Abstract
CARMILs are a conserved family of large multidomain proteins that regulate and target actin assembly by interacting with actin capping protein (CP). Vertebrates contain three highly conserved CARMIL isoforms encoded by three genes, whereas lower organisms contain only one isoform and gene. In order to investigate the functions of vertebrate CARMILs, we identified and characterized the three CARMIL genes in zebrafish (Danio rerio). We isolated and sequenced complete and partial cDNAs from embryos. The three genes display distinct spatial and temporal expression patterns during development. Sequence and phylogenetic analyses of cDNAs and predicted protein sequences reveal that the three zebrafish genes fall into the three conserved isoform groups previously defined for other vertebrates, which have isoform-specific and overlapping functions in human cultured cells. These results provide new tools and offer insight into understanding the role of the regulation of actin assembly dynamics during embryonic development and tissue morphogenesis.
Keywords: lrrc16a, rltpr, lrrc16b, capping protein, actin
Introduction
The CARMIL family of proteins are highly conserved, large (~1400-aa), multidomain regulators of heterodimeric actin capping protein (CP). CARMIL was first identified as Acan125 in Acanthamoeba by its ability to bind the SH3 domain of a class-I myosin [Xu et al., 1995]. The name CARMIL was coined later, based on the biochemical ability of the Dictyostelium homologue, p116, to serve as a direct molecular linker between CP, Arp2/3 complex, and myosin-I [Jung et al., 2001]. Amoebae and invertebrates have one gene encoding CARMIL, while all vertebrates studied to date have three CARMIL genes, which encode proteins that fall into three highly conserved and distinct isoforms termed CARMIL1, CARMIL2 and CARMIL3 [Edwards et al., 2014].
Biochemical studies reveal that CARMILs allosterically regulate CP through the direct interaction of an intrinsically disordered domain of CARMIL, which contains two CP-binding motifs in tandem in all multicellular and some unicellular organisms. One motif, called CP-interacting (CPI), is found in many proteins other than CARMILs. The second motif, called CARMIL-specific interaction (CSI), is found only in CARMILs, and it is not present in CARMILs from amoebae (Figure 1A) [Edwards et al., 2013; Kim et al., 2012; Lanier et al., 2015]. The interaction of CP with CPI-motif containing proteins is essential for the localization and function of CP in human cultured cells [Edwards et al., 2015].
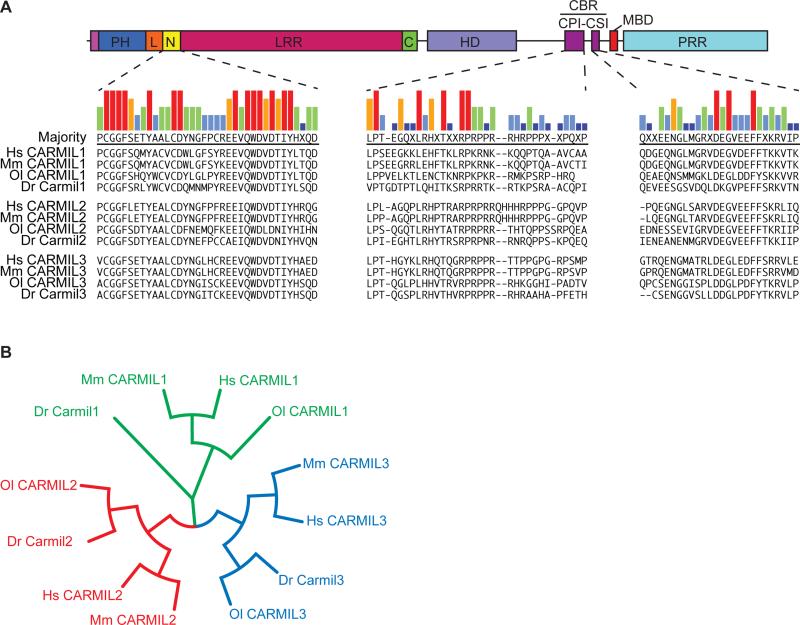
Figure 1. Identification and Sequence Analysis of Zebrafish CARMIL Proteins.
A) Diagram of CARMIL domain architecture, adapted from Zwolak and colleagues [Zwolak et al., 2013], with alignment of sequences from the highly conserved N-cap (N), CP-Interaction (CPI) and CARMIL-specific Interacting (CSI) regions. Species abbreviations: Hs, human; Mm, mouse; Ol, medaka; Dr, zebrafish. B) Cladogram created using amino-acid sequences of full-length CARMILs.
