Summary
The efficiency of gene expression in all organisms depends on the nucleotide composition of the coding region. GC content and codon usage are the two key sequence features known to influence gene expression, but the underlying molecular mechanisms are not entirely clear. Here we have determined the relative contributions of GC content and codon usage to the efficiency of nuclear gene expression in the unicellular green alga Chlamydomonas reinhardtii. By comparing gene variants that encode an identical amino acid sequence but differ in their GC content and/or codon usage, we show that codon usage is the key factor determining translational efficiency and, surprisingly, also mRNA stability. By contrast, unfavorable GC content affects gene expression at the level of the chromatin structure by triggering heterochromatinization. We further show that mutant algal strains that permit high-level transgene expression are less susceptible to epigenetic transgene suppression and do not establish a repressive chromatin structure at the transgenic locus. Our data disentangle the relationship between GC content and codon usage, and suggest simple strategies to overcome the transgene expression problem in Chlamydomonas.
Keywords: Chlamydomonas reinhardtii, codon usage, GC content, translation, RNA stability, post-transcriptional regulation, reporter gene, YFP, epigenetics, chromatin structure, histone modification
Introduction
The green alga Chlamydomonas reinhardtii has become an invaluable model system in many research areas, including photosynthesis and chloroplast biology, genome evolution, environmental sensing and signal transduction as well as cell motility and flagellar function in eukaryotes (e.g., Weeks, 1992; Harris, 2001; Merchant et al., 2007). The unicellular, vegetatively haploid alga reproduces sexually or asexually and can be grown under photoautotrophic, mixotrophic or heterotrophic conditions (using acetate as the sole source of reduced carbon; Harris, 1989). Its three genomes in the nucleus, the plastid and the mitochondria are sequenced (Merchant et al., 2007) and amenable to genetic manipulation by stable transformation (Kindle et al., 1991; Remacle et al., 2006; Neupert et al., 2012). Moreover, excellent genetic and genomic resources are available in Chlamydomonas, with large strain collections and mutant collections that can be explored and screened for desired phenotypes (e.g., Gonzalez-Ballester et al., 2011; Terashima et al., 2015).
Over the years, the community also has assembled a large toolbox for post-genomics research in Chlamydomonas, including various reporter genes (Fuhrmann et al., 1999; Fuhrmann et al., 2004; Shao and Bock, 2008; Neupert et al., 2009; Rasala et al., 2013), cassettes for transgene expression (Schroda et al., 2000; Fischer and Rochaix, 2001), protocols for chemical mutagenesis, UV mutagenesis and insertional mutagenesis (Dent et al., 2005; Neupert et al., 2009; Gonzalez-Ballester et al., 2011; Shao et al., 2013; Zhang et al., 2014; Dent et al., 2015), and gene silencing methods employing RNA interference (RNAi; Rohr et al., 2004) or artificial microRNAs (amiRNAs; Molnar et al., 2009; Zhao et al., 2009). However, until recently, the poor expression of transgenes from the nuclear genome of Chlamydomonas (Fuhrmann et al., 1999; Schroda et al., 2000) as well as loss of transgene expression over time (Cerutti et al., 1997; Yamasaki et al., 2008) represented major limitations to both functional genomic research and the exploitation of the alga as a production platform for biofuels, green chemicals, industrial enzymes and biopharmaceuticals (Scaife et al., 2015). While transgene introduction into the nuclear genome by stable genetic transformation is efficient, the isolation of transgenic algal clones that express the foreign gene to reasonably high levels has been very difficult (in particular when expression of cDNAs was attempted) or failed altogether.
Since promoter choice clearly has an impact on the probability of successful transgene expression in Chlamydomonas (Fischer and Rochaix, 2001) and the construction of strong chimeric promoters reduced the severity of the problem in some cases (Schroda et al., 2000), it was hypothesized that epigenetic silencing at the transcriptional level is causally responsible for inefficient transgene expression. However, since also resynthesis of the coding region and adjustment of the codon usage to that of the extremely GC-rich nuclear genome of the alga can improve transgene expression (Fuhrmann et al., 1999; Fuhrmann et al., 2004; Shao and Bock, 2008), there is clearly an additional translational component influencing expression levels. Both translation initiation and translation elongation efficiencies are important determinants of protein synthesis rates in eukaryotes (Xia, 2015) and, very recently, translation rates have also been shown to have a strong impact on mRNA stability (Presnyak et al., 2015).
To overcome the serious limitation posed by poor transgene expression in Chlamydomonas, we recently developed a genetic screen that allowed the selection of algal mutants that express introduced transgenes to high levels. From UV mutagenesis experiments, two such expression strains, termed UVM4 and UVM11, were isolated (Neupert et al., 2009). Their superior transgene expression properties correlated with greatly increased mRNA levels pointing to transcriptional gene silencing as the likely epigenetic cause of poor transgene expression in the wild type (Neupert et al., 2009). The two expression strains have become widely used tools for efficient reporter gene expression and expression of transgenes that are of biotechnological interest (e.g., Karcher et al., 2009; Zäuner et al., 2012; Bohne et al., 2013; Lauersen et al., 2013; reviewed, e.g., in Jinkerson and Jonikas, 2015).
Expectedly, the isolation of the expression strains does not solve all problems with transgene expression in Chlamydomonas. As pointed out already in the initial report on the identification of the UVM4 and UVM11 strains (Neupert et al., 2009), if poor expression of a particular transgene has an additional post-transcriptional component, such as mRNA instability or inefficient translation, further optimization work is needed to improve expression levels. In this regard, two factors are frequently discussed as important determinants of transgene expression efficiency: codon usage and the unusually high GC content of the Chlamydomonas nuclear genome (Merchant et al., 2007). Although codon usage can be influenced by selective pressure on nucleotide composition in the genome, the codon usage of an organism is shaped by many additional factors most of which are related to the translation process (e.g., tRNA pool sizes, efficiency of wobbling and superwobbling; Alkatib et al., 2012). However, thus far, the relative importance of GC content and codon usage to gene expression efficiency has not been rigorously investigated in any system.
Here, we report a systematic comparison of gene variants that encode an identical amino acid sequence and differ only in their GC content and/or codon usage. This allowed us to determine the influence of nucleotide composition versus codon usage to the efficiency of gene expression in Chlamydomonas reinhardtii. We find that (i) codon usage rather than GC content is the key determinant of gene expression efficiency, (ii) high translation rates strongly enhance mRNA stability, (iii) the nucleotide composition of the coding region can feed back onto the chromatin status in the promoter region, and (iv) the use of codon-optimized transgenes in our UVM expression strains facilitates transgene expression to up to more than 1% of the total soluble protein, which remains stable over time. This opens up new applications in cell biology and algal biotechnology.
Results
Design of YFP gene variants for expression in Chlamydomonas
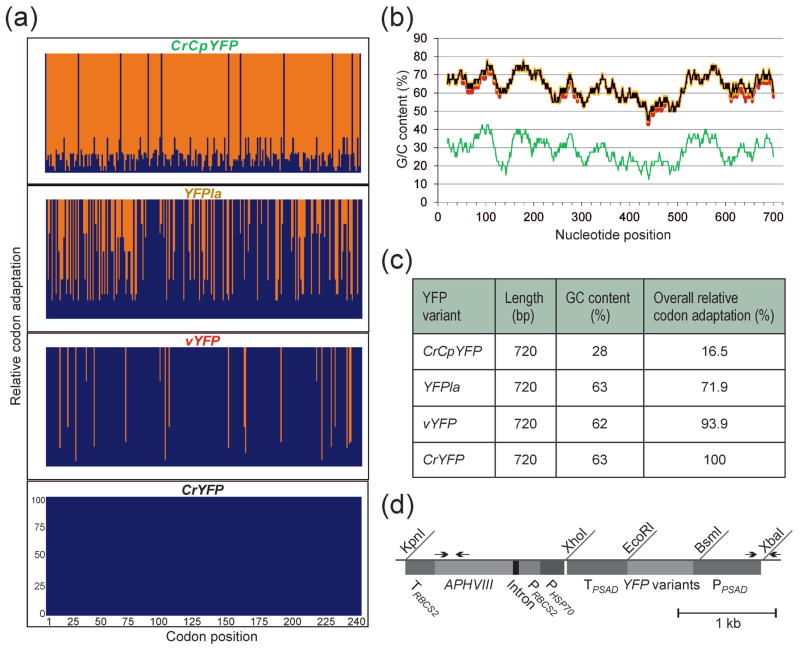
To disentangle the contributions of GC content and codon usage to the efficiency of gene expression in wild-type strains and expression strains of Chlamydomonas reinhardtii, we designed four versions of the gene encoding the yellow fluorescence protein (YFP). All four versions are identical with respect to the amino acid sequence they encode, but differ in their GC content and codon usage (Figure 1a–c). The codon usage was assessed by the relative codon adaptation (RCA), a reference set-based index that has been shown to outperform the codon adaptation index (CAI) as a predictor of gene expression efficiency (Fox and Erill, 2010).
Figure 1.
YFP gene variants and their properties, and physical map of the expression vector used for nuclear transformation.
