Abstract
The polymorphism of the Apolipoprotein E (APOE) promoter rs405509 can regulate the transcriptional activity of the APOE gene and is related to Alzheimer's disease (AD). However, its effects on cognitive performance and the underlying brain mechanisms remain unknown. Here, we performed a battery of neuropsychological tests in a large sample (837 subjects) of nondemented elderly Chinese people, and explored the related brain mechanisms via the construction of diffusion MRI‐based structural connectome and graph analysis from a subset (84 subjects) of the sample. Cognitively, the rs405509 risk allele (TT) carriers showed decreased attention and execution functions compared with noncarriers (GG/GT). Regarding the topological alterations of the brain connectome, the risk allele group exhibited reduced global and local efficiency of white matter structural networks, mainly in the left anterior and posterior cingulate cortices (PCC). Importantly, the efficiency of the left PCC is correlated with the impaired attention function and mediates the impacts of the rs405509 genotype on attention. These results demonstrated that the rs405509 polymorphism affects attention function in nondemented elderly people, possibly by modulating brain structural connectivity of the PCC. This polymorphism may help us to understand the neural mechanisms of cognitive aging and to serve as a potential marker assessing the risk of AD. Hum Brain Mapp 36:4847–4858, 2015. © 2015 Wiley Periodicals, Inc.
Keywords: APOE promoter, brain connectome, cognition, diffusion MRI, graph theory
INTRODUCTION
Genes are a major factor influencing human behavior and cognition; this is reportedly even more so the case for the elderly [Harris and Deary, 2011; McClearn et al., 1997]. One of the most studied genes is the Apolipoprotein E (ApoE, protein; APOE, gene). The ɛ4 allele of APOE is associated with higher risks of late‐onset Alzheimer's disease (AD) [Liu et al., 2013] and with earlier cognitive decline with aging in those who are cognitively normal [Caselli et al., 2009; Deary et al., 2002; Wisdom et al., 2009]. With the advanced neuroimaging techniques, evidence has also shown that the APOE ɛ4 allele is associated with brain structural and functional changes, which may underlie the reported cognitive impairment [Heise et al., 2011; Machulda et al., 2011; Reiman et al., 2004; Shaw et al., 2007].
Recently, the roles of polymorphisms of other genes, especially polymorphisms of the APOE promoter, have been unearthed [Poirier et al., 1993]. The APOE promoter rs405509, also known as −219T/G or Th1/E47cs, is a DNA sequence that acts as a transcriptional factor‐binding site and has been strongly suggested to control APOE expression such as influencing the parenchymal Aβ deposition [Lambert et al., 1998b, 2005]. The TT allele of the APOE promoter has been reported to be a possible risk factor for AD [Beyer et al., 2002; Lambert et al., 1998b, 2002]. However, the effects of the rs405509 variation (TT vs. GG/GT) on cognitive performances in healthy elderly people and the related neural mechanisms remain largely unexplored.
Neuroimaging techniques are tools for bridging the gap in our understanding of how genetic risks impact cognition by modifying the underlying brain circuitry. Given that high‐level cognitive processes depend on interaction among distributed brain regions, MRI‐based brain connectome analyses can be used to explore the possible neural mechanisms of brain diseases and related cognitive alterations. Using the network model and graph‐theory approaches, several recent studies have shown that the topological efficiency of white matter (WM) structural networks constructed from diffusion MRI (dMRI) was decreased in AD [Lo et al., 2010] and mild cognitive impairment (MCI) [Bai et al., 2012; Shu et al., 2012], and was sensitive to cognitive decline during normal aging [Wen et al., 2011]. Importantly, Brown et al. found different age‐related trajectories in the local clustering of WM structural networks for carriers of APOE ɛ4 allele compared with noncarriers [Brown et al., 2011]. Recently, our group has revealed the modulation effect of rs405509 genotype on cortical thickness of parahippocampus during nondemented aging [Chen et al., 2015]. However, in contrast to these advances, the impact of the APOE promoter (rs405509) variants on the topological organization of brain connectome is still unclear, and the influence of the brain endophenotypes on genotype‐related cognitive differences is unknown.
The present study is primarily interested in nondemented older subjects. First, the effects of the APOE promoter rs405509 risk variants on cognitive performance were assessed using a battery of neuropsychological tests in a large Chinese population. Second, to identify the promoter polymorphism that is related to brain alterations, we used dMRI and fiber tractography techniques to construct the WM structural networks and investigate the alterations of the topological organization of brain connectome in the risk allele carrier group compared with the noncarriers. Finally, we explored the relationship between the disrupted cognitive functions and altered topological metrics of the brain structural connectome.
MATERIALS AND METHODS
Participants
A total of 837 nondemented subjects (age range 55–80 years, mean age 65.1 ± 7.1, 534 females) were included in the present study. All participants were right‐handed and native Chinese speakers from the BABRI (Beijing Aging Brain Rejuvenation Initiative) database. The inclusion criteria of the participants were as follows: (a) a score of at least 24 on the Mini‐Mental Status Examination (MMSE); (b) no history of neurologic, psychiatric, or systemic illnesses known to influence cerebral function, including serious vascular diseases, head trauma, tumor, current depression, alcoholism, and epilepsy; and (c) no prior history of taking psychoactive medications; and (d) able to cope with the physical demands of the MRI scanning. The exclusion criteria were as follows: (a) structural abnormalities other than cerebrovascular lesions, such as tumors, subdural hematomas, and contusions due to previous head trauma that could impair cognitive function; (b) history of addictions, neurologic or psychiatric diseases, or treatments that would affect cognitive function; (c) large vessel disease, such as cortical or subcortical infarcts and watershed infarcts; and (d) diseases with white matter lesions according to their medical history and the age‐related white matter changes (ARWMC) rating scale [Wahlund et al., 2001 ]. This study was approved by the Institutional Review Board of the State Key Laboratory of Cognitive Neuroscience and Learning, Beijing Normal University. Written informed consent was obtained from each participant. The demographic information of all participants is presented in Table 1.
Table 1.
