Abstract
Alzheimer’s disease (AD) is a common and complex neurodegenerative disease. Age at onset (AAO) of AD is an important component phenotype with a genetic basis, and identification of genes in which variation affects AAO would contribute to identification of factors that affect timing of onset. Increase in AAO through prevention or therapeutic measures would have enormous benefits by delaying AD and its associated morbidities. In this paper, we performed a family-based genome wide association study for AAO of late-onset AD in whole exome sequence data generated in multigenerational families with multiple AD cases. We conducted single marker and gene-based burden tests for common and rare variants, respectively. We combined association analyses with variance component linkage analysis, and with reference to prior studies, in order to enhance evidence of the identified genes. For variants and genes implicated by the association study, we performed a gene-set enrichment analysis to identify potential novel pathways associated with AAO of AD. We found statistically significant association with AAO for three genes (WRN, NTN4, and LAMC3) with common associated variants, and for four genes (SLC8A3, SLC19A3, MADD, and LRRK2) with multiple rare associated variants that have a plausible biological function related to AD. The genes we have identified are in pathways that are strong candidates for involvement in the development of AD pathology and may lead to a better understanding of AD pathogenesis.
Keywords: DNA sequencing, Linear Mixed Model, Pedigree, Family-based association
INTRODUCTION
Alzheimer’s disease (AD: MIM104300) is a common and complex neurodegenerative disease. In the US it is reported as the 6th leading cause of death (Murphy et al., 2013), with direct costs in caring for subjects estimated as >200 billion dollars annually (Thies et al., 2013). Unlike a number of other common diseases associated with aging for which death rates have declined, including heart disease, stroke, and some cancers, the death rate attributable to AD has substantially increased since 2000 (Thies et al., 2013). These issues lead to a projection of substantial rising costs in the near future, not only in the US, but in other countries as well (Banerjee, 2014, Thies et al., 2013). Even a modest increase in age-at-onset (AAO) of AD through prevention or therapeutic measures would have enormous benefits as it would delay the disease and associated morbidities. To date, there have been no successful pharmacological or other therapies that achieve this goal (Thies et al., 2013).
AD risk has a genetic basis, with evidence for familial aggregation first noted more than 60 years ago (Sjögren et al., 1952). Evidence for a genetic basis was later strengthened with results from twin (Gatz & Pedersen, 2013, Gatz et al., 1997, Pedersen et al., 2004) and family (Mayeux et al., 1991, Van Duijn et al., 1991) studies. Identification of four genes with variation that contributes to AD risk provided definitive confirmation (Corder et al., 1993, Goate et al., 1991, Levy-Lahad et al., 1995, Sherrington et al., 1995). Rare mutations in APP, PSEN1, and PSEN2 are typically characterized by highly-penetrant early-onset disease (EOAD, age < 65 yrs) (Bertram et al., 2008, Bird et al., 1996), while common variation in APOE is associated with altered AD risk in more typical and common late-onset AD (LOAD, age ≥ 65 yrs). Risk increases as a function of number of APOE ε4 alleles, and decreases as a function of number of APOE ε2 alleles, relative to the baseline ε3 allele (Corder et al., 1994, Corder et al., 1993). A recent study employing targeted high-throughput sequencing also implicates rare variation in APP, PSEN1, and PSEN2 in risk of LOAD (Cruchaga et al., 2012) and rare variants in TREM2 and PLD3 genes have been implicated in LOAD as well (Benitez et al., 2013, Cruchaga et al., 2014, Guerreiro et al., 2013, Jonsson et al., 2013). Finally, large genome wide studies (GWAS) have recently implicated multiple additional risk loci (Harold et al., 2009, Lambert et al., 2009, Lambert et al., 2013, Naj et al., 2011, Seshadri et al., 2010). Currently, efforts are under way to obtain additional evidence for involvement of the genes proposed in GWAS studies (Holton et al., 2013, Lord et al., 2014).
In AD, age is an important factor. While AD is very rare in younger individuals, especially below age 60 yrs, mutations in the known early onset genes are believed to account for ~50% of early-onset cases (Finckh et al., 2005, Ikeuchi et al., 2008, Lleo et al., 2002, Tandon & Fraser, 2002). Incidence of AD increases with age in all populations surveyed (Fratiglioni et al., 2000, Hebert et al., 2003, Lobo et al., 2000, Rocca et al., 1991, Sosa-Ortiz et al., 2012), with annual incidence in the US increasing from ~1% at ages 65–70 yrs, to 6–8% by age 85 yrs and up (Mayeux, 2003). As a result of this high annual incidence rate, the prevalence of AD among individuals of age 85 yrs and above is ~32% (Thies et al., 2013). In the study of AD, age may be used either as a covariate, or as a phenotype of direct interest. As a phenotype that is directly relevant to AD, AAO is correlated among family-members, with wide variability among families (Bird et al., 1996). Transmission models of AAO in family-based samples support a genetic basis (Daw et al., 2000). Also consistent with a genetic basis, AAO differs among APOE genotypes, with genotype-specific risk inversely proportional to genotype-specific AAO. Differences among APOE genotype-specific onset-distributions are consistent across studies, whether measured on a genetic background of an EOAD mutation in one of the presenilin genes (Pastor et al., 2003, Wijsman et al., 2005) or in more typical LOAD (Farrer et al., 1997). These observations all suggest that AAO may be a useful phenotype for study of the genetic basis of AD.
To date AAO as the phenotype of interest has been used in only a few genome scans of LOAD. Family-based linkage-analysis is the primary approach that has been used to identify regions of interest (Choi et al., 2011, Dickson et al., 2008, Holmans et al., 2005, Lee et al., 2008, Zhao et al., 2013b), with more limited recent use of GWAS in samples of unrelated, affected subjects (Kamboh et al., 2012, Naj et al., 2014). Together with regional analyses (Wijsman et al., 2004), these genome scans implicate regions containing AAO loci. In particular, regions on chromosome 6 and 19p replicated across independent sets of pedigrees. No inclusion of sequence data in AAO studies has yet been reported. There are two main issues with use of AAO. First, study subjects typically are identified through retrospective sampling designs. Analysis of AAO as a continuous variable from a GWAS study can give biased or misleading results when analyses are conducted as if a prospective sampling design had been used (Lin & Zeng, 2009). This contrasts with logistic regression typically used for analysis of case-control studies, which provides equivalent estimates of relative risk for both prospective and retrospective studies (Prentice & Pyke, 1979). Second, with AAO it is necessary to address age-censoring. As a result, analysis has either been carried out using only affected subjects (Holmans et al., 2005, Kamboh et al., 2012, Lee et al., 2008, Naj et al., 2014), or with a model that accounts for age-censoring (Choi et al., 2011, Dickson et al., 2008, Zhao et al., 2013b). Restricting analysis of AAO to AD cases leads to different interpretation of results than does analysis that includes unaffected subjects and incorporates age censoring. Analysis of AAO only in AD cases provides information about the genetic basis only of AAO modification, given predisposition to AD. Inclusion of unaffected subjects and incorporation of age-censoring allows a broader interpretation. In this context, reduced age-at-onset in cases compared to controls as inferred from age-censored data may lead to the inference of increased age-specific risk that is a function of genotype in a region of interest.
In our study here, we performed a family-based GWAS for AAO of LOAD in whole exome sequencing (WES) data in families with multiple AD cases. Our WES data consisted of only AD cases, so our analysis is a case-only analysis of AAO. While GWAS SNP chips provide a relatively high genomic coverage of the common (Minor Allele Frequency, MAF>0.05) genetic variation (>80% (Li et al., 2008)), a portion of common variation and all rare genetic variation is poorly covered. The sequence data allow access to both rare and common variants. Therefore, the use of WES family data allows the study of both common and rare variants, which are likely to be enriched in families (Wijsman, 2012). In order to identify new loci (both rare and common variants) and genes that may be a modifying factor of AAO, we conducted a classical single marker association analysis for common variants, and a gene-based burden association analysis for multiple rare variants in genes. We combined association analyses with variance component linkage analysis, in addition to referring to previous analyses of AAO in other family-based samples, in order to enhance evidence of the identified genes. We also performed a gene-set enrichment analysis on the list of identified genes and explored the biological function of these genes to help understanding AAO/AD pathogenesis.