(a) Relative codon adaptation (RCA) of the different YFP gene variants compared to the nuclear genome of Chlamydomonas reinhardtii. Blue bars indicate the relative adaptation (in %) of each individual codon in the reading frames of the four gene variants. The x-axis indicates the codon numbers within the gene.
(b) GC content (in %) and its distribution over the reading frames of the four YFP gene variants. The values were determined in a sliding window of 40 bp. CrYFP is indicated in black, vYFP in red, YFPla in orange and the A/T-rich variant CrCpYFP in green (cf. colors in panel a).
(c) Overall properties of the different YFP gene variants. All variants have the same size and identical amino acid sequence, but they are highly variable in their GC content and codon usage (RCA).
(d) Physical map of the transformation vector used for expression of the YFP gene variants in Chlamydomonas. The YFP genes were cloned into a PSAD expression cassette (Fischer and Rochaix, 2001) using the restriction sites BsmI and EcoRI. The paromomycin resistance gene APHVIII serves as selectable marker and is driven by a hybrid promoter consisting of fused expression elements from the HSP70A gene (PHSP70) and the RBCS2 gene (PRBCS2). Arrows indicate primers used for PCR analysis of transformed strains. PPSAD: promoter from the PSAD gene; TPSAD: terminator from the PSAD gene.
The Venus variant of YFP (vYFP) was originally developed as a sensitive reporter for mammalian cells (Nagai et al., 2002). It is relatively GC-rich (62%) and has an overall RCA of 93.9%. As a presumably worst-case gene version, we derived a chloroplast codon-optimized sequence from the vYFP gene, subsequently referred to as CrCpYFP (for Chlamydomonas reinhardtii chloroplast YFP gene). Like the plastid genomes of vascular plants, the chloroplast genome of Chlamydomonas is highly AT-rich (Shimada and Sugiura, 1991; Maul et al., 2002) and has a strong bias towards codons with A or T in the third position. Consequently, the CrCpYFP version has a very low GC content (of only 28%) and an extremely low RCA of 16.5% (Figure 1a–c).
We also synthesized a fully codon-optimized YFP gene version (CrYFP) that has the same GC content as the coding sequences in the Chlamydomonas nuclear genome (Merchant et al., 2007) and an RCA of 100%. To distinguish between the impact of GC content and that of codon usage, we took advantage of the presence of family boxes in the genetic code that specify a single amino acid and, therefore, allow synonymous substitutions of NNC triplets with NNG triplets and vice versa (Table 1). For example, the amino acid valine, specified by the four GTN triplets, is preferentially encoded by GTG in Chlamydomonas (RCA=1). The synonymous valine codon GTC has the same GC content but is much less frequently used (RCA=0.33). By systematically introducing such synonymous substitutions into the coding region of CrYFP (and, additionally, exchanging AGC Ser codons by TCG; Table 1), we lowered the RCA of the gene from 100% to 71.9% while retaining the GC content of 63%. This gene variant was named YFPla (for “lowly adapted”; Figure 1a–c).
Table 1.
Codon replacement list for the generation of the YFPla gene variant from CrYFP.
| Selected codon | ||
|---|---|---|
|
|
||
| Amino acid | CrYFP | YFPla |
|
| ||
| Ala | GCC | GCG |
|
| ||
| Gly | GGC | GGG |
|
| ||
| Leu | CTG | CTC |
|
| ||
| Pro | CCC | CCG |
|
| ||
| Arg | CGC | CGG |
|
| ||
| Thr | ACC | ACG |
|
| ||
| Val | GTG | GTC |
|
| ||
| Ser | AGC | TCG |
The four YFP versions were placed into the same expression cassette and the same transformation vector for nuclear transformation (Figure 1d). The constructs were then introduced into the nuclear genome of strains Elow47 (a wild type-like strain and immediate progenitor strain of the expression strains UVM4 and UVM11; Neupert et al., 2009) and UVM11 by glass bead-assisted transformation. This resulted in altogether eight sets of transgenic clones that could be compared with each other with respect to their efficiency of expressing a protein of identical amino acid sequence.
Establishment of an in-gel fluorescence assay for YFP expression in Chlamydomonas
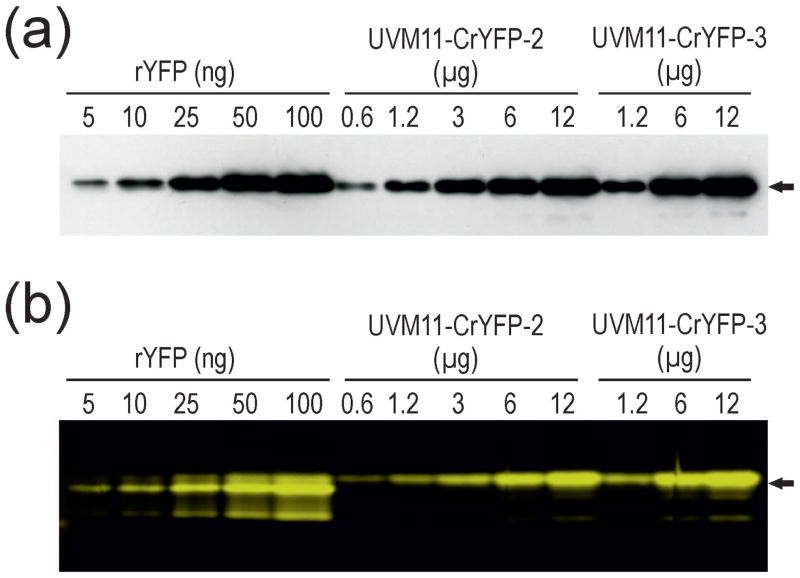
To be able to quantitate the YFP expression levels in the various transgenic strains, we expressed the His-tagged protein in Escherichia coli and purified it to homogeneity. We then sought to develop an immunoblot-independent quantitative assay that would allow us to directly visualize and quantitate YFP accumulation by measuring fluorescence emission. To this end, we selected two strongly YFP-expressing strains identified by a preliminary fluorescence microscopy-based screening of UVM11 transformants generated with the CrYFP construct and optimized electrophoretic separation under non-denaturing conditions to retain full fluorescence of YFP. We then analyzed dilution series of extracted total soluble protein samples by immonoblot assays with an anti-GFP antibody (that recognizes YFP at high sensitivity, because YFP is a mutant version of GFP; Kato et al., 2002) and compared the data with identical dilution series assayed by in-gel fluorescence imaging of protein extracts separated under non-denaturing conditions (Figure 2). No differences were detected between the two methods, indicating that detection of in-gel fluorescence represents a simpler, faster and much less laborious procedure than immunoblotting.
Figure 2.
Semi-quantitative analysis of YFP accumulation in transgenic Chlamydomonas strains. Immunoblot analysis is compared with imaging of in-gel YFP fluorescence.
(a) Immunoblot analysis of dilution series of recombinant His-tagged YFP (rYFP) purified from Escherichia coli and total soluble protein of two Chlamydomonas UVM11 strains transformed with the CrYFP construct (transformants number 2 and 3).
(b) In-gel fluorescence analysis of the same samples analyzed under non-denaturing electrophoresis conditions followed by fluorescence imaging. The gel was scanned using a Typhoon scanner applying the 526 SP Fluorescein filter. The amount of protein loaded in each lane is given in ng (rYFP) or μg (algal strains). Arrows indicate the expected molecular mass of YFP of 26.8 kDa. Note that, according to both methods, YFP accumulation in strain number 3 is greater than 1% of the total soluble protein, the highest expression level of a recombinant protein in Chlamydomonas reported to date.
One of the two strongly YFP-expressing strains, UVM11-CrYFP2, was included as an internal standard in all subsequent analyses of mRNA and protein accumulation in the transgenic algal strains generated with the different YFP gene constructs.
YFP accumulation from the different gene variants in wild-type and expression strains
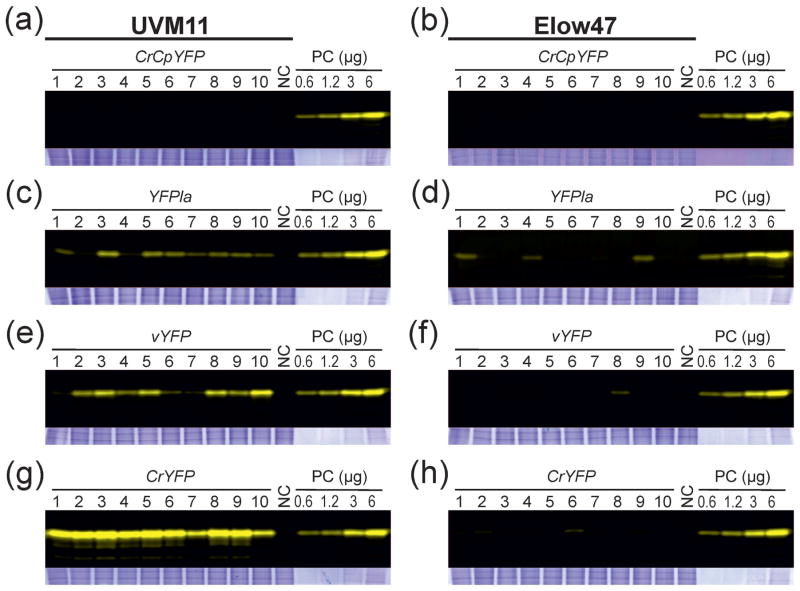
Next, we randomly picked transformants from each of the eight transformation experiments and verified integration of the complete YFP cassette into the nuclear genome by PCR assays (see Experimental Procedures). For each construct and algal strain, ten transgenic algal clones that harbored the complete cassette were then analyzed with respect to YFP accumulation levels using our established in-gel fluorescence assay (Figure 3). The transformed clones of the wild type-like strain Elow47 all showed no or barely detectable YFP expression (Figure 3b,d,f,h). In all the CrCpYFP transformants, YFP expression was below the detection limit (Figure 3b), suggesting that the AT-rich gene with very low RCA is not suitable for expression in Chlamydomonas. For each of the other three gene variants, a few clones displaying weakly fluorescing YFP bands could be identified, indicating that upon integration into a favorable genomic location, expression is possible. However, compared to the standard (PC; strain UVM11-CrYFP2), the YFP accumulation level in these clones was more than tenfold lower (Figure 3d,f,h).