Demographic information and neuropsychological characterization of all participants
| TT carriers (n = 415) | GG/GT carriers (n = 422) | F(t/χ 2) value | P | |
|---|---|---|---|---|
| Age (yr) | 65.39 (6.99) | 64.79 (7.26) | 1.21 | 0.23 |
| Gender (M/F) | 170/245 | 133/289 | 5.28 | 0.033 |
| APOE ɛ4 (carrier/noncarrier) | 95/320 | 31/391 | 39.5 | <0.001 |
| General cognition | ||||
| Mini‐Mental Status Examination (MMSE) | 27.33 (2.21) | 27.75 (1.91) | 5.61 | 0.018* |
| Episodic Memory | ||||
| Auditory Verbal Learning Test (AVLT) delayed‐recall | 5.37 (2.61) | 5.75 (2.66) | 2.13 | 0.15 |
| Rey‐Osterrieth Complex Figure test (ROCF) recall | 12.83 (6.53) | 13.18 (6.58) | 0.09 | 0.77 |
| Attention | ||||
| Symbol Digit Modalities Test (SDMT) | 33.29 (11.57) | 35.03 (11.87) | 3.92 | 0.048* |
| Trail Making Test‐A (TMT‐A) (s) | 62.78 (32.03) | 61.24 (30.76) | 0.06 | 0.81 |
| Executive Function | ||||
| Stroop Color and Word Test (SCWT)‐C Time | 77.39 (23.82) | 78.33 (23.85) | 2.15 | 0.14 |
| SCWT‐C Number | 44.77 (6.86) | 44.57 (4.59) | 0.35 | 0.56 |
| Trail Making Test‐B (TMT‐B) (s) | 190.2 (86.06) | 178.94 (70.28) | 5.24 | 0.022* |
The values are represented with mean (standard deviation) of each group. For the details of the neuropsychological tests, see Materials and Methods.
*P < 0.05.
Neuropsychological Testing
All participants received a battery of neuropsychological tests assessing general mental status and other cognitive domains, such as memory, attention and executive function. The general mental status was assessed with MMSE [Crum et al., 1993], which served as exclusion criteria for subjects scoring ≤23 who were considered to have possible dementia [Zhang, 1993]. The comprehensive neuropsychological battery comprised the following three cognition domains (and the included test in each): (a) memory [Auditory Verbal Learning Test (AVLT) [Guo et al., 2007], Rey‐Osterrieth Complex Figure test (ROCF) (recall) [Rey, 1941]]; (b) attention [Trail Making Test‐A (TMT‐A) [Reitan, 1958] and Symbol Digit Modalities Test (SDMT) [Sheridan et al., 2006]]; and (c) executive function [Trail Making Test‐B (TMT‐B) [Reitan, 1958] and Stroop Color and Word Test (SCWT)‐C [Guo, 2005]].
Genetic Analysis
Genomic DNA for each participant was extracted from peripheral blood (200 μl) using the QuickGene‐Mini80 equipment and QuickGene DNA whole blood kit S (Fujifilm, Tokyo, Japan). We genotyped the rs405509 polymorphism using the Custom Taqman® SNP Genotyping Assays (Applied Biosystems, Foster City). An additional two SNPs, rs429358, and rs7412, which collectively form the APOE ɛ2 (with haplotype of rs429358‐rs7412: T‐T), ɛ3 (T‐C), and ɛ4 alleles (C‐C), were also genotyped. The sample success rates for all three SNPs were 100%, and the reproducibility of all the genotyping was 100% according to a duplication analysis of at least 10% of the genotypes. According to the rs405509 genotyping, all subjects were divided into two groups: 415 TT homozygotes and 422 G‐allele carriers (including 58 GG and 364 GT carriers).
MRI Data Acquisition
The MRI data were acquired using a SIEMENS TRIO 3T scanner at the Imaging Center for Brain Research, Beijing Normal University. Among the participants, 84 subjects underwent high‐quality MRI scanning, which included a 3D T1‐weighted MRI scan and a dMRI scan. A T1‐weighted image was acquired by using magnetization‐prepared rapid gradient‐echo (MPRAGE) sequence: 176 sagittal slices, slice thickness = 1 mm, repetition time (TR) = 1,900 ms, echo time (TE) = 3.44 ms, field of view (FOV) = 256 × 256 mm2, acquisition matrix = 256 × 256. The diffusion‐weighted images were acquired by a single‐shot EPI sequence: 70 axial slices, slice thickness = 2 mm, no gap, 30 diffusion directions with b = 1,000 s/mm2, and an additional image with b = 0, TR = 9,500 ms, TE = 92 ms, FOV = 256 × 256 mm2, acquisition matrix = 128 × 128, average = 3. Through genetic analysis, 40 TT allele carriers and 44 GG/GT allele carriers with MRI data were identified. Demographic information for the participants with MRI data is presented in Supporting Information Table S1.
MRI Data Preprocessing
The preprocessing of dMRI data included eddy current and motion artifact correction, estimation of the diffusion tensor, and calculation of the fractional anisotropy (FA). Briefly, the eddy current distortions and motion artifacts in the dMRI data were corrected by applying an affine alignment of each diffusion‐weighted image to the b = 0 image. After this process, the diffusion tensor elements were estimated by solving the Stejskal and Tanner equation, and the FA value of each voxel was calculated [Basser et al., 1994; Basser and Pierpaoli, 1996]. All the preprocessing procedure of dMRI data was performed with the FDT toolbox in FSL (http://www.fmrib.ox.ac.uk/fsl).
Brain Connectome Construction
Network node definition
For a brain network at the macroscale, the entire gray matter (GM) was typically parcellated into a number of regions of interest (ROIs), each representing a network node. The procedure of defining the nodes has been previously described [Gong et al., 2009; Shu et al., 2011]. Briefly, the individual FA image was first coregistered to the T1‐weighted image. The T1‐weighted image was then non‐linearly normalized to the ICBM‐152 template in MNI space using FMRIB's Non‐linear Image Registration Tool (FNIRT, FSL, http://www.fmrib.ox.ac.uk/fsl/). Finally, the inverse transformations were applied to the Automated Anatomical Labeling (AAL) atlas [Tzourio‐Mazoyer et al., 2002] with 90 cortical and subcortical brain regions in the MNI space, resulting in native‐space GM parcelletions for each subject. The transforming procedures were implemented by using PANDA (http://www.nitrc.org/projects/panda/) (Fig. 1).
Figure 1.