MATERIALS AND METHODS
Subjects and phenotyping
Our data consisted of 77 subjects diagnosed with AD. These subjects were selected from pedigrees with multiple cases of late onset AD (> 60 yrs onset age) in multigenerational families from public repositories: 58 subjects from the NIA-LOAD/NCRAD collection (Wijsman et al., 2011) and 19 subjects from the NIMH (Blacker et al., 1997) collection. Single subjects, but not families, from the NIA-LOAD/NCRAD families have been incorporated into GWAS studies of AAO, and the NIMH families have previously been used for a linkage-analysis genome scan of AAO (Choi et al., 2011). Neither set of families has been used for evaluation of sequence-based variants as contributors to AAO, which was the goal of the current study. Pedigrees ranged in size from 10 to 25 subjects, and consisted of 3–4 generations/pedigree. The assumption was that use of families increases ability to detect effects of (potentially) rare alleles. Selection of specific families additionally required availability of DNA from at least two relatively distantly-related LOAD cases (e.g., avuncular to second cousin relationships), thus minimizing their relatedness relative to subjects from other available pedigrees, and reducing the sizes of regions of interest identified in the families. DNA samples from up to four cases per family were used when they were available, therefore also including additional, closer relationships. For inclusion in the current analysis, subjects also were required to be of European ancestry. Additional Hispanic pedigrees selected at the same time with the same criteria were only used for Principal Component Analysis in order to ensure a homogenous group of subjects. They were not used for association analysis because of analytical complications of joint analysis of an admixed set of pedigrees with European- and Hispanic-descent families. The AD affection status was defined as meeting NINCDS-ADRDA criteria for definite, probable, or possible AD (Mckhann et al., 1984, Wijsman et al., 2011). In both the NIMH and NIA-LOAD/NCRAD samples, AAO for AD cases was defined as the age at which first symptoms of AD were reported (Choi et al., 2011, Wijsman et al., 2011). In the subjects used for our analysis, the mean of AAO and its standard deviation are 70.6 and 9.24, respectively. Our study was approved by the University of Washington institutional review board. All samples used were collected with appropriate consent for this study.
Whole Exome Sequencing data
The gene coding sequences were captured using Nimblegen SeqCap EZ Human Exome Library v2.0 kit (Roche, Basel, Switzerland) following the manufacturer’s recommended instruction. The capture kit targets 28,858 genes with total size of the target regions 36.5 Mbp. The sequencing library clusters were generated on Illumina flowcells using cBlot (Illumina, Inc.) and pair-end 101bp sequencing was performed on the Illumina HIseq2000 sequencing platform at the Department of Genome Sciences, University of Washington. The raw base calling was performed with CASAVA (Illumina, Inc.). Sequenced reads were aligned to NCBI human reference genome GRCh37 (hg19) using the Burrows-Wheeler Aligner (Li & Durbin, 2010). BAM files were generated using SAMtools (Li et al., 2009). PCR duplicates were marked using Picard (http://broadinstitute.github.io/picard/). After base recalibration the sequence reads were realigned around indels and mapped. For single nucleotide variant calling, the Genome Analyzer Toolkit (GATK) was utilized (Mckenna et al., 2010). The average read depth for called positions was 70.89.
In all analyses, we considered autosomal di-allelic polymorphic variants (i.e. 102,603 SNPs). Quality control steps were performed to filter out possible sequencing errors. We used the following exclusion criteria: ABHet (Allele balance for heterozygotes) > 0.75, HRun (Largest Contiguous Homopolymer Run of Variant Allele In Either Direction) > 4.0, QUAL (Phred-scaled quality score) ≤ 50, QD (Variant Confidence/Quality by Depth) < 5, or SB (Strand Bias) ≥ 0.10. We also excluded SNPs with evidence of Hardy-Weinberg (HW) disequilibrium (p-value<10−3). HW testing was based on the 32 unrelated individuals in our WES dataset. QC procedures led to the exclusion of 15,937 SNPs (i.e. 14,948 due to sequence call quality filters and 989 due to HW disequilibrium). The number of remaining SNPs was 86,666.
Statistical Analysis
Principal Component Analysis
Families in the NIA-LOAD/NCRAD sample were recruited by multiple sites across the US (Wijsman et al., 2011) and families in the NIMH sample were recruited by three different sites (Blacker et al., 1997). As a result, subjects may be drawn from different genetic backgrounds even within subjects declared as of European ancestry. For the NIA-LOAD/NCRAD sample, several families are known to be Caribbean Hispanic. As described previously, there was strong evidence for population stratification in the NIA-LOAD/NCRAD European-American sample (Wijsman et al., 2011), and an indication of possible stratification in the NIMH sample (Choi et al., 2011). To account and correct for population stratification in the full sample used here, we performed a supervised principal component analysis (PCA) using EIGENSTRAT (Price et al., 2006) using SNPs from the WES data. For the PCA, we added to our AD cases the 1000 Genomes (1KG) project (Abecasis et al., 2010) subjects (release November 2010) of European (EUR), African (AFR), and Asian (ASN) descent. PCA was performed using common variants (Minor allele frequency (MAF) >0.05) with low pairwise linkage disequilibrium (LD), and with available genotypes in both our data and the 1KG data. We used PLINK (Purcell et al., 2007) to select SNPs as follows: in a window of 150 SNPs, we estimated LD for all pairs of SNPs and filtered out one of each pair having an r2 > 0.2. We used overlapping sliding windows with a step-size of five SNPs. The procedure led to a set of 8,822 SNPs. Our PCA results identified 16 WES subjects who do not cluster with the remaining subjects of European descent (Fig. S1 in supplementary material). These subjects were all of known Caribbean Hispanic descent and were excluded from subsequent analysis, which led to exclusion of entire pedigrees, with no exclusion of any of the European-descent subjects. Among the remaining subjects, 56 subjects had available AAO information, and out of these 56 subjects, APOE genotypes were available for 47 subjects.
Imputation Analysis for APOE
As recently shown (Radmanesh et al., 2014), missing APOE genotypes can now be accurately imputed using the very dense 1000 Genomes data as a reference. To avoid decrease of the sample size due to missing APOE genotypes for nine subjects, we performed an imputation analysis to infer the missing APOE genotypes using the 1000 Genomes data (August 2010). Briefly, we used SHAPEIT2 (Delaneau et al., 2012) to phase the NIA-LOAD/NCRAD GWAS subjects and minimac (http://genome.sph.umich.edu/wiki/Minimac) to impute the missing genotypes. Then we extracted the allelic dosages for the two APOE SNPs (i.e., rs7412 and rs429358) in the case of missing APOE genotypes and we used them along with the known APOE genotypes in subsequent association analyses. With these procedures, all 56 subjects were used.
Association Analysis
We conducted two family-based genome wide association analyses for the log-transformed AAO. The first analysis used a single marker test where one SNP was tested at a time. The second analysis used a gene-based association burden test (Weighted Sum approach (Madsen & Browning, 2009)). The aim of the first analysis was to find evidence of association between AAO and SNPs with MAF>0.05. The aim of the second analysis was to find evidence of association between AAO and multiple rare variants in a gene (SNPs with MAF≤0.05). All association analyses were performed using the statistical package R (http://www.r-project.org/). To account for family relationship among subjects, we used a linear mixed model (LMM) implemented in the R-package “kinship”. In all models, we used the theoretical kinship matrix obtained from the pedigree structure information.
Single Marker Association Test for Common Variants
For the single marker association test, assuming an additive model, we considered 39,993 SNPs with MAF > 0.05 and with at least 50 non-missing genotypes. Among these SNPs, 30,384, 20,574, and 13,590 have minor allele frequencies greater than 0.1, 0.2, and 0.3, respectively. We used the model: log(AAO) = β × X + δZ + ε, where X is the vector of genotypes coded additively as 0, 1, or 2 copies of the minor allele, Z is the vector of observed covariates (e.g., APOE genotypes when used), β and δ are the marker and covariate fixed effect coefficients, respectively, and where Φ is a matrix of twice the coefficient of kinship between pairs of subjects, I is an identity matrix, and and are the polygenic and residual variances, respectively. To test for association, we used the Wald test (H1 : β ≠ 0 vs H0 : β = 0).
Burden Association Test for Multiple Rare Variants
We performed the weighted sum burden association test (Madsen & Browning, 2009) for rare variants (MAF≤0.05). This approach collapses rare variants within genes by giving them weights that are inversely proportional to minor allele frequency. SNPs were annotated using ANNOVAR (Wang et al., 2010). A gene was tested if it had at least two non-synonymous SNPs with MAF < 0.05 and if the sum of the MAFs was greater than 0.05. The association model we used was: where √wi ~ dBeta(5,25) (Wu et al., 2011), where Z and ε are defined above. Again, we used the Wald test to test for association.
Variance Component Linkage Analysis
In order to determine which of our association signals are also supported by evidence for linkage and to incorporate this evidence into the overall evaluation of the signals, we performed a variance component linkage analysis using SOLAR (Almasy & Blangero, 1998) for all 26 NIA-LOAD pedigrees, which have GWAS SNP data. The small number of NIMH pedigrees had a different, microsatellite marker scan, which could not be combined with the NIA-LOAD SNP markers for joint analysis. These pedigrees were therefore not used for the linkage analysis since the 19 available subjects is too small a sample to support estimation of the multiple parameters needed for a separate VC linkage analysis. For each chromosome, we first selected a set of equally-spaced SNPs (~0.5 centiMorgan) with relatively high MAF (>0.4) and in linkage equilibrium. Then, we estimated the Identity-By-Descent (IBD) distribution at each marker position in a full multipoint computation using a Markov Chain Monte Carlo (MCMC) method for pedigrees that have more than 15 transmitted meioses and exact computation for the remaining pedigrees. This analysis was performed using the program gl_auto in the MORGAN package (Thompson, 2011). IBD estimates at 5 cM intervals were converted to SOLAR input-format and a model with additive variance components only was fitted and compared to a model with polygenic variance, only, in a likelihood ratio test. We ran two versions of this model: the first one adjusted for APOE genotypes (VC wApoe) and the second one did not (VC sansApoe).