Figure 3.
In-gel fluorescence analysis of YFP expression in algal transformants generated with the different YFP gene variants. A comparison of 10 randomly picked transgenic clones (with verified full-length integration of the YFP cassette; see Experimental Procedures) generated with strains UVM11 (a, c, e, g) and Elow47 (b, d, f, h) is shown. Samples of 12 μg total soluble protein (TSP) were separated by non-denaturing SDS-PAGE. Protein samples isolated from the non-transformed strains was used as negative control (NC). A dilution series (0.6, 1.2, 3 and 6 μg TSP) of a YFP-expressing strain (CrYFP transformant number 2 in strain UVM11; cf. Figure 2) was loaded onto each gel as a positive control (PC). Gels were scanned using a Typhoon scanner applying the 526 SP Fluorescein filter. To control for equal loading, the upper (high molecular weight) part of the gel was stained with Coomassie and is shown below each blot.
(a) YFP accumulation in 10 independent clones of expression strain UVM11 transformed with gene variant CrCpYFP.
(b) YFP accumulation in control strain Elow47 transformed with CrCpYFP.
(c) YFP accumulation in strain UVM11 transformed with YFPla.
(d) YFP accumulation strain Elow47 transformed with YFPla.
(e) YFP accumulation strain UVM11 transformed with vYFP.
(f) YFP accumulation in strain Elow47 transformed with vYFP.
(g) YFP accumulation in strain UVM11 transformed with CrYFP.
(h) YFP accumulation in strain Elow47 transformed with CrYFP.
The CrCpYFP gene also conferred no YFP accumulation in UVM11 (Figure 3a), indicating that, even in the expression strains, highly AT-rich and very lowly codon-adapted transgenes cannot be expressed to appreciable levels. By contrast, YFP accumulation occurred in nearly all transgenic clones generated with the other three YFP gene variants (Figure 3c,e,g), confirming the superior expression properties of the UVM11 strain (Neupert et al., 2009). The fully codon-optimized gene version, CrYFP, yielded the by far highest YFP expression levels, with very little variation between the ten randomly selected transgenic clones (Figure 3g). The YFPla variant (that has an identical GC content but a low RCA; Figure 1a–c) conferred much lower YFP accumulation, again with little variation between clones (Figure 3c). This result strongly suggests that codon usage is an important determinant of gene expression efficiency, independent of the GC content of the coding region. This conclusion was further substantiated by analysis of the vYFP transformants. Although the vYFP gene has a slightly lower GC content than YFPla and CrYFP, it triggered, on average, significantly higher YFP accumulation than the YFPla variant (Figure 3c,e), most probably due to its higher RCA (93.9% vs. 71.9%; Figure 1c).
Correlation between YFP mRNA and protein accumulation
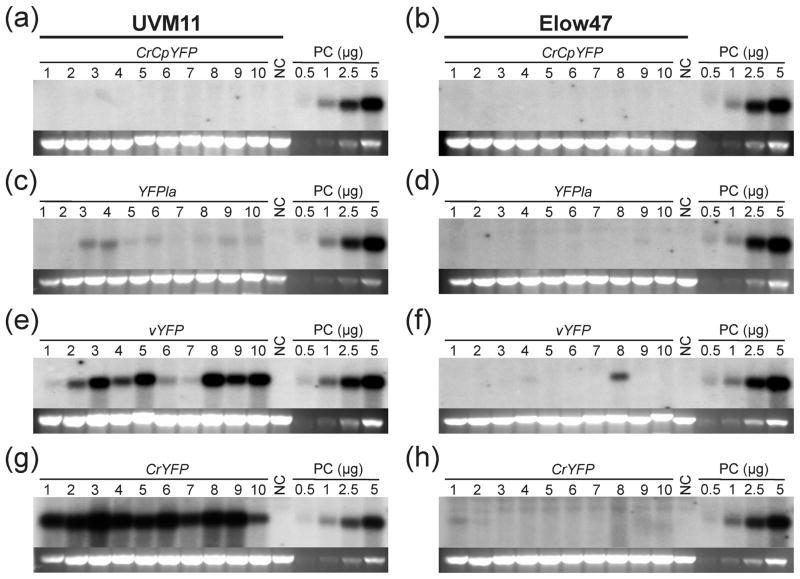
Having seen strong differences in protein accumulation between constructs and algal strains, we next examined whether these expression differences can be explained with different mRNA accumulation levels. We, therefore, performed RNA gel blot analyses to determine YFP mRNA abundance in all 80 transgenic clones (Figure 4).
Figure 4.
RNA gel blot analysis of YFP transcript accumulation in Chlamydomonas strains UVM11 and Elow47 transformed with different YFP gene constructs. Samples of 10 μg total RNA extracted from the same transformed clones that had been analyzed by in-gel fluorescence (cf. Figure 3) were electrophoretically separated in denaturing 1.2% agarose gels. The blots were hybridized to a radiolabeled probe comprising a mixture of the complete coding regions of all four YFP variants at equimolar ratios. RNA samples isolated from the non-transformed strains were used as negative control (NC). A dilution series (0.5, 1.0, 2.5 and 5.0 μg RNA) of a YFP-expressing strain (CrYFP transformant number 2 in strain UVM11; cf. Figure 2) was loaded onto each gel as a positive control (PC) that also allows comparison between blots. The band representing the 25S rRNA (of the cytosolic 80S ribosome) in the ethidium bromide-stained gel prior to blotting is shown below each blot as a loading control.
(a) YFP mRNA accumulation in 10 independent clones of expression strain UVM11 transformed with gene variant CrCpYFP.
(b) YFP mRNA accumulation in control strain Elow47 transformed with CrCpYFP.
(c) YFP mRNA accumulation in strain UVM11 transformed with YFPla.
(d) YFP mRNA accumulation in strain Elow47 transformed with YFPla.
(e) YFP mRNA accumulation strain UVM11 transformed with vYFP.
(f) YFP mRNA accumulation in strain Elow47 transformed with vYFP.
(g) YFP mRNA accumulation in strain UVM11 transformed with CrYFP.
(h) YFP mRNA accumulation in strain Elow47 transformed with CrYFP.
Overall, the analysis of RNA accumulation revealed a strong correlation between RNA abundance and protein abundance (Figures 3 and 4). Most transformed clones generated in strain Elow47 displayed undetectably low YFP mRNA levels. Hybridization signals were detected only in some of the exceptional transformants that showed some YFP accumulation in the in-gel fluorescence assays (Figures 3 and 4). Transformants generated in the UVM11 genetic background accumulated YFP mRNA to levels that were largely proportional to the protein accumulation levels. Interestingly, transformants obtained with YFPla, even though having the same GC content as the fully codon-optimized YFP gene (CrYFP), accumulated only relatively low mRNA levels. By contrast, the vYFP variant, which has a significantly higher RCA than the YFPla variant, gave rise to much higher mRNA (and protein) levels. Given that all transgenes are expressed from identical cassettes in identical transformation vectors, the observed correlation between RCA and mRNA accumulation strongly suggests that translating ribosomes stabilize the mRNA.
UVM11 transformants do not have higher transgene copy numbers
As transgene expression levels also can be influenced by copy number of the transgenes integrated into the genome, we wanted to exclude the possibility that transformation of the UVM11 strain results in higher transgene copy numbers. Since transforming DNA is believed to integrate into preexisting double-strand breaks in the genome (Salomon and Puchta, 1998), this could, for example, be the case if the UVM11 mutation affects DNA repair in the nucleus.
