Summary
During pancreas development, epithelial buds undergo branching morphogenesis to form an exocrine and endocrine gland. Proper morphogenesis is necessary for correct lineage allocation of pancreatic progenitors; however, the cellular events underlying pancreas morphogenesis are unknown. Here, we employed time-lapse microscopy and fluorescent labeling of cells to analyze cell behaviors associated with pancreas morphogenesis. We observed that outer bud cells adjacent to the basement membrane are pleomorphic and rearrange frequently; as well, they largely remain in the outer cell compartment even after mitosis. These cell behaviors and pancreas branching depend on cell contacts with the basement membrane, which induce actomyosin cytoskeleton remodeling via integrin-mediated activation of FAK/Src signaling. We show that integrin signaling reduces E-cadherin-mediated cell-cell adhesion in outer cells, and provide genetic evidence that this regulation is necessary for initiation of branching. Our study suggests that regulation of cell motility and adhesion by local niche cues initiates pancreas branching morphogenesis.
Keywords: pancreas, morphogenesis, branching, progenitor, integrin, time-lapse imaging, adhesion, E-cadherin, Src, FAK, extracellular matrix
Introduction
Branch formation is a morphogenetic process to construct organs comprised of elaborate epithelial networks. Branching allows organs to maximize their surface area, which is critical for absorptive and secretory functions. Recent advances in live imaging and the advent of fluorescent reporter strategies have begun to reveal the cellular behaviors used to create the unique branching patterns of the salivary glands, mammary gland, and kidney (Chi et al., 2009; Ewald et al., 2008; Larsen et al., 2006). However, the mechanisms underlying pancreas branching morphogenesis are still unknown.
The pancreas develops as ventral and dorsal evaginations of the endodermal epithelium into the surrounding mesenchyme (Shih et al., 2013). The earliest sign of pancreas branching becomes apparent around embryonic day (E) 11.5, when the initially smooth epithelial surface begins to form stubby outgrowths that subsequently elongate into branches (Villasenor et al., 2010). Careful analysis of pancreatic sections throughout development shows that pancreas branching is associated with the formation of a multi-lumen tubular plexus, which then extends and remodels into a single-lumen ductal system (Kesavan et al., 2009; Petzold et al., 2013; Villasenor et al., 2010). Failure to organize pancreatic epithelial progenitors into tubes causes a defect in their lineage allocation, suggesting a link between morphogenesis and cell specification (Kesavan et al., 2009).
Time-lapse imaging studies have provided insight into global patterns of pancreas branching (Puri and Hebrok, 2007). However, previous imaging studies in the pancreas were not designed to follow the behavior of individual cells. Hence, the cellular mechanisms by which the pancreatic epithelium transforms into a highly branched organ remain unclear.
The cellular behaviors that drive tissue morphogenesis require the actomyosin network to change cell shape and cell contacts (Guillot and Lecuit, 2013; Munjal and Lecuit, 2014). The forces generated by such networks govern cellular behaviors through coupling to the plasma membrane by E-cadherin complexes or the basement membrane by integrins (Legate et al., 2009). Crossregulation between E-cadherin-mediated cell-cell adhesions and integrin-mediated cell-extracellular matrix (ECM) contacts has been demonstrated at a cellular level, in particular in the context of tumor cells (Canel et al., 2013). Yet, the role of cell-cell and cell-ECM contacts in pancreatic organ morphogenesis is unknown.
Here, we used genetic strategies to mosaically label pancreatic epithelial cells with fluorescent proteins, allowing us to follow the behaviors of individual cells by time-lapse microscopy in pancreas explants.
Results
Live imaging of Developing Pancreas Explants Reveals Dynamic Cellular Behaviors of Progenitor Cells
Because pancreas branching is a dynamic process involving complex cellular movements, we sought to establish a live-imaging platform that would allow us to monitor individual cell behaviors during pancreas morphogenesis. To achieve this, we crossed Pdx1-CreER mice with Rosa26mTomato/mGFP/+ (R26mT/mG) mice to generate mice in which individual pancreatic progenitor cells are labeled by different fluorochromes. Upon tamoxifen administration, Cre recombinase is activated in progenitor cells, resulting in the excision of the membrane-bound mTomato (mT) gene and permitting the expression of membrane-bound green fluorescent protein (mG) (Figure 1A). By titrating the tamoxifen dosage administered to pregnant dams, we can achieve mosaic labeling of the pancreatic progenitor cell population with mT and mG (Figure 1B). In combination with time-lapse confocal microscopy, this approach allows us to follow the division and movement of pancreatic progenitor cells at single cell resolution in explant cultures for up to 3 days (Figure 1B). By analyzing sequential image frames, we used this platform to define fundamental cellular processes that underlie pancreas branching morphogenesis.
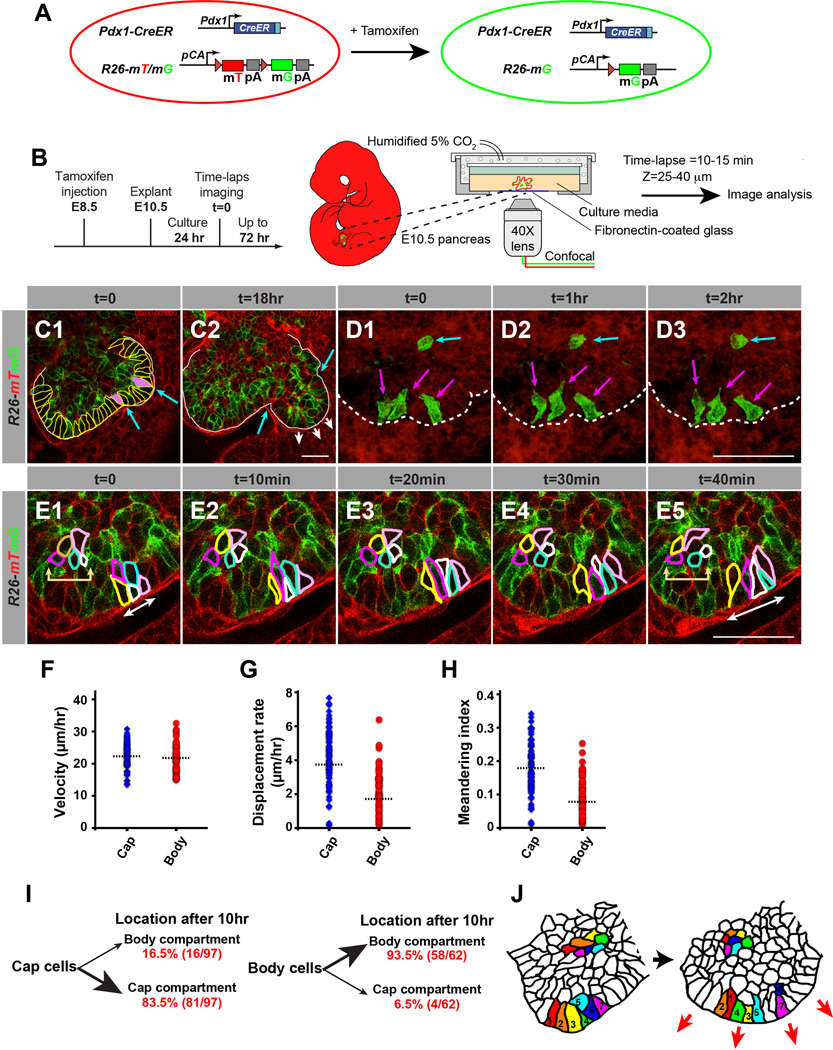
Figure 1. Time-Lapse Confocal Microscopic Analysis of Progenitor Cell Behaviors in Pancreas Explants.