RESULTS
Single Marker Test
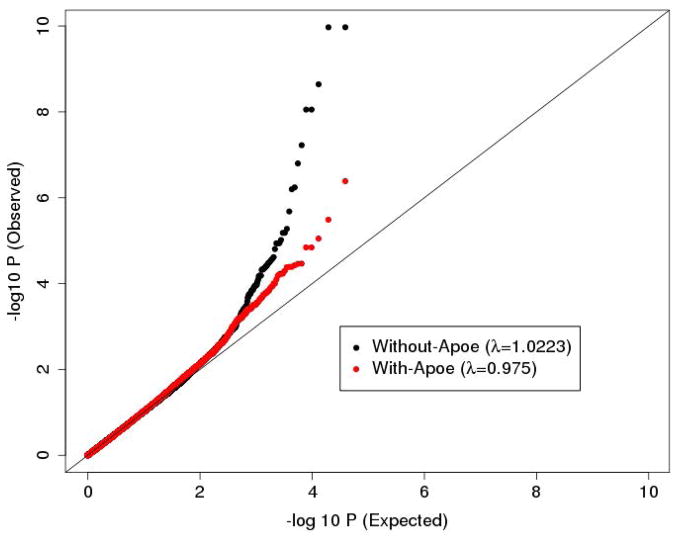
We first ran association analyses without any covariates. The QQ-plot and genomic control coefficient (Devlin & Roeder, 1999) (λ= 1.022) showed slight inflation in the statistical test distribution (Fig. 1). This excess of significant results may be driven by the effect of APOE on sample ascertainment, and thus on sample structure as suggested in (Wijsman et al., 2011). To reduce the observed inflation, and to also avoid detecting signals driven by APOE (a very well established associated factor with AD (Corder et al., 1994, Farrer et al., 1997)), we ran a second association analysis adjusting for APOE as covariate in the LMM. The statistical distribution obtained by this analysis was better controlled, as both the genomic control coefficient (λ=0.975) and QQ-plot showed (Fig. 1).
Fig. 1.
QQ-plot of the single marker test analysis that adjusts (red) and does not adjust (black) for APOE. The plot is based on all 39,993 tested SNPs.
As expected, we observed significant evidence of association for SNPs in the APOE region on chromosome 19 in the first analysis without adjustment for APOE (from 44 Mbp to 47 Mbp). None of the SNPs tested are the two SNPs that define the three critical APOE alleles, as those SNPs fail QC analysis because of low read depth. At a nominal threshold (α=0.05), 10 SNPs were significant in this region. The SNP rs11879355 provided the strongest evidence for association with a p-value of 6.2×10−3 (Table S1 in supplementary material). After adjusting for APOE, all 10 significant SNPs were no longer significant at a nominal threshold of α =0.05 (Table S1 in supplementary material).
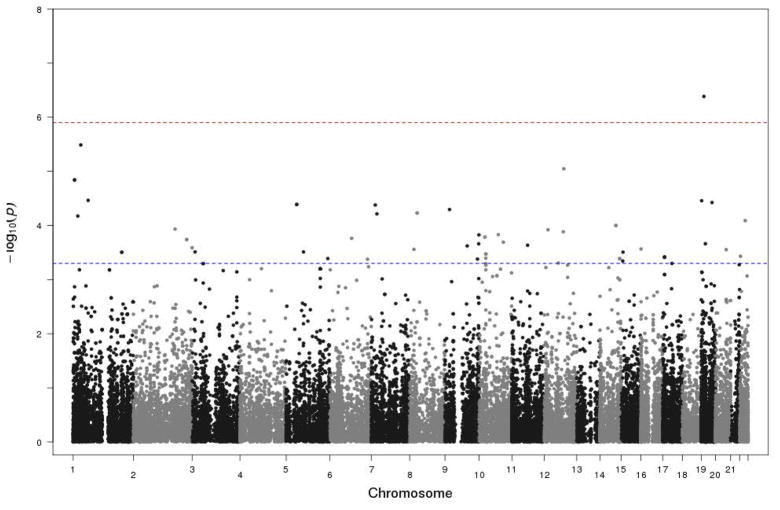
Our main results from single SNP analyses are based on the second analysis, which adjusts for APOE, and are shown in the Manhattan plot in Fig. 2. Our strategy was to focus on the most significant non-synonymous SNPs with p-values less than 5×10−4 (i.e. 15 SNPs). The results of these SNPs are shown in Table 1. All these SNPs, except rs1800378, had negative effect sizes, indicating that they decrease AAO. The MAF of 13 of these SNPs was less than 0.1. The significance of association tests of the 15 SNPs ranged from 4.99×10−4 to 4.12×10−7. We identified one SNP on chromosome 19 (rs2291516, MAF=0.08, p-value=4.12×10−7) with Bonferroni-corrected significance (0.05/39993 = 1.25×10−6). This SNP is in the gene RGL3. The remaining 14 SNPs are located in 11 different genes on nine different chromosomes.
Fig. 2.
Manhattan plot of the single marker test analysis that adjusts for APOE. The plot is based on all 39,993 tested SNPs. The horizontal blue line is the threshold we used to decide the most significant SNPs. The horizontal red line is the genome-wide significance threshold.
Table 1.
Most significant SNPs obtained by the single marker test
| Chr | Position | Gene | rsname | Minor | Major | MAF | F | β | P | βa | Pa |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 201,180,100 | IGFN1 | - | T | C | 0.071 | - | −0.147 | 1.18E-03 | −0.144 | 3.12E-04 |
| 1 | 201,180,340 | IGFN1 | rs139390045 | G | A | 0.071 | 0.084 | −0.147 | 1.18E-03 | −0.144 | 3.12E-04 |
| 5 | 43,613,046 | NNT | rs35201656 | G | A | 0.054 | 0.042 | −0.116 | 2.59E-02 | −0.157 | 4.08E-05 |
| 5 | 43,653,243 | NNT | rs41271083 | T | C | 0.054 | 0.042 | −0.116 | 2.59E-02 | −0.157 | 4.08E-05 |
| 8 | 30,921,935 | WRN | rs2230009 | A | G | 0.054 | 0.056 | −0.346 | 1.07E-10 | −0.296 | 5.88E-05 |
| 9 | 133,963,008 | LAMC3 | rs4740412 | A | G | 0.214 | 0.266 | −0.077 | 1.31E-02 | −0.089 | 4.17E-04 |
| 9 | 139,118,673 | QSOX2 | rs12380852 | C | T | 0.107 | 0.117 | −0.132 | 1.71E-04 | −0.113 | 2.17E-04 |
| 10 | 23,729,362 | OTUD1 | - | T | A | 0.063 | - | −0.102 | 8.68E-02 | −0.203 | 1.63E-04 |
| 15 | 28,230,318 | OCA2 | rs1800407 | T | C | 0.054 | 0.078 | −0.142 | 4.34E-03 | −0.157 | 3.11E-04 |
| 16 | 615,048 | C16orf11 | rs113068385 | A | G | 0.055 | 0.036 | −0.053 | 3.05E-01 | −0.151 | 2.71E-04 |
| 17 | 7,324,788 | SPEM1 | rs33989543 | A | G | 0.054 | 0.070 | −0.258 | 6.57E-06 | −0.151 | 2.71E-04 |
| 17 | 7,735,063 | DNAH2 | rs57985356 | G | T | 0.071 | 0.085 | −0.141 | 3.84E-03 | −0.138 | 3.82E-04 |
| 17 | 7,735,934 | DNAH2 | rs61745181 | A | G | 0.071 | 0.076 | −0.141 | 3.84E-03 | −0.138 | 3.82E-04 |
| 19 | 11,508,177 | RGL3 | rs2291516 | A | G | 0.080 | 0.101 | −0.155 | 3.60E-04 | −0.168 | 4.12E-07 |
| 19 | 17,361,116 | USHBP1 | rs1043963 | A | G | 0.063 | 0.086 | −0.133 | 8.46E-03 | −0.157 | 2.17E-04 |
MAF= Minor Allele Frequency in the sample; F= Minor allele frequency in the ESP project data of European-American sample; b = Effect size not adjusted for APOE; P= P-value not adjusted for APOE;
Analysis that adjusts for APOE. All analyses were based on N=56 subjects.
Burden Test
The number of tested genes with at least two non-synonymous SNPs with MAF < 0.05 and the sum of the MAFs greater than 0.05 was 1,949. The first quartile, median, mean, and third quartile of the number of rare variants in a gene were 3, 4, 4.96, and 6, respectively. We ran two versions of burden tests. The first one adjusts for APOE (wApoe) and the second one does not (sansApoe). Genomic control coefficients (0.776 for wApoe and 0.996 for sansApoe) and QQ-plots showed better properties of the burden test obtained by sansApoe. This trend was different from what we obtained in the single marker test. This is due to the fact that the weighted sum of variants in a gene is likely to be less correlated with APOE than the alleles of each marker alone. Therefore, our results were based on the sansApoe analysis. Manhattan and QQ-plots can be found in Fig. S2 and Fig. S3 in supplementary material. In the burden test analysis, we focused on the 10 most significant genes (Table 2) from our genome scan, with p-values ranging from 3.11×10−4 to 5.3×10−6. Again, to ensure that our most significant results were not driven by the effect of APOE, we explored the wApoe analysis of the 10 selected genes. The significance of all genes decreased slightly in the wApoe analysis.