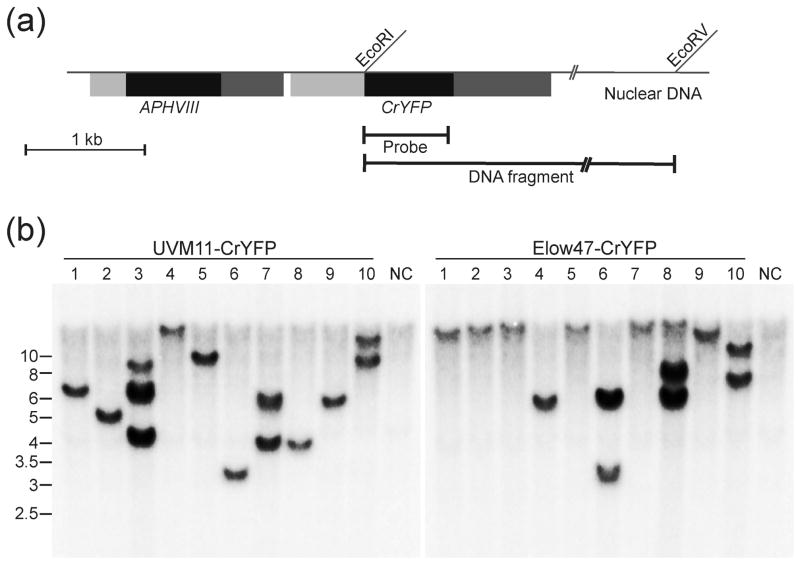
To test for possible copy number effects, we compared ten transformed UVM11 clones generated with the CrYFP construct with ten clones generated in strain Elow47 by restriction fragment length polymorphism (RFLP) analysis. In both strains, most transformants (7 out of 10) produced a single hybridizing band in the Southern blot (Figure 5) and the remaining three transgenic clones produced two or three bands. This result suggests that, in both strains, most transformants represent single-copy transgene insertions. Importantly, no correlation between transgene copy numbers and transgene expression levels was seen (Figures 3–5), indicating that codon usage is a much more important determinant of gene expression than copy number.
Figure 5.
Southern blot analysis of CrYFP transformants of strains UVM11 and Elow47.
(a) Schematic map of the transformation vector integrated into the nuclear genome of Chlamydomonas. The restriction sites used for RFLP analysis by Southern blotting are indicated. The location of the EcoRV site in the flanking nuclear DNA is hypothetical (and variable depending on the integration site in the genome). The hybridization probe (Probe) and the detected hybridizing restriction fragment (DNA fragment) are also indicated. Coding regions are represented as block boxes, promoters as dark grey boxes and terminators as light grey boxes.
(b) Southern blot analysis of ten randomly picked CrYFP transformants of expression strain UVM11 (left panel) and ten randomly picked CrYFP transformants of control strain Elow47 (right panel). Samples of 20 μg total DNA from each transformed clone were digested with the restriction enzymes EcoRI and EcoRV and separated by agarose gel electrophoresis. DNA samples extracted from non-transformed UVM11 and Elow47 were digested with the same restriction enzymes and used as negative control (NC). The entire coding region of CrYFP was used as a template for probe generation. Fragment sizes of the molecular weight marker are given at the left in kb.
Stability of transgene expression
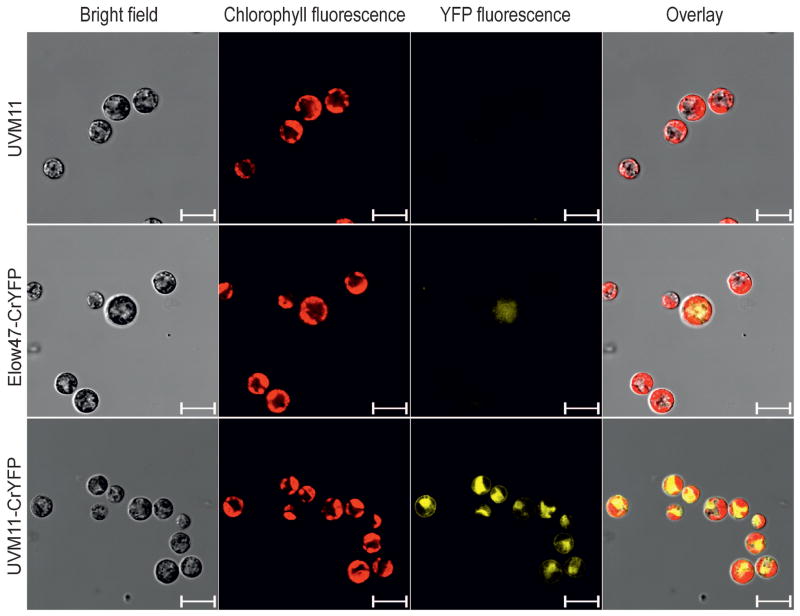
Combined with the UVM11 expression strain, the fully codon optimized YFP variant should provide a new powerful tool enabling cell biological research in Chlamydomonas at high sensitivity, including subcellular localization analyses and in vivo protein-protein interaction studies using FRET, BRET, BiFC and similar techniques. Analysis of transformed algal clones by fluorescence microscopy confirmed that the attained YFP levels are readily detectable. The UVM11 transformants carrying the CrYFP cassette show extremely bright yellow fluorescence in the cytosol (Figure 6). By contrast, YFP fluorescence in those exceptional Elow47 transformants that showed some recombinant protein accumulation was barely detectable (Figure 6).
Figure 6.
Analysis of YFP accumulation in CrYFP transformants of strains UVM11 and Elow47 by confocal laser-scanning microscopy. Bright-field images, chlorophyll fluorescence, YFP fluorescence and the overlay of all images are shown. A 510–535 nm filter was used for detection of YFP fluorescence, and a 630–670 nm filter for detection of chlorophyll fluorescence. The non-transformed strain UVM11 was included as a negative control. Scale bars: 10 μm.
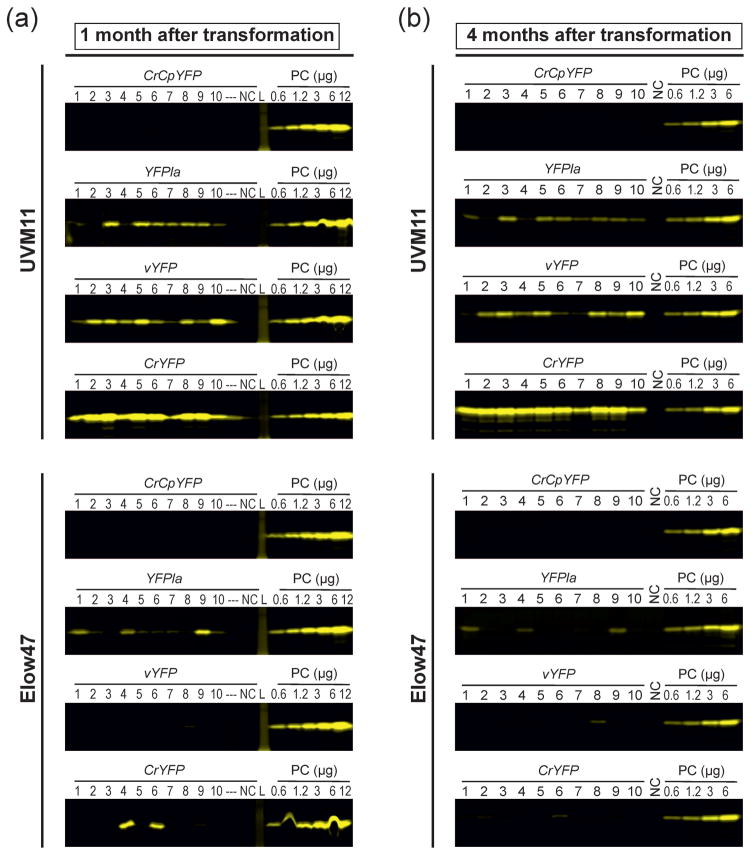
The utility of a transgene expression system also depends on the stability of expression over time. Epigenetic silencing can lead to instability and eventually complete loss of transgene expression (Cerutti et al., 1997). To examine the stability of YFP expression in our transgenic algal strains, all 80 transgenic clones were vegetatively propagated for four months under non-selective conditions and then reinvestigated using our in-gel fluorescence assay (Figure 7). When YFP accumulation levels one month after transformation were compared with those four months after transformation, no significant change was seen in any of the UVM11 transformants. Most YFP-positive Elow47 transformants also maintained expression, although a few exceptions were observed in which the expression level was strongly reduced after 4 months or expression was lost altogether (e.g., CrYFP strains number 4 and 6; Figure 7).
Figure 7.
Stability of transgene expression over time. In-gel fluorescence analyses of YFP expression in the different transformants of strains UVM11 and Elow47 are shown. Samples of 12 μg total soluble protein were separated by non-denaturing SDS-PAGE. Protein samples extracted from non-transformed UVM11 or Elow47 were used as negative control (NC). A dilution series (0.6, 1.2, 3, 6 and 12 μg TSP) of a YFP-expressing strain (CrYFP transformant number 2 in UVM11) was loaded onto each gel as a positive control (PC) and a standard for comparisons between gels. Gels were scanned using a Typhoon scanner and applying the 526 SP Fluorescein filter. L: protein ladder (molecular weight marker); ---: empty lane in the gel.
(a) YFP accumulation in transformants generated with CrCpYFP, YFPla, vYFP and CrYFP in strains UVM11 and Elow47 one month after transformation.
(b) YFP accumulation in the same transformants four months after transformation. The transformed strains were maintained under non-selective conditions. See also Figure 3 which shows the same fluorescence scans and, additionally, loading controls for all gels.