(A) Schematic of the dual-color transgenic mouse model (Pdx1-CreER; Rosa(R)26mTomato/mGFP/+) for mosaic labeling of pancreatic progenitor cells with membrane-bound green fluorescent protein (mG) and membrane-bound tomato protein (mT). (B) Experimental design for the analysis of embryonic pancreas organ cultures by time-lapse fluorescence microscopy. (C-E) Optical sections (C,E) or 3D projection images (D) through the center of a pancreatic bud at several time points (t). t=0 demarcates the beginning of the imaging period, 24 hours after starting the organ culture. (C) Cell shape change of cap cells coincides with site of branch formation. (C1) At t=0 cap cells (outlined in yellow) are of columnar shape. Some cap cells (filled in purple) exhibit basal constriction and a wide apical surface. The location of these cells coincides with the appearance of ‘dips’ in the epithelial surface (indicated by blue arrows). (C2) After 18 hours, cells between these ‘dips’ (indicated by blue arrows) branch out and form a new lobe (indicated by white arrows). (D) Time-lapse 3D reconstructed images of clonally labeled progenitor cells undergoing shape changes. Cap cells (magenta arrows) display more dynamic shape changes than body cells (blue arrow). (E) Clusters of progenitor cells undergoing cell rearrangement. The labeled cap cell cluster (white two-headed arrow) displays more dynamic cell rearrangement than the body cell cluster (beige two-headed arrow). (F-H) Cell movement indices, including velocity (F), displacement rate (G), and meandering index (H). (I) Cell counting and time-lapse cell location analysis show that, after 10 hours, the majority of cap cells remain in the cap cell layer and the majority of body cells remain in the body cell compartment. (J) Graphical summary showing the different cell behaviors of cap and body cells. Scale bar = 25 µm. See also Figure S1 and Movie S1.
We injected pregnant dams at embryonic day (E) 8.5 with tamoxifen, dissected pancreatic buds from Pdx1-CreER; R26mT/mG/+ embryos at E10.5, cultured the explants for 24 hours, and captured images at 10–15 minute intervals over 24 hours. In these movies, we analyzed parameters such as cell shape changes, cell rearrangements, migratory patterns, and cell divisions. After the initial 24-hour culture period (defined as time (t) 0), the surface of the pancreatic epithelium was largely smooth with the exception of a few areas where sites of future invagination were discernable (Figure 1C1). Consistent with in vivo findings (Villasenor et al., 2010), clear epithelial invaginations indicative of branching morphogenesis became apparent during the subsequent 18 hours in culture (Figure 1C2). At the beginning of the imaging period, two major domains could be distinguished: an outer pseudostratified columnar epithelial layer of ‘cap’ cells and an inner compartment of ‘body’ cells (Figure 1C1) (Villasenor et al., 2010). The majority of cap cells display a wide basal surface and constricted apical side (Figure 1C1; Movie S1). However, we also observed sporadic cap cells with a constricted basal side and wide apical surface (Figure 1C1, blue arrows; Movie S1). Time-lapse analysis revealed that those cap cells demarcate sites of future epithelial invaginations (Figure 1C2; Movie S1), indicating that branch formation is preceded by a cell shape change of cap cells. Analysis of individual cap cells over a time span of 2 hours revealed dramatic and rapid cell shape changes (Figure 1D, magenta arrows; Movie S1). Furthermore, we observed dynamic cell intercalations, or position rearrangements, among neighboring cap cells, resulting in the widening of defined segments within the epithelial surface (Figure 1E, white arrows; Movie S1). In contrast to cap cells, body cells maintained their shape and position during the same time period (Figure 1D, cyan arrows; Figure 1E, beige arrows). Together, these results show that cap cells are more pleiomorphic and dynamic than body cells.
Based on the distinctive cell shape changes and dynamic rearrangements observed in cap cells, we postulated that cap and body cells exhibit differences in cell motility. To track the movement of individual cells in space and time, we performed time-lapse microscopy of pancreatic explants from transgenic mice expressing nuclear green fluorescent protein in pancreatic progenitor cells (Figure S1A–C; Movie S1). These time-lapse movies allowed us to quantify individual cell movement parameters, such as velocity (distance over time), displacement rate (distance traveled from origin in a set time) and meandering index (a ratio of displacement from origin to track length). While the velocity of cap and body cell movements was similar (Figure 1F), cap cells exhibited a higher displacement rate and meandering index than body cells (Figure 1G,H). These findings show that cap cells move with more directionality than body cells.
To determine whether cap and body cells change location between the two compartments, we tracked the location of individual cap and body cells over a period of 10 hours. We found that 83.5% (81/97) of cap cells stayed in the cap cell compartment, while 93.5% (58/62) of body cells remained in the body cell compartment (Figure 1I). Together, these results suggest that cap and body cell location is largely pre-determined early and that cap cells could play an important role in driving the changes in organ shape associated with the initiation of pancreas branching (Figure 1J).