Table 2.
Burden test results for the ten most significant genes
| Chr | Gene | Pos first SNP | Pos last SNP | V0.05 | S_MAF | β | P | βa | Pa |
|---|---|---|---|---|---|---|---|---|---|
| 2 | SLC19A3 | 228,552,234 | 228,563,911 | 3 | 0.054 | −0.266 | 5.85E-06 | −0.166 | 2.77E-03 |
| 11 | MADD | 47,296,533 | 47,315,499 | 2 | 0.054 | −0.254 | 2.00E-05 | −0.169 | 2.85E-03 |
| 11 | C11orf82 | 82,625,814 | 82,644,904 | 4 | 0.080 | −0.208 | 3.08E-05 | −0.105 | 2.71E-02 |
| 11 | KDELC2 | 108,352,777 | 108,357,137 | 2 | 0.054 | −0.241 | 5.24E-05 | −0.219 | 5.21E-04 |
| 12 | LRRK2 | 40,619,082 | 40,758,652 | 4 | 0.054 | −0.223 | 2.84E-04 | −0.133 | 2.09E-02 |
| 14 | SLC8A3 | 70,527,576 | 70,634,200 | 3 | 0.071 | −0.196 | 2.56E-04 | −0.049 | 3.68E-01 |
| 14 | SLC24A4 | 92,909,807 | 92,959,940 | 5 | 0.063 | −0.177 | 5.30E-06 | −0.288 | 1.00E-04 |
| 15 | STARD9 | 42,930,972 | 43,011,009 | 8 | 0.080 | −0.109 | 3.11E-04 | −0.053 | 3.45E-01 |
| 19 | GRIN3B | 1,000,785 | 1,009,585 | 3 | 0.063 | −0.190 | 9.00E-05 | −0.119 | 3.47E-02 |
| 19 | LENG8 | 54,966,557 | 54,968,038 | 3 | 0.063 | −0.202 | 2.35E-04 | −0.112 | 3.52E-02 |
V0.05 = SNPs with MAF<0.05; S_MAF= Sum of the V0.05 minor allele frequencies; β= Effect size not adjusted for APOE; P= P-value not adjusted for APOE.
Analysis that adjusts for APOE. All analyses were based on N=56 subjects.
Variance Component Linkage Analysis
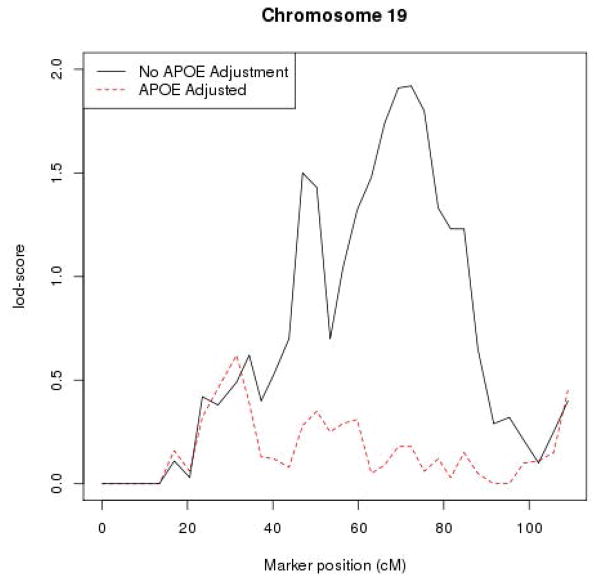
In the VC sansApoe, we identified five regions with lod-scores ≥1.5 (Chromosome 2, 4, 9, 13, and 19) (Supplementary material). The region on chromosome 19 is the longest one and has the highest maximum lod-score, reaching ~2, out of these five regions (Fig. 3). The relevant gene in this region is most likely APOE since the lod-score maximizes at approximately the location of APOE on chromosome 19. Indeed, the VC wApoe analysis showed a drastic decrease of lod-scores in this region (i.e., lod-scores drop from ~2 for VC sansApoe to <0.2 for VC wApoe at the position of APOE at ~72 cM, Fig. 3). In the same analysis, most, but not all, lod-scores of other regions also decreased. The region on chromosome 9 was the most robust to APOE adjustment with its lod-score decreasing modestly from 1.47 to 1. New regions appeared when adjusting for APOE: 1) one around 210 cM on chromosome 2 (lod-score adjusted for APOE = 1.1) and one around 50 cM on chromosome 4 (lod-score adjusted for APOE = 1.4). Moreover, a modest signal on chromosome 19p around 30 – 40 cM (lod ~0.6) is effectively immune to whether or not there is adjustment for APOE.
Fig. 3.
Lod-score plot for chromosome 19. The red dashed line represents the lod-scores of VC analysis adjusting for APOE. The black line represents the lod-scores of VC analysis without adjustment for APOE.
Analysis of known genes from the literature
We explored our association results for the SNPs and genes reported in a previous AAO GWAS (Naj et al., 2014). The authors considered previously identified genes associated with AD (i.e.; CR1, BIN1, CD2AP, EPHA1, CLU, MS4A4A, PICALM, ABCA7, and CD33). They identified association between AAO and SNPs in CR1 (rs6701713, p-value=7.2×10−4), BIN1 (rs7561528, p-value=4.8×10−4), and PICALM (rs561655, p-value=2.2×10−3). From the list of SNPs reported in this previous study, only one SNP was found in our WES data (rs3752246, ABCA7). This SNP was not significant in both our and their study (p-value=0.69 and 0.064, respectively). Note that the remaining SNPs were located in introns. Nonetheless, several SNPs with MAF greater than 0.05, in CR1 and ABCA7, were nominally significant in our single marker test analysis of AAO. In CR1, two SNPs (rs2274567 and rs3811381) had p-values of 0.034. In ABCA7, two SNPs (rs3764645 and rs3752234) were nominally significant with p-values equal to 0.018 and 0.023, respectively. These results are shown in Table S2 in supplementary material. Using the burden test, three genes (i.e.; BIN1, EPHA1, and ABCA7) out of the nine considered in (Naj et al., 2014) had at least two SNPs with MAF less than 0.05, which means that they were tested. However, none of them was significant (Table S3 in supplementary material).
Bioinformatic enrichment analysis
We ran a gene-set enrichment analysis (GSEA) using a user-friendly web-based tool, WebGestalt (http://bioinfo.vanderbilt.edu/webgestalt/, (Wang et al., 2013)). We performed GSEA enrichment analysis using both GO and KEGG databases on the list of: 1) the 16 most significant genes obtained by the single marker test, 2) the ten most significant genes obtained by the burden test, and 3) the combined list of 26 genes from both tests. We focused on pathways with p-values less than 0.05 after a Bonferroni correction for multiple testing (Table 3). Only the burden test achieved this threshold, and using the list of “burden test genes”, six GO pathways containing five genes (SLC8A3, SLC24A4, SLC19A3, GRIN3B, and LRRK2) from the burden list showed significant results after Bonferroni correction (adjusted p-value range=[0.0096 – 0.0288]) (Table 3). It is notable that identified genes belong to multiple associated pathways.
Table 3.
Gene-set enrichement analysis using WebGestalt for the most significant genes obtained by the burden test
| Pathway ID | Pathway name | Database | C | O | E | R | rawP | adjP |
|---|---|---|---|---|---|---|---|---|
| GO:0015291 | Secondary active transmembrane transporter activity | GO molecular function | 192 | 3 (SLC24A4, SLC8A3, SLC19A3 ) | 0.11 | 26.53 | 2.0E-04 | 9.6E-03 |
| GO:0043025 | Neuronal cell body | GO cellular component | 291 | 3 (SLC8A3, GRIN3B, LRRK2 ) | 0.15 | 19.53 | 4.0E-04 | 1.5E-02 |
| GO:0044297 | Cell body | GO cellular component | 312 | 3 (SLC8A3, GRIN3B, LRRK2 ) | 0.16 | 18.22 | 5.0E-04 | 1.9E-02 |
| GO:0015297 | Antiporter activity | GO molecular function | 60 | 2 (SLC24A4, SLC8A3 ) | 0.04 | 56.61 | 5.0E-04 | 2.4E-02 |
| GO:0006816 | Calcium ion transport | GO biological process | 216 | 3 (SLC24A4, SLC8A3, GRIN3B ) | 0.13 | 22.58 | 2.0E-04 | 2.7E-02 |
| GO:0022804 | Active transmembrane transporter activity | GO molecular function | 309 | 3 (SLC24A4, SLC8A3, SLC19A3 ) | 0.18 | 16.49 | 6.0E-04 | 2.9E-02 |
C= The number of reference genes in the pathway; O= The number of genes in the gene set and also in the pathway; E= The expected number in the pathway; R= Ratio of enrichment; rawP= P-value from hypergeometric test; adjP= Bonferroni-corrected P-value.