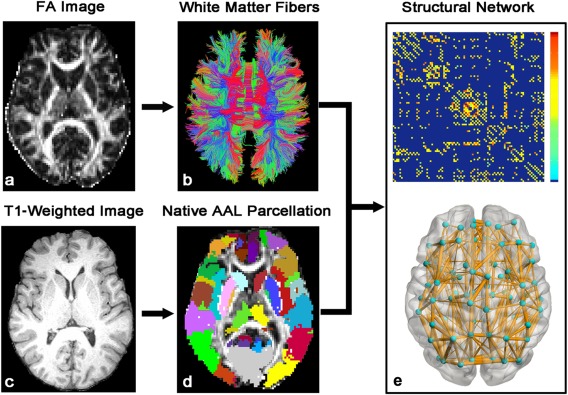
The flowchart of WM network construction by diffusion MRI. The reconstruction of whole‐brain WM fibers was performed using dMRI deterministic tractography with the Diffusion toolkit (http://trackvis.org). The network nodes were defined by parcellating the brain into 90 regions of AAL template through a T1‐weighted image. Two regions were considered structurally connected if at least three fiber streamlines with two endpoints was located in these two regions. Then, the weighted networks of each subject were created by computing the mean FA along the fiber pathways (FA‐weighted) that connect each pair of brain regions. The matrix and three‐dimensional representation (axial view) of the WM network of a normal subject are shown in the right panel. The nodes and connections were mapped onto the cortical surfaces using the in‐house BrainNet viewer software (http://www.nitrc.org/projects/bnv/). For further details, see Materials and Methods. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
WM tractography
Diffusion tensor tractography was implemented with the Diffusion toolkit (http://trackvis.org) [Wang et al., 2007] by using the “fiber assignment by continuous tracking” method [Mori et al., 1999]. The tracking procedure was seeded from the center of each voxel with a FA >0.2 across the entire brain and was terminated if the turning angle was greater than 45 degrees or the fiber entered a voxel with a FA lower than 0.2. For each subjects, tens of thousands of streamlines were generated to etch out all of the major WM tracts.
Network edge definition
For the network edges, two GM regions were considered structurally connected if there was at least three fiber streamlines with its two end‐points located in these two regions [Shu et al., 2011]. We defined the average FA along the pathways of the interconnecting streamlines between two regions as the weights of the network edges. As a result, the FA‐weighted WM network for each participant was constructed, which was represented by a symmetric 90 × 90 matrix (Fig. 1).
Graph Analysis
To characterize the topological organization of WM structural networks, several common graph measures were considered: network strength (S p), global efficiency (E glob), local efficiency (E loc), shortest path length (L p), clustering coefficient (C p), and small‐world parameters (λ, γ, and σ) [Rubinov and Sporns, 2010]. For regional characteristics, we investigated the nodal efficiency [Achard and Bullmore, 2007]. The detailed definitions of these graph metrics are described below. All network analyses were performed using in‐house GRETNA software (http://www.nitrc.org/projects/gretna/) and were visualized using BrainNet Viewer software (http://www.nitrc.org/projects/bnv).
Network strength
For a network (graph) G with N nodes and K edges, we calculated the strength of G as:
where S(i) is the sum of the edge weights wij linking to node i. The strength of a network is the average of the strengths across all of the nodes in the network.
Small‐world properties
Small‐world network parameters (clustering coefficient, C p, and shortest path length, L p) were originally proposed by Watts and Strogatz [1998]. In this study, we investigated the small‐world properties of the weighted brain networks. The clustering coefficient of a node i, C(i), which was defined as the likelihood whether the neighborhoods were connected with each other or not, is expressed as follows:
where ki is the degree of node i, and is the weight, which is scaled by the mean of all weights to control each participant's cost at the same level. The clustering coefficient is zero, C(i) = 0, if the nodes are isolated or with just one connection, i.e., ki = 0 or ki = 1. The clustering coefficient, C p, of a network is the average of the clustering coefficient over all nodes, which indicates the extent of local interconnectivity or cliquishness in a network [Watts and Strogatz, 1998].
The path length between any pair of nodes (e.g., node i and node j) is defined as the sum of the edge lengths along this path. For weighted networks, the length of each edge was assigned by computing the reciprocal of the edge weight, 1/wij. The shortest path length, Lij, is defined as the length of the path for node i and node j with the shortest length. The shortest path length of a network is computed as follows:
where N is the number of nodes in the network. The L p of a network quantifies the ability for information propagation in parallel.
To examine the small‐world properties, the clustering coefficient, C p, and shortest path length, L p, of the brain networks were compared with those of random networks. In this study, we generated 100 matched random networks, which had the same number of nodes, edges, and degree distribution as the real networks [Maslov and Sneppen, 2002]. Of note, we retained the weight of each edge during the randomization procedure such that the weight distribution of the network was preserved. Furthermore, we computed the normalized shortest path length (lambda), , and the normalized clustering coefficient (gamma), , where and are the mean shortest path length and the mean clustering coefficient of 100 matched random networks. Of note, the two parameters correct the differences in the edge number and degree distribution of the networks across individuals. A real network would be considered small‐world if and [Watts and Strogatz, 1998]. In other words, a small‐world network has not only the higher local interconnectivity but also the approximately equivalent shortest path length compared with the random networks. These two measurements can be summarized into a simple quantitative metric, small‐worldness,
which is typically greater than 1 for small‐world networks.
Network efficiency
The global efficiency of G measures the global efficiency of the parallel information transfer in the network [Latora and Marchiori, 2001], which can be computed as:
where Lij is the shortest path length between node i and node j in G.
The local efficiency of G reveals how much the network is fault tolerant, showing how efficient the communication is among the first neighbors of the node i when it is removed. The local efficiency of a graph is defined as:
where Gi denotes the subgraph composed of the nearest neighbors of node i.
Regional nodal characteristics
To determine the nodal (regional) characteristics of the WM networks, we computed the regional efficiency, E nodal(i) [Achard and Bullmore, 2007]:
where Lij is the shortest path length between node i and node j in G. E nodal(i) measures the average shortest path length between a given node i and all of the other nodes in the network.
Statistical Analysis
Demographic factors, including age, gender and APOE ɛ4 carrier distribution of the rs405509 genotypes were compared using either two‐sample t‐test or the χ 2 test. For the genotype effects on the neuropsychological tests and the graph metrics of brain structural networks, comparisons were performed with a multiple linear regression analysis, with age, gender, and APOE ɛ4 effects as covariates of no interest. For regional differences, the false discovery rate (FDR) correction for multiple comparisons was performed. Finally, we investigated the relationship between the altered network metrics and the neuropsychological scores by partial correlation, with removal of the effects of age, gender, APOE ɛ4 and rs405509 genotype. All of the above statistical analyses were performed with SPSS software (version 17.0; SPSS, Chicago, IL). The genotype distribution in our samples was tested with the Hardy‐Weinberg test by the software PLINK [Purcell et al., 2007].