Cap Cells Exhibit Mitosis-Associated Cell Dispersal
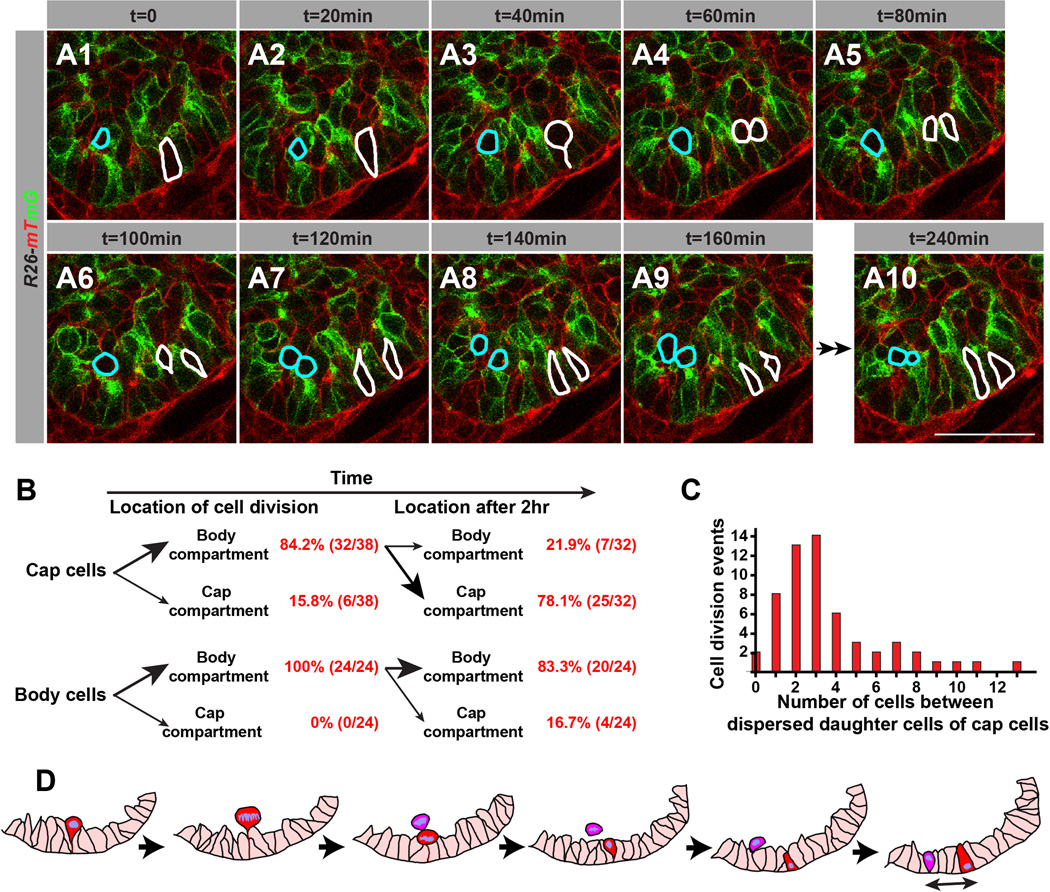
By tracking individual cells, we also observed distinct cellular behaviors in mitotic cap and body cells (Figure 2A; Movie S2). Body cells (outlined in blue in Figure 2A) divided within the body cell compartment with the two daughter cells typically remaining adjacent to each other after cytokinesis (Figure 2A7–10,B). In contrast, the majority of cap cells (84.2%; n=38, outlined in white in Figure 2A; Movie S2) moved away from the outer cell layer (Figure 2A1–3) to the body cell compartment where cell division occurred (Figure 2A4). After cytokinesis, most of the daughter cells (78.1%; n=32) immediately separated and migrated back to the cap cell layer (Figure 2A5–9, 2B). We found that the daughter cap cells often reinserted back into the cap cell compartment in noncontiguous positions. The space between two daughter cells ranged widely from zero to more than 10 cells, the majority of cells being separated by one to four cells (Figure 2C; Movie S2). This behavior, known as mitosis-associated cell dispersal (Grosse et al., 2011; Packard et al., 2013), was not observed in body cells, and thus appears to be a unique feature of cap cells. Given the high division rate of cap cells, this cellular behavior together with rapid cell shape changes, cell intercalation, and directional cell migration would lead to extensive epithelial cell rearrangements within the cap cell compartment at the onset of pancreas branching morphogenesis (Figure 2D).
Figure 2. Time-Lapse Analysis of Labeled Pancreatic Progenitor Cells Undergoing Mitosis and Cell Migration.
(A) Time-lapse fluorescence microscopy of mosaically labeled pancreatic progenitor cells in pancreas explants from Pdx1-CreER; R26mT/mG/+ embryos. Cap cells undergoing mitosis during the 4-hour imaging period are outlined in white and body cells in blue. (B) Time-lapse cell location analysis and cell counting of dividing cap and body cells at the time of and 2 hours after cell division. (C) The number of cells between two dispersed daughter cells after reinsertion into the cap cell layer were counted (cell division events, n=95). (D) Graphical summary of the mitosis-associated cell dispersal of cap cells. Scale bar = 25 µm. See also Movie S2.
ECM-Integrin Signaling Controls Pancreas Branching and Cap Cell Behavior
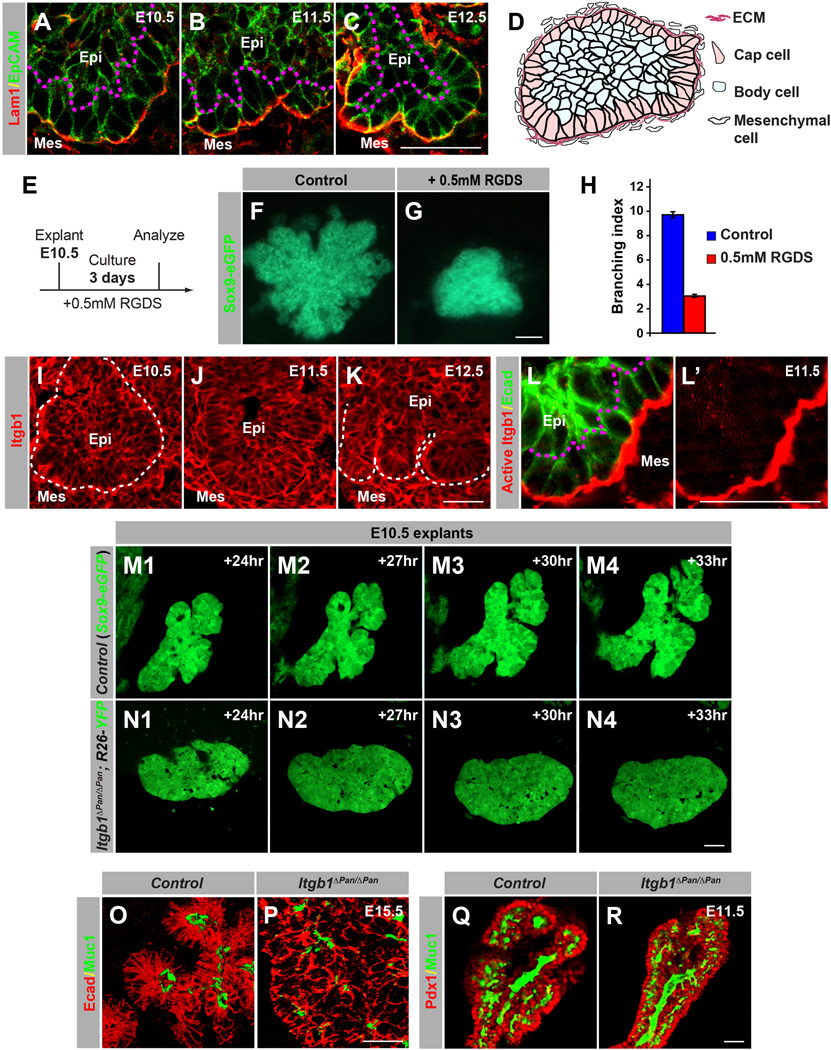
Our findings have revealed distinct cellular behaviors in cap and body cells; however, the signals regulating these cellular behaviors are unclear. As cap and body cell location is largely pre-determined at the beginning of branching (Figure 1I), non-diffusible local cues might establish cap and body cell compartments and regulate their distinct cellular behaviors. Given evidence that the basal side of cap cells, but not body cells, directly contacts the ECM (Figure 3A–D), we hypothesized that microenvironmental signals communicated via the ECM could mediate these localized cellular behaviors. To determine whether the interaction of cap cells with the ECM is necessary for pancreas morphogenesis, we disrupted cap cell interaction with the ECM in pancreas explants with arginyl-glycyl-aspartyl-serine (RGDS), a tetrapeptide that competitively inhibits ECM-cell interactions (Figure 3E). Treatment of pancreas explants with RGDS peptide inhibited the formation of branches (Figure 3F–H), suggesting that early progenitor cells need to interact with the ECM for branching morphogenesis to occur.
Figure 3. ECM-Integrin Signaling Controls Pancreas Branching.