DISCUSSION
In this paper, we presented the result of a (WES) family-based association study for AAO of LOAD subjects. Risk of AD and its AAO are related traits, as shown by overlap on both traits of effects of known genetic factors, such as APOE (Corder et al., 1993, Farrer et al., 1997). Nonetheless, the interpretation and implication of results focused on AD risk vs. AAO is different. A focus on AAO among AD cases, as in our study, may bring new insights into factors affecting onset of AD in those that are at risk. This allows a more nuanced and useful measure than simple risk of AD, and is highly pertinent to downstream investigation. Even a modest increase in AAO of AD through prevention or therapeutic measures would have enormous benefits by simply delaying the onset of disease.
In our study design, WES was carried on two to three affected subjects per family that has multiple affected subjects. This family-based design might be more efficient than a population-based design, especially for rare variants that are enriched in pedigrees (Wijsman, 2012). In addition, the rare variants that might be implicated in AD or the AAO of AD are likely to be shared by these affected subjects. Despite the modest size of our WES dataset, our results suggest that a good design that uses a carefully selected set of subjects can provide promising results. This is demonstrated by our replication of several signals from (Naj et al., 2014), a study based on more than 9,000 unrelated subjects, by our identification of several additional candidate genes for AD, and also finding that some of the identified genes are located in regions with evidence of linkage for AAO in this study, as well as in other studies.
There are also several aspects of both the design and the analysis that should contribute to robust results. First, as is common in similar studies, we took a number of steps to make sure that our results are statistically-robust and not explained by artifacts of confounders (e.g.; poor SNP quality and population stratification). Second, by focusing on AAO in affected subjects, only, we avoid the problem posed by the censored age data in unaffected subjects, for which there are not yet analytical methods that give statistically-robust results for variance-components analysis in pedigree samples. Third, our choices of analysis details were chosen to make the results robust to small sample size. By using direct sequence data and methods that allow for the possibility of multiple variants within relevant genes while also capitalizing on the increased information that can be obtained from a continuous trait, we eliminate many of the reasons that very large sample sizes became necessary during the era of GWAS case-control studies.
The use of WES data permits the evaluation of rare genetic variations in the functional parts of the genome (exons), which are not genotyped or well-tagged in classical GWAS SNP chips (Li et al., 2008). In addition, WES may provide a direct observation of common variants that are not well-tagged in GWAS SNP chips. Using a single marker test analysis for common variants and a burden test analysis for multiple rare variants have permitted us to detect novel candidate genes that may play a functional role in modifying AAO of AD. Based on literature review, among genes we have identified, three (WRN, OMIM 604611; NTN4, OMIM 610401; and LAMC3, OMIM 604349) with common associated SNPs and four with multiple rare variants associated with AAO (SLC8A3, OMIM 607991; SLC19A3, OMIM 606152; MADD, OMIM 603584; and LRRK2, OMIM 609007) have strong prior evidence for involvement in AD (http://www.genecards.org/).
Two genes, NTNA4 and LAMC3, belong to a family of proteins related to laminins. The gene Netrin 4 (NTN4) (rs17288108, p-value=3.48×10−5, MAF=0.125, p.(Y205H)) was originally found to have a role in neuronal axon migration and may play an important role in development (Cirulli & Yebra, 2007). Shen et al (Shen et al., 2012) recently showed that NTN4 expression is up-regulated during β-amyloid induced injury of neurite outgrowth, an effect that is reversed with addition of acetylcholinestarase inhibitors. This is consistent with the possibility that NTN4 might play a role in the development of AD pathology (Shen et al., 2012). The gene LAMC3 (rs4740412, p-value=1.44×10−4, MAF= 0.214, p.(R1459Q)) is a member of the Laminin family of heterotrimeric molecules that function in stabilization of epithelial structures. LAMC3 is strongly expressed in developing human fetal brain with highest expression in temporo-occipital regions. Recessive mutations in LAMC3 cause a syndrome with cortical malformations and seizures (Barak et al., 2011) further underlining its importance in brain development. Laminin interacts with β-amyloid supporting its role in Alzheimer’s disease (Morgan & Inestrosa, 2001). In addition, LAMC3 is a part of network that includes PICALM, a gene previously reported as associated with AD (Carrasquillo et al., 2010, Harold et al., 2009, Lambert et al., 2013).
Among genes identified by the burden test, there was LRRK2 (leucine-rich repeat serine/threonine-protein kinase 2, p-value=2.84×10−4), which is the most common cause of dominant inherited Parkinson’s disease (PARK8) (Lesage & Brice, 2009). Common variants in this gene have been also found to increase the susceptibility to Parkinson’s disease (MIM168600) (Gilks et al., 2005, Nalls et al., 2011, Simon-Sanchez et al., 2009). Several LRRK2 protein functions might lead to its effects in AD. LRRK2 regulates autophagy through a calcium-dependent activation of the CaMKK/AMPK signaling pathway (Gomez-Suaga et al., 2012) and mediates the synaptotoxic effects of Amyloid beta oligomers through tau phosphorylation (Mairet-Coello et al., 2013). LRRK2 might also contribute to Lewy Body pathology in Alzheimer’s disease (Linnertz et al., 2014). The remaining four genes that have functions related to AD can be found in the supplementary material.
Finally, the VC linkage analysis we performed gives strength to some genes identified in our single marker and burden association analyses. An interesting gene is RGL3, which both gave the strongest single-variant results in the current analysis, for SNP rs2291516, and is in a region with evidence of linkage on chromosome 19p located upstream of APOE. Even though the evidence for linkage in the current sample is moderate, it is interesting because at the same position, a strong signal was identified when adjusting for APOE in an earlier evaluation of regions containing AAO loci (Wijsman et al., 2004) with later confirmatory evidence provided by two other independent samples (Choi et al., 2011, Zhao et al., 2013a). The pedigrees used in these studies do not overlap with pedigrees used here. The region with evidence for linkage to AAO also includes SNP rs1043963 in USHBP1, which also gave positive results in the single marker association test performed here. Another gene, SLC19A3, which was identified by the burden test, is also located in a region with evidence of linkage on chromosome 2. This gene has a biological function that might be related to AD (supplementary material).
Current advances in sequencing allow for whole exome and genome sequencing in tens of thousands of samples. However, in the context of sequencing data, replication is not simple. Combining or replicating studies that use next-generation sequence data has new challenges. Read depth and analytical procedures for alignment and variant calling affect results, requiring both re-calling of the sequence data, and use of methods that include modeling read depth to avoid spurious results (Derkach et al., 2014). Such procedures do not yet include related subjects. In addition, even such large samples might not have sufficient power to account for multiple testing and large samples introduce additional complications of aggregating large datasets, including variation in phenotypic measurements, such as AAO across studies.
In conclusion, with our statistical approach that uses a family-based association study design, we have identified several candidate genes that have additional functional evidence for association with AD. Our family-based design and focus on coding regions of the genome attenuates the issues of multiple testing that complicate classic case-control designs. The identification of candidate genes is important from the perspective of a follow-up in larger case-control samples or in other family-based samples. Our approach that identifies a focused list of candidate genes, specific types of variants that are responsible for association (single low frequency variant vs. burden of rare variants) and the specific AAO phenotype allows for increase in power and replication study in smaller datasets.
Supplementary Material
Fig. S1 Principal Component Analysis results. Our WES subjects and subjects from the three populations of the 1000 genomes project are plotted on the principal component 1 (PC1) vs. principal component 2 (PC2). Our WES subjects are represented by red crosses, and the EUR, AFR, and ASN subjects are represented by blue, cyan, and pink circles, respectively.
Fig. S2 Manhattan plot of the burden test analysis (Weighted Sum Approach) that does not adjust for APOE. The plot is based on all 1,949 tested genes. The horizontal blue line represents the least significant gene among the 10 most significant genes. The horizontal red line is the genome-wide significance threshold.
Fig. S3 QQ-plot of the burden test analysis (Weighted Sum Approach) that adjusts (red) or does not adjust (black) for APOE. The plot is based on all 1,949 tested genes.
Acknowledgments
This work was partially supported by grant funding from NIH R01 AG039700 and NIH P50 AG005136. Subjects and samples used here were originally collected with grant funding from NIH U24 AG026395, U24 AG021886, P50 AG008702, P01 AG007232, R37 AG015473, P30 AG028377, P50 AG005128, P50 AG016574, P30 AG010133, P50 AG005681, P01 AG003991, U01 MH046281, U01 MH046290, and U01 MH046373. The funders had no role in study design, analysis, or preparation of the manuscript.
Footnotes
COMPETING IN INTERESTS
The authors declare no competing interests.
AUTHORS’ CONTRIBUTIONS
All authors participated in study design and analysis and drafted the manuscript.
Contributor Information
Mohamad Saad, Email: mhsaad@uw.edu.
Zoran Brkanac, Email: zbrkanac@uw.edu.
Ellen M. Wijsman, Email: wijsman@uw.edu.
