Supplemental Digital Content is available in the text.
Key Words: breast cancer, coring method, molecular pathology, tissue microarray
Abstract
Sampling of formalin-fixed paraffin-embedded (FFPE) tissue blocks is a critical initial step in molecular pathology. Image-guided coring (IGC) is a new method for using digital pathology images to guide tissue block coring for molecular analyses. The goal of our study is to evaluate the use of IGC for both tissue-based and nucleic acid–based projects in molecular pathology. First, we used IGC to construct a tissue microarray (TMA); second, we used IGC for FFPE block sampling followed by RNA extraction; and third, we assessed the correlation between nuclear counts quantitated from the IGC images and RNA yields. We used IGC to construct a TMA containing 198 normal and breast cancer cores. Histopathologic analysis showed high accuracy for obtaining tumor and normal breast tissue. Next, we used IGC to obtain normal and tumor breast samples before RNA extraction. We selected a random subset of tumor and normal samples to perform computational image analysis to quantify nuclear density, and we built regression models to estimate RNA yields from nuclear count, age of the block, and core diameter. Number of nuclei and core diameter were the strongest predictors of RNA yields in both normal and tumor tissue. IGC is an effective method for sampling FFPE tissue blocks for TMA construction and nucleic acid extraction. We identify significant associations between quantitative nuclear counts obtained from IGC images and RNA yields, suggesting that the integration of computational image analysis with IGC may be an effective approach for tumor sampling in large-scale molecular studies.
Over the past 2 decades, there have been major advances in the development of methods for the analysis of DNA, RNA, and protein expression from clinical cancer samples. Recently, many of these methods have been optimized for analyses of archival patient tissue samples stored as formalin-fixed paraffin-embedded (FFPE) tissue blocks. The development of tissue microarray (TMA)1 technology, which permits the analysis of hundreds of patient samples on a single glass slide, has further advanced the field by enabling the relatively high-throughput analysis of cancer biomarkers on large patient cohorts. Performing well-powered studies are essential for the validation of tissue-based biomarkers for clinical care. Thus, these technologies will play an important role in enabling the clinical translation of new molecular technologies for personalized medicine.
Although there have been major advances in the development of methods for the high-throughput analysis of DNA, RNA, and protein expression from FFPE tissue samples, there has been relatively less development of technologies to enable rapid, accurate sampling of FFPE tissue blocks for downstream molecular analyses. Three commonly used techniques for FFPE tissue sampling are laser-capture microdissection (LCM),2 macrodissection (MD) of tissue sections from unstained slides,3 and direct coring (DC) of a tissue block using a coring needle. Each of these 3 approaches (LCM, MD, DC) offers a range of strengths and weaknesses, in terms of precision and efficiency, with DC representing the most efficient but least precise method and LCM representing the most precise but least efficient method. Of these 3 approaches, only DC can be used for TMA construction. Given the limitations of existing approaches, there would be value in developing a new method that is both precise and efficient.
In this study, we evaluated a new alternative method, IGC, for TMA construction and for sampling FFPE tissue blocks for large-scale projects in molecular pathology. IGC is a semiautomated method involving the visualization of a digital whole-slide image overlaid on top of an image of the FFPE tissue block to guide the coring of the FFPE block. The goal of IGC is to balance the increased precision of LCM-based and MD-based methods with the increased efficiency of DC.
In this study, we pursued 3 aims; first we constructed a TMA consisting of normal breast tissue and invasive breast cancer to assess the accuracy of IGC for obtaining targeted lesions. Second, we performed a large-scale study that involved IGC followed by RNA extraction and quantification on a total of 377 normal breast tissue samples and 386 tumor samples. Third, we applied computational image analysis to IGC images from 50 normal cases and 50 tumor cases to estimate the number of nuclei, and we assessed the association of number of nuclei, diameter of coring needle, number of cores, and block age with RNA yields in normal and tumor breast tissue.