These results suggest that expression of the YFP reporter in strain UVM11 is stable and does not suffer from epigenetic transgene inactivation. Whether or not this is causally related to the mutation in UVM11 that confers the expression trait (in that the same mechanism is responsible for epigenetic transgene inactivation upon transformation and epigenetic transgene silencing over time) remains to be determined.
Chromatin basis of the high transgene expression capacity in strain UVM11
Our previous characterization of the UVM4 and UVM11 strains had suggested a transcriptional cause of the much higher transgene expression capacity in the UVM strains compared to wild-type strains (Neupert et al., 2009). It, therefore, seemed reasonable to assume that the expression differences between the strains are due to epigenetic differences in the chromatin status established at the transgenic locus after incorporation of the transgene into the nuclear genome (Casas-Mollano et al., 2007; Casas-Mollano et al., 2008; Strenkert et al., 2011a; Strenkert et al., 2013). To examine the molecular basis of the differences in transgene expression capacity between the wild type and the expression strain UVM11, chromatin immunoprecipitation (ChIP) experiments were performed. To correct for expression differences between individual transformed clones (as caused, for example, by position effects or the occasional integration of more than one copy of the transgene into the genome; Figure 5), at least 200 independent clones from each transformation experiment were pooled. Thus, each pool of transformants represents both the average expression level conferred by a particular gene variant in a particular strain and the typical chromatin state at the transgenic locus.
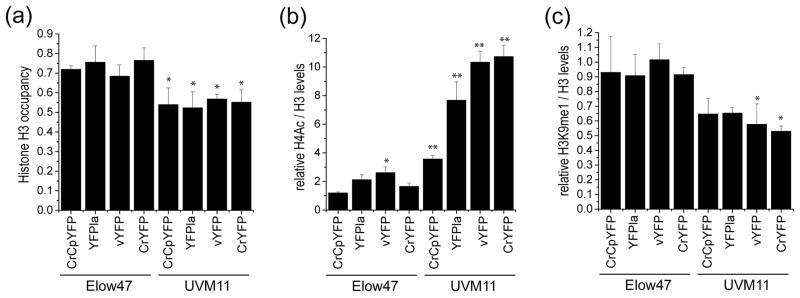
To compare nucleosome occupancy in UVM11 and the corresponding wild type-like strain Elow47, histone H3 abundance at the promoter region of the YFP loci was measured for all YFP variants and in all pools of transformants (Figure 8a). As an internal genomic control, the CYC6 promoter region (of the gene encoding cytochrome c6) was also analyzed (Strenkert et al., 2013). On average, H3 occupancy at the YFP loci was approximately 25% lower in strain UVM11 than in Elow47, suggesting that the promoter region of the transgene is loaded with fewer nucleosomes in the expression strain. Interestingly, this effect was independent of the YFP variant in that also the AT-rich gene variant that expressed poorly in UVM11 (CrCpYFP; Figures 3 and 4) displayed reduced nucleosome loading (Figure 8a).
Figure 8.
Comparative analysis of histone occupancy and histone modification in strains UVM11 and Elow47. Independent pools consisting of at least 200 transformants each strain were analyzed. * p < 0.05; * p < 0.001 (one-way ANOVA, with Holm–Sidak post hoc test).
(a) Comparison of histone H3 occupancy.
(b) Comparison of relative histone H4 acetylation (H4ac). H4ac levels were determined relative to the levels of histone H3.
(c) Comparison of relative histone H3 monomethylation at lysine 9 (H3K9me1). H3K9me1 levels were determined relative to the levels of histone H3.
Next, we investigated two histone modification marks as proxies for active and repressive chromatin, respectively. Acetylation of histone H4 (H4Ac) correlates with active chromatin states that are permissive for gene transcription, whereas monomethylation of lysine residue 9 in histone H3 (H3K9me1) correlates with repressive chromatin states and low transcriptional activity (Strenkert et al., 2013). As internal control regions, the CYC6 promoter was used for H4Ac and the telomere flanking region was analyzed for H3K9me1 (Strenkert et al., 2013). Consistent with the high transgene expression capacity in strain UVM11, the level of H4Ac was significantly higher in all pools of transformants generated with this strain than in the corresponding pools of Elow47 transformants (Figure 8b). Interestingly, there was a pronounced correlation between the level of H4Ac and the expression efficiency of the YFP gene variants in strain UVM11. While the lowly expressing CrCpYFP showed only a moderate increase in H4Ac, the two highly expressing variants vYFP and CrYFP displayed strongly enhanced levels of H4 acetylation (Figure 8b).
When H3K9me1 levels were determined, they were found to be lower in all pools of UVM11 transformants, in line with H3K9me1 being a mark of repressive chromatin. The negative correlation of H3K9me1 levels with the expression strength of the different YFP variants was less pronounced than the (positive) correlation seen with H4Ac (Figure 8c). Taken together, the analysis of histone occupancy and histone modifications strongly suggest that the superior expression capacity of strain UVM11 is due to a reduced sensitivity to heterochromatization of the transgenic locus after transgene integration into the nuclear genome.
Discussion
In this work, we have dissected components involved in efficient nuclear gene expression in the model alga Chlamydomonas. Analysis of different variants of a gene encoding a reporter protein of identical amino acid sequence allowed us to determine the relative contributions of nucleotide composition and codon usage to the efficiency of transgene expression. Comparison of gene variants with similar GC content but different codon usage (YFPla versus vYFP and CrYFP; Figure 1) revealed that optimum codon usage is an important determinant of expression efficiency. We propose that it might even be the key factor and that adjusting the GC content to that of the highly GC-rich nuclear genome of Chlamydomonas improves expression efficiency only indirectly, by having an (on average positive) effect on codon optimality.
Comparative analysis of protein accumulation and mRNA accumulation in transgenic algal strains revealed that both are very strongly correlated (cf. Figures 3 and 4). This observation can have two possible causes: (i) high mRNA accumulation allows for higher translation, or conversely, (ii) high translation rates condition high mRNA stability. Since all transgenes are driven by exactly the same expression cassettes (promoter and terminator), it seems reasonable to propose that translational efficiency directly impacts RNA stability in that translating ribosomes protect the mRNA from endoribonucleolytic degradation and, in this way, increase mRNA stability. This conclusion is in line with recent findings that, in budding yeast, mRNA stability is strongly influenced by codon usage and, hence, translation rates (Presnyak et al., 2015).
In many organisms, codon usage bias is positively correlated with expression level in that highly transcribed genes tend to have a more extreme preference for the most frequently used codons than lowly expressed genes (Wang and Roossinck, 2006). However, this does not affect the strategy for codon usage optimization in synthetic gene design as long as the most frequently occurring triplet is used for each amino acid. When the relative synonymous codon usage (RSCU) values were determined for highly expressed Chlamydomonas genes and compared to the overall RSCU values in all genes, the most frequently used codons were identical for all amino acids, with the sole exception that the AGC serine codon was slightly more frequently used in highly transcribed genes than the otherwise most frequently occurring serine codons UCC and UCG (Wang and Roossinck, 2006).
Our analyses reported here also confirm the vastly superior expression properties of the UVM expression strain over wild-type strains (Figures 3, 4 and 6–8; Neupert et al., 2009). However, it is important to note that transgenes with extremely unfavorable codon usage (i.e., the highly AT-rich gene variant CrCpYFP; Figure 1) cannot even be expressed in our expression strains. It is also noteworthy that all transformed UVM11 clones that contained the complete CrYFP cassette also expressed it to high levels (Figures 3 and 4). This observation indicates that (i) in strain UVM11, the major obstacle to successful transgene expression in Chlamydomonas is overcome by the mutation(s) this expression strain harbors, and (ii) transgene expression levels in UVM11 transformants are not much dependent on the insertion site in the genome. Thus, it seems reasonable to conclude that the gene(s) knocked out in our expression strains encode key regulator(s) of transgene suppression in Chlamydomonas. A drawback of the original UVM4 and UVM11 strains was that they were generated in a cell wall-deficient strain and, therefore, were not motile and could not be used to study flagellar biology or tactic movements. In our ongoing efforts to identify the mutated genes by mapping and genome resequencing, we managed to cross one of the strains to a wild-type strain which allowed us to recover walled and motile progeny that retains the favorable expression properties.
Our ChIP experiments revealed that all transformed clones in the background of the Elow47 strain show very low levels of H4Ac, consistent with efficient transgene silencing in wild-type strains of the alga. By contrast, H4Ac levels were strongly increased in the mutant strain, again pointing to a key role of the mutated gene in chromatin modification and the establishment of a repressive chromatin state at incoming (naked) DNA sequences. This conclusion would be compatible with a role of the transgene inactivation mechanism in defense against invading foreign nucleic acids from viruses and other pathogens, as proposed previously (Casas-Mollano et al., 2008).