References
- Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. American Journal of Human Genetics. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee S. The macroeconomics of dementias - Will the world economy get Alzheimer’s disease? Archives of Medical Research. 2014;43:705–709. doi: 10.1016/j.arcmed.2012.10.006. [DOI] [PubMed] [Google Scholar]
- Barak T, Kwan KY, Louvi A, Demirbilek V, Saygi S, Tuysuz B, Choi M, Boyaci H, Doerschner K, Zhu Y, Kaymakcalan H, Yilmaz S, Bakircioglu M, Caglayan AO, Ozturk AK, Yasuno K, Brunken WJ, Atalar E, Yalcinkaya C, Dincer A, Bronen RA, Mane S, Ozcelik T, Lifton RP, Sestan N, Bilguvar K, Gunel M. Recessive LAMC3 mutations cause malformations of occipital cortical development. Nat Genet. 2011;43:590–594. doi: 10.1038/ng.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez BA, Cooper B, Pastor P, Jin SC, Lorenzo E, Cervantes S, Cruchaga C. TREM2 is associated with the risk of Alzheimer’s disease in Spanish population. Neurobiol Aging. 2013;34:1711 e1715–1717. doi: 10.1016/j.neurobiolaging.2012.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, Lange C, Mullin K, Parkinson M, Hsiao M, Hogan MF, Schjeide BMM, Hooli B, DiVito J, Ionita L, Jiang HY, Laird N, Moscarillo T, Ohlsen KL, Elliott K, Wang X, Hu-Lince D, Ryder M, Murphy A, Wagner SL, Blacker D, Becker KD, Tanzi RE. Genome-wide Association Analysis Reveals Putative Alzheimer’s Disease Susceptibility Loci in Addition to APOE. American Journal of Human Genetics. 2008;83:623–632. doi: 10.1016/j.ajhg.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird TD, Levy-Lahad E, Poorkaj P, Sharma V, Nemens E, Lampe T, Schellenberg GD. Wide range in age of onset for chromosome 1-related familial Alzheimer’s disease. Annals of Neurology. 1996;40:932–936. doi: 10.1002/ana.410400619. [DOI] [PubMed] [Google Scholar]
- Blacker D, Haines JL, Rodes L, Terwedow H, Go RC, Harrell LE, Perry RT, Bassett SS, Chase G, Meyers D, Albert MS, Tanzi R. ApoE-4 and age at onset of Alzheimer’s disease: the NIMH genetics initiative. Neurology. 1997;48:139–147. doi: 10.1212/wnl.48.1.139. [DOI] [PubMed] [Google Scholar]
- Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F, Crook JE, Pankratz VS, Dickson DW, Graff-Radford NR, Petersen RC, Morgan K, Younkin SG. Replication of CLU, CR1, and PICALM Associations With Alzheimer Disease. Archives of Neurology. 2010;67:961–964. doi: 10.1001/archneurol.2010.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Marchani EE, Bird TD, Steinbart EJ, Blacker D, Wijsman EM. Genome Scan of Age-at-Onset in the NIMH Alzheimer Disease Sample Uncovers Multiple Loci, Along With Evidence of Both Genetic and Sample Heterogeneity. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2011;156B:785–798. doi: 10.1002/ajmg.b.31220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirulli V, Yebra M. Netrins: beyond the brain. Nat Rev Mol Cell Biol. 2007;8:296–306. doi: 10.1038/nrm2142. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schmechel DE, Gaskell PC, Rimmler JB, Locke PA, Conneally PM, Schmader KE, Small GW, Roses AD, Haines JL, Pericak Vance MA. Protective Effect of Apolipoprotein-E Type-2 Allele for Late-Onset Alzheimer-Disease. Nature Genetics. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Cruchaga C, Chakraverty S, Mayo K, Vallania FL, Mitra RD, Faber K, Williamson J, Bird T, Diaz-Arrastia R, Foroud TM, Boeve BF, Graff-Radford NR, St Jean P, Lawson M, Ehm MG, Mayeux R, Goate AM. Rare Variants in APP, PSEN1 and PSEN2 Increase Risk for AD in Late-Onset Alzheimer’s Disease Families. Plos One. 2012;7:e31039. doi: 10.1371/journal.pone.0031039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruchaga C, Karch CM, Jin SC, Benitez BA, Cai Y, Guerreiro R, Harari O, Norton J, Budde J, Bertelsen S, Jeng AT, Cooper B, Skorupa T, Carrell D, Levitch D, Hsu S, Choi J, Ryten M, Hardy J, Trabzuni D, Weale ME, Ramasamy A, Smith C, Sassi C, Bras J, Gibbs JR, Hernandez DG, Lupton MK, Powell J, Forabosco P, Ridge PG, Corcoran CD, Tschanz JT, Norton MC, Munger RG, Schmutz C, Leary M, Demirci FY, Bamne MN, Wang X, Lopez OL, Ganguli M, Medway C, Turton J, Lord J, Braae A, Barber I, Brown K, Passmore P, Craig D, Johnston J, McGuinness B, Todd S, Heun R, Kolsch H, Kehoe PG, Hooper NM, Vardy ER, Mann DM, Pickering-Brown S, Kalsheker N, Lowe J, Morgan K, David Smith A, Wilcock G, Warden D, Holmes C, Pastor P, Lorenzo-Betancor O, Brkanac Z, Scott E, Topol E, Rogaeva E, Singleton AB, Kamboh MI, St George-Hyslop P, Cairns N, Morris JC, Kauwe JS, Goate AM. Rare coding variants in the phospholipase D3 gene confer risk for Alzheimer’s disease. Nature. 2014;505:550–554. doi: 10.1038/nature12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daw E, Bird T, Nemens E, Nochlin D, Schellenberg G, Wijsman E. ApoE and other loci affect age of Alzheimer’s disease onset in families with PS2 mutation. American Journal of Human Genetics. 2000;67(Suppl):192. doi: 10.1002/ajmg.b.30087. [DOI] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods. 2012;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- Derkach A, Chiang T, Gong J, Addis L, Dobbins S, Tomlinson I, Houlston R, Pal DK, Strug LJ. Association analysis using next generation sequence data from publicly available control groups: The robust variance score statistics. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btu196. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- Dickson MR, Li J, Wiener HW, Perry RT, Blacker D, Bassett SS, Go RCP. A genomic scan for age at onset of Alzheimer’s disease in 437 families from the NIMH genetic initiative. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2008;147B:784–792. doi: 10.1002/ajmg.b.30689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, PericakVance MA, Risch N, van Duijn CM. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease - A meta-analysis. JAMA-Journal of the American Medical Association. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Finckh U, Kuschel C, Anagnostouli M, Patsouris E, Pantes GV, Gatzonis S, Kapaki E, Davaki P, Lamszus K, Stavrou D, Gal A. Novel mutations and repeated findings of mutations in familial Alzheimer disease. Neurogenetics. 2005;6:85–89. doi: 10.1007/s10048-005-0211-x. [DOI] [PubMed] [Google Scholar]
- Fratiglioni L, Launer LJ, Andersen K, Breteler MMB, Copeland JRM, Dartigues JF, Lobo A, Martinez-Lage J, Soininen H, Hofman A Neurologic Dis Elderly Res G. Incidence of dementia and major subtypes in Europe: A collaborative study of population-based cohorts. Neurology. 2000;54:S10–S15. [PubMed] [Google Scholar]
- Gatz M, Pedersen NL. Study of Dementia in Swedish Twins. Twin Research and Human Genetics. 2013;16:313–316. doi: 10.1017/thg.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatz M, Pedersen NL, Berg S, Johansson B, Johansson K, Mortimer JA, Posner SF, Viitanen M, Winblad B, Ahlbom A. Heritability for Alzheimer’s disease: The study of dementia in Swedish twins. Journals of Gerontology Series a-Biological Sciences and Medical Sciences. 1997;52:M117–M125. doi: 10.1093/gerona/52a.2.m117. [DOI] [PubMed] [Google Scholar]
- Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, Shaw K, Bhatia KP, Bonifati V, Quinn NP, Lynch J, Healy DG, Holton JL, Revesz T, Wood NW. A common LRRK2 mutation in idiopathic Parkinson’s disease. Lancet. 2005;365:415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- Goate A, Chartier-Harlin MC, Mullan M, Brown J, Crawford F, Fidani L, Giuffra L, Haynes A, Irving N, James L, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- Gomez-Suaga P, Luzon-Toro B, Churamani D, Zhang L, Bloor-Young D, Patel S, Woodman PG, Churchill GC, Hilfiker S. Leucine-rich repeat kinase 2 regulates autophagy through a calcium-dependent pathway involving NAADP. Hum Mol Genet. 2012;21:511–525. doi: 10.1093/hmg/ddr481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JS, Younkin S, Hazrati L, Collinge J, Pocock J, Lashley T, Williams J, Lambert JC, Amouyel P, Goate A, Rademakers R, Morgan K, Powell J, St George-Hyslop P, Singleton A, Hardy J. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nature Genetics. 2009;62:1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert LE, Scherr PA, Bienias JL, Bennett DA, Evans DA. Alzheimer disease in the US population - Prevalence estimates using the 2000 census. Archives of Neurology. 2003;60:1119–1122. doi: 10.1001/archneur.60.8.1119. [DOI] [PubMed] [Google Scholar]