(A-C) Immunofluorescence staining of pancreatic sections from E10.5 (A), E11.5 (B), and E12.5 (C) embryos for the ECM marker Laminin1 (Lam1) and the epithelial marker EpCAM. (D) Graphical summary showing localization of the ECM in the early embryonic pancreas. (E-H) RGDS peptide treatment of pancreatic explants blocks ECM-integrin signaling and perturbs branching morphogenesis. (E) Experimental design. (F,G) Pancreas explants from Sox9-eGFP mice untreated (F) or treated with RGDS peptide (G). (H) Quantification of the branching index by counting the number of lobes per explant (n=6). (I-L) Cap cells display activated integrin signaling. Immunofluorescence staining of pancreatic sections at E10.5 (I), E11.5 (J), and E12.5 (K) for Integrin-β1 (Itgb1) (I–K), or activated Itgb1 (L,L’). E-cadherin (Ecad) (L) visualizes the pancreatic epithelium. (M,N) Time-lapse images of control Sox9-eGFP (M) and Itgb1 mutant (Itgb1ΔPan/ΔPan;R26YFP/+, N) pancreatic explants. (O,P) 3D reconstructed images of staining for Ecad and Mucin1 (Muc1) shows absence of branching in Itgb1ΔPan/ΔPan embryos at E15.5. (Q,R) Staining at E11.5 for Muc1 and Pdx1 shows micro-lumen formation in Itgb1ΔPan/ΔPan embryos. ECM, extracellular matrix; RGDS, Arginine-Glycine-Aspartate-Serine tetra-peptide; Epi, epithelium; Mes, mesenchyme. Scale bar = 25 µm (A-C, I-R), 50 µm (F,G). See also Figure S2, S3, S4 and Movie S3.
Integrin family receptors function as transmembrane linkers that connect ECM molecules to actin filaments in the cell cortex, thereby regulating the shape, orientation, and movement of cells (Guo and Giancotti, 2004; Legate et al., 2009). Among all integrin subunits, the β1 integrin subunit (Itgb1) is most central to signaling via the ECM (Hynes, 2002). Itgb1 was ubiquitously expressed in pancreatic epithelial and surrounding mesenchymal cells from E10.5 to E12.5 (Figure 3I–K). However, the activated form of Itgb1 was mainly detected at the basal surface of cap cells, indicating that ECM-integrin signaling is predominately activated in cap cells (Figure 3L,L’).
To investigate whether integrin signaling is required for pancreas branching, we deleted Itgb1 specifically in pancreatic progenitors by generating Pdx1-Cre; Itgb1f/f mice (hereafter abbreviated as Itgb1ΔPan/ΔPan) (Figure S2A,B). To verify that Itgb1 deletion perturbs cell-ECM interactions, we performed in vitro cell adhesion assays to measure the ECM binding capacity of Itgb1-deficient pancreatic progenitors. As expected, the binding of Itgb1-deficient pancreatic progenitors to ECM substrates was drastically diminished (Figure S2C,D). Interestingly, Laminin1 staining also revealed thinning of the basement membrane in Itgb1ΔPan/ΔPan embryos (Figure S2E,F), suggesting that loss of Itgb1 also affects ECM remodeling.
By imaging pancreatic explants in real time, we next determined whether loss of Itgb1 affects the initiation of branching morphogenesis. In control embryos, we observed the formation of ‘tips’ and ‘dips’ characteristic of early pancreas branching after 24 hours of culture (Figure 3M; Figure S2G; Movie S3). By contrast, pancreata from Itgb1 ΔPan/ΔPan embryos maintained a round shape and failed to exhibit signs of branching even after an extended culture period for more than three days (Figure 3N; Figure S2H; Movie S3). Absence of branching was also evident in sections from Itgb1ΔPan/ΔPan embryos at E12.5 and E15.5 (Figure S2B,F; Figure 3O,P). Notably, the numbers of apoptotic cells were similar in Itgb1ΔPan/ΔPan and control pancreata at E11.5 (Figure S2I–L), indicating that the branching defect is not a mere consequence of programmed cell death.
Pancreas branching defects have also been observed in mouse mutants with defects in microlumen formation, which occurs as a result of polarization and apical constriction of body cells (Kesavan et al., 2009; Villasenor et al., 2010). Interestingly, microlumen formation was not perturbed in Itgb1ΔPan/ΔPan embryos (Figure 3Q,R), suggesting that early branching morphogenesis and microlumen formation can be uncoupled.
Downstream Pathways of ECM-Integrin Signaling Control Actomyosin Dynamics and Pancreas Branching
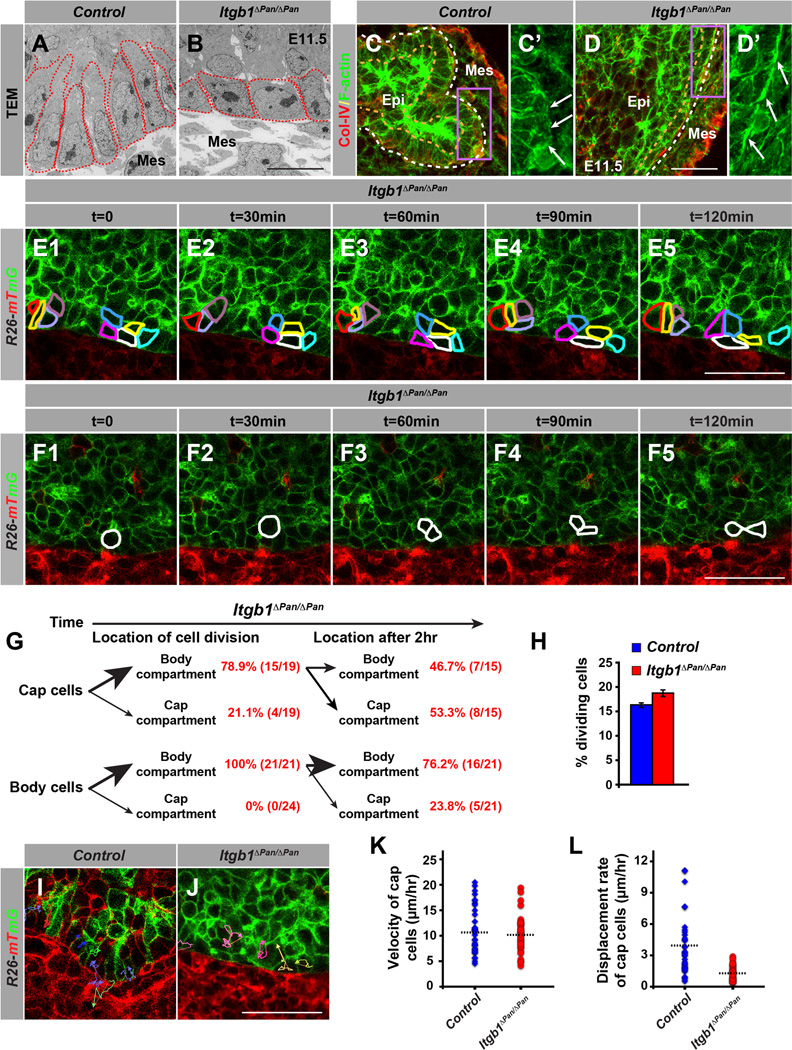
To determine whether the lack of branching morphogenesis in Itgb1ΔPan/ΔPan embryos is associated with aberrant cellular behaviors, we examined cap cell dynamics after Itgb1 deletion. Itgb1-deficient cap cells exhibited a shape change from columnar (Figures 1C and 4A) to cuboidal (Figure 4B). As cell shape is maintained by the actin cytoskeleton (Legate et al., 2009), we hypothesized that the loss of Itgb1 perturbs organization of the actin cytoskeleton. Indeed, Itgb1ΔPan/ΔPan embryos displayed aberrant organization of F-actin fibers, such that the F-actin fibers were mostly aligned parallel to the basal side of cap cells instead of perpendicular as in control embryos (Figure 4C–D’, white arrows). To determine whether other actomyosin-dependent cellular processes, such as cell rearrangements, mitosis, and migration, are affected by Itgb1 deletion, we used live cell imaging to generate time-lapse movies from Itgb1ΔPan/ΔPan; R26mT/mG/+ pancreatic explants. In contrast to cap cells in control embryos, which exhibited dynamic cell rearrangements (Figure 1E; Movie S1), Itgb1-deficient cap cells maintained their relative positions over time (Figure 4E; Movie S4). To determine whether cap cells require Itgb1 expression for mitosis-associated cell dispersal, we followed cap cells and their daughters during cell division. Similar to control embryos (Figure 2A–C; Movie S2), cap cells in Itgb1ΔPan/ΔPan embryos moved into the body cell compartment prior to cell division (Figure 4F1,2 and G; Movie S4). However, unlike controls, most of the daughter cells remained closely associated with each other (Figure 4F3–5) and often did not migrate back into the cap cell compartment (Figure 4G; Movie S4). Notably, the overall mitotic rate of pancreatic progenitors was similar in Itgb1ΔPan/ΔPan and control embryos at E11.5 (Figure 4H). This suggests that during initiation of pancreas branching Itgb1 deletion does not affect epithelial cell proliferation, but is required for daughter cell dispersion following mitosis.