MATERIALS AND METHODS
IGC
We used the IGC procedure for TMA construction and for sampling FFPE tissue blocks for RNA extraction (Fig. 1). Hematoxylin and eosin (H&E)-stained and FFPE blocks containing both tumor and normal epithelium breast tissue were selected. The H&E-stained slides were scanned by the whole-slide digital Pannoramic scanner (3D-Histech, Budapest, Hungary). Then, pathologists (E.-Y.O, A.H.B) annotated the digital images to indicate 4 circular regions of interest from each whole-slide digital image (1 for tumor in center, 1 for tumor in periphery, and the other 2 for normal epithelium and stroma) using the Pannoramic Viewer Software package (3D-Histech). Circles for the selected areas may range in size from 0.6 to 2 mm in diameter, based on the desired size of the sample core. We selected different color cores to indicate whether the core was targeting tumor or normal tissue. In this study, we used a 1 or 1.5 mm diameter coring needle. The annotated whole-slide digital image was overlaid onto an image of the corresponding FFPE block using the software provided by the TMA-Master (3D-Histech). After selecting 2 reference points, the 2 images (from slide and block) were aligned and a computer-guided robotic arm obtained the selected core from the FFPE block (Fig. 1).
FIGURE 1.

Overview of the image-guided coring (IGC) procedure.
TMA
Study Population
For TMA construction, we evaluated 50 FFPE breast tissue blocks with the diagnosis of invasive ductal carcinoma from the archives of the Department of Pathology at the Beth Israel Deaconess Medical Center under an institutionally approved IRB protocol. For all cases except one, we obtained 4 cores per each case consisting of a central region of invasive tumor, a region of invasive cancer in the periphery of the tumor, normal terminal duct lobular units (TDLUs), and stroma. For the 1 remaining case, we were unable to obtain a core from normal breast, due to insufficient normal tissue.
Image-guided TMA Construction
To construct the TMA using IGC, we designed a TMA map for the recipient block. The TMA machine drilled holes (gap between holes: 800 µm, depth of holes: 7.5 mm) based on the map using a 1.5-mm drill needle. Then we exchanged the drill needle with a core needle and a robotic arm was triggered to obtain the cylindrical tissue core from the “donor” block and transferred it into the prepared hole on the “recipient” block. For morphologic evaluation of each core, we prepared an H&E-stained histologic section from the TMA.
Obtaining Tissue Cores for RNA Extraction
Study Population
We selected 386 tumor and 377 normal adjacent breast tissue samples from the Nurses’ Health Study (NHS) breast tissue archive. This study was approved by the Committee on the Use of Human Subjects in Research at Brigham and Women’s Hospital.
Obtaining Cores
After procuring the annotated tissue using the IGC technique, the obtained cores were inserted into a safe-lock 0.2-mL tube via the robotic arm. We applied IGC to a total of 763 samples from the NHS (386 tumor and 377 normal adjacent breast tissue samples). We obtained a median of 3 cores from tumor cases and 5 cores from normal cases. The core diameter used ranged from 1 to 1.5 mm depending on core availability and sample characteristics. The core length was 7.5 mm. Subsequently, RNA was extracted from the tumor and normal cores using the semiautomated AllPrep FFPE DNA/RNA kit (Qiagen), and the concentration of extracted RNA was measured via the Nano-Drop ND-2000 spectrophotometer. The RNA extraction protocol is presented as Supplementary Information (Supplemental Digital Content 1, http://links.lww.com/AIMM/A82).
Image Analysis
For image analysis, we captured images corresponding to each core taken from a randomly sampled set of 50 samples of normal breast tissue and 50 samples of invasive breast cancer. Overall, we acquired 248 images of normal breast and 149 images of breast cancer, corresponding to one image per core of tissue taken from these 100 cases. For each image, we used image analysis software (version 3.6.1, Definiens Tissue Studio) to perform nuclear identification, segmentation, and counting. For each of the 100 samples, we collected information on: age of the block, size of cores, number of cores, number of nuclei per image, and RNA yield (ng/µL).
Statistical Analyses
To assess the association of block age, core diameter, number of cores, and epithelial nuclear count with RNA yields, we performed rank-based regression, using the Rfit package4 in R to estimate RNA concentration from a nonparametric model considering the number of nuclei per image, number of cores, core diameter, and age of the block.
RESULTS
IGC for TMA Construction
To assess the accuracy of IGC for sampling the desired tissue of interest, we constructed a TMA and reviewed the accuracy of the resulting tissue sampling. The constructed TMA contained a total of 198 tissue cores, each with a diameter of 1.5 mm. The overall accuracy for obtaining cores from the tumor central area was 49/50 (98%), from the tumor periphery was 49/50 (98%), from normal stroma was 49/49 (100%), and from normal TDLUs was 41/49 (84%) (Table 1). These data show that the IGC method obtains very high accuracy for sampling tumor in the center and periphery and normal stroma, with a lower (but still substantial) accuracy for sampling normal TDLUs. In the instances where sampling of normal TDLUs failed, the tissue contained normal stroma only.