Mediation Analysis
After examining rs405509 genotype effects on cognitive performance and network metrics, we performed a series of mediation analyses [Hayes, 2013] to identify if the topological metrics of brain connectome could mediate the role of risk allele in cognitive decline. A mediator model typically consists of one (or more) independent (or exogenous) variable(s), a mediator variable, and a dependent variable. Both the independent and the mediator variables are assumed to influence the dependent variable. The independent variable can influence the dependent variable both directly and indirectly (i.e., through its effect on the mediating variable). Accordingly, in the present study, the rs405509 risk allele carrying status (TT: 1; GG/GT: 2) constituted the independent variable, disrupted cognitive performance was the dependent variable and altered network measures were the mediator variables. Therefore, we would assume that the association between genotype and cognitive performance is mediated by the altered brain connectome. The mediation analyses were performed with SPSS software.
Reproducibility Analysis
Previous studies have shown that brain graph metrics are highly dependent on the resolution of the network (i.e., number of nodes) [Zalesky et al., 2010]. In addition to coarse AAL parcellation (L‐AAL), we also use the high‐resolution template (H‐1024) [Zalesky et al., 2010] with 1,024 ROIs with equal size to define network nodes. For each participant, the H‐1024 FA‐weighted WM network was constructed and the same procedure of network analysis was performed as in the L‐AAL network. We calculated the network strength, efficiency metrics and small‐world parameters for global network properties and the nodal efficiency for regional network properties. Statistical analyses were used for both global and regional network properties to investigate the genotype effects on the topological organization of the H‐1024 WM networks.
RESULTS
Effects of rs405509 Polymorphism on the Cognitive Performance
Demographic information and neuropsychological characteristics for the participants are presented in Table 1 and Supporting Information Table S1. For the genotype distribution in our samples, no deviation from the Hardy‐Weinberg equilibrium was found (P > 0.05). In the large samples (415 TT allele carriers and 422 G‐allele carriers), the TT allele carriers showed significantly decreased MMSE (P = 0.018), decreased SDMT (P = 0.048) and increased TMT‐B time (P = 0.022) compared with the GG/GT allele carrier (Table 1), suggesting a decline in the general cognition, attention and executive functions in the risk allele group. Based on the relatively small samples with available MRI data (40 TT allele carriers and 44 GG/GT carriers), similar findings were obtained (MMSE: P = 0.044; SDMT: P = 0.0001; TMT‐B: P = 0.001; TMT‐A: P = 0.004) (Supporting Information Table S1).
Effects of rs405509 Polymorphism on the Topological Organization of Brain Structural Connectome
First, the WM networks of both the TT allele carriers and G‐allele carriers showed prominent small‐world properties (λ ≈ 1, γ > 1), which suggests a balance of information integration and segregation of the human brain. However, for the global network metrics, the TT allele carriers exhibited decreased global efficieny (P = 0.007) and local efficiency (P = 0.044), decreased clustering coefficient (P = 0.045), increased absolute and normalized shortest path length (L p: P = 0.006; λ: P = 0.003) than the G‐allele carriers (Fig. 2). For the regional alterations, the most significantly disrupted regions in the TT allele group were located in the left anterior cingulate cortex (ACC), left posterior cingulate cortices (PCC) and right superior temporal gyrus (P < 0.05, FDR corrected) (Fig. 3A,B), and a trend of differences in left middle cingulate cortex and left middle temporal gyrus were also identified (P < 0.01, uncorrected) (Fig. 3A). No regions with increased efficiency in the TT allele carriers relative to the G‐allele carriers were found.
Figure 2.
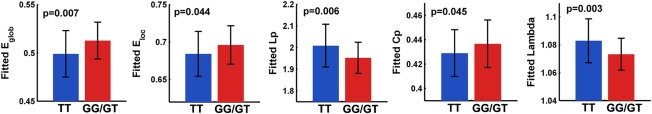
Group differences in global metrics of WM structural connectome. The bar and error bar represent the mean values and standard deviations of the network properties in each group after removing the effects of age, gender and APOE ɛ4, respectively. Significantly reduced global efficiency, local efficiency, clustering coefficient and increased Lp and lambda of WM networks in risk allele carriers relative to the noncarriers were observed (all P < 0.05). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Figure 3.
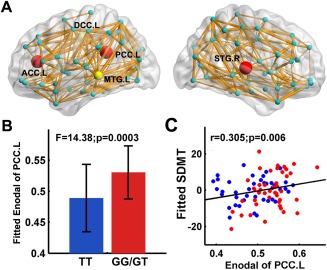
The distribution of brain regions with decreased nodal efficiency in the TT allele group. (A) The node sizes indicate the significance of between‐group differences in the regional efficiency. Nodes in yellow and red showed reduced efficiency in risk allele carriers versus noncarriers (P < 0.01); nodes in red represent regions with group differences remained after FDR corrections. The network shown here was constructed by averaging the structural connection matrices across all subjects and thresholded with a sparsity of 10%. The nodal regions are located according to their centroid stereotaxic coordinates. ACC: anterior cingulate cortex; PCC: posterior cingulate cortex; DCC: dorsal middle cingulate cortex; MTG: middle temporal gyrus; STG: superior temporal gyrus. (B) The bar and error bar represent the mean values and standard deviations of the nodal efficiency of the left PCC in each group after removing the effects of age, gender and APOE ɛ4, respectively. (C) Plots showing the decrease in nodal efficiency of the left PCC with SDMT scores. The blue and red dots represent the adjusted values of TT allele carriers and GG/GT carriers after controlling for age, gender, APOE ɛ4 and rs405509 genotype, respectively. [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Relationship Between Altered Network Topology and Disrupted Cognition
We correlated the altered global and regional network metrics with the disrupted general cognition (MMSE), attention (SDMT), and executive (TMT‐B time) functions across all subjects, while removing the effects of age, gender, APOE ɛ4 and rs405509 genotype. We found that the nodal efficiency of the left PCC was positively correlated with the SDMT score (r = 0.31 P = 0.006) (Fig. 3C). No other correlations between the network metrics and neuropsychological scores were found.
Mediation Analysis
In the mediation analysis, the independent factor was rs405509 variants and dependent variables were cognitive measures that showed significant genotype differences, such as MMSE, SDMT and TMT‐B. The proposed mediators were the altered global and nodal network metrics, which showed significant genotype effects in the above regression analyses. As shown in Figure 4, mediation analysis indicated that the nodal efficiency of the left PCC mediated the effect of rs405509 variants on SDMT score (Z = 2.01, P = 0.045). No other significant mediation effects were found.
Figure 4.