Figure 4. ECM-Integrin Signaling Regulates Cap Cell Behaviors.
(A,B) Transmission electron microscope (TEM) images of pancreata from Itgb1ΔPan/ΔPan and control embryos at E11.5. Deletion of Itgb1 alters the shape of cap cells (outlined by red dotted line) from columnar (A) to cuboidal (B). (C,D) Itgb1 deletion results in accumulation of filamentous actin (F-actin) at the basal side of cap cells. Staining for the actomyosin marker F-actin and collagen-IV (Col-IV) in control (C) and Itgb1ΔPan/ΔPan embryos (D) at E11.5. The white and beige dotted lines in C,D delineate the basal side and apical side of the cap cells, respectively. Fields demarcated by purple boxes in C,D are shown at higher magnification in C’,D’. Arrows in C’ and D’ point to the basal side of cap cells. (E-L) Time-lapse analysis of cap cells in pancreatic explants from Itgb1ΔPan/ΔPan; R26mT/mG/+ embryos for cell shape changes (E), mitosis (F) and cell migration (I–L). (E,F) Optical sections through the center of a pancreatic bud at several time points (t). t=0 demarcates the beginning of the imaging period 24 hours after starting the organ culture at E10.5. (E) Cap cells (cells in two different clusters are outlined) display little cell shape change and rearrangement during a 2-hour period, see Movie S4, also see Figure 2A and Movie S2 for comparison to controls. (F) Daughter cells stay associated with each other. A dividing cap cell and its daughter cells are outlined in white. (G) Time-lapse cell location analysis and cell counting of dividing cap and body cells. In Itgb1ΔPan/ΔPan embryos, cap cells migrate into the body cell compartment to divide, but in contrast to controls (see Figure 2B), the daughter cells frequently stay in the body cell compartment after mitosis. (H) Percentage of dividing cells was calculated by counting PHH3+ cells relative to Ecad+ cells (n=3). (I,J) Cell migration tracking of representative cap cells in control (I) and Itgb1ΔPan/ΔPan explants (J). (K,L) Cap cells in Itgb1ΔPan/ΔPan and control embryos exhibit similar velocity (K) but the displacement rate is reduced in Itgb1ΔPan/ΔPan embryos (L). ECM, extracellular matrix; Epi, epithelium; Mes, mesenchyme. Scale bar = 10 µm (A,B); 25 µm (C-F, I,J). See also Figure S3, S4 and Movie S4.
We next tracked the migration of individual cap cells in pancreas explants from Itgb1ΔPan/ΔPan embryos (Figure 4I,J). While we observed little effect of Itgb1 deletion on the velocity of cap cells (Figure 4K), Itgb1-deficient cap cells exhibited a significantly lower displacement rate than Itgb1-expressing cap cells (Figure 4L). Hence, loss of Itgb1 does not affect the ability of cap cells to move but impairs directionality of their migration. Together, these findings support the conclusion that the observed cap cell behaviors are induced by local cues from the ECM via integrin-mediated remodeling of the actin cytoskeleton.
Cytoskeleton organization is controlled by multiple kinase pathways including the Src, FAK, and ERK signaling pathways (Guo and Giancotti, 2004; Legate et al., 2009), which are all regulated by ECM-integrin signaling (Figure S3A). In agreement with ltgb1 signaling being specifically active in cap cells (Figure 3L), the phosphorylated and activated forms of Src, FAK, and ERK were expressed predominantly in cap cells (Figure S3B–D). Furthermore, the expression of active Src, FAK, and ERK was reduced in ltgb1-deficient cap cells (Figure S3E–G), suggesting that ECM-integrin signaling is required for the activation of these downstream kinase pathways. To investigate whether these signaling pathways control pancreas branching, we treated pancreas explants with pharmaceutical inhibitors that block Src and FAK activity. Unlike control explants (Figure S3H,I,W,X), explants treated with the inhibitors failed to develop branches (Figure S3M,N,R,S,W,X). In addition, we observed altered F-actin organization and irregular cell shapes in Src and FAK inhibitor-treated explants (Figure S3O–Q,T–V). To further determine the requirement of actomyosin dynamics for pancreas branching, we treated explants with the inhibitors of actomyosin contractility, Blebbistatin and Cytochalasin D, or activated Rac1 or Rho GTPases. These treatments blocked branching morphogenesis and disrupted cap cell organization (Figure S4A–W). Together, these data suggest that ECM-integrin signaling initiates pancreas branch formation by regulating actomyosin dynamics through the Src, FAK, and ERK signaling pathways.