TABLE 1.
Accuracy of Obtaining Cores From Tumor in Center, Tumor in Periphery, Normal Epithelium, and Stroma in Tissue Microarray

IGC for Obtaining Tissue Cores for RNA Extraction
Although TMAs enable the targeted in situ analysis of a small set of biomarkers across large patient populations, there are no methods yet available to perform broad profiling assays on TMAs. These assays (e.g., microarrays, next-generation sequencing-based assays) require nucleic acid extraction before downstream molecular analysis. To evaluate the utility of IGC for these types of studies, we next used IGC for tissue sampling, before RNA extraction for a large-scale gene expression microarray study. Following RNA extraction, the median concentration of extracted RNA in 386 tumor samples was 125.56 and was 23.31 ng/μL in 377 normal tissue samples. Thus, we obtained sufficient RNA for downstream molecular analyses for the vast majority of tumor samples (RNA concentration of at least 20 ng/μL for 89% of tumor samples), and for just over half of the normal breast tissue samples (RNA concentration of at least 20 ng/μL for 55% of normal samples). These data show that tissue sampling with IGC is highly effective for tumor samples, and although the method was effective in most cases of normal samples, a significant number of cases (∼45%) still failed to obtain adequate RNA yields, suggesting that additional tissue sampling and/or extraction method optimization may be warranted for the normal breast tissue FFPE samples. The only moderate level of success in obtaining at least 20 ng/µL of RNA from the normal samples reflects the challenge of obtaining adequate RNA from normal tissue in FFPE, which are far less cellular than neoplastic areas of tissue.
IGC for Estimating RNA Yields From Quantitative Characteristics of the Histopathologic Image
In contrast to methods which do not involve digital acquisition of histopathologic images before FFPE tissue block sampling, a potential advantage of the IGC method is that it may enable the use of computational image analysis of histopathologic images to inform tumor sampling for downstream molecular analysis. For example, computational image analysis of nuclear density may predict RNA yields and potentially could be used to guide the extent of sampling.
To begin to test this approach, we used computational image analysis to quantify nuclear density in IGC images from a set of 50 cases of normal breast and 50 cases of breast cancer. For each case, we measured block age, number of cores acquired, core diameter, and computed the sum of the number of nuclei per IGC image per case (descriptive statistics of the study set presented in Table 2).
TABLE 2.
Descriptive Statistics of Study Set for Correlating Block Characteristics With RNA Yield

In normal and tumor tissue, the total number of nuclei was significantly associated with RNA yield in both normal and tumor tissues [Spearman ρ=0.52 (P=0.0002) in normal tissue; and Spearman ρ=0.56 (P=3.66e−5) in tumor tissues; Fig. 2]. To determine the association of number of nuclei with RNA yields, when adjusting for block age, coring needle diameter, and number of cores obtained, we performed rank-based linear regression.4
FIGURE 2.

Scatterplot of number of nuclei and RNA yields in normal (top) and breast cancer tissue (bottom). Each point represents a case [N=50 normal (top), N=50 breast cancer (bottom)]. The line in each scatterplot is the rank-based linear regression line of number of nuclei to RNA concentration.
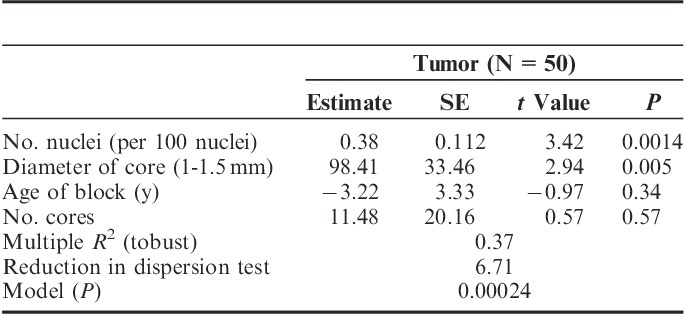
In the regression analyses, we find a significant association of number of nuclei with RNA yield in both the normal and tumor tissues, when adjusting for age of block, number of cores, and core diameter (Tables 3 and 4). Core diameter was also independently associated with RNA yields in both normal and tumor tissues. In both normal and tumor tissue, no significant independent predictive information was provided by the age of the block and number of cores. These data suggest that quantification of number of nuclei using computational image analysis can provide additional independent information for predicting RNA yields, beyond that provided by number of cores, size of cores, and age of block.
