Summary
In an RNAi library screen for loss of kinetoplast DNA (kDNA), we identified an uncharacterized Trypanosoma brucei protein, named TbLOK1, required for maintenance of mitochondrial shape and function. We found the TbLOK1 protein located in discrete patches in the mitochondrial outer membrane. Knockdown of TbLOK1 in procyclic trypanosomes caused the highly interconnected mitochondrial structure to collapse, forming an unbranched tubule remarkably similar to the streamlined organelle seen in the bloodstream form. Following RNAi, defects in mitochondrial respiration, inner membrane potential, and mitochondrial transcription were observed. At later times following TbLOK1 depletion, kDNA was lost and a more drastic alteration in mitochondrial structure was found. Our results demonstrate the close relationship between organelle structure and function in trypanosomes.
Keywords: trypanosomes, DNA replication, kDNA loss, mitochondrial shape and morphology
Introduction
Trypanosoma brucei is a single-celled parasite that causes devastating diseases in sub-Saharan Africa, such as sleeping sickness or Human African Trypanosomiasis, and a cattle disorder called nagana. African trypanosomes are transmitted by the tsetse fly, with one stage of its life cycle, the procyclic form (PCF), living in the midgut of the insect. After migration to the salivary gland, a fly bite transmits the metacyclic form into the mammalian host, where it differentiates into still another life cycle stage called the bloodstream form (BSF). T. brucei has a single mitochondrion with distinct morphologies in its various stages, reflecting different metabolic activities (Priest & Hajduk, 1994, Vickerman, 1985). Two of the T. brucei life cycle stages, the PCF and the BSF, are readily cultured in the lab and studies have shown that PCFs largely depend upon mitochondrial oxidative phosphorylation for their ATP production. Consequently, PCFs contain an elaborated organelle with many branches and interconnections, as well as abundant cristae. On the other hand, BSFs get their energy from glucose in the mammalian blood and make most of their ATP by substrate level phosphorylation. BSFs lack a functional electron transport chain, and therefore contain a much simpler mitochondrion in the form of a single, unbranched tubule with fewer cristae.
The mitochondrial genome, known as kinetoplast DNA or kDNA, is a complex network of interlocked DNA circles, including several thousand minicircles and a few dozen maxicircles (Liu et al., 2005, Lukes et al., 2005, Shlomai, 2004). Each cell has a single kDNA network that is condensed into a disk-shaped structure in the mitochondrial matrix, adjacent to the flagellar basal body. Maxicircles, like the mitochondrial DNAs of most eukaryotes, encode subunits of the respiratory complexes. However, in T. brucei most maxicircle transcripts must be extensively edited to form functional open reading frames (Stuart et al., 2005)]. Guide RNAs, the templates for editing, are primarily encoded on a complex array of minicircles. Thus, it is critical that during cell division each daughter cell receives not only maxicircles, but also a complete repertoire of minicircles. To help ensure proper partitioning, the kDNA network is linked to the basal body and flagellum by a transmembrane filament system known as the tripartite attachment complex (TAC)(Ogbadoyi et al., 2003). After DNA replication, division and separation of the flagellar basal body is thought to drag the attached sister networks to daughter cells.
Measuring kDNA loss, we screened an RNAi library for genes involved in kDNA replication and segregation (Englund et al., 2005, Motyka et al., 2006, Zhao et al., 2008). We identified a gene essential for PCF growth that we call TbLOK1 (for loss of kDNA). While RNAi-mediated knockdown of the TbLOK1 protein did cause kinetoplast loss, we found that the disappearance of the kDNA occurred well after cell division ceased, arguing that the requirement of TbLOK1 for kDNA maintenance is indirect. In contrast, the earliest defects following induction of TbLOK1 RNAi were a dramatic alteration of mitochondrial structure and a concomitant loss of organelle function. Our results raise the intriguing possibility that the TbLOK1 protein plays a direct role in the control of mitochondrial shape and indicates that normal mitochondrial structure is critical for full activity of this crucial organelle.
Results
Identification of TbLOK1
During an RNAi library screen of PCF trypanosomes for kDNA replication proteins, we identified a previously unstudied gene (GeneDB: Tb09.211.1940) required for kinetoplast maintenance. We named this new gene TbLOK1 for loss of kDNA. TbLOK1 encodes a 19 kDa (168 amino acid residues) basic protein (pI 10.0) with no obvious domains, motifs, or subcellular localization signals, other than two regions (residues 21 to 42 and 77 to 99) that are predicted to be transmembrane segments (although they are of moderate hydrophobicity). Homologs to the TbLOK1 protein exist in other kinetoplastids, including Leishmania major, L. infantum and T. cruzi (Fig. S1), but we could not identify counterparts in more distantly-related organisms.