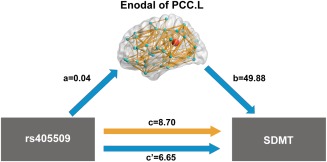
Mediation analysis. Independent factor was rs405509 variants, dependent variables were SDMT scores and the mediator was the nodal efficiency of the left PCC. Path a shows coefficient for the effect of genotype on the nodal efficiency of left PCC. Path b shows the coefficient for the effect of the nodal efficiency of left PCC on SDMT. Paths c and c′show coefficients for the total (yellow) and direct (blue) effects of genotype on SDMT, respectively. The nodal efficiency of the left PCC in the WM structural network mediated the effect of rs405509 variants on SDMT. All coefficients standardized. Sobel test was performed for mediation analysis (Z = 2.01, P = 0.045). [Color figure can be viewed in the online issue, which is available at http://wileyonlinelibrary.com.]
Reproducibility of Findings
For the high‐resolution network, similar group differences in global and regional network metrics were found as those from the L‐AAL network. Compared with G‐allele carriers, TT allele carriers showed decreased global and local efficiency (E glob: P = 0.008; E loc: P = 0.030), increased L p (P = 0.007), increased λ (P = 0.003) and increased γ (P = 0.031). For the regional alterations, TT group showed reduced nodal efficiency, mainly located in the bilateral posterior and middle cingulate cortices, bilateral putamen, right middle temporal gyrus, left hippocampus, left cuneus and right insula (P < 0.05, corrected). No regions with increased efficiency in the TT allele carriers relative to the G‐allele carriers were found.
DISCUSSION
This present study revealed significant effects of the APOE promoter (rs405509) variation on cognitive performance in nondemented elderly people and explored the related topological alterations in the brain structural connectome, which are independent of the effects of the APOE ɛ4 polymorphism. The risk allele carriers showed decreased cognitive attention and execution functions compared with the noncarriers. With respect to brain alterations, the risk allele group exhibited reduced global and local efficiency of WM networks, primarily located in the left anterior and posterior cingulate cortices. Importantly, the decreased efficiency in the left PCC was correlated with the impaired attention function, and the genotype impact on attention was mediated by the efficiency of left PCC. The results suggested that the rs405509 polymorphism targets a specific pattern of cognitive decline in healthy elderly people and demonstrated the effects of rs405509 variations on brain structural connectivity of the PCC. This polymorphism may help to understand the neural mechanisms of cognitive decline in nondemented elderly people and serve as a potential marker to assess the preclinical abnormal changes related to AD.
rs405509 Polymorphism Affects Attention and Executive Functions
According to previous studies, normal aging and AD‐related decline in cognitive performance mainly affects attention, memory and execution functions; a high heritability of cognitive aging was established by the behavioral genetic studies [Grady, 2012]. Until now, the only confirmed genetic variant that influenced cognitive ability in normal elderly was the APOE ɛ4 allele [Wisdom et al., 2009]. APOE ɛ4 carriers performed significantly worse than noncarriers when testing for episodic memory, executive functioning, global cognitive functioning and perceptual speed [Deary et al., 2002; Schiepers et al., 2011]. In the present study, we found similar effects of the risk variant of APOE promoter rs405509 on global cognition, attention and execution functions, but not on memory. Neuropathologically defined subtypes of AD with distinct clinical characteristics, hippocampal sparing and limbic‐predominant AD subtypes might account for approximately 25% of cases. Various domains of cognitive disruption may be related to different subtypes of AD [Murray et al., 2011; Wolk and Dickerson, 2010]. Therefore, we hypothesized that risk allele carriers of rs400509 may interact with APOE ɛ4 on pathological lesions of AD, because of this their effects may not be the same, which would lead to different influences on cognition and even different subtypes of AD.
rs405509 Polymorphism Affects the Topological Organization of Brain Structural Connectome
Brain network analysis has provided a new way to quantify the information integration and segregation of brain systems. The risk allele group showed decreased global and local efficiency of WM networks, suggesting less efficient information transfer across different regions, thus providing possible substrate for the disrupted attention and executive functions [Wen et al., 2011], although we have not found a direct relationship here. The topological alterations were similar to the findings of the DTI network studies in AD, MCI, and normal aging [Bai et al., 2009; Lo et al., 2010; Shu et al., 2012]. Importantly, a recent study has investigated the effects of a common risk allele for AD, APOE ɛ4, on the brain WM connectome [Brown et al., 2011]. They found different age‐related trajectories in the local clustering of brain structural networks for carriers of APOE ɛ4 allele compared with noncarriers. And the present study is the first time to investigate the effects of APOE promoter rs405509 genotype on the topology of WM structural connectome. Some similar and specific findings were found in different studies. Specifically, decreased clustering of WM networks and accelerated disruption in some limbic regions, such as ACC, PCC/Precuneus in the risk allele group were found in both studies. The disrupted architecture of WM networks in the TT allele group is supposed to be related to more rapid degeneration in structural connectivity during aging, which may be due to neuronal shrinkage, loss of small axon fibers and WM demyelination [Bartzokis et al., 2012; Terry et al., 1987; Yeatman et al., 2014], which is also supported by our recent finding of accelerated age‐related reduction of cortical thickness in the risk allele group [Chen et al., 2015]. Some other possible factors, such as invisible vascular lesions, cerebral atrophy, or the age differences between two groups may also contribute to the reduced global connectivity in the risk allele group. Interestingly, most of the regions affected by the APOE promoter, such as the ACC, PCC and superior temporal gyrus, are located in the higher‐order association cortices that are responsible for information integration [Mesulam, 1998 ]. Moreover, these regions have been implicated as key components of the default‐mode network (DMN) [Raichle et al., 2001], and consistently reported to be affected in early AD and APOE ɛ4 carriers [Buckner et al., 2005; Machulda et al., 2011].
PCC Mediates the Effects of rs405509 Polymorphism on Attention Function
The most disrupted brain region in the T homozygotes is found to be located in the PCC. Previous neuroimaging studies have consistently revealed both structural and functional alterations in the PCC and related connectivity in APOE ɛ4 carrier, normal aging and AD [Buckner et al., 2005; Heise et al., 2011; Machulda et al., 2011; Tomasi and Volkow, 2011; Zhang et al., 2007]. As a central part of the DMN [Buckner et al., 2009], the PCC has a high baseline metabolic rate [Raichle et al., 2001] and wide structural and functional connections, responsible for multimodal information integration [Hagmann et al., 2008]. As a highly connected hub region of brain networks, the PCC is more vulnerable to normal aging and disease processes compared with other brain regions [Crossley et al., 2014; Stam, 2014; van den Heuvel and Sporns, 2013], thus providing a potential imaging‐based biomarker for early disease detection and prevention. Specifically, the PCC has been consistently implicated in the pathology of early AD, including cortical atrophy, metabolic disruption, and amyloid deposition [Buckner et al., 2005]. Reduced nodal efficiency in the PCC reflected disrupted interactions with other regions, which suggests decreased structural connectivity within the DMN or between the DMN and other systems. This result is consistent with several other studies that report effects of the APOE ɛ4 allele on WM integrity in brain tracts of the DMN, such as cingulum bundles [Ryan et al., 2011], parahippocampal WM [Honea et al., 2009], and the corpus callosum [Westlye et al., 2012]. According to previous genetic studies, the rs405509 can regulate the transcriptional activity and expression levels of the APOE gene [Artiga et al., 1998; Campillos et al., 2003; Ramos et al., 2005], such as influencing parenchymal Aβ deposition [Lambert et al., 1998b, 2005]. However, the relationship between the biological changes and macroscale alterations in the brain connectome need to be further studied.