CARMILs have multiple protein domains. Structural and biochemical analysis of mouse CARMIL1 revealed an N-terminal, non-canonical, pleckstrin homology (PH) domain, which is connected to a horseshoe-shaped leucine-rich region (LRR) by a conserved and structured linker [Zwolak et al., 2013]. The LRR is capped on both ends by helix-loop-helix motifs that are also conserved among CARMILs; the N-terminal cap (N-cap) is especially well-conserved [Liang et al., 2009; Zwolak et al., 2013]. The LRR is followed by a helical dimerization domain (HD) [Zwolak et al., 2013]. Vertebrate isoforms readily form homodimers, but heterodimers are not observed, even in cells expressing multiple isoforms [Liang et al., 2009]. Heterodimers can be induced to form, but only when expression levels are artificially high (Edwards, M, Lanier, H, and Cooper, JA unpublished data).
The C-terminal half of CARMIL is intrinsically disordered and includes several functional domains. The CP-binding region (CBR) contains the CPI and CSI motifs, which adopt specific structures when bound to the surface of CP, as revealed by structural and biochemical studies [Hernandez-Valladares et al., 2010; Kim et al., 2012]. The interaction of CARMILs with CP, mediated by the CBR, is known to be necessary for the function of CARMIL1 and CARMIL2 in human cultured cells [Edwards et al., 2015; Lanier et al., 2015]. The direct binding of CP by CARMIL targets CP to membranes, and it decreases, but does not abolish, the actin capping activity of CP [Edwards et al., 2013].
The C-terminal half also includes an unstructured domain of basic and hydrophobic residues, which targets CARMIL to membranes [Brzeska et al., 2010; Lanier et al., 2015], along with a proline-rich domain capable of binding to SH3 domains found in the tails of certain class-I myosins. Only vertebrate CARMILs have the membrane-binding domain [Lanier et al., 2015], and CARMILs from all organisms have proline-rich domains [Edwards et al., 2014].
Among vertebrate isoforms, CARMIL1 has been the most widely studied, in humans and mice. CARMIL1 localizes to the plasma membrane at the leading edge of migrating cells via its PH and membrane-binding domains, both of which can interact with liposomes [Edwards et al., 2013; Lanier et al., 2015; Liang et al., 2009; Zwolak et al., 2013]. In cultured cells, CARMIL1 is necessary for lamellipodia formation, membrane ruffling and macropinocytosis, as well as integrin-mediated Rac1 activation [Liang et al., 2009]. In contrast, CARMIL2 localizes prominently to vimentin intermediate filaments [Lanier et al., 2015; Liang et al., 2009], and this interaction is necessary for all CARMIL2 functions in cultured human cells [Lanier et al., 2015]. CARMIL2 also localizes to membranes, and it is important for lamellipodia formation, membrane ruffling and macropinocytosis [Lanier et al., 2015]. CARMIL2 differs from CARMIL1 in that loss-of-function mutant cells adopt a distinctive multipolar cell shape [Liang et al., 2009]. Evidence that CARMIL1 and CARMIL2 have distinct functions includes the observation that expression of one isoform cannot rescue loss-of-function phenotypes of the other [Liang et al., 2009]. Less is known about CARMIL3, which was identified as a human oncofetal gene (LRRC16B) with effects on cell proliferation [Hsu et al., 2010].
Levels of vertebrate CARMIL isoform expression have not been well studied, either among tissues or during development and differentiation. Previous work has been limited to a set of specific tissues or a single stage of development. In an RT-PCR study of five mouse tissues, CARMIL2 showed high-level expression in thymus and spleen with lower levels in bone marrow and testis and no detectable expression in brain [Liang et al., 2013]. In the same study, CARMIL1 was expressed highly in testis with weak expression in spleen, brain and bone marrow, but not in thymus; CARMIL3 was expressed in brain and testis at moderate levels, but not in thymus, spleen or bone marrow [Liang et al., 2013]. In a study of normal human tissues, using bioinformatics and RT-PCR, CARMIL3 expression was highest in brain and testis, with lesser amounts in many other tissues in adult samples. In fetal tissue samples, CARMIL3 expression was more widespread across tissues [Hsu et al., 2010]. While expression of CARMIL genes in whole animals has not been studied systematically; Carmil2 (rltpr) expression was observed in spinal cord and ganglia of adult zebrafish in a gene-trapping study [Trinh et al., 2011]. In that study, fusion alleles for the other two CARMIL genes were not obtained.
Zebrafish is a powerful model system for the study of developmental, physiological, and pathological processes in vertebrates. During development, cell migration is essential for the morphogenesis of tissues and organs in vertebrates [Solnica-Krezel, 2005]. These cellular movements rely on the actin cytoskeleton, and regulation of actin assembly dynamics is likely to play an important role [Daggett et al., 2004]. Since CP is the major regulator of fast-growing actin barbed ends, its regulation by CARMILs likely plays a significant role during these cellular movements and processes. Here we identify the three CARMIL-encoding genes in zebrafish and examine their expression during early zebrafish development.