ECM-Integrin Initiates Pancreas Branching by Modulating Cell Adhesion
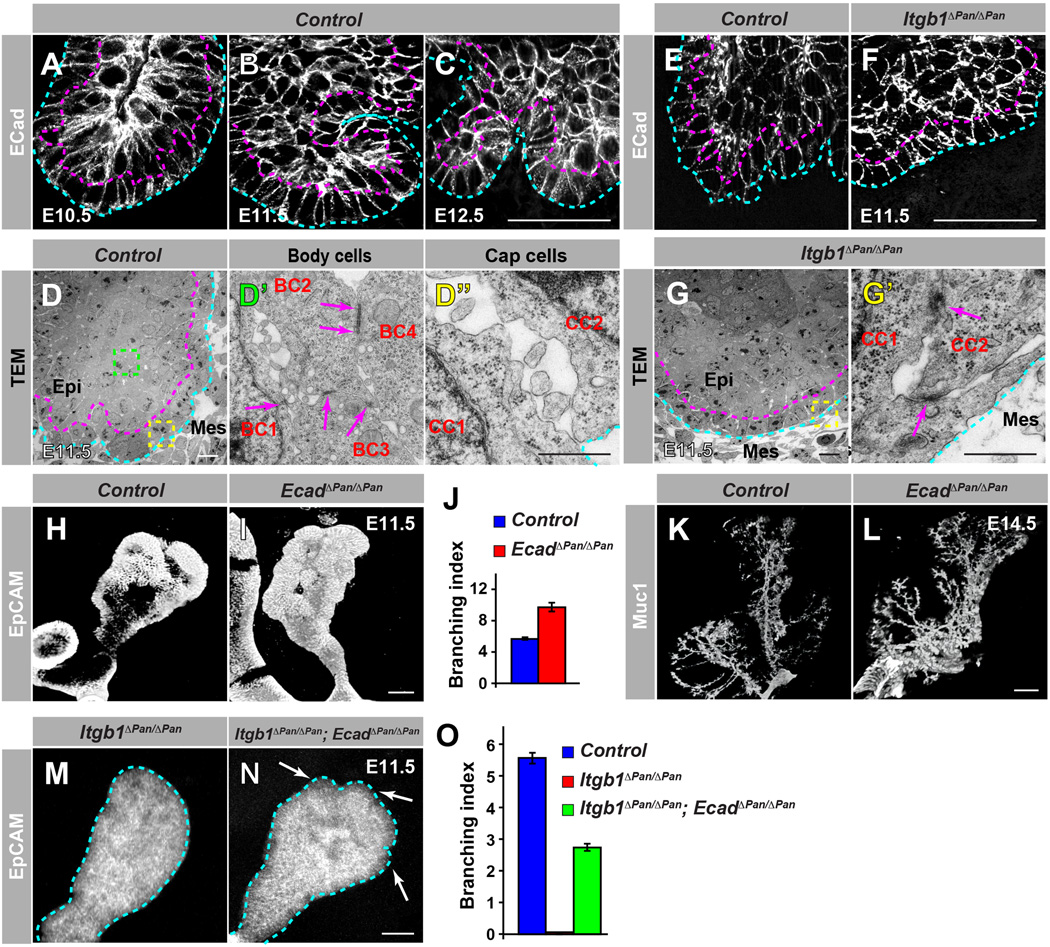
As both the cell shape changes and cell migration require local changes in cell-cell junctions between neighboring cells (Guillot and Lecuit, 2013), we sought to determine whether cell adhesive properties contribute to the distinct behaviors of cap and body cells. Cell-cell interactions, mediated by adherens junctions, are established by homophilic interactions of cadherins (Halbleib and Nelson, 2006). At the onset of pancreas branching, E-cadherin, the best-characterized cadherin family member, was highly expressed in body cells but weakly in cap cells (Figure 5A,B). However, as branching morphogenesis progressed, E-cadherin expression increased in cap cells (Figure 5C), suggesting that E-cadherin downregulation in cap cells is limited to the onset of branching. We then used transmission electron microscopy (TEM) to further characterize junction complexes and cellular ultra-structures in cap and body cells. In body cells, adherens junctions can be recognized as electron-dense structures between membranes of adjacent cells (Figure 5D,D’, magenta arrows in D’). These electron-dense structures were less frequent in cap cells at E11.5, suggesting that cap cells are not connected by mature adherens junctions at this stage. Instead, we saw junctions that were less electron-dense and observed extensive membrane protrusions connecting cap cells (Figure 5D,D”), indicating that cap cells are more loosely connected than body cells. These observations are consistent with the notion that low adhesion between cap cells facilitates cap cell motility to initiate branching.
Figure 5. Initiation of Pancreas Branching Morphogenesis Depends on Regulation of Cell Adhesion by ECM-Integrin Signaling.
(A-C) Immunofluorescence staining of pancreatic sections from E10.5 (A), E11.5 (B), and E12.5 (C) embryos for E-cadherin (Ecad) shows lower Ecad levels in cap cells than in body cells at E10.5 and E11.5. (E,F) Ecad staining intensity in cap cells is higher in Itgb1ΔPan/ΔPan than in control embryos at E11.5. The blue dotted lines in A-F delineate the basal side and the magenta dotted lines the apical side of the cap cell layer. (D,G) Deletion of Itgb1 alters cell-cell junctions between cap cells. TEM images of pancreata from control (D) and Itgb1ΔPan/ΔPan (G) embryos at E11.5. Mature adherens junctions (magenta arrows in D’) are identified in body cells (D,D’) but not in cap cells of control embryos (D,D”). Mature adherens junctions (magenta arrows in G’) are also seen in cap cells of Itgb1ΔPan/ΔPan embryos (G,G’). D’ and D” show magnifications of the areas boxed in green and yellow, respectively, in D. G’ shows a magnification of the area boxed in yellow in G. The blue dotted lines in D,D”,G,G’ delineate the basal side and the magenta dotted lines in D,G the apical side of the cap cell layer. (H-L) Ecad deletion in pancreatic progenitors causes hyperbranching. Whole mount immunofluorescence staining for EpCAM in control (H) and EcadΔPan/ΔPan (I) embryos at E11.5. (J) Quantification of the branching index by counting the number of lobes at E11.5 (n=4). Whole mount immunofluorescence staining for Mucin1 (Muc1) in control (K) and EcadΔPan/ΔPan (L) embryos reveals a hyperbranched ductal network at E14.5. (M-O) Ecad inactivation partially rescues branching in the Itgb1-deficient pancreas. Whole mount immunofluorescence staining for EpCAM in Itgb1ΔPan/ΔPan (M) and Itgb1ΔPan/ΔPan; EcadΔPan/ΔPan (N) embryos at E11.5. The arrows in N point to early branches in the pancreas of Itgb1ΔPan/ΔPan; EcadΔPan/ΔPan embryos. (O) Quantification of the branching index at E11.5 (n=3). Epi, epithelium; Mes, mesenchyme; BC, body cell; CC, cap cell. Scale bar = 25 µm (A-C, H,I,M,N), 10 µm (D,G), 200 nm (D’,D”,C’), 125 µm (K,L). See also Figure S5.
We next investigated whether cell-cell adhesion in cap cells is under the control of ECM-integrin signaling in the early pancreas. Compared to controls, cap cells in Itgb1ΔPan/ΔPan embryos displayed higher E-cadherin staining intensity (Figure 5E–F). TEM analysis further revealed electron-dense structures similar to adherens junctions between adjacent cap cells in Itgb1ΔPan/ΔPan embryos (Figure 5G,G’, magenta arrows in G’). This indicates that loss of integrin signaling leads to E-cadherin upregulation and premature formation of adherens junctions in cap cells.
To determine whether E-cadherin regulation is important for pancreas branching, we generated mice with a pancreas-specific deletion of E-cadherin (Pdx1-Cre; Ecadf/f mice, hereafter abbreviated as EcadΔPan/ΔPan). TEM analysis showed that E-cadherin deletion results in a loss of adherens junctions in both cap and body cells (Figure S5A). This change in cell-cell junctions in EcadΔPan/ΔPan mice was associated with increased numbers of branches at E11.5 (Figure 5H–J) and a hyperbranched ductal tree at E14.5 (Figure 5K–L). These findings suggest that a reduction in cell-cell adhesion through down-regulation of E-cadherin plays a role in the initiation of pancreas branching.