- Holmans P, Hamshere M, Hollingworth P, Rice F, Tunstall N, Jones S, Moore P, DeVrieze FW, Myers A, Crook R, Compton D, Marshall H, Meyer D, Shears S, Booth J, Ramic D, Williams N, Norton N, Abraham R, Kehoe P, Williams H, Rudrasingham V, O’Donovan M, Jones L, Hardy J, Goate A, Lovestone S, Owen M, Williams J. Genome screen for loci influencing age at onset and rate of decline in late onset Alzheimer’s disease. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2005;135B:24–32. doi: 10.1002/ajmg.b.30114. [DOI] [PubMed] [Google Scholar]
- Holton P, Ryten M, Nalls M, Trabzuni D, Weale ME, Hernandez D, Crehan H, Gibbs JR, Mayeux R, Haines JL, Farrer LA, Pericak-Vance MA, Schellenberg GD, Ramirez-Restrepo M, Engel A, Myers AJ, Corneveaux JJ, Huentelman MJ, Dillman A, Cookson MR, Reiman EM, Singleton A, Hardy J, Guerreiro R Alzheimer’s Dis Genetics C. Initial Assessment of the Pathogenic Mechanisms of the Recently Identified Alzheimer Risk Loci. Annals of Human Genetics. 2013;77:85–105. doi: 10.1111/ahg.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi T, Kaneko H, Miyashita A, Nozaki H, Kasuga K, Tsukie T, Tsuchiya M, Imamura T, Ishizu H, Aoki K, Ishikawa A, Onodera O, Kuwano R, Nishizawa M. Mutational analysis in early-onset familial dementia in the Japanese population. Dementia and Geriatric Cognitive Disorders. 2008;26:43–49. doi: 10.1159/000141483. [DOI] [PubMed] [Google Scholar]
- Jonsson T, Stefansson H, Steinberg S, Jonsdottir I, Jonsson PV, Snaedal J, Bjornsson S, Huttenlocher J, Levey AI, Lah JJ, Rujescu D, Hampel H, Giegling I, Andreassen OA, Engedal K, Ulstein I, Djurovic S, Ibrahim-Verbaas C, Hofman A, Ikram MA, van Duijn CM, Thorsteinsdottir U, Kong A, Stefansson K. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamboh MI, Barmada MM, Demirci FY, Minster RL, Carrasquillo MM, Pankratz VS, Younkin SG, Saykin AJ, Sweet RA, Feingold E, Dekosky ST, Lopez OL The Alzheimer’s Disease Neuroimaging Initiative. Genome-wide association analysis of age-at-onset in Alzheimer’s disease. Molecular Psychiatry. 2012;17:1340–1346. doi: 10.1038/mp.2011.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nature Genetics. 2009;41:1094–1099. doi: 10.1038/ng.439. [DOI] [PubMed] [Google Scholar]
- Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, Jun G, DeStefano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thornton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Hollingworth P, Ramirez A, Hanon O, Fitzpatrick AL, Buxbaum JD, Campion D, Crane PK, Baldwin C, Becker T, Gudnason V, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MJ, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuinness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Naranjo MCD, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nature Genetics. 2013;45:1452–U1206. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Barral S, Cheng R, Chacon I, Santana V, Williamson J, Lantigua R, Medrano M, Jimenez-Velazquez IZ, Stern Y, Tycko B, Rogaeva E, Wakutani Y, Kawarai T, St George-Hyslop P, Mayeux R. Age-at-onset linkage analysis in Caribbean Hispanics with familial late-onset Alzheimer’s disease. Neurogenetics. 2008;9:51–60. doi: 10.1007/s10048-007-0103-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage S, Brice A. Parkinson’s disease: from monogenic forms to genetic susceptibility factors. Hum Mol Genet. 2009;18:R48–59. doi: 10.1093/hmg/ddp012. [DOI] [PubMed] [Google Scholar]
- Levy-Lahad E, Wasco W, Poorkaj P, Romano DM, Oshima J, Pettingell WH, Yu CE, Jondro PD, Schmidt SD, Wang K, Crowley AC, Fu YH, Guenette SY, Galas D, Nemens E, Wijsman EM, Bird TD, Schellenberg GD, Tanzi RE. Candidate gene for the chromosome 1 familial Alzheimer’s disease locus. Science. 1995;269:973–977. doi: 10.1126/science.7638622. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Li C, Guan W. Evaluation of coverage variation of SNP chips for genome-wide association studies. Eur J Hum Genet. 2008;16:635–643. doi: 10.1038/sj.ejhg.5202007. [DOI] [PubMed] [Google Scholar]
- Lin DY, Zeng D. Proper analysis of secondary phenotype data in case-control association studies. Genet Epidemiol. 2009;33:256–265. doi: 10.1002/gepi.20377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linnertz C, Lutz MW, Ervin JF, Allen J, Miller NR, Welsh-Bohmer KA, Roses AD, Chiba-Falek O. The genetic contributions of SNCA and LRRK2 genes to Lewy Body pathology in Alzheimer’s disease. Hum Mol Genet. 2014;23:4814–4821. doi: 10.1093/hmg/ddu196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lleo A, Blesa R, Queralt R, Ezquerra M, Molinuevo JL, Pena-Casanova J, Rojo A, Oliva R. Frequency of mutations in the presenilin and amyloid precursor protein genes in early-onset Alzheimer disease in Spain. Archives of Neurology. 2002;59:1759–1763. doi: 10.1001/archneur.59.11.1759. [DOI] [PubMed] [Google Scholar]
- Lobo A, Launer LJ, Fratiglioni L, Andersen K, Di Carlo A, Breteler MMB, Copeland JRM, Dartigues JF, Jagger C, Martinez-Lage J, Soininen H, Hofman A Neurologic Dis Elderly Res G. Prevalence of dementia and major subtypes in Europe: A collaborative study of population-based cohorts. Neurology. 2000;54:S4–S9. [PubMed] [Google Scholar]
- Lord J, Lu AJ, Cruchaga C. Identification of rare variants in Alzheimer’s disease. Frontiers in Genetics. 2014;5:369. doi: 10.3389/fgene.2014.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mairet-Coello G, Courchet J, Pieraut S, Courchet V, Maximov A, Polleux F. The CAMKK2-AMPK kinase pathway mediates the synaptotoxic effects of Abeta oligomers through Tau phosphorylation. Neuron. 2013;78:94–108. doi: 10.1016/j.neuron.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeux R. Epidemiology of neurodegeneration. Annual Review of Neuroscience. 2003;26:81–104. doi: 10.1146/annurev.neuro.26.043002.094919. [DOI] [PubMed] [Google Scholar]
- Mayeux R, Sano M, Chen J, Tatemichi T, Stern Y. Risk of Dementia in 1st-Degree Relatives of Patients with Alzheimers-Disease and Related Disorders. Archives of Neurology. 1991;48:269–273. doi: 10.1001/archneur.1991.00530150037014. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer’s disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer’s Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- Morgan C, Inestrosa NC. Interactions of laminin with the amyloid beta peptide. Implications for Alzheimer’s disease. Braz J Med Biol Res. 2001;34:597–601. doi: 10.1590/s0100-879x2001000500006. [DOI] [PubMed] [Google Scholar]
- Murphy SL, Xu J, Kochanek KD. Prevention, C.f.D.C.a, editor. Deaths: Final data for 2010. 2013. pp. 1–118. [PubMed] [Google Scholar]
- Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JSK, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nature Genetics. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naj AC, Jun G, Reitz C, Kunkle BW, Perry W, Park YS, Beecham GW, Rajbhandary RA, Hamilton-Nelson KL, Wang LS, Kauwe JS, Huentelman MJ, Myers AJ, Bird TD, Boeve BF, Baldwin CT, Jarvik GP, Crane PK, Rogaeva E, Barmada MM, Demirci FY, Cruchaga C, Kramer PL, Ertekin-Taner N, Hardy J, Graff-Radford NR, Green RC, Larson EB, St George-Hyslop PH, Buxbaum JD, Evans DA, Schneider JA, Lunetta KL, Kamboh MI, Saykin AJ, Reiman EM, De Jager PL, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Martin ER, Haines JL, Mayeux RP, Farrer LA, Schellenberg GD, Pericak-Vance MA, Albert MS, Albin RL, Apostolova LG, Arnold SE, Barber R, Barnes LL, Beach TG, Becker JT, Beekly D, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Cantwell LB, Cao C, Carlson CS, Carney RM, Carrasquillo MM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cribbs DH, Crocco EA, DeCarli C, DeKosky ST, Dick M, Dickson DW, Duara R, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, et al. Effects of multiple genetic Loci on age at onset in late-onset Alzheimer disease: a genome-wide association study. JAMA Neurol. 2014;71:1394–1404. doi: 10.1001/jamaneurol.2014.1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls MA, Plagnol V, Hernandez DG, Sharma M, Sheerin UM, Saad M, Simon-Sanchez J, Schulte C, Lesage S, Sveinbjornsdottir S, Stefansson K, Martinez M, Hardy J, Heutink P, Brice A, Gasser T, Singleton AB, Wood NW. Imputation of sequence variants for identification of genetic risks for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet. 2011;377:641–649. doi: 10.1016/S0140-6736(10)62345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor P, Roe C, Villegas A, Bedoya G, Chakraverty S, Garcia G, Tirado V, Norton J, Rios S, Martinez M, Kosik K, Lopera F, Goate A. Apolipoprotein E ε4 modifies Alzheimer’s disease onset in an E280A PS1 Kindred. Annals of Neurology. 2003;54:163–169. doi: 10.1002/ana.10636. [DOI] [PubMed] [Google Scholar]