Results
Identification of CARMIL family members in zebrafish
To identify genes encoding CARMILs, we searched the zebrafish genome with an amino-acid sequence from the most highly conserved CARMIL-specific region, termed N-cap (Fig. 1A). tBLASTn searches identified three putative genes. Analysis of the predicted protein sequences available from the genome database for the three genes indicated that each gene encodes a specific CARMIL isoform. Carmil1 is encoded by lrrc16a on chromosome 19, Carmil2 by rltpr on chromosome 18, and Carmil3 by lrrc16b on chromosome 2.
To extend the sequence analysis, we examined cDNA sequences for the three genes. For the carmil2 and carmil3 genes, the annotated genome sequence (zv9) predicts full-length transcripts. To test the validity of these predictions, we prepared cDNAs from total RNA isolated from 24 hours post fertilization (hpf) embryos. Using oligonucleotide primers based on the predicted transcript, we obtained overlapping cDNA sequences that correspond well to the prediction for the full-length transcript (rltpr (carmil2): ENSDART00000141051; lrrc16b (carmil3): ENSDART00000127934). The resulting cDNA sequences were submitted to GenBank with accession numbers KR912199 for carmil2 and KR912200 for carmil3.
For Carmil1, the gene is close to a telomere, and the longest predicted transcript in the annotated genome is only 316 nucleotides long (lrrc16a: ENSDART00000141918). This transcript is predicted to encode a polypeptide of 106 residues containing the CARMIL-specific N-cap domain. To identify larger transcripts, we used the predicted protein sequence of medaka (Oryzias latipes) CARMIL1 as a query for tBLASTn searches of zebrafish ESTs. Six ESTs were identified that did not match either carmil2 or carmil3. These sequences were aligned, and oligonucleotide primers based on the EST sequences were used to amplify first-strand cDNA from total RNA as above. A partial cDNA transcript of 4,449 nucleotides was obtained and submitted to GenBank with accession number KR912198. This cDNA includes the 5’ UTR and the start site, and it encodes a predicted protein of 1,435 amino acids, which includes all of the CARMIL domains depicted in Figure 1A but lacks the STOP codon and 3’ UTR.
The domain structures and lengths of the three predicted protein sequences are characteristic of other vertebrate CARMILs (Fig. 1A). Each isoform displays a high degree of sequence similarity when compared to other vertebrate CARMIL isoforms, throughout their entire sequence, including the most highly conserved domains (Fig. 1A). We used the zebrafish and medaka CARMIL sequences to extend the phylogenetic analysis of vertebrate CARMILs. In phylogenetic trees, the zebrafish CARMILs fall into the three groups that define the conserved isoforms CARMIL1, CARMIL2 and CARMIL3 of other vertebrates (Fig. 1B) [Edwards et al., 2014].
CARMIL gene expression during early development
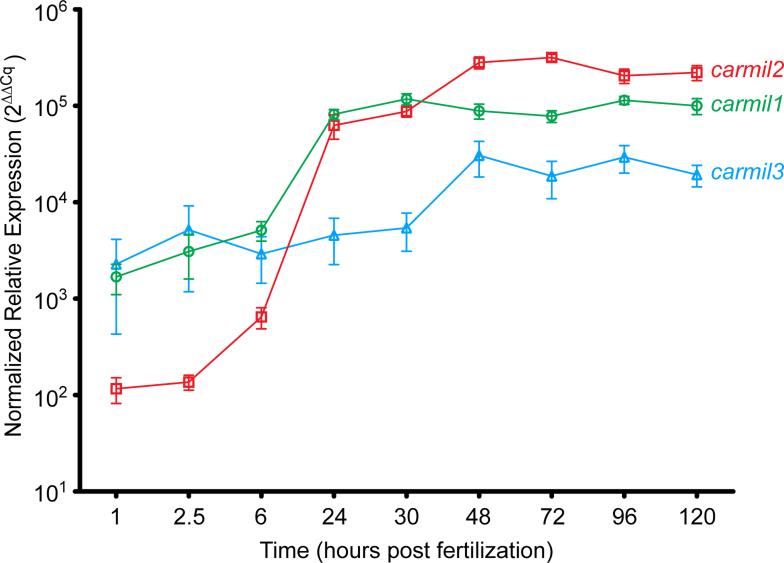
We used our cDNA sequences to determine the level and spatial distribution of CARMIL gene expression during development. First, to measure relative expression levels over time, we performed qRT-PCR analysis on total RNA isolated from whole embryos (Fig. 2). The three CARMIL genes were expressed maternally (1 and 2.5 hpf) and zygotically (6 through 120 hpf); however, their relative levels of expression differed from one another. At maternal stages (1 and 2.5 hpf), carmil1 and carmil3 were expressed at similar levels. carmil2 was expressed but at >10-fold lower levels (Fig. 2 - note the log scale). From 6 to 24 hpf, carmil1 and carmil2 expression levels increased by large amounts (100- and 1000-fold, respectively), and carmil3 expression increased by a smaller amount (<10-fold). Between 30 and 48 hpf, expression levels of carmil1 decreased slightly, whereas those of carmil2 and carmil3 continued to increase. From 72 through 120 hpf, none of the three genes showed large changes in gene expression; carmil2 continued to be expressed at levels greater than either carmil1 or carmil3. These results show that the levels and temporal expression of the zebrafish CARMIL genes are distinct from one another.