Furthermore, we found that the reduced efficiency of the left PCC is correlated with a decreased attention/processing speed in healthy elderly people, and also revealed a significant mediation effect of the PCC nodal efficiency on the association between genotype and attention function. The result may have important implications for the mechanisms underlying individual differences in cognitive performance and may be explained from a neuroanatomical perspective: the information transfer efficiency of the PCC with other regions accounts for attention function, especially processing speed in the elderly. This theory is also supported by a systematic review that showed that the PCC plays an important role in attention function in both cognition and disease [Leech and Sharp, 2013]. The mediation effect of WM provides new insight into understanding the mechanisms of genotype‐related cognitive differences, which may be related to the role of the APOE promoter in the WM development process of the human brain, including dendritic and axon growth and synaptogenesis that underlie circuit development [Judson et al., 2011].
Currently, the interaction between the APOE promoter and the APOE ɛ4 allele in the development of AD remains unclear. According to previous genetic studies, the rs405509 was shown to have a major effect on APOE expression levels [Artiga et al., 1998; Campillos et al., 2003; Ramos et al., 2005]. However, whether this gene is an auxiliary factor that augments the influence of the APOE ɛ4 allele or is an independent risk factor for AD is still being debated. Based on large samples and well controlled studies, as well as a strict meta‐analysis [Lambert et al., 2002], the important and significant association of rs405509 with AD is widely accepted. Several epidemiological studies have consistently shown that the frequency of rs405509 TT genotype in AD patients is significantly higher than that of healthy controls [Lambert et al., 1998a,b,2004], and the genotype has a significant odds ratio in AD [Heijmans et al., 2002]. Further studies are needed to elucidate the relationship of APOE promoter polymorphisms and the APOE ɛ4 allele [Poovindran Anada et al., 2012].
Methodological Issues
Several methodological issues should be addressed. First, although these findings are useful for developing a more mechanistic understanding of the neurobiology of cognitive aging and risk of AD, the present study focuses on common variations in a single candidate gene. Future work should characterize the additive effects of, and interactions between, multiple risk alleles, such as APOE ɛ4 and TT allele of the APOE promoter, in the context of both normal and abnormal aging. Second, longitudinal studies will be important in determining the time course of any alterations of structural and functional connectivity for at‐risk populations. Large population samples with the neuropathological/biochemical examination would be needed to study the contribution of other genes to accelerated cognitive aging and risk of AD. As this investigation was an exploratory study of the APOE promoter, replication in other independent dataset with larger samples is also required to further establish the gene‐imaging phenotype association.
Supporting information
Supporting Information Table 1.
REFERENCES
- Achard S, Bullmore E (2007): Efficiency and cost of economical brain functional networks. PLoS Comput Biol 3:e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artiga MJ, Bullido MJ, Sastre I, Recuero M, Garcia MA, Aldudo J, Vazquez J, Valdivieso F (1998): Allelic polymorphisms in the transcriptional regulatory region of apolipoprotein E gene. FEBS Lett 421:105–108. [DOI] [PubMed] [Google Scholar]
- Bai F, Shu N, Yuan Y, Shi Y, Yu H, Wu D, Wang J, Xia M, He Y, Zhang Z (2012): Topologically convergent and divergent structural connectivity patterns between patients with remitted geriatric depression and amnestic mild cognitive impairment. J Neurosci 32:4307–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai F, Watson DR, Yu H, Shi Y, Yuan Y, Zhang Z (2009): Abnormal resting‐state functional connectivity of posterior cingulate cortex in amnestic type mild cognitive impairment. Brain Res 1302:167–174. [DOI] [PubMed] [Google Scholar]
- Bartzokis G, Lu PH, Heydari P, Couvrette A, Lee GJ, Kalashyan G, Freeman F, Grinstead JW, Villablanca P, Finn JP, Mintz J, Alger JR, Altshuler LL (2012): Multimodal magnetic resonance imaging assessment of white matter aging trajectories over the lifespan of healthy individuals. Biol Psychiatry 72:1026–1034. [DOI] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D (1994): MR diffusion tensor spectroscopy and imaging. Biophys J 66:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Pierpaoli C (1996): Microstructural and physiological features of tissues elucidated by quantitative‐diffusion‐tensor MRI. J Magn Reson 111:209–219. [DOI] [PubMed] [Google Scholar]
- Beyer K, Lao JI, Gomez M, Riutort N, Latorre P, Mate JL, Ariza A (2002): The Th1/E47cs‐G apolipoprotein E (APOE) promoter allele is a risk factor for Alzheimer disease of very later onset. Neurosci Lett 326:187–190. [DOI] [PubMed] [Google Scholar]
- Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KJ, Small GW, Bookheimer SY (2011): Brain network local interconnectivity loss in aging APOE‐4 allele carriers. Proc Natl Acad Sci USA 108:20760–20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA (2005): Molecular, structural, and functional characterization of Alzheimer's disease: Evidence for a relationship between default activity, amyloid, and memory. J Neurosci 25:7709–7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, Andrews‐Hanna JR, Sperling RA, Johnson KA (2009): Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci 29:1860–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campillos M, Lamas JR, Garcia MA, Bullido MJ, Valdivieso F, Vazquez J (2003): Specific interaction of heterogeneous nuclear ribonucleoprotein A1 with the −219T allelic form modulates APOE promoter activity. Nucleic Acids Res 31:3063–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caselli RJ, Dueck AC, Osborne D, Sabbagh MN, Connor DJ, Ahern GL, Baxter LC, Rapcsak SZ, Shi J, Woodruff BK, Locke DE, Snyder CH, Alexander GE, Rademakers R, Reiman EM (2009): Longitudinal modeling of age‐related memory decline and the APOE epsilon4 effect. New Engl J Med 361:255–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Li P, Gu B, Liu Z, Li X, Evans AC, Gong G, Zhang Z (2015): The effects of an APOE promoter polymorphism on human cortical morphology during nondemented aging. J Neurosci 35:1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, Bullmore ET (2014): The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain 137:2382–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crum RM, Anthony JC, Bassett SS, Folstein MF (1993): Population‐based norms for the Mini‐Mental State Examination by age and educational level. JAMA 269:2386–2391. [PubMed] [Google Scholar]