The TbLOK1 protein is essential for trypanosome growth
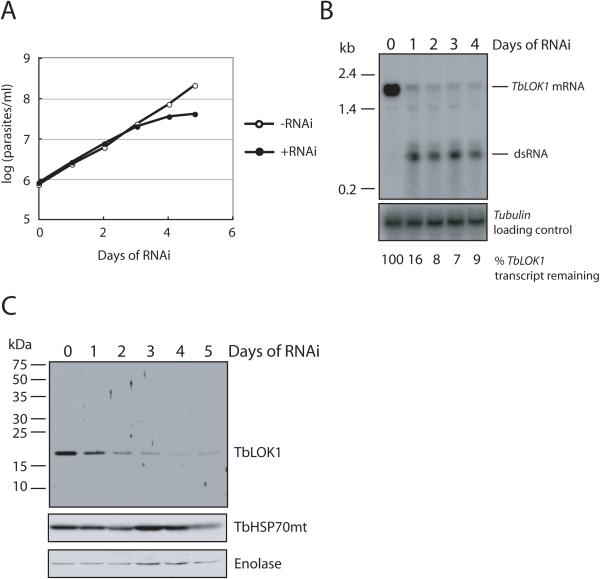
RNAi-mediated depletion of the TbLOK1 protein had a strong effect on trypanosome viability (Fig. 1A). Following induction of RNAi, cells began to slow in their growth after 3 days, and completely stopped dividing by day 4. The decrease in the TbLOK1 protein and mRNA levels closely paralleled the cell growth defect. The 19 kDa TbLOK1 protein was greatly reduced by day 3 and virtually undetectable by day 4 (Fig. 1B), while the levels of mitochondrial HSP70 (Effron et al., 1993, Guler et al., 2008) and the cytosolic enolase protein (Bakshi & Shapiro, 2004) did not change. Northern blots showed that RNAi significantly decreased TbLOK1 mRNA by day 1, with maximal depletion (> 85%) occurring by day 4 (Fig. 1C).
Figure 1. The TbLOK1 protein is essential for trypanosome growth.
A. Knockdown of TbLOK1 stops cell division. Cells were grown without (−RNAi) or with (+RNAi) induction of TbLOK1 RNAi, and then parasites were counted. Values on the Y-axis are the measured cell number times the dilution factor. B. TbLOK1 mRNA levels decrease following RNAi induction. At the indicated times, total RNA was electrophoresed, blotted and probed with TbLOK1 or the tubulin gene (load control). RNA from equivalent number of cells was loaded in each lane. C. The TbLOK1 protein levels drop following RNAi. At the indicated times, cells were isolated and extracted proteins were electrophoresed and western blotted with antibodies to TbLOK1, TbHSP70mt and enolase (a load control). Proteins from equivalent numbers of cells were loaded in each lane.
Knockdown of the TbLOK1 protein causes a striking defect in kDNA segregation, but a delayed and incomplete loss of kDNA
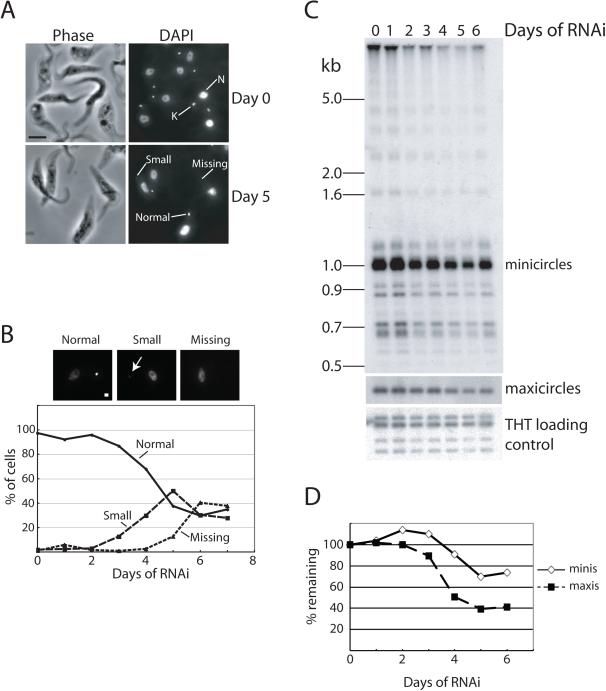
Although we identified TbLOK1 in a screen for trypanosomes that lose their kDNA, we found that the kDNA loss following RNAi was modest compared to the growth defect (Fig. 2). After 4 days of TbLOK1 RNAi, when cells had completely stopped dividing, DAPI (4′, 6-diamidino-2-phenylindole) staining showed that about 70% of trypanosomes still had normal amounts of kDNA. Even after 6 days of RNAi, about a third of the cells contained normal-sized kDNA networks, another third had a small kinetoplast, and only a third of the cells lacked any detectable kDNA (Fig. 2A, B). Southern blot analysis confirmed our microscope-based assays, showing an incomplete loss of both minicircles and maxicircles (Fig. 2C). By day 5 of RNAi, minicircle content decreased only ~ 30%, while maxicircles dropped by ~ 60%. Moreover, free minicircle analysis failed to show significant accumulation of any minicircle intermediates that would indicate a specific block in replication (Fig. S2). With TbLOK1 RNAi, loss of kDNA occurred after the inhibition of growth. In contrast, loss of kDNA preceded or was simultaneous with the cessation of cell division when known replication proteins, such as mitochondrial topoisomerase II (Wang & Englund, 2001) and DNA ligase kα (Downey et al., 2005), were depleted.