Figure 2. Levels of Zebrafish CARMIL Gene Expression During Development.
Normalized relative expression levels, calculated as described in Methods, over time. Each plotted point is the mean of three separate experiments containing two replicates of each gene. Error bars are SEM.
Tissue specificity of expression
Next, we examined CARMIL family gene expression in developing zebrafish by whole-mount in situ hybridization with DIG-labeled RNA probes. The three genes showed distinct expression patterns at all stages examined (Fig. 3).
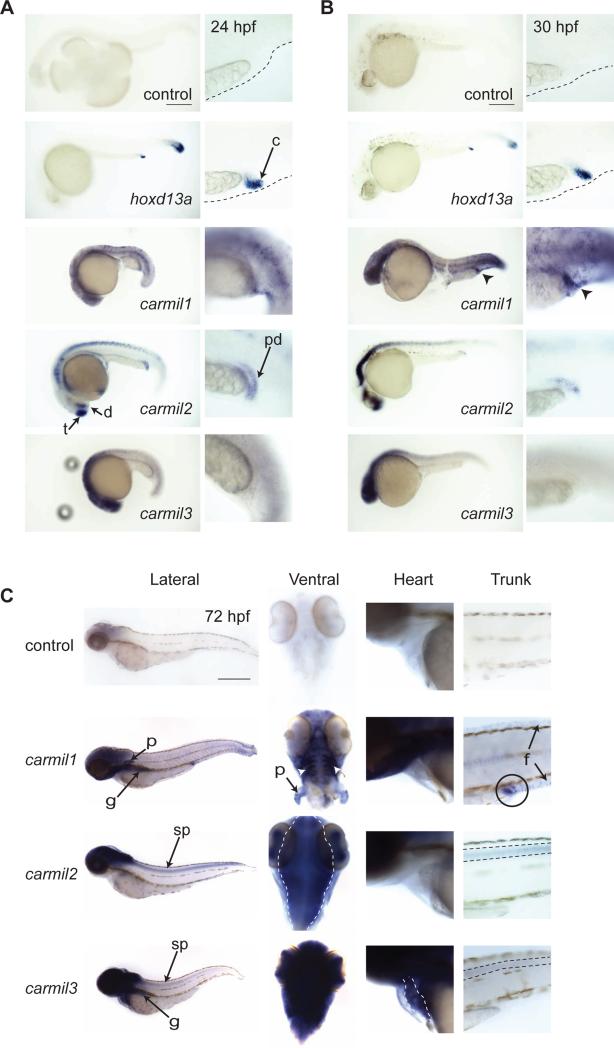
Figure 3. Whole-mount RNA in situ Hybridization for CARMIL Gene Expression.
Lateral view of 24 hpf (A) and 30 hpf (B) embryos treated with RNA probes for the indicated genes. Adjacent panels show increased magnification of the distal end of the yolk extension and the cloaca. hoxd13a was used to identify the cloaca [Pyati et al., 2006; Thisse and Thisse, 2005]. Arrowheads in B indicate increased carmil1 staining at the cloaca. C) 72 hpf embryos treated with RNA probes for the indicated genes. Lateral view of whole fish along with higher magnification images of the ventral view of the head, with yolk removed, lateral view of the developing heart, and lateral view of the trunk. Within the embryo head, carmil1 is concentrated in the branchial arches (white arrow heads) and pectoral fins (p). Both carmil2 and carmil3 are expressed in the embryo eyes. carmil2 expression is concentrated within the brain (dotted outline), and carmil3 is expressed throughout the head and brain. carmil3 shows specific staining in the developing heart (dotted outline). In lateral views of the embryo's trunk, carmil1 is expressed in the cloaca (circle) and fin folds (f). carmil2 and carmil3 expression is seen in the spinal cord (dotted line). Scale: 250 μm for 24 hpf (A) and 30 hpf (B); 500 μm for 72 hpf (C). t – telencephalon; d – diencephalon; c – cloaca; pd – pronephric duct; p – pectoral fin; g – gut; f – fin folds; sp – spinal cord.