- Deary IJ, Whiteman MC, Pattie A, Starr JM, Hayward C, Wright AF, Carothers A, Whalley LJ (2002): Cognitive change and the APOE epsilon 4 allele. Nature 418:932 [DOI] [PubMed] [Google Scholar]
- Gong G, He Y, Concha L, Lebel C, Gross DW, Evans AC, Beaulieu C (2009): Mapping anatomical connectivity patterns of human cerebral cortex using in vivo diffusion tensor imaging tractography. Cereb Cortex 19:524–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady C (2012): The cognitive neuroscience of ageing. Nat Rev Neurosci 13:491–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo QH, Hong Z, Lv CZ (2005) Application of Stroop colorword test on Chinese elderly patients with mild cognitive impairment and mild Alzheimer's dementia. Chin J Neuromed 4:701–704. [Google Scholar]
- Guo Q, Yu P, Hong Z, Lv C (2007): Norm of auditory verbal learning test in the normal aged in Chinese community. Chin J Clin Psychol 15:132–135. [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, Sporns O (2008): Mapping the structural core of human cerebral cortex. PLoS Biol 6:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SE, Deary IJ (2011): The genetics of cognitive ability and cognitive ageing in healthy older people. Trends Cognit Sci 15:388–394. [DOI] [PubMed] [Google Scholar]
- Hayes AF (2013) Introduction to Mediation, Moderation, and Conditional Process Analysis: A Regression‐Based Approach. New York: Guilford Press. [Google Scholar]
- Heijmans BT, Slagboom PE, Gussekloo J, Droog S, Lagaay AM, Kluft C, Knook DL, Westendorp RG (2002): Association of APOE epsilon2/epsilon3/epsilon4 and promoter gene variants with dementia but not cardiovascular mortality in old age. Am J Med Genet 107:201–208. [DOI] [PubMed] [Google Scholar]
- Heise V, Filippini N, Ebmeier KP, Mackay CE (2011): The APOE varepsilon4 allele modulates brain white matter integrity in healthy adults. Mol Psychiatry 16:908–916. [DOI] [PubMed] [Google Scholar]
- Honea RA, Vidoni E, Harsha A, Burns JM (2009): Impact of APOE on the healthy aging brain: A voxel‐based MRI and DTI study. J Alzheimers Dis 18:553–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judson MC, Eagleson KL, Levitt P (2011): A new synaptic player leading to autism risk: Met receptor tyrosine kinase. J Neurodev Disord 3:282–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Pasquier F, Delacourte A, Frigard B, Cottel D, Perez‐Tur J, Mouroux V, Mohr M, Cecyre D, Galasko D, Lendon C, Poirier J, Hardy J, Mann D, Amouyel P, Chartier‐Harlin MC. (1998a): Pronounced impact of Th1/E47cs mutation compared with −491 AT mutation on neural APOE gene expression and risk of developing Alzheimer's disease. Hum Mol Genet 7:1511–1516. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Pasquier F, Cottel D, Frigard B, Amouyel P, Chartier‐Harlin MC (1998b): A new polymorphism in the APOE promoter associated with risk of developing Alzheimer's disease. Hum Mol Genet 7:533–540. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Araria‐Goumidi L, Myllykangas L, Ellis C, Wang JC, Bullido MJ, Harris JM, Artiga MJ, Hernandez D, Kwon JM, Frigard B, Petersen RC, Cumming AM, Pasquier F, Sastre I, Tienari PJ, Frank A, Sulkava R, Morris JC, St Clair D, Mann DM, Wavrant‐DeVrieze F, Ezquerra‐Trabalon M, Amouyel P, Hardy J, Haltia M, Valdivieso F, Goate AM, Perez‐Tur J, Lendon CL, Chartier‐Harlin MC (2002): Contribution of APOE promoter polymorphisms to Alzheimer's disease risk. Neurology 59:59–66. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Berr C, Cottel D, Amouyel P, Helbecque N (2004): APOE promoter polymorphisms and dementia in the elderly. Neurosci Lett 365:116–119. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Mann D, Richard F, Tian J, Shi J, Thaker U, Merrot S, Harris J, Frigard B, Iwatsubo T, Lendon C, Amouyel P (2005): Is there a relation between APOE expression and brain amyloid load in Alzheimer's disease? J Neurol Neurosurg Psychiatry 76:928–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latora V, Marchiori M (2001): Efficient behavior of small‐world networks. Phys Rev Lett 87:198701. [DOI] [PubMed] [Google Scholar]
- Leech R, Sharp DJ (2013): The role of the posterior cingulate cortex in cognition and disease. Brain 137:12–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Kanekiyo T, Xu H, Bu G (2013): Apolipoprotein E and Alzheimer disease: Risk, mechanisms and therapy. Nat Rev Neurol 9:106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo CY, Wang PN, Chou KH, Wang J, He Y, Lin CP (2010): Diffusion tensor tractography reveals abnormal topological organization in structural cortical networks in Alzheimer's disease. J Neurosci 30:16876–16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machulda MM, Jones DT, Vemuri P, McDade E, Avula R, Przybelski S, Boeve BF, Knopman DS, Petersen RC, Jack CR Jr. (2011): Effect of APOE epsilon4 status on intrinsic network connectivity in cognitively normal elderly subjects. Arch Neurol 68:1131–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslov S, Sneppen K (2002): Specificity and stability in topology of protein networks. Science 296:910–913. [DOI] [PubMed] [Google Scholar]
- McClearn GE, Johansson B, Berg S, Pedersen NL, Ahern F, Petrill SA, Plomin R (1997): Substantial genetic influence on cognitive abilities in twins 80 or more years old. Science 276:1560–1563. [DOI] [PubMed] [Google Scholar]
- Mesulam MM (1998) From sensation to cognition. Brain 121:1013–1052. [DOI] [PubMed] [Google Scholar]
- Mori S, Crain BJ, Chacko VP, van Zijl PC (1999): Three‐dimensional tracking of axonal projections in the brain by magnetic resonance imaging. Ann Neurol 45:265–269. [DOI] [PubMed] [Google Scholar]