Figure 2. Knockdown of the TbLOK1 protein causes an incomplete loss of kDNA.
A. TbLOK1-depleted cells lose kDNA. Uninduced cells (day 0) and cells induced with tetracycline to produce double-stranded RNA corresponding to TbLOK1 for 5 days were DAPI-stained and examined by phase contrast (left panels) and fluorescence (right panels) microscopy. K, kinetoplast. N, nucleus. B. Kinetics of kDNA loss. At indicated time of TbLOK1 RNAi, trypanosomes were fixed, stained with DAPI, and at least 100 cells at each time point were scored for either normal, small or absent kDNA. A typical fluorescence image of each category is shown above graph. White arrow points to a small kinetoplast. C. Minicircle and maxicircles decrease following LOK1 RNAi. Total trypanosome DNA was isolated at the indicated times following RNAi, digested with HindIII and XbaI, electrophoresed and Southern blotted. The nuclear-encoded trypanosome hexose transporter (THT) was the loading control.
In contrast to its moderate effect on kDNA loss, we found that TbLOK1 RNAi caused a stronger defect in normal, symmetrical kDNA segregation (Fig. 3A). After 4 days of TbLOK1 depletion, about 85% of trypanosomes with two kDNAs divided their kinetoplasts asymmetrically, so that the sister kDNA networks differed significantly in size (Fig. 3B). Using fluorescence in situ hybridization (FISH) with specific minicircle and maxicircle probes, we found that both minicircles and maxicircles divided asymmetrically (Fig. 3D). Also indicative of a problem in kDNA separation, FISH showed that TbLOK1 RNAi produced an increase in the number of long ‘threads’ of maxicircles connecting the two segregated minicircle networks (Fig. 3C & D). Recent studies have shown that maxicircle segregation is a very late step in the kinetoplast duplication cycle (Gluenz et al., 2011, Gluenz et al., 2007), and although ‘threads’ are occasionally seen in normal trypanosomes, their accumulation in TbLOK1 RNAi cells indicates difficulty in partitioning maxicircles between the progeny kinetoplasts.
Figure 3. TbLOK1 depletion leads to asymmetric kDNA division.
A. Examples of cells undergoing symmetric (sym, left panels) or asymmetric (asym, right panels) kDNA division. Arrows indicate kDNA. B. Quantitation of cells undergoing symmetric (gray bars) or asymmetric (black bars) kDNA division during RNAi-mediated knockdown of TbLOK1. C. Quantitation of the number of cells with two kDNAs, in which the two kinetoplasts are connected by a maxicircle thread or nabelschnur (Gluenz et al., 2011, Gluenz et al., 2007) after RNAi of TbLOK1 as visualized by FISH with specific probes. D. Examples of FISH analysis with minicircle and maxicircle probes in cells containing maxicircle threads after knockdown of TbLOK1. Arrows indicate thread structures. K, kinetoplast; N, nucleus.