carmil1 expression was seen throughout the embryo at 24 hpf and 30 hpf, with increased staining of the epidermis (Fig. 3A-B). At 30 hpf, carmil1 expression became prominent at the presumptive site of the cloaca (circle in Fig. 3B) similar to staining seen for hoxd13a, a marker for the cloaca. Later in development, carmil1 expression persisted throughout the epidermis, at the cloaca and fin folds (Fig. 3C), and appeared prominently in the branchial arches and pectoral fins (ventral view in Fig. 3C). At 72 hpf, both carmil1 and carmil3 appear to stain the developing gut (Fig. 3C).
carmil2 expression at 24 hpf was seen in the central nervous system, including the telencephalon, diencephalon, hindbrain and spinal cord (Fig. 3A). carmil2 staining was also seen at the distal end of the pronephric duct at this time [Wingert et al., 2007]. At 30 hpf, the pattern was similar to the pattern at 24 hpf, with a distinct lack of staining in the midbrain region. Pronephric duct staining was diminished at 30 hpf (Fig. 3B) and completely absent at 72 hpf (Fig. 3C). At 72 hpf, carmil2 staining was now present throughout the brain (outline in the ventral panel Fig. 3C), eyes, and spinal cord (trunk Fig. 3C).
For carmil3, at 24 and 30 hpf, expression was observed throughout the brain and in the eyes (Fig. 3A and B). At 72 hpf, expression remained strong within the brain, head and eyes; with the addition of strong staining to the guy and faint staining of the spinal cord. At 72 hpf, carmil3 was the only CARMIL gene strongly expressed within the heart (Fig. 3C).
Overall, while the three CARMIL genes displayed overlapping expression in certain tissues, they displayed specific expression patterns in different tissues and at different stages of development. Most notably, at 72 hpf, carmil1 expression was seen within the cloaca, fin folds, and branchial arches; carmil2 was expressed within the brain, eyes, and spinal cord; and carmil3 was expressed throughout the head, brain, and eyes, and it was the only isoform observed within the heart.
Discussion
We discovered that, similar to other vertebrates, the zebrafish genome has three genes that encode three proteins of the CARMIL family. Sequence analysis places the proteins into the highly conserved phylogenetic groups of CARMIL isoforms found in all vertebrate genomes. The three genes have distinct expression patterns during zebrafish development, both temporarily and spatially. Both the sequence analysis and the expression patterns support the notion that the three genes have distinct functions during development.
We identified cDNA sequences for the three CARMIL genes, based on analysis of the current genome sequence and DNA sequencing of cDNAs prepared from fish embryos. The three predicted proteins contain all of the domains found in vertebrate CARMILs, including the conserved N-cap and CSI domains that are highly characteristic of CARMILs. Despite the duplication of the zebrafish genome following their ancestral split from mammals [Amores et al., 1998; Gates et al., 1999; Meyer and Schartl, 1999], we found only single representatives of each of the three genes [Edwards et al., 2014].
Previous studies of CARMIL isoform expression have been limited to cultured cells [Liang et al., 2009], selected tissues [Hsu et al., 2010; Liang et al., 2013; Matsuzaka et al., 2004] or a specific isoform at a single stage of development [Trinh et al., 2011]. Here we examined expression in the whole organism over time during development. We found that all three genes are expressed at all times assayed, but with distinct changes in expression levels and tissue distributions over time.
carmil1 is the most widely expressed of the three genes in zebrafish tissues, and it is the only CARMIL isoform observed within the epidermis, fins, and the branchial arches (Fig. 3 B and D). Interestingly, both carmil1 and carmil2 expression is seen near the site of the presumptive cloaca. carmil2 was seen at the pronephric duct at 24 hpf, with staining diminishing at 30 hpf and completely absent at 72 hpf (Fig. 3). carmil1 expression was not observed at the cloaca until 30 hpf (Fig. 3B) persists through 120 hpf (not shown). The timing of the expression of these two CARMIL genes is interesting in that formation of the zebrafish kidney occurs before the formation and opening of the cloaca [Parkin et al., 2009; Pyati et al., 2006; Wallace and Pack, 2003; Wingert et al., 2007].
carmil2 and carmil3 display similar in situ staining patterns, reflecting expression within the brain and spinal cord of developing embryos. The actin cytoskeleton has important roles in neuronal development and neuritogenesis. For example, migration of glial cells requires activation and modulation of Arp2/3-nucleated actin assembly in response to external cues [Gilmour et al., 2002; Klämbt, 2009]. Dendritic spines contain a highly dynamic pool of actin [Star et al., 2002], and spine formation requires CP [Fan et al., 2011]. In addition, Arp2/3-mediated actin polymerization is important for growth cone motility and filopodia formation, as well as neuritogenesis [Korobova and Svitkina, 2008; 2010]. Thus, regulators of CP activity are likely to play a prominent role for morphogenesis of these tissues.
carmil3 is the only isoform for which expression is observed within the developing heart (Fig. 3C). Expression appears at 72 hpf and is not seen at earlier time points, including 48 hpf. The zebrafish heart undergoes a morphological and functional change from a simple two-chambered tube-shaped organ at 48 hpf into a multi-layered and mature functioning organ by 120 hpf [Matrone et al., 2015].