- Murray ME, Graff‐Radford NR, Ross OA, Petersen RC, Duara R, Dickson DW (2011): Neuropathologically defined subtypes of Alzheimer's disease with distinct clinical characteristics: A retrospective study. Lancet Neurol 10:785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier J, Davignon J, Bouthillier D, Kogan S, Bertrand P, Gauthier S (1993): Apolipoprotein E polymorphism and Alzheimer's disease. Lancet 342:697–699. [DOI] [PubMed] [Google Scholar]
- Poovindran Anada R, Ganesan D, Ramahsamay N, Wong KT (2012): A prevalence study of single nucleotide polymorphisms in the promoter of the apolipoprotein E gene in different ethnic groups in Malaysia. Neurol Asia 17:341–346. [Google Scholar]
- Purcell S, Neale B, Todd‐Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC (2007): PLINK: A tool set for whole‐genome association and population‐based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL (2001): A default mode of brain function. Proc Natl Acad Sci USA 98:676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos MC, Matias S, Artiga MJ, Pozueta J, Sastre I, Valdivieso F, Bullido MJ (2005): Neuronal specific regulatory elements in apolipoprotein E gene proximal promoter. Neuroreport 16:1027–1030. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J (2004): Functional brain abnormalities in young adults at genetic risk for late‐onset Alzheimer's dementia. Proc Natl Acad Sci USA 101:284–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reitan R (1958): Validity of the trail making test as an indicator of organic brain damage. Percept Motor Skills 8:271–276. [Google Scholar]
- Rey A (1941): L‐examen psychologique dans les cas d'encephalopathie traumatique. Arch Psychol 1941:286–340. [Google Scholar]
- Rubinov M, Sporns O (2010): Complex network measures of brain connectivity: Uses and interpretations. Neuroimage 52:1059–1069. [DOI] [PubMed] [Google Scholar]
- Ryan L, Walther K, Bendlin BB, Lue LF, Walker DG, Glisky EL (2011): Age‐related differences in white matter integrity and cognitive function are related to APOE status. Neuroimage 54:1565–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiepers OJ, Harris SE, Gow AJ, Pattie A, Brett CE, Starr JM, Deary IJ (2011): APOE E4 status predicts age‐related cognitive decline in the ninth decade: Longitudinal follow‐up of the Lothian Birth Cohort 1921. Mol Psychiatry 17:315–324. [DOI] [PubMed] [Google Scholar]
- Shaw P, Lerch JP, Pruessner JC, Taylor KN, Rose AB, Greenstein D, Clasen L, Evans A, Rapoport JL, Giedd JN (2007): Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: An observational study. Lancet Neurol 6:494–500. [DOI] [PubMed] [Google Scholar]
- Sheridan LK, Fitzgerald HE, Adams KM, Nigg JT, Martel MM, Puttler LI, Wong MM, Zucker RA (2006): Normative symbol digit modalities test performance in a community‐based sample. Arch Clin Neuropsychol 21:23–28. [DOI] [PubMed] [Google Scholar]
- Shu N, Liang Y, Li H, Zhang J, Li X, Wang L, He Y, Wang Y, Zhang Z (2012): Disrupted topological organization in white matter structural networks in amnestic mild cognitive impairment: Relationship to subtype. Radiology 265:518–527. [DOI] [PubMed] [Google Scholar]
- Shu N, Liu Y, Li K, Duan Y, Wang J, Yu C, Dong H, Ye J, He Y (2011): Diffusion tensor tractography reveals disrupted topological efficiency in white matter structural networks in multiple sclerosis. Cereb Cortex 21:2565–2577. [DOI] [PubMed] [Google Scholar]
- Stam CJ (2014): Modern network science of neurological disorders. Nat Rev Neurosci 15:683–695. [DOI] [PubMed] [Google Scholar]
- Terry RD, DeTeresa R, Hansen LA (1987): Neocortical cell counts in normal human adult aging. Ann Neurol 21:530–539. [DOI] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND (2011): Aging and functional brain networks. Mol Psychiatry 17:471, 549–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, Joliot M (2002): Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. Neuroimage 15:273–289. [DOI] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O (2013): Network hubs in the human brain. Trends Cognit Sci 17:683–696. [DOI] [PubMed] [Google Scholar]
- Wahlund LO, Barkhof F, Fazekas F, Bronge L, Augustin M, Sjogren M, Wallin A, Ader H, Leys D, Pantoni L, Pasquier F, Erkinjuntti T, Scheltens P (2001): A new rating scale for age‐related white matter changes applicable to MRI and CT. Stroke 32:1318–1322. [DOI] [PubMed] [Google Scholar]
- Wang R, Benner T, Sorensen A, Wedeen V (2007): Diffusion Toolkit: A Software Package for diffusion Imaging Data Processing and Tractography. ISMRM abstract Proc Intl Soc Mag Reson Med 15:3720.
- Watts DJ, Strogatz SH (1998): Collective dynamics of 'small‐world' networks. Nature 393:440–442. [DOI] [PubMed] [Google Scholar]
- Wen W, Zhu W, He Y, Kochan NA, Reppermund S, Slavin MJ, Brodaty H, Crawford J, Xia A, Sachdev P (2011): Discrete neuroanatomical networks are associated with specific cognitive abilities in old age. J Neurosci 31:1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westlye LT, Reinvang I, Rootwelt H, Espeseth T (2012): Effects of APOE on brain white matter microstructure in healthy adults. Neurology 79:1961–1969. [DOI] [PubMed] [Google Scholar]
- Wisdom NM, Callahan JL, Hawkins KA (2009): The effects of apolipoprotein E on non‐impaired cognitive functioning: A meta‐analysis. Neurobiol Aging 32:63–74. [DOI] [PubMed] [Google Scholar]
- Wolk DA, Dickerson BC (2010): Apolipoprotein E (APOE) genotype has dissociable effects on memory and attentional‐executive network function in Alzheimer's disease. Proc Natl Acad Sci USA 107:10256–10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeatman JD, Wandell BA, Mezer AA (2014): Lifespan maturation and degeneration of human brain white matter. Nat Commun 5:4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Yucel M, Pantelis C, Bullmore ET (2010): Whole‐brain anatomical networks: Does the choice of nodes matter? Neuroimage 50:970–983. [DOI] [PubMed] [Google Scholar]
- Zhang M (1993): Guideline of Psychiatrical Scales. Changsha: Human Sciences Technology Press; pp 185–189. [Google Scholar]
- Zhang Y, Schuff N, Jahng GH, Bayne W, Mori S, Schad L, Mueller S, Du AT, Kramer JH, Yaffe K, Chui H, Jagust WJ, Miller BL, Weiner MW (2007): Diffusion tensor imaging of cingulum fibers in mild cognitive impairment and Alzheimer disease. Neurology 68:13–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Table 1.