All YFP transgenes tested here are driven by identical expression signals and three of the gene variants (YFPla, vYFP and CrYFP; Figure 1) have nearly identical nucleotide composition (GC content). Interestingly, the highly AT-rich gene variant CrCpYFP showed considerably lower levels of histone H4 acetylation (H4Ac; Figure 8). This may indicate that, in addition to secondarily impacting codon usage and, thus, translational efficiency, differences in GC content also have a direct effect on the chromatin established at the transgenic locus after its insertion into the genome. While nucleosome occupancy appears to be independent of the GC content, highly AT-rich regions are preferentially heterochromatinized both in wild-type strains and in our expression strains (Figure 8). This finding points to distinct roles of GC content and codon usage in determining the efficiency of (trans)gene expression. While codon usage acts at the translational level (and secondarily at the level of mRNA stability), the GC content influences chromatin modification and, at least if extremely unfavorable (as in CrCpYFP), triggers transcriptional repression through heterochromatinization. The latter finding points to an interesting influence of the nucleotide composition of the coding region on the chromatin state at the promoter. A possible mechanism how this could be brought about is that low translation triggers mRNA degradation the breakdown products of which then trigger heterochromatinization through a small RNA-mediated silencing mechanism.
Quantitation of recombinant protein accumulation in the expression strains producing the fully codon-optimized CrYFP revealed that YFP accumulated in the transformants to more than 1% of the total soluble protein. To our knowledge, this is the highest expression level of a recombinant protein made from a single-copy nuclear transgene in Chlamydomonas reported to date. Importantly, expression in the UVM strains is stable over time and does not suffer from frequent epigenetic transgene silencing (Figure 7). Thus, the combination of our expression strains with codon optimization of the transgene provides a powerful strategy to maximize foreign protein accumulation and, in this way, will help to remove the arguably most serious bottleneck in basic research and biotechnology with Chlamydomonas. This will considerably expand the range of cell biological methods applicable to Chlamydomonas, facilitate new approaches in functional genomics (Jinkerson and Jonikas, 2015) and also stimulate novel applications in algal biotechnology (Scaife et al., 2015).
In sum, our work reported here has dissected factors involved in successful transgene expression and disentangled the impact of GC content and codon usage. We have demonstrated that, while GC content exerts an influence on local chromatin structure, codon usage is the key parameter determining translational efficiency, and secondarily, also mRNA stability. Finally, our systematic comparison of the expression strain UVM11 with the corresponding wild type-like strain revealed an epigenetic basis of the low transgene expression capacity in wild-type strains of Chlamydomonas which probably operates via the efficient establishment of a repressive chromatin structure at any incoming naked piece of DNA.
Experimental procedures
Algal strains and culture conditions
Chlamydomonas reinhardtii strains Elow47 and UVM11 (Neupert et al., 2009) were used for transformation experiments. Both strains are derived from the cell wall-deficient, arginine auxotrophic strain cw15-302 (cwd mt+ arg7). Elow47 was obtained after co-transformation of cw15 arg- with the CRY1-1 emetine resistance gene and the ARG7 gene providing arginine prototrophy. UVM11 was obtained by UV light-induced mutagenesis of Elow47 and selection for high transgene expression (Neupert et al., 2009). Algal cells were cultivated mixotrophically either in liquid Tris-acetate-phosphate (TAP) medium (Harris, 1989) or on agar-solidified TAP medium at 22°C under continuous light (50–100 μE m−2 s−1).
Construction of transformation vectors
Four different YFP gene variants were generated that are identical in their amino acid sequence, but vary in GC content and relative codon adaptation to the nuclear genome of Chlamydomonas reinhardtii. Plasmid pJR39 (Neupert et al., 2009) was used as transformation vector harbouring the gene for the Venus variant of YFP (vYFP). The CrCpYFP gene variant was synthesized based on the codon usage of the chloroplast genome of Chlamydomonas (GeneArt, Regensburg, Germany). An overlap extension PCR was conducted to introduce the codon substitutions K207A and Y238I. The first PCR was performed with primers PsaD-CrCpYFPfw (5′-GTGCATTCTAGGACCCCACTGCTACTCACAACAAGCCCCATGGTTTCAAAAGGTG AAG-3′; restriction sites underlined, start codon in bold) and CrCpYFP-B-rev2 (5′-GATAAagcTGATTGATATGATAA-3′; nucleotide replacements indicated by lowercase letters) introducing a BsmI restriction site at the 5′ end and replacing an AAA Lys codon by a GCT Ala triplet (K207A). The second PCR was performed with primers CrcpYFP-B-fw2 (5′-CAATCAgctTTATCAAAAGATCC-3′) and CrCpYFP-Brev (5′-TGAATTCTTATTTAatTAATTCATCCATAC-3′; stop codon in bold) to introduce an EcoRI site and replace a TAT Tyr codon by an ATT Ile codon (Y238I). In the final PCR, gel-purified amplicons from PCRs 1 and 2 were used as templates and amplified using primers PsaD-CrCpYFPfw and CrCpYFP-Brev. The final full-length amplicon was cloned into pCR™ 2.1-TOPO™ TA, and after sequence confirmation, cloned into pJR39 as a BsmI/EcoRI restriction fragment, generating vector pRMB8.
The CrYFP gene variant was designed to have 100% relative codon adaptation to the nuclear genome of Chlamydomonas (http://www.kazusa.or.jp/codon/). The gene was synthesized (GeneCust, Dudelange, Luxemburg) with BsmI and EcoRI restriction sites at the 5′ and 3′ ends, respectively. The fragment was cloned into the similarly digested vector pJR39, resulting in plasmid pRMB12.
The YFPla variant was designed to have the same GC content and GC distribution as the fully adapted variant CrYFP, but with a lower level of codon optimization for the Chlamydomonas nuclear genome. To this end, codons of CrYFP were replaced with synonymous codons of the same GC content (but lower relative adaptation) in all possible cases (Table 1). The YFPla gene variant was synthesized (GeneCust) and cloned into pJR39 as BsmI/EcoRI restriction fragment, generating plasmid pRMB13.
To be able to quantify YFP expression levels, recombinant YFP was produced that has the same amino acid sequence as the YFP gene variants. The AT-rich YFP variant was amplified using primers seq-F (5′-GGTTTCCTCGCCGAGCAAGCC-3′) and YFP-His-rv (5′-TCTCGAGTTTAATTAATTCATCCATACC-3′), cloned into vector pCR™ 2.1-TOPO™ TA, and subsequently cloned as NcoI/XhoI fragment into the similarly digested vector pET28a(+) (Novagen), resulting in E. coli expression vector pRMB25.
Transformation of Chlamydomonas reinhardtii
Nuclear transformation of the Elow47 and UVM11 strains of C. reinhardtii was performed using the glass bead method (Kindle, 1990; Neupert et al., 2012). All transformation plasmids were linearized with the restriction enzyme AhdI that has a unique recognition site in the vector backbone. 1 μg of linearized plasmid DNA was used for transformation. Selection was performed on TAP plates supplemented with 10 μg mL−1 paromomycin.
DNA isolation, Southern blot analysis and PCR
Genomic DNA from Chlamydomonas was extracted according to published protocols (Schroda et al., 2001) with the following modifications. The cell pellet was resuspended in 200 μL extraction buffer (2% cetyltrimethylammonium bromide, 100 mM Tris-HCl pH 8.0, 20 mM EDTA pH 8.0, 1.4 M NaCl, 2% β-mercaptoethanol). 200 μL chloroform/isoamyl alcohol (24:1) was added, the suspension was incubated at 65°C for 20 min under shaking (1,400 rpm) for 30 s every 2 min, and then centrifuged at 10°C for 10 min at 12,500 rpm. DNA was precipitated from the supernatant with 0.7 volume isopropanol followed by washing of the pellet with 70% (v/v) ethanol. The air-dried pellet was resuspended in 50–100 μL water with 1 μL (10 mg mL−1) RNase A and incubated for 15 min at 37°C.
To identify transformants with the complete transformation cassette integrated into their genome (i.e., the selectable marker gene and the entire YFP cassette), PCR analyses were performed with primer pairs amplifying sequences upstream and downstream of the YFP gene. Primers PPsaDrev (5′-CGAGCCCTTCGAACAGCCAGGCCG-3′) and M13for (5′-GTAAAACGACGGCCAGT-3′) were used to amplify the 5′ end of the PsaD promoter upstream of the YFP coding region (380 bp amplicon) and primers APHVIII.rev (5′-CCTCAGAAGAACTCGTCCAACAGCC-3′) and APHVIII.fw (5′-GGAGGATCTGGACGAGGAGCGGAAG-3′) were used to amplify the 3′ end of the APHVIII marker gene (Figure 1d). Transformants yielding both PCR products were selected as positive clones.
For Southern blot analysis, samples of 20 μg total DNA were digested with the restriction enzymes EcoRI and EcoRV, separated in 1% agarose gels, and transferred onto Hybond™ N+ nylon membranes (GE Healthcare) by capillary blotting. The CrYFP coding region was amplified, purified by agarose gel electrophoresis and labeled with [α-32P]dCTP by random priming (Megaprime™ DNA labeling system; GE Healthcare). Hybridization took place at 65°C using standard protocols.
RNA extraction and RNA gel blot analysis
Total RNA was isolated from algal cultures grown to cell densities of 5×106 to 1×107 cells mL−1 in TAP medium using the Direct-Zol™ RNA MiniPrep kit (Zymo Research). Samples of 10 μg RNA were separated in formaldehyde-containing 1.2% agarose gels followed by transfer onto Hybond™ N+ nylon membranes by capillary blotting. Hybridization was performed at 65°C using [α-32P]dCTP-labeled probes produced by random priming (Megaprime™ DNA labeling system). A mixture of equimolar ratios of the complete coding regions of all four YFP variants, amplified from their respective plasmids, was used as template for probe generation.