- Pedersen NL, Gatz M, Berg S, Johansson B. How heritable is Alzheimer’s disease late in life? Findings from Swedish twins. Annals of Neurology. 2004;55:180–185. doi: 10.1002/ana.10999. [DOI] [PubMed] [Google Scholar]
- Prentice RL, Pyke R. Logistic disease incidence models and case-control studies. Biometrika. 1979;66:403–411. [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radmanesh F, Devan WJ, Anderson CD, Rosand J, Falcone GJ. Accuracy of imputation to infer unobserved APOE epsilon alleles in genome-wide genotyping data. Eur J Hum Genet. 2014;22:1239–1242. doi: 10.1038/ejhg.2013.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocca WA, Hofman A, Brayne C, Breteler MMB, Clarke M, Copeland JRM, Dartigues JF, Engedal K, Hagnell O, Heeren TJ, Jonker C, Lindesay J, Lobo A, Mann AH, Molsa PK, Morgan K, Oconnor DW, Droux AD, Sulkava R, Kay DWK, Amaducci L. Frequency and Distribution of Alzheimers-Disease in Europe - a Collaborative Study of 1980–1990 Prevalence Findings. Annals of Neurology. 1991;30:381–390. doi: 10.1002/ana.410300310. [DOI] [PubMed] [Google Scholar]
- Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, Bis JC, Smith AV, Carassquillo MM, Lambert JC, Harold D, Schrijvers EMC, Ramirez-Lorca R, Debette S, Longstreth WT, Janssens A, Pankratz VS, Dartigues JF, Hollingworth P, Aspelund T, Hernandez I, Beiser A, Kuller LH, Koudstaal PJ, Dickson DW, Tzourio C, Abraham R, Antunez C, Du YC, Rotter JI, Aulchenko YS, Harris TB, Petersen RC, Berr C, Owen MJ, Lopez-Arrieta J, Varadarajan BN, Becker JT, Rivadeneira F, Nalls MA, Graff-Radford NR, Campion D, Auerbach S, Rice K, Hofman A, Jonsson PV, Schmidt H, Lathrop M, Mosley TH, Au R, Psaty BM, Uitterlinden AG, Farrer LA, Lumley T, Ruiz A, Williams J, Amouyel P, Younkin SG, Wolf PA, Launer LJ, Lopez OL, van Duijn CM, Breteler MMB. Genome-wide Analysis of Genetic Loci Associated With Alzheimer Disease. Journal of the American Medical Association. 2010;303:1832–1840. doi: 10.1001/jama.2010.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JN, Wang DS, Wang R. The protection of acetylcholinesterase inhibitor on beta-amyloid-induced the injury of neurite outgrowth via regulating axon guidance related genes expression in neuronal cells. Int J Clin Exp Pathol. 2012;5:900–913. [PMC free article] [PubMed] [Google Scholar]
- Sherrington R, Rogaev EI, Liang Y, Rogaeva EA, Levesque G, Ikeda M, Chi H, Lin C, Li G, Holman K, et al. Cloning of a gene bearing missense mutations in early-onset familial Alzheimer’s disease. Nature. 1995;375:754–760. doi: 10.1038/375754a0. [DOI] [PubMed] [Google Scholar]
- Simon-Sanchez J, Schulte C, Bras JM, Sharma M, Gibbs JR, Berg D, Paisan-Ruiz C, Lichtner P, Scholz SW, Hernandez DG, Kruger R, Federoff M, Klein C, Goate A, Perlmutter J, Bonin M, Nalls MA, Illig T, Gieger C, Houlden H, Steffens M, Okun MS, Racette BA, Cookson MR, Foote KD, Fernandez HH, Traynor BJ, Schreiber S, Arepalli S, Zonozi R, Gwinn K, van der Brug M, Lopez G, Chanock SJ, Schatzkin A, Park Y, Hollenbeck A, Gao J, Huang X, Wood NW, Lorenz D, Deuschl G, Chen H, Riess O, Hardy JA, Singleton AB, Gasser T. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat Genet. 2009;41:1308–1312. doi: 10.1038/ng.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjögren T, Sjögren H, Lindgren AG. Morbus Alzheimer and morbus Pick: a genetic, clinical and patho-anatomical study. Munksgaard, Copenhagen: 1952. [PubMed] [Google Scholar]
- Sosa-Ortiz AL, Acosta-Castillo I, Prince MJ. Epidemiology of Dementias and Alzheimer’s Disease. Archives of Medical Research. 2012;43:600–608. doi: 10.1016/j.arcmed.2012.11.003. [DOI] [PubMed] [Google Scholar]
- Tandon A, Fraser P. The presenilins. Genome Biology. 2002:3. doi: 10.1186/gb-2002-3-11-reviews3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies W, Bleiler L, Alzheimer’s A. 2013 Alzheimer’s disease facts and figures Alzheimer’s Association. Alzheimers & Dementia. 2013;9:208–245. doi: 10.1016/j.jalz.2013.02.003. [DOI] [PubMed] [Google Scholar]
- Thompson E. The structure of genetic linkage data: from LIPED to 1M SNPs. Hum Hered. 2011;71:86–96. doi: 10.1159/000313555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Duijn CM, Clayton D, Chandra V, Fratiglioni L, Graves AB, Heyman A, Jorm AF, Kokmen E, Kondo K, Mortimer JA, Rocca WA, Shalat SL, Soininen H, Hofman A. Familial Aggregation of Alzheimers-Disease and Related Disorders - a Collaborative Reanalysis of Case-Control Studies. International Journal of Epidemiology. 1991;20:S13–S20. doi: 10.1093/ije/20.supplement_2.s13. [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM. The role of large pedigrees in an era of high-throughput sequencing. Human Genetics. 2012;131:1555–1563. doi: 10.1007/s00439-012-1190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM, Daw EW, Yu CE, Payami H, Steinbart EJ, Nochlin D, Conlon EM, Bird TD, Schellenberg GD. Evidence for a novel late-onset Alzheimer’s disease locus on chromosome 19p13.2. American Journal of Human Genetics. 2004;75:398–409. doi: 10.1086/423393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman EM, Daw EW, Yu X, Steinbart EJ, Nochlin D, Bird TD, Schellenberg G. APOE and other loci affect age-at-onset in Alzheimer’s disease families with PS2 mutation. American Journal of Medical Genetics Part B-Neuropsychiatric Genetics. 2005;132B:14–20. doi: 10.1002/ajmg.b.30087. [DOI] [PubMed] [Google Scholar]
- Wijsman EM, Pankratz ND, Choi Y, Rothstein JH, Faber KM, Cheng R, Lee JH, Bird TD, Bennett DA, Diaz-Arrastia R, Goate AM, Farlow MR, Ghetti B, Sweet RA, Foroud TM, Mayeux R. Genome wide association of familial late onset Alzheimer’s disease replicates BIN1 and CLU, and nominates CUGBP2 in interaction with APOE. PLoS Genetics. 2011;7:e1001308. doi: 10.1371/journal.pgen.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu MC, Lee S, Cai TX, Li Y, Boehnke M, Lin XH. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Marchani EE, Cheung CY, Steinbart EJ, Schellenberg GD, Bird TD, Wijsman EM. Genome scan in familial late-onset Alzheimer’s disease: a locus on chromosome 6 contributes to age-at-onset. Am J Med Genet B Neuropsychiatr Genet. 2013a;162B:201–212. doi: 10.1002/ajmg.b.32133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W, Marchani EE, Cheung CYK, Steinbart EJ, Schellenberg GD, Bird TD, Wijsman EM. Genome scan in familial late-onset Alzheimer’s disease: a locus on chromosome 6 contributes to age at onset. American Journal of Medical Genetics - Neuropsychiatric Genetics. 2013b;162B:201–212. doi: 10.1002/ajmg.b.32133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Principal Component Analysis results. Our WES subjects and subjects from the three populations of the 1000 genomes project are plotted on the principal component 1 (PC1) vs. principal component 2 (PC2). Our WES subjects are represented by red crosses, and the EUR, AFR, and ASN subjects are represented by blue, cyan, and pink circles, respectively.
Fig. S2 Manhattan plot of the burden test analysis (Weighted Sum Approach) that does not adjust for APOE. The plot is based on all 1,949 tested genes. The horizontal blue line represents the least significant gene among the 10 most significant genes. The horizontal red line is the genome-wide significance threshold.
Fig. S3 QQ-plot of the burden test analysis (Weighted Sum Approach) that adjusts (red) or does not adjust (black) for APOE. The plot is based on all 1,949 tested genes.