CARMILs regulate actin dynamics indirectly via interactions with actin-binding proteins, notably CP. While CARMILs bind Arp2/3 in lower eukaryotes [Jung et al., 2001, Xu et al., 1995], vertebrate CARMILs appear to have lost this ability [Liang et al., 2009; Yang et al., 2005]. In human cultured cells, the interaction with CP is necessary for the function of CARMIL1 [Edwards et al., 2013] and CARMIL2 [Lanier et al., 2015]. Furthermore, the function of CP in cells appears to require interaction with a CPI-motif protein, such as CARMIL [Edwards et al., 2015]. CARMILs also interact with SH3-containing myosin-Is, and the physiological significance of this interaction has not been examined in detail. Myosin-I, as an actin-based motor, may actively transport CARMIL to the leading edge of migrating cells where CARMIL then interacts with CP [Fujiwara et al., 2014]. Finally, the morphological changes involved in cell migratory events, and the role of regulators of actin dynamics has not been extensively studied. Studying the regulation of CP by CARMILs in specific tissues will allow us to gain further insight into the role CP-mediated actin dynamics play during tissue morphogenesis and development.
Methods
Zebrafish husbandry
Zebrafish of the AB* strain were maintained at 28.5°C following standard operating procedures and guidelines established by the Washington University Zebrafish Facility, described in detail at zebrafishfacility.wustl.edu/documents.html. Embryos were obtained by natural mating and staged according to Kimmel et al. [Kimmel et al., 1995].
CARMIL gene identification
Zebrafish CARMIL1 (lrrc16a), CARMIL2 (rltpr) and CARMIL3 (lrrc16b) family members were identified via tBLASTn using queries with amino-acid sequence from the N-cap domain of human CARMIL2b. cDNA was generated from oligo(d)T-primed total RNA harvested from 24 hours post-fertilization (24 hpf) embryos. For carmil2 and carmil3, first-strand cDNA was sequenced with primers chosen from predictions of full-length transcripts from the annotated genome (zv9).
For carmil1, the annotated genome predicted two partial-length transcripts. We confirmed the existence of the predicted transcripts and used predicted primers to extend the sequence using first-strand cDNA. One transcript contained the N-cap domain, and a second one contained the 5’ UTR and start codon. PCR with other primers predicted to span exon-exon boundaries generated a cDNA sequence with both predicted transcripts and an intervening 125-nucleotide sequence. The genome database is incomplete downstream of the predicted N-cap-containing transcript; sequence between contigs CU611039.6 and CABZ01067085.1 is lacking in the zv9 and GRCz10 genome assemblies. To obtain cDNA sequences for this region, we identified EST sequences by tBLASTn with amino-acid sequence from the predicted medaka carmil1 cDNA. The identified ESTs were GenBank EB911600.1, AL927635.1, BE017584.1, CK146524.1, AL914428.1, and CR926826.1. Using primer walking, we filled the gaps between the predicted carmil1 cDNAs and the ESTs, ultimately generating a cDNA of 4449 nucleotides representing a near-complete transcript lacking only the extreme C-terminus of the predicted polypeptide and 3’-untranslated region.
Our cDNA sequences were submitted to GenBank with accession numbers KR912198 for carmil1, KR912199 for carmil2, and KR912120 for carmil3. Sequence alignments were constructed with the MUSCLE algorithm [Edgar, 2004] and analysis performed with MegAlign Pro (DNASTAR); cladograms were generated with Evolview [Zhang et al., 2012].
qRT-PCR Analysis
Total RNA was isolated using TRIzol reagent (Life Technologies) from whole embryos at specific time points following fertilization. First-strand cDNA was synthesized from 1 μg total RNA for each sample with random hexamers (Roche) and Superscript III reverse transcriptase (Invitrogen). qPCR analysis was performed with Precision Melt Supermix (Bio-Rad) on a Bio-Rad CFX Connect real-time system run for 40 cycles, with actb1 and gapdh as reference genes. Cq values were calculated with Bio-Rad CFX Manager software. Expression values (ΔΔCq) were calculated and normalized by defining the corrected Cq value for 40 cycles as equal to zero.