Based on these results, we postulated that the observed up-regulation of E-cadherin and premature formation of adherens junctions in cap cells in Itgb1ΔPan/ΔPan embryos could contribute to the branching defect. To test this, we generated Itgb1ΔPan/ΔPan; EcadΔPan/ΔPan embryos and examined pancreas morphology. Confirming our earlier findings, Itgb1ΔPan/ΔPan embryos displayed a smooth outer cap cell epithelium at E11.5 and showed no branching morphogenesis (Figure 5M; Figure S5B,C). By contrast, Itgb1ΔPan/ΔPan; EcadΔPan/ΔPan embryos exhibited ‘dips’ on the pancreatic epithelial surface, signifying the initiation of pancreas branching (Figure 5N,O, white arrows in N; Figure S5D, white arrows). Thus, can partially restore branching in Itgb1ΔPan/ΔPan embryos. Together, our results support a model whereby pancreas branching morphogenesis is initiated via ECM-integrin signaling-mediated regulation of cell adhesion in cap cells.
Discussion
The Importance of Cellular Behaviors for Pancreas Organ Development
Our 3D analysis of confocal time-lapse sequences of cultured embryonic pancreata revealed region-specific cell behaviors of pancreatic epithelial cells. We show that cap cells are more pleomorphic and motile than body cells and rearrange more freely. Our findings suggest that these cap cell behaviors are controlled through remodeling of the actomyosin cytoskeleton by local cues from the basement membrane. Disrupting integrin-mediated cell interactions with the basement membrane rendered cap cells more similar to body cells with regard to their morphology and movements. Moreover, in the absence of integrin signaling, cap cells tended to stay in the body cell compartment after their division, indicating that the basement membrane provides instructive cues for segregating the two compartments. Our data suggest that these unique behaviors of cap cells are likely important for the initiation of pancreas branching. Moreover, the early segregation of cap and body cell compartments could help establish differences in cell fate, as outer cells later differentiate into acinar cells, whereas inner cells give rise to endocrine and ductal cells (Shih et al., 2013). The concept that morphogenesis and cell fate choices could be coordinated by spatially-restricted ECM cues is in accordance with previous observations that exposure to ECM biases progenitors toward the acinar cell fate (Gittes et al., 1996; Kesavan et al., 2009).
A previous study showed that Ptf1aCre-mediated deletion of Itgb1 has no effect on pancreas morphogenesis (Bombardelli et al., 2010). The discrepancy with our findings is likely explained by the slightly later expression of Ptf1aCre compared to Pdx1-Cre (Seymour et al., 2012). This suggests that the ECM and integrin signaling are particularly critical for the initiation of branching morphogenesis during a short early time window of pancreas development.
Unique Features of Pancreas Branching Morphogenesis
At a macroscopic level, there are similarities between pancreas and other branched organs, in particular mammary and salivary glands. The mammary and salivary glands are also transiently organized into an outer cap cell and inner body cell compartment (Huebner and Ewald, 2014). As in the pancreas, outer layer cells of salivary glands differentiate into acinar cells, and their behavior is similarly controlled by integrin-mediated cell interactions with the basement membrane (Hsu et al., 2013). However, there are clear differences between salivary gland and pancreas morphogenesis at both the cellular and molecular levels. In developing salivary glands, branching is initiated by the formation of shallow clefts, which subsequently deepen to subdivide the single bud into multiple smaller buds (Harunaga et al., 2011). Our live imaging analysis did not reveal formation of clefts during initiation of pancreas branching. Instead, we observed formation of small epithelial invaginations, accompanied by collective outgrowth of epithelial cells between two invagination sites.
In addition to morphogenetic differences, the patterns of E-cadherin regulation are distinct in salivary glands and pancreas. In salivary glands, cap cells lining the clefts selectively downregulate E-cadherin (Sakai et al., 2003), while pancreatic cap cells have uniformly low E-cadherin levels. Furthermore, E-cadherin inhibition in salivary glands has been reported to not affect (Hsu et al., 2013) or to inhibit (Walker et al., 2008) branch initiation, whereas we observed hyperbranching in the pancreas after E-cadherin deletion.
Our data also suggest differences between salivary glands and pancreas in how E-cadherin is regulated at the molecular level. In salivary glands, E-cadherin is transcriptionally repressed by Snail2 and Btbd7, which are expressed in cells at the bottom of the clefts (Onodera et al., 2010). In contrast, we were unable to detect Btbd7 or Snail2 expression in pancreatic cap cells (data not shown), suggesting alternative mechanisms for E-cadherin regulation. In addition to transcriptional mechanisms, E-cadherin can be regulated post-translationally by stimulation of E-cadherin endocytosis through the integrin-Src axis (Martinez-Rico et al., 2010). Our finding that E-cadherin regulation in cap cells appeared to be Src-dependent (Figure S3P), suggests that this mechanism might be employed in the pancreas to globally reduce E-cadherin levels in cells contacting the basement membrane. Of note, high integrin signaling activity and low E-cadherin levels are also a key feature of pancreatic ductal adenocarcinoma (Grzesiak et al., 2007; von Burstin et al., 2009). Hence, it is possible that mature acinar cells co-opt mechanisms used during embryogenesis to gain motility during malignant transformation. Employing explant systems and 3D–organoids, the here described time-lapse imaging technology will allow us to systematically study how niche cues affect cell behaviors in development and disease.
Experimental Procedures
Mouse Strains
Mouse strains are described in Supplemental Experimental Procedures. All animal experiments described herein were approved by the University of California San Diego Institutional Animal Care and Use Committees.
Live Imaging
Pancreas explants are described in Supplemental Experimental Procedures. After 24 hours of culture, the explants were placed on the microscope stage in 37 °C culture chambers with a controlled atmosphere of humidified 5% CO2. Time-lapse imaging was performed using Zeiss LSM780 inverted confocal microscope with C-Apochromat 40×/1.20 W objective lens (Figures 1, 2, and 4) and C-Apochromat 20×/0.75 objective lens (Figure S1) and Olympus FV1000 inverted confocal microscope with Uapo 40×/1.15 W objective lens (Figure 3 and S2). Explants were optically sectioned every 0.5 µm in a 512 × 512 format with up to 40 µm Z-stacks every 10 min for 48 hours. Images were acquired with Zen software and then reconstructed in 3D with Volocity™ software. The contrast was adjusted and selected optical planes or z-projections of sequential optical sections were used to assemble time-lapse movies.
A detailed description of all experimental procedures, including ex vivo pancreas explants, cell tracking, and electron microscopy, is available in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgements
We thank R. MacDonald and D. Melton for mouse strains. We acknowledge the support of the University of California, San Diego Microscopy and Electron Microscopy Core Facilities, and the Sanford Consortium Histology and Imaging Core Facility. This work was supported by the NIH/NIDDK awards DK078803 and DK68471 to M.S., JDRF grants 1-2005-1084 and 1-2004-13, and the WA State Life Sciences Discovery Fund Program grant 4553677 to V.C., and JDRF postdoctoral fellowship 3-2009-161 to H.P.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.P.S. and M.S. conceived the project, designed the experiments and analyzed the data. H.P.S. and D.P. performed all analyses of mouse genetic models, pancreatic explant culture systems and live-imaging experiments. V.C. performed the cell-adhesion assay. H.P.S. and M.S. wrote the manuscript.