Recombinant protein expression and purification from Escherichia coli
The YFP expression vector pRMB25 was transformed into E. coli strain BL21 Tuner™ DE3 (Novagen). The YFP-expressing bacterial strain was grown in LB medium overnight at 37°C under continuous shaking at 200 rpm. YFP expression was induced by addition of 1 mM IPTG and continued incubation under shaking for 5 h. Cells were collected by centrifugation for 15 min at 3,000g and 4°C. The cell pellet was resuspended in 5 mL lysis buffer [50 mM Tris/HCl pH 8.0, 300 mM NaCl, 10 mM imidazole, 1x EDTA-free cOmplete protease inhibitor (Roche, Mannheim, Germany)] per gram of cells. Subsequently, 1 mg mL−1 lysozyme, 10 mg mL−1 RNase A and 5 mg mL−1 DNase I were added, and the solution was incubated on ice for 30 min. Cells were disrupted by sonication (Sonifier®, W-250 D, G. Heinemann Ultraschall- und Labortechnik) six times at 10% amplitude for 10 s on ice with pauses of 10 s, and three times at 20% amplitude for 20 s with pauses of 10 s. Samples were then centrifuged for 30 min at 8,000g and 4°C. The supernatant was filtered through a 0.4 μm filter and purified by FPLC (ÄKTAexplorer 100 Air, GE Healthcare) under native conditions using the HisTrap HP column 1 ml (GE Healthcare) according to the manufacturer’s protocol. The protein concentration was measured using the Bradford assay, and the purity was assessed by gel electrophoretic protein separation followed by staining with Coomassie Brilliant Blue.
Protein extraction and immunoblot analyses
Total soluble protein was extracted by resuspending Chlamydomonas cell pellets in 200 μl lysis buffer [50 mM HEPES/KOH pH 7.5, 10 mM KAc, 5 mM MgAc, 1 mM EDTA, 1mM DTT, 1x protease inhibitor cocktail cOmplete (Roche)], followed by disruption of cells by sonication (Sonifier®, W-250 D) at 10% amplitude for 15 s on ice. Proteins were quantified using the Bradford assay (Roti®Quant, Roth, Karlsruhe). For immunoblotting, protein samples were denatured at 95°C for 3 min in 1x sample buffer, separated in denaturing 15% SDS–PAA gels and transferred onto PVDF (polyvinylidene difluoride) membranes (Hybond™ P, GE Healthcare) using a Trans-Blot® electrophoretic transfer cell (Bio-Rad) in standard Tris-glycine transfer buffer (25 mM Tris/HCl, 192 mM glycine, pH 8.3). Immunobiochemical protein detection was performed using a 1:20,000 dilution of monoclonal anti-GFP primary antibody (Clontech) and a 1:20,000 dilution of anti-mouse HRP-conjugated secondary antibody (Agrisera). Hybridization signals were visualized by the ECL™ Prime detection system (GE Healthcare).
In-gel fluorescence assays
Total soluble protein was isolated under non-denaturing conditions using the standard extraction protocol described above with the following modifications. DTT was omitted from the lysis buffer, the final SDS concentration was decreased to 1% in the sample buffer, the protein samples were directly loaded onto the PAA gel without prior incubation at 95°C, and the electrophoretic separation was performed at 4 °C. The gels were scanned with a Typhoon™ Trio+ scanner (GE Healthcare) using green laser light for excitation and the 526 SP emission filter.
Microscopy
Fluorescence of YFP was detected in living cells with a confocal laser-scanning microscope (TCS SP5; Leica, Wetzlar) using an argon laser for excitation (514 nm), a 510–535 nm filter for detection of YFP fluorescence, and a 630–720 nm filter for detection of chlorophyll fluorescence.
Chromatin immuno-precipitation (ChIP)
For ChIP, 108 cells were collected and cross-linked with 0.35% formaldehyde at room temperature. The reaction was quenched using a final concentration of 125 mM glycine. Cross-linked material was resuspended in 1.2 mL lysis buffer and fragmented with a Branson sonifier and a MICRO TIP, 1/4 inch, tapered, for volumes of 10–50 mL (Branson Ultrasonics). After sonication, the lysate was precleared of starch and debris by centrifugation for 2 min at 16,100g and 4°C. The lysate was then aliquoted, frozen in liquid nitrogen and stored at −80°C until further use. ChIP was performed as described previously (Strenkert et al., 2011; Strenkert et al., 2011a; Strenkert et al., 2013b). Briefly, aliquots corresponding to 2×107 cells were diluted 1:10 with ChIP buffer (1.1% Triton X-100, 1.2 mM EDTA, 167 mM NaCl, and 16.7 mM Tris/HCl, pH 8) and supplemented with BSA at a final concentration of 100 mg/mL. Antibodies specific for the following epitopes were used: histone H3 (ab1791; Abcam), histone H3K9me1 (ab9045; Abcam) and histone H4Ac (06–866). Antibody-protein-DNA complexes were allowed to form during a 1 h incubation at 4°C, then complexed with 6 mg presoaked protein A Sepharose beads (Sigma-Aldrich) during a 2 h incubation at 4°C, and precipitated by a 20 s centrifugation at 12,000g. Sepharose beads were washed once with washing buffer 1 (0.1% SDS, 1% TritonX-100, and 2 mM EDTA, pH 8) containing 150 mM NaCl, once with washing buffer 1 containing 500 mM NaCl, once with washing buffer 2 (250 mM LiCl, 1% Nonidet P-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8), and twice with TE (1 mM EDTA and 10 mM Tris-Cl, pH 8 ). Protein-DNA complexes were eluted by two incubations for 15 min each at 65°C in elution buffer (1% SDS and 0.1 M NaHCO3), and cross-links were reverted by an overnight incubation at 65°C after addition of NaCl to a final concentration of 0.5 M. Proteins were digested by incubation for 1 h at 55°C after addition of proteinase K (3.5 mg/mL), EDTA (8 mM) and Tris-HCl, pH 8.0 (32 mM). DNA was extracted once with phenol/chloroform/isoamyl alcohol (25:24:1), once with chloroform/isoamyl alcohol (24:1), and precipitated by incubation with 2 volumes of ethanol after addition of 0.3 M sodium acetate pH 5.2 and 10 mg/mL glycogen for at least 3 h at −20°C. Precipitated DNA was collected by a 20 min centrifugation at 4°C and 16,000g, washed with 70% ethanol, air-dried and resuspended in Tris pH 8 (10 mM). Controls ChIP experiments in the absence of an antibody were included in every batch. The amount of precipitated DNA was quantified by qPCR using the amount of input DNA as a reference as well as internal control regions: the CYC6 promoter for histone H3 and H4Ac, and the telomere flanking region for H3K9me1 (Strenkert et al., 2013). The following primers were used: PPSADrev (5′ AGCAAGCCAGGGTTAGGTGTT 3′), PPSADforCrCp (5′ TTTGTGACCATTTACATCACCATCT 3′), PPSADforvYFP (5′ CTTGTGGCCGTTTACGTCG 3′), PPSADforCryfp (5′ CTTGTGGCCGTTCACGTCG 3′), PPSADforYFPla (5′ CTTGTGCCCGTTGACGTCC3′), PCYC6for (5′ ACACGCCCCTCATTCACAGA 3′), PCYC6rev (5′ GCACACGAGACACTCCGAGC 3′), TFR1for (5′ GGGTTTTGCAGGGTTTGGA 3′) and TFR1rev (5′ CCTCATCATGGTCACCCACA 3′).
Acknowledgments
We thank Dr. Marc Lohse for help with codon optimization and Dr. Gal Wittenberg for help with purification of recombinant YFP. This work was supported by the Max Planck Society, a PhD fellowship from the Ministry of Science, Research and Technology of Iran to R. Barahimipour, an EMBO fellowship to D. Strenkert, grants from the Deutsche Forschungsgemeinschaft and the Bundesministerium für Bildung und Forschung to R. Bock, and a grant from the National Institutes of Health (GM42143) to S. Merchant.
Footnotes
The authors declare no conflict of interest.