Whole mount RNA in situ hybridization
Embryos were fixed in 4% paraformaldehyde overnight at 4°C, dechorionated by hand and dehydrated with a series of methanol washes. Probe constructs were amplified and TA-cloned into pCR4-TOPO (Invitrogen). Forward and reverse primers were as follows: carmil1 – CGCTCGTCTGCAGCTAGTCCTGTT and TGATGGCCAGCAGATCCCTGTT; carmil2 – ACCCATTCGTCGCTCGCTAAGA and GATTTGCGCAGGGTCAGTCCAT; carmil3 – GTGATGAAATCAAGAGCAGTCCTGG and TAGCACAATCGTCTCACACAAAAA; hoxd13a – GAGCCCATAACAGACATGAG and GGATCCATTAACCCTCACTAAAGGGAAAAGAGAGGCGAGGAGTGAG [Thisse and Thisse, 2005]. Antisense probes were generated by in vitro transcription with DIG-labeled ribonucleotides (Roche). The carmil2 probe sense strand served as a negative control. Whole-mount in situ hybridization was performed as described [Thisse and Thisse, 2008].
Acknowledgements
We are very grateful to Drs. Liliana Solnica-Krezel and Steve Johnson, the Solnica-Krezel laboratory and the Washington University Zebrafish Facility, for advice and assistance with the project and the manuscript. We are particularly indebted to Drs. Ryan Gray, Jade Li and Diane Sepich of the Solnica-Krezel group and Mr. Stephen Canter of the zebrafish facility. We are grateful to our colleagues Michael Onken, Hunter Lanier and Marc Edwards for discussions and advice. This work was supported by NIH grant GM95509 to JAC. BCS was supported by T32 CA 113275.
References
- Amores A, Force A, Yan YL, Joly L, Amemiya C, Fritz A, Ho RK, Langeland J, Prince V, Wang YL, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Brzeska H, Guag J, Remmert K, Chacko S, Korn ED. An experimentally based computer search identifies unstructured membrane-binding sites in proteins: application to class I myosins, PAKS, and CARMIL. Journal of Biological Chemistry. 2010;285:5738–5747. doi: 10.1074/jbc.M109.066910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daggett DF, Boyd CA, Gautier P, Bryson-Richardson RJ, Thisse C, Thisse B, Amacher SL, Currie PD. Developmentally restricted actin-regulatory molecules control morphogenetic cell movements in the zebrafish gastrula. Curr Biol. 2004;14:1632–1638. doi: 10.1016/j.cub.2004.08.024. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Liang Y, Kim T, Cooper JA. Physiological role of the interaction between CARMIL1 and capping protein. Mol Biol Cell. 2013;24:3047–3055. doi: 10.1091/mbc.E13-05-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, McConnell P, Schafer DA, Cooper JA. CPI motif interaction is necessary for capping protein function in cells. Nat Commun. 2015:1–10. doi: 10.1038/ncomms9415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards M, Zwolak A, Schafer DA, Sept D, Dominguez R, Cooper JA. Capping protein regulators fine-tune actin assembly dynamics. Nat Rev Mol Cell Biol. 2014:1–13. doi: 10.1038/nrm3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Tang X, Vitriol E, Chen G, Zheng JQ. Actin Capping Protein is required for Dendritic Spine Development and Synapse Formation. J. Neurosci. 2011;31:10228–10233. doi: 10.1523/JNEUROSCI.0115-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara I, Remmert K, Piszczek G, Hammer JA. Capping protein regulatory cycle driven by CARMIL and V-1 may promote actin network assembly at protruding edges. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1313738111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates MA, Kim L, Egan ES, Cardozo T. A genetic linkage map for zebrafish: comparative analysis and localization of genes and expressed sequences. Genome. 1999 [PubMed] [Google Scholar]
- Gilmour DT, Maischein H-M, Nüsslein-Volhard C. Migration and function of a glial subtype in the vertebrate peripheral nervous system. Neuron. 2002;34:577–588. doi: 10.1016/s0896-6273(02)00683-9. [DOI] [PubMed] [Google Scholar]
- Hernandez-Valladares M, Kim T, Kannan B, Tung A, Aguda AH, Larsson M, Cooper JA, Robinson RC. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat Struct Mol Biol. 2010;17:497–503. doi: 10.1038/nsmb.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C-C, Chiang C-W, Cheng H-C, Chang W-T, Chou C-Y. Identifying LRRC16B as an oncofetal gene with transforming enhancing capability using a combined bioinformatics and experimental approach. Oncogene. 2010 doi: 10.1038/onc.2010.451. [DOI] [PubMed] [Google Scholar]