TABLE 3.
Multivariate Rank-based Regression of Coring Parameters and RNA Concentration in Normal Breast Tissue Samples

TABLE 4.
Multivariate Rank-based Regression of Coring Parameters and RNA Concentration in Breast Cancer Samples
DISCUSSION
In this study, we evaluated the performance of IGC, which is a new approach for sampling FFPE tissue for large-scale studies in molecular pathology. We evaluated the efficacy of IGC for TMA construction and for tissue sampling before RNA extraction. In the present study, histopathologic evaluation of H&E-stained TMAs constructed by IGC showed that the targeted area was sampled in 98% of tumor cores and in 91.8% of the cores from the normal breast tissue, including an accuracy of 84% for the normal TDLU and 100% for normal stroma. In a previous study by Collins et al,5 the accuracy of sampling normal TDLUs for a TMA using a conventional manual coring approach (core size=0.6 mm) was reported to be 50%. Taken together, these data suggest that IGC shows higher accuracy for sampling of tumor and normal stroma, and that IGC improves on manual DC-based approaches for sampling of normal TDLUs, likely due to both the increased core diameter as well as the increased precision due to using the histopathologic image to directly guide the coring.
In the second part of our study, we used IGC in the setting of a large-scale project within the NHS to sample FFPE blocks before RNA extraction. Using the IGC sampling approach and applying a standard protocol for RNA extraction from FFPE tissue, we showed moderate success rates for obtaining adequate RNA yields from tumor tissue (>20 ng/µL in 88.8% of cases), with somewhat less success in the normal samples (>20 ng/µL in only 55.4% of cases).
Little is currently known of the factors that predict RNA yields from FFPE pathologic specimens. A potential use of IGC in this regard could be to enable predictions of RNA yield from pathologic attributes of the specimen, including nuclear count estimated directly from the histopathologic image of the region of tissue undergoing coring. In this study, we performed nonparametric regression to assess whether nuclear count on IGC images as determined by computational image analysis was predictive of RNA yields. In both tumor and normal tissue, we find that number of nuclei is a significant predictor of RNA yield, even when adjusting for other factors (number of cores, core diameter, and age of the block). These data suggest that IGC coupled with computational image analysis may enable more efficient use of valuable and limited tissue specimens by enabling the construction of statistical models to predict RNA yields based on quantitative features of the histopathologic specimen. These models could then be used to inform decisions as to the extent of sampling required for a given molecular assay.
Supplementary Material
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website, www.appliedimmunohist.com.
ACKNOWLEDGMENTS
The authors thank the participants and staff of the Nurses’ Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY.
Footnotes
Supported by the National Library of Medicine of the National Institutes of Health under Award Number K22LM011931, the National Cancer Institute of the National Institutes of Health by the NIH NCI U19/GAME-ON DRIVE (CA148065) initiative, NIH P01 CA87969, NIH UM1 CA186107, and SPORE grant P50CA168504 Career Development Award. The content is solely the responsibility of the authors and does not necessarily represent the views of the National Institutes of Health.
A.H.B. is on the Scientific Advisory Board of Definiens. The remaining authors declare no conflict of interest.
REFERENCES
- 1.Kallioniemi OP, Wagner U, Kononen J, et al. Tissue microarray technology for high-throughput molecular profiling of cancer. Hum Mol Genet. 2001;10:657–662. [DOI] [PubMed] [Google Scholar]
- 2.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science. 1996;274:998–1001. [DOI] [PubMed] [Google Scholar]
- 3.Kim HK, Kim J, Korolevich S, et al. Distinctions in gastric cancer gene expression signatures derived from laser capture microdissection versus histologic macrodissection. BMC Med Genomics. 2011;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kloke J, McKean J. Rfit: rank-based estimation for linear models. 2012. Available at: http://rjournal.github.io/archive/2012-2/RJournal_2012-2_Kloke+McKean.pdf. Accessed May 12, 2015.
- 5.Collins LC, Wang Y, Connolly JL, et al. Potential role of tissue microarrays for the study of biomarker expression in benign breast disease and normal breast tissue. Appl Immunohistochem Mol Morphol. 2009;17:438–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Digital Content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal's Website, www.appliedimmunohist.com.