References
- Alkatib S, Scharff LB, Rogalski M, Fleischmann TT, Matthes A, Seeger S, Schöttler MA, Ruf S, Bock R. The contributions of wobbling and superwobbling to the reading of the genetic code. PLoS Genet. 2012;8:e1003076. doi: 10.1371/journal.pgen.1003076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne A-V, Schwarz C, Schottkowski M, Lidschreiber M, Piotrowski M, Zerges W, Nickelsen J. Reciprocal regulation of protein synthesis and carbon metabolism for thylakoid membrane biogenesis. PLoS Biol. 2013;11:e1001482. doi: 10.1371/journal.pbio.1001482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, van Dijk K, Eisenhart J, Cerutti H. SET3p monomethylates histone H3 on lysine 9 and is required for the silencing of tandemly repeated transgenes in Chlamydomonas. Nucleic Acids Res. 2007;35:939–950. doi: 10.1093/nar/gkl1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, Jeong B-r, Xu J, Moriyama H, Cerutti H. The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc Natl Acad Sci USA. 2008;105:6486–6491. doi: 10.1073/pnas.0711310105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Johnson AM, Gillham NW, Boynton JE. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell. 1997;9:925–945. doi: 10.1105/tpc.9.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent RM, Sharifi MN, Malnoe A, Haglund C, Calderon RH, Wakao S, Niyogi KK. Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 2015;82:337–351. doi: 10.1111/tpj.12806. [DOI] [PubMed] [Google Scholar]
- Fischer N, Rochaix J-D. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genom. 2001;265:888–894. doi: 10.1007/s004380100485. [DOI] [PubMed] [Google Scholar]
- Fox JM, Erill I. Relative codon adaptation: a generic codon bias index for prediction of gene expression. DNA Res. 2010;17:185–196. doi: 10.1093/dnares/dsq012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19:353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M, Hausherr A, Ferbitz L, Schödl T, Heitzer M, Hegemann P. Monitoring dynamic expression of nuclear genes in Chlamydomonas reinhardtii by using a synthetic luciferase reporter gene. Plant Mol Biol. 2004;55:869–881. doi: 10.1007/s11103-004-2150-6. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Ballester D, Pootakham W, Mus F, Yang W, Catalanotti C, Magneschi L, de Montaigu A, Higuera JJ, Prior M, Galván A, Fernandez E, Grossman AR. Reverse genetics in Chlamydomonas: a platform for isolating insertional mutants. BMC Plant Methods. 2011;7:24. doi: 10.1186/1746-4811-7-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas Sourcebook. Academic Press; San Diego, CA: 1989. [Google Scholar]
- Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- Jinkerson RE, Jonikas MC. Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 2015;82:393–412. doi: 10.1111/tpj.12801. [DOI] [PubMed] [Google Scholar]
- Karcher D, Köster D, Schadach A, Klevesath A, Bock R. The Chlamydomonas chloroplast HLP protein is required for nucleoid organization and genome maintenance. Mol Plant. 2009;2:1223–1232. doi: 10.1093/mp/ssp083. [DOI] [PubMed] [Google Scholar]
- Kato N, Pontier D, Lam E. Spectral profiling for the simultaneous observation of four distinct fluorescent proteins and detection of protein-protein interaction via fluorescence resonance energy transfer in tobacco leaf nuclei. Plant Phyisol. 2002;129:931–942. doi: 10.1104/pp.005496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL, Richards KL, Stern DB. Engineering the chloroplast genome: Techniques and capabilities for chloroplast transformation in Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1991;88:1721–1725. doi: 10.1073/pnas.88.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauersen KJ, Berger H, Mussgnug JH, Kruse O. Efficient recombinant protein production and secretion from nuclear transgenes in Chlamydomonas reinhardtii. J Biotechnol. 2013;167:101–110. doi: 10.1016/j.jbiotec.2012.10.010. [DOI] [PubMed] [Google Scholar]
- Lauersen KJ, Kruse O, Mussgnug JH. Targeted expression of nuclear transgenes in Chlamydomonas reinhardtii with a versatile, modular vector toolkit. Appl Microbiol Biotechnol. 2015;99:3491–3503. doi: 10.1007/s00253-014-6354-7. [DOI] [PubMed] [Google Scholar]
- Maul JE, Lilly JW, Cui L, dePamphilis CW, Miller W, Harris EH, Stern DB. The Chlamydomonas reinhardtii plastid chromosome: islands of genes in a sea of repeats. Plant Cell. 2002;14:2659–2679. doi: 10.1105/tpc.006155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Grigoriev IV, Rokhsar DS, Grossman AR Chlamydomonas Annotation Team, JGI Annotation Team. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–251. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Bassett A, Thuenemann E, Schwach F, Karkare S, Ossowski S, Weigel D, Baulcombe D. Highly specific gene silencing by artificial microRNAs in the unicellular alga Chlamydomonas reinhardtii. Plant J. 2009;58:165–174. doi: 10.1111/j.1365-313X.2008.03767.x. [DOI] [PubMed] [Google Scholar]
- Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nature Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Neupert J, Karcher D, Bock R. Generation of Chlamydomonas strains that efficiently express nuclear transgenes. Plant J. 2009;57:1140–1150. doi: 10.1111/j.1365-313X.2008.03746.x. [DOI] [PubMed] [Google Scholar]
- Neupert J, Shao N, Lu Y, Bock R. Genetic transformation of the model green alga Chlamydomonas reinhardtii. Meth Mol Biol. 2012;847:35–47. doi: 10.1007/978-1-61779-558-9_4. [DOI] [PubMed] [Google Scholar]
- Presnyak V, Alhusaini N, Chen YH, Martin S, Morris N, Kline N, Olson S, Weinberg D, Baker KE, Graveley BR, Coller J. Codon optimality is a major determinant of mRNA stability. Cell. 2015;160:1111–1124. doi: 10.1016/j.cell.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala BA, Barrera DJ, Ng J, Plucinak TM, Rosenberg JN, Weeks DP, Oyler GA, Peterson TC, Haerizadeh F, Mayfield SP. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013;74:545–556. doi: 10.1111/tpj.12165. [DOI] [PubMed] [Google Scholar]
- Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc Natl Acad Sci USA. 2006;103:4771–4776. doi: 10.1073/pnas.0509501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohr J, Sarkar N, Balenger S, Jeong B-r, Cerutti H. Tandem inverted repeat system for selection of effective transgenic RNAi strains in Chlamydomonas. Plant J. 2004;40:611–621. doi: 10.1111/j.1365-313X.2004.02227.x. [DOI] [PubMed] [Google Scholar]
- Salomon S, Puchta H. Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 1998;17:6086–6095. doi: 10.1093/emboj/17.20.6086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaife MA, Nguyen GTDT, Rico J, Lambert D, Helliwell KE, Smith AG. Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant J. 2015;82:532–546. doi: 10.1111/tpj.12781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M, Blöcker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Schroda M, Vallon O, Whitelegge JP, Beck CF, Wollman FA. The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing. Plant Cell. 2001;13:2823–2839. doi: 10.1105/tpc.010202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N, Bock R. A codon-optimized luciferase from Gaussia princeps facilitates the in vivo monitoring of gene expression in the model alga Chlamydomonas reinhardtii. Curr Genet. 2008;53:381–388. doi: 10.1007/s00294-008-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao N, Duan GY, Bock R. A mediator of singlet oxygen responses in Chlamydomonas reinhardtii and Arabidopsis identified by a luciferase-based genetic screen in algal cells. Plant Cell. 2013;25:4209–4226. doi: 10.1105/tpc.113.117390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Sugiura M. Fine structural features of the chloroplast genome: comparison of the sequenced chloroplast genomes. Nucleic Acids Res. 1991;19:983–995. doi: 10.1093/nar/19.5.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Sommer F, Schulz-Raffelt M, Schroda M. Transcription factor-dependent chromatin remodeling at heat shock and copper-responsive promoters in Chlamydomonas reinhardtii. Plant Cell. 2011a;23:2285–2301. doi: 10.1105/tpc.111.085266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Schroda M. Protocol: methodology for chromatin immunoprecipitation (ChlP) in Chlamydomonas reinhardtii. Plant Methods. 2011b;7:35. doi: 10.1186/1746-4811-7-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strenkert D, Schmollinger S, Schroda M. Heat shock factor 1 counteracts epigenetic silencing of nuclear transgenes in Chlamydomonas reinhardtii. Nucleic Acids Res. 2013;41:5273–5289. doi: 10.1093/nar/gkt224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terashima M, Freeman ES, Jinkerson RE, Jonikas MC. A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. 2015;81:147–159. doi: 10.1111/tpj.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Roossinck MJ. Comparative analysis of expressed sequences reveals a conserved pattern of optimal codon usage in plants. Plant Mol Biol. 2006;61:699–710. doi: 10.1007/s11103-006-0041-8. [DOI] [PubMed] [Google Scholar]
- Weeks DP. Chlamydomonas: an increasingly powerful model plant cell system. Plant Cell. 1992;4:871–878. [Google Scholar]
- Xia X. A major controversy in codon-anticodon adaptation resolved by a new codon usage index. Genetics. 2015;199:573–579. doi: 10.1534/genetics.114.172106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki T, Miyasaka H, Ohama T. Unstable RNAi effects through epigenetic silencing of an inverted repeat transgene in Chlamydomonas reinhardtii. Genetics. 2008;180:1927–1944. doi: 10.1534/genetics.108.092395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zäuner S, Jochum W, Bigorowski T, Benning C. A cytochrome b5-containing plastid-located fatty acid desaturase from Chlamydomonas reinhardtii. Eukaryot Cell. 2012;11:856–863. doi: 10.1128/EC.00079-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Patena W, Armbruster U, Gang SS, Blum SR, Jonikas MC. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell. 2014;26:1398–1409. doi: 10.1105/tpc.114.124099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Wang W, Bai X, Qi Y. Gene silencing by artificial micro RNAs in Chlamydomonas. Plant J. 2009;58:157–164. doi: 10.1111/j.1365-313X.2008.03758.x. [DOI] [PubMed] [Google Scholar]