- Jung G, Remmert K, Wu X, Volosky JM, Hammer JA. The Dictyostelium CARMIL protein links capping protein and the Arp2/3 complex to type I myosins through their SH3 domains. J Cell Biol. 2001;153:1479–1498. doi: 10.1083/jcb.153.7.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Ravilious GE, Sept D, Cooper JA. Mechanism for CARMIL protein inhibition of heterodimeric actin-capping protein. Journal of Biological Chemistry. 2012;287:15251–15262. doi: 10.1074/jbc.M112.345447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Klämbt C. Modes and regulation of glial migration in vertebrates and invertebrates. Nat. Rev. Neurosci. 2009;10:769–779. doi: 10.1038/nrn2720. [DOI] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Arp2/3 complex is important for filopodia formation, growth cone motility, and neuritogenesis in neuronal cells. Mol Biol Cell. 2008;19:1561–1574. doi: 10.1091/mbc.E07-09-0964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Svitkina T. Molecular architecture of synaptic actin cytoskeleton in hippocampal neurons reveals a mechanism of dendritic spine morphogenesis. Mol Biol Cell. 2010;21:165–176. doi: 10.1091/mbc.E09-07-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier H, Kim T, Cooper JA. CARMIL2 is a Novel Molecular Connection between Vimentin and Actin Essential for Cell Invasion. 2015 doi: 10.1091/mbc.E15-08-0552. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier H, McConnell P, Cooper JA. Cell Migration and Invasion Require a Membrane-Binding Domain of CARMIL2. 2015 doi: 10.1074/jbc.M115.676882. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Niederstrasser H, Edwards M, Jackson CE, Cooper JA. Distinct Roles for CARMIL Isoforms in Cell Migration. Mol. Biol. Cell. 2009;20:5290–5305. doi: 10.1091/mbc.E08-10-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Cucchetti M, Roncagalli R, Yokosuka T, Malzac A, Bertosio E, Imbert J, Nijman IJ, Suchanek M, Saito T, et al. The lymphoid lineage-specific actin-uncapping protein Rltpr is essential for costimulation via CD28 and the development of regulatory T cells. Nat Immunol. 2013 doi: 10.1038/ni.2634. [DOI] [PubMed] [Google Scholar]
- Matrone G, Wilson KS, Mullins JJ, Tucker CS, Denvir MA. Temporal cohesion of the structural, functional and molecular characteristics of the developing zebrafish heart. Differentiation. 2015 doi: 10.1016/j.diff.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Matsuzaka Y, Okamoto K, Mabuchi T, Iizuka M, Ozawa A, Oka A, Tamiya G, Kulski JK, Inoko H. Identification, expression analysis and polymorphism of a novel RLTPR gene encoding a RGD motif, tropomodulin domain and proline/leucine-rich regions. Gene. 2004;343:291–304. doi: 10.1016/j.gene.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol. 1999;11:699–704. doi: 10.1016/s0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- Parkin CA, Allen CE, Ingham PW. Hedgehog signalling is required for cloacal development in the zebrafish embryo. Int. J. Dev. Biol. 2009;53:45–57. doi: 10.1387/ijdb.082669cp. [DOI] [PubMed] [Google Scholar]
- Pyati UJ, Cooper MS, Davidson AJ, Nechiporuk A, Kimelman D. Sustained Bmp signaling is essential for cloaca development in zebrafish. Development. 2006;133:2275–2284. doi: 10.1242/dev.02388. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L. Conserved patterns of cell movements during vertebrate gastrulation. Curr Biol. 2005;15:R213–R228. doi: 10.1016/j.cub.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Star EN, Kwiatkowski DJ, Murthy VN. Rapid turnover of actin in dendritic spines and its regulation by activity. Nature Neuroscience. 2002;5:239–246. doi: 10.1038/nn811. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. High Throughput Expression Analysis of ZF-Models Consortium Clones. ZFIN Direct Data Submission. 2005 [Google Scholar]
- Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
- Trinh LA, Hochgreb T, Graham M, Wu D, Ruf-Zamojski F, Jayasena CS, Saxena A, Hawk R, Gonzalez-Serricchio A, Dixson A, et al. A versatile gene trap to visualize and interrogate the function of the vertebrate proteome. Genes & Development. 2011;25:2306–2320. doi: 10.1101/gad.174037.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Developmental Biology. 2003 doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- Wingert RA, Selleck R, Yu J, Song H-D, Chen Z, Song A, Zhou Y, Thisse B, Thisse C, McMahon AP, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–1938. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Zot AS, Zot HG. Identification of Acan125 as a myosin-I-binding protein present with myosin-I on cellular organelles of Acanthamoeba. J Biol Chem. 1995;270:25316–25319. doi: 10.1074/jbc.270.43.25316. [DOI] [PubMed] [Google Scholar]
- Yang C, Pring M, Wear MA, Huang M, Cooper JA, Svitkina TM, Zigmond SH. Mammalian CARMIL inhibits actin filament capping by capping protein. Dev Cell. 2005;9:209–221. doi: 10.1016/j.devcel.2005.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gao S, Lercher MJ, Hu S, Chen W-H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Research. 2012;40:W569–W572. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwolak A, Yang C, Feeser EA, Michael Ostap E, Svitkina T, Dominguez R. CARMIL leading edge localization depends on a non-canonical PH domain and dimerization. Nat Commun. 2013;4 doi: 10.1038/ncomms3523. [DOI] [PMC free article] [PubMed] [Google Scholar]