References
- Bombardelli L, Carpenter ES, Wu AP, Alston N, DelGiorno KE, Crawford HC. Pancreas-specific ablation of beta1 integrin induces tissue degeneration by disrupting acinar cell polarity. Gastroenterology. 2010;138:2531–2540. doi: 10.1053/j.gastro.2010.02.043. 2540 e2531–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canel M, Serrels A, Frame MC, Brunton VG. E-cadherin-integrin crosstalk in cancer invasion and metastasis. Journal of cell science. 2013;126:393–401. doi: 10.1242/jcs.100115. [DOI] [PubMed] [Google Scholar]
- Chi X, Michos O, Shakya R, Riccio P, Enomoto H, Licht JD, Asai N, Takahashi M, Ohgami N, Kato M, et al. Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev Cell. 2009;17:199–209. doi: 10.1016/j.devcel.2009.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald AJ, Brenot A, Duong M, Chan BS, Werb Z. Collective epithelial migration and cell rearrangements drive mammary branching morphogenesis. Dev Cell. 2008;14:570–581. doi: 10.1016/j.devcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gittes GK, Galante PE, Hanahan D, Rutter WJ, Debase HT. Lineage-specific morphogenesis in the developing pancreas: role of mesenchymal factors. Development. 1996;122:439–447. doi: 10.1242/dev.122.2.439. [DOI] [PubMed] [Google Scholar]
- Grosse AS, Pressprich MF, Curley LB, Hamilton KL, Margolis B, Hildebrand JD, Gumucio DL. Cell dynamics in fetal intestinal epithelium: implications for intestinal growth and morphogenesis. Development. 2011;138:4423–4432. doi: 10.1242/dev.065789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiak JJ, Ho JC, Moossa AR, Bouvet M. The integrin-extracellular matrix axis in pancreatic cancer. Pancreas. 2007;35:293–301. doi: 10.1097/mpa.0b013e31811f4526. [DOI] [PubMed] [Google Scholar]
- Guillot C, Lecuit T. Mechanics of epithelial tissue homeostasis and morphogenesis. Science. 2013;340:1185–1189. doi: 10.1126/science.1235249. [DOI] [PubMed] [Google Scholar]
- Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5:816–826. doi: 10.1038/nrm1490. [DOI] [PubMed] [Google Scholar]
- Halbleib JM, Nelson WJ. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev. 2006;20:3199–3214. doi: 10.1101/gad.1486806. [DOI] [PubMed] [Google Scholar]
- Harunaga J, Hsu JC, Yamada KM. Dynamics of salivary gland morphogenesis. J Dent Res. 2011;90:1070–1077. doi: 10.1177/0022034511405330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu JC, Koo H, Harunaga JS, Matsumoto K, Doyle AD, Yamada KM. Region-specific epithelial cell dynamics during branching morphogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:1066–1077. doi: 10.1002/dvdy.24000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner RJ, Ewald AJ. Cellular foundations of mammary tubulogenesis. Semin Cell Dev Biol. 2014;31:124–131. doi: 10.1016/j.semcdb.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes RO. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- Kesavan G, Sand FW, Greiner TU, Johansson JK, Kobberup S, Wu X, Brakebusch C, Semb H. Cdc42-mediated tubulogenesis controls cell specification. Cell. 2009;139:791–801. doi: 10.1016/j.cell.2009.08.049. [DOI] [PubMed] [Google Scholar]
- Larsen M, Wei C, Yamada KM. Cell and fibronectin dynamics during branching morphogenesis. Journal of cell science. 2006;119:3376–3384. doi: 10.1242/jcs.03079. [DOI] [PubMed] [Google Scholar]
- Legate KR, Wickstrom SA, Fassler R. Genetic and cell biological analysis of integrin outside-in signaling. Genes & development. 2009;23:397–418. doi: 10.1101/gad.1758709. [DOI] [PubMed] [Google Scholar]
- Martinez-Rico C, Pincet F, Thiery JP, Dufour S. Integrins stimulate E-cadherin-mediated intercellular adhesion by regulating Src-kinase activation and actomyosin contractility. Journal of cell science. 2010;123:712–722. doi: 10.1242/jcs.047878. [DOI] [PubMed] [Google Scholar]
- Munjal A, Lecuit T. Actomyosin networks and tissue morphogenesis. Development. 2014;141:1789–1793. doi: 10.1242/dev.091645. [DOI] [PubMed] [Google Scholar]
- Onodera T, Sakai T, Hsu JC, Matsumoto K, Chiorini JA, Yamada KM. Btbd7 regulates epithelial cell dynamics and branching morphogenesis. Science. 2010;329:562–565. doi: 10.1126/science.1191880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Packard A, Georgas K, Michos O, Riccio P, Cebrian C, Combes AN, Ju A, Ferrer-Vaquer A, Hadjantonakis AK, Zong H, et al. Luminal mitosis drives epithelial cell dispersal within the branching ureteric bud. Dev Cell. 2013;27:319–330. doi: 10.1016/j.devcel.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold KM, Naumann H, Spagnoli FM. Rho signalling restriction by the RhoGAP Stard13 integrates growth and morphogenesis in the pancreas. Development. 2013;140:126–135. doi: 10.1242/dev.082701. [DOI] [PubMed] [Google Scholar]
- Puri S, Hebrok M. Dynamics of embryonic pancreas development using real-time imaging. Developmental biology. 2007;306:82–93. doi: 10.1016/j.ydbio.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Larsen M, Yamada KM. Fibronectin requirement in branching morphogenesis. Nature. 2003;423:876–881. doi: 10.1038/nature01712. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Shih HP, Patel NA, Freude KK, Xie R, Lim CJ, Sander M. A Sox9/Fgf feed-forward loop maintains pancreatic organ identity. Development. 2012;139:3363–3372. doi: 10.1242/dev.078733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih HP, Wang A, Sander M. Pancreas organogenesis: from lineage determination to morphogenesis. Annual review of cell and developmental biology. 2013;29:81–105. doi: 10.1146/annurev-cellbio-101512-122405. [DOI] [PubMed] [Google Scholar]
- Villasenor A, Chong DC, Henkemeyer M, Cleaver O. Epithelial dynamics of pancreatic branching morphogenesis. Development. 2010;137:4295–4305. doi: 10.1242/dev.052993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Burstin J, Eser S, Paul MC, Seidler B, Brandl M, Messer M, von Werder A, Schmidt A, Mages J, Pagel P, et al. E-cadherin regulates metastasis of pancreatic cancer in vivo and is suppressed by a SNAIL/HDAC1/HDAC2 repressor complex. Gastroenterology. 2009;137:361–371. doi: 10.1053/j.gastro.2009.04.004. 371 e361–365. [DOI] [PubMed] [Google Scholar]
- Walker JL, Menko AS, Khalil S, Rebustini I, Hoffman MP, Kreidberg JA, Kukuruzinska MA. Diverse roles of E-cadherin in the morphogenesis of the submandibular gland: insights into the formation of acinar and ductal structures. Developmental dynamics : an official publication of the American Association of Anatomists. 2008;237:3128–3141. doi: 10.1002/dvdy.21717. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.