Y, van Iperen EP, Sivapalaratnam S, Tragante V, Lanktree MB, Lange LA, Almoguera B, Appelman YE, et al. Large-scale gene-centric meta-analysis across 32 studies identifies multiple lipid loci. Am J Hum Genet. 2012;91:823–38. doi: 10.1016/j.ajhg.2012.08.032. doi: 10.1016/j.ajhg.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiadis G, Sabater-Lleal M, Buil A, Souto JC, Borrell M, Lathrop M, Watkins H, Almasy L, Hamsten A, Soria JM. Genetic determinants of plasma β2-glycoprotein I levels: a genome-wide association study in preliminary pedigrees from Spain. J Thromb Haemostn. 2013;11:521–528. doi: 10.1111/jth.12120. doi: 10.1111/jth.12120. [DOI] [PubMed] [Google Scholar]
- Benedict C, Axelsson T, Söderberg S, Larsson A, Ingelsson E, Lind L, Schiöth HB. Fat mass and obesity-associated gene (FTO) is linked to higher plasma levels of the hunger hormone ghrelin and lower serum levels of the satiety hormone leptin in older adults. Diabetes. 2014;63:3955–3959. doi: 10.2337/db14-0470. doi: 10.2337/db14-0470. [DOI] [PubMed] [Google Scholar]
- Castro A, Lázaro I, Selva DM, Céspedes E, Girona J, NúriaPlana, Guardiola M, Cabré A, Simó R, Masana L. APOH is increased in the plasma and liver of type 2 diabetic patients with metabolic syndrome. Atherosclerosis. 2010;209:201–205. doi: 10.1016/j.atherosclerosis.2009.09.072. doi: 10.1016/j.atherosclerosis.2009.09.072. [DOI] [PubMed] [Google Scholar]
- Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingue BW, Belsky DW, Harris KM, Smolen A, McQueen MB, Boardman JD. Polygenic risk predicts obesity in both white and black young adults. PLoS One. 2014 doi: 10.1371/journal.pone.0101596. doi: 10.1371/journal.pone.0101596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouni-Berthold I, Berthold HK. PCSK9 Antibodies for the treatment of hypercholesterolemia. Nutrients. 2014;6:5517–5533. doi: 10.3390/nu6125517. doi: 10.3390/nu6125517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Yin RX, Li H, Wang YM, Wu JZ, Yang DZ. Association of the Trp316Ser variant (rs1801690) near the apolipoprotein H (β2-glycoprotein-I) gene and serum lipid levels. Int J Clin Exp Pathol. 2015;8:7291–304. [PMC free article] [PubMed] [Google Scholar]
- Hasstedt SJ. jPAP: Document-driven software for genetic analysis. Genet Epidemiol. 2005;29:255. Abstract. [Google Scholar]
- Hasstedt SJ, Thomas A. Detecting pleiotropy and epistasis using variance components linkage analysis in jPAP. Hum Hered. 2011;72:258–263. doi: 10.1159/000331690. doi: 10.1159/000331690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helder SG, Collier DA. The genetics of eating disorders. Curr Top Behav Neurosci. 2011;6:157–175. doi: 10.1007/7854_2010_79. doi: 10.1007/7854_2010_79. [DOI] [PubMed] [Google Scholar]
- Hunt SC, Hasstedt SJ, Xin Y, Dalley BK, Milash BA, Yakobson E, Gress RE, Davidson LE, Adams TD. Polymorphisms in the NPY2R gene show significant associations with BMI that are additive to FTO, MC4R, and NPFFR2 gene effects. Obesity. 2011;19:2241–2247. doi: 10.1038/oby.2011.239. doi: 10.1038/oby.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuonen D. Saddlepoint approximations for distributions of quadratic forms in normal variables. Biometrika. 1999;86:929–935. doi: 10.1093/biomet/86.4.929. [Google Scholar]
- McDowell MA, Fryar CD, Hirsch R, Ogden CL. Anthropometric reference data for children and adults: U.S. population, 1999-2002. Adv Data. 2005;7:1–5. [PubMed] [Google Scholar]
- Oka T, Nishimura Y, Zang L, Hirano M, Shimada Y, Wang Z, Umemoto N, Kuroyanagi J, Nishimura N, Tanaka T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010 doi: 10.1186/1472-6793-10-21. doi: 10.1186/1472-6793-10-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponsuksili S, Murani E, Schellander K, Schwerin M, Wimmers K. Identification of functional candidate genes for body composition by expression analyses and evidencing impact by association analysis and mapping. Biochim Biophys Acta. 2005;1730:31–40. doi: 10.1016/j.bbaexp.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao JM, He ML, McAllister TA, Guan LL. Effect of age on bovine subcutaneous fat proteome: molecular mechanisms of physiological variations during beef cattle growth. J Anim Sci. 2014;92:3316–3327. doi: 10.2527/jas.2013-7423. doi: 10.2527/jas.2013-7423. [DOI] [PubMed] [Google Scholar]
- Ruaño G, Bernene J, Windemuth A, Bower B, Wencker D, Seip RL, Kocherla M, Holford TR, Petit WA, Hanks S. Physiogenomic comparison of edema and BMI in patients receiving rosiglitazone or pioglitazone. Clin Chim Acta. 2009;400:48–55. doi: 10.1016/j.cca.2008.10.009. doi: 10.1016/j.cca.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Sandholt CH, Hansen T, Pedersen O. Beyond the fourth wave of genome-wide obesity association studies. Nutr Diabetes. 2012 doi: 10.1038/nutd.2012.9. doi: 10.1038/nutd.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Self SG, Liang K-Y. Asymptotic Properties of Maximum Likelihood Estimators and Likelihood Ratio Tests under Nonstandard Conditions. J Amer Stat Assoc. 1987;82:605–610. [Google Scholar]
- Tan LJ, Liu SL, Lei SF, Papasian CJ, Deng HW. Molecular genetic studies of gene identification for sarcopenia. Hum Genet. 2012;131:1–31. doi: 10.1007/s00439-011-1040-7. doi: 10.1007/s00439-011-1040-7. [DOI] [PubMed] [Google Scholar]
- Tanaka T, Ngwa JS, van Rooij FJ, Zillikens MC, Wojczynski MK, Frazier-Wood AC, et al. Genome-wide meta-analysis of observational studies shows common genetic variants associated with macronutrient intake. Am J Clin Nutr. 2013;97:1395–1402. doi: 10.3945/ajcn.112.052183. doi: 10.3945/ajcn.112.052183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung YC, Yeo GS, O'Rahilly S, Coll AP. Obesity and FTO: changing focus at a complex locus. Cell Metab. 2014;20:710–771. doi: 10.1016/j.cmet.2014.09.010. doi: 10.1016/j.cmet.2014.09.010. [DOI] [PubMed] [Google Scholar]
- Williams RR, Hunt SC, Barlow GK, Chamberlain RM, Weinberg AD, Cooper HP, Carbonari JP, Gotto AM., Jr. Health family trees: a tool for finding and helping young family members of coronary and cancer prone pedigrees in Texas and Utah. Am J Public Health. 1988;78:1283–1286. doi: 10.2105/ajph.78.10.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]