Loss of the TbLOK1 protein leads to a rapid collapse of mitochondrial shape
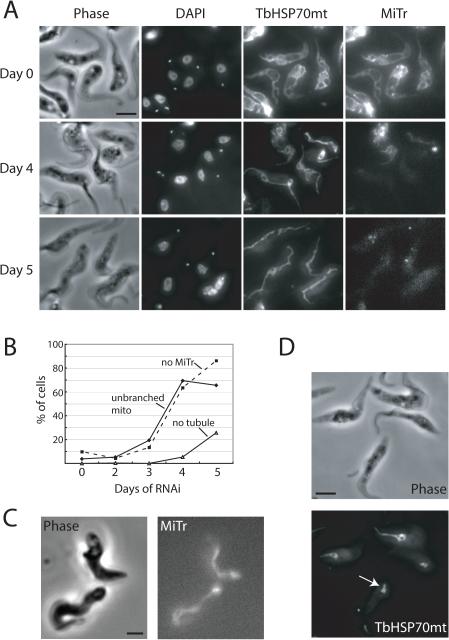
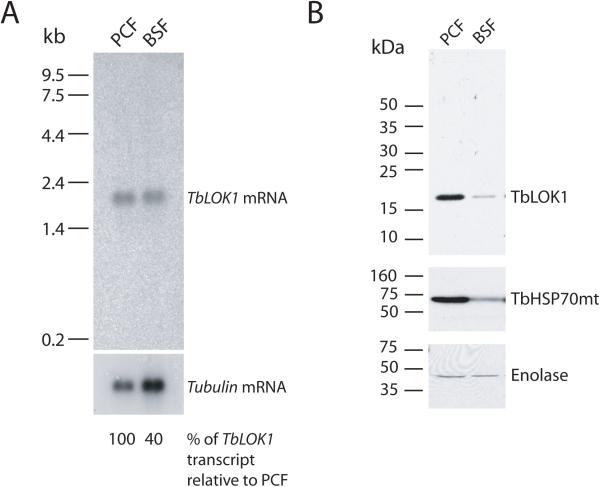
Coincident with the cell growth defect, knockdown of the TbLOK1 protein caused a striking change in mitochondrial morphology. Using the matrix-localized TbHSP70mt protein to track mitochondrial shape during our immunofluorescence studies, we found the typical branched, interconnected organization of tubular-shaped organelles in cells prior to RNAi induction (Fig. 4A, day 0). The first noticeable change following TbLOK1 knockdown was a dramatic loss of the mitochondrial branches and connections. At day 3, the mitochondrion in ~ 20% of the cells appeared to be a thick, unbranched tubule, and this number rose to ~ 70% by day 4 and 5 (Fig. 4A & B). We note that many of the cells at day 4 and 5 still have kDNA, indicating that loss of the kinetoplast is not the cause of the morphology defect (more on this below). The simple tubule seen in PCF cells depleted for TbLOK1 is strikingly similar to the mitochondrion found in BSF trypanosomes (Fig. 4C). Interestingly, we found that levels of the TbLOK1 protein in BSFs were reduced compared to PCF trypanosomes; however, as TbHSP70mt is also less abundant in bloodstream form cells, this difference could be due to reduced mitochondrial volume in this life cycle stage (Fig. 5).
Figure 4. TbLOK1 is necessary for normal mitochondrial shape.
A. TbLOK1 RNAi causes loss of mitochondrial structure. Uninduced cells (day 0), or those induced for TbLOK1 RNAi for 4 or 5 days were stained with MitoTracker Red (MiTr) and DAPI, decorated with antibodies to TbHSP70mt, and then examined by phase and fluorescence microscopy. B. Kinetics of mitochondrial changes following TbLOK1 depletion. Images from RNAi-treated cells (~100 cells from each time point) were examined as in A, and those with abnormal mitochondria (either unbranched organelles, or those that lost tubular mitochondria shape) were counted. The dotted line on the graph indicates the number of cells whose mitochondrion failed to stain with MitoTracker. C. Image of wild-type (strain 927) BSF cells stained with MitoTracker. D. Collapse of mitochondrial structure after prolonged TbLOK1 knockdown. After 5 days of TbLOK1 RNAi, trypanosomes were stained using antibodies to TbHSP70mt and imaged. The complete loss of a tubular-shaped mitochondrion in one cell is shown (white arrow).
Figure 5. Levels of TbLOK1 in different life cycle stages.
A. Transcript abundance in wild-type 927 PCF and 427 BSF cells. RNA from equivalent number of cells was loaded in each lane, and tubulin was used as a loading control for Northern blots. B. Western blot comparing levels of TbLOK1, TbHSP70mt and enolase in PCF and BSF cells. Proteins from equivalent numbers of cells were loaded in each lane.
At later times following TbLOK1 knockdown, we observed a more severe disruption in mitochondrial shape. ~ 5% of the cells at day 4 and ~ 26% on day 5 had lost all extended tubular structure, and instead only a small mitochondrial ‘blob’ (possibly a short tubule) persisted, usually at the posterior end of the cell (Fig. 4D). Our results therefore suggest that RNAi-mediated reduction in the TbLOK1 protein causes a progressive change in mitochondrial shape. First, coinciding with the cessation of cell growth is the collapse of mitochondrial connections and branches. Later, at about the time cells begin to lose their kDNA, the organelle becomes much more disorganized and loses nearly all of its tubular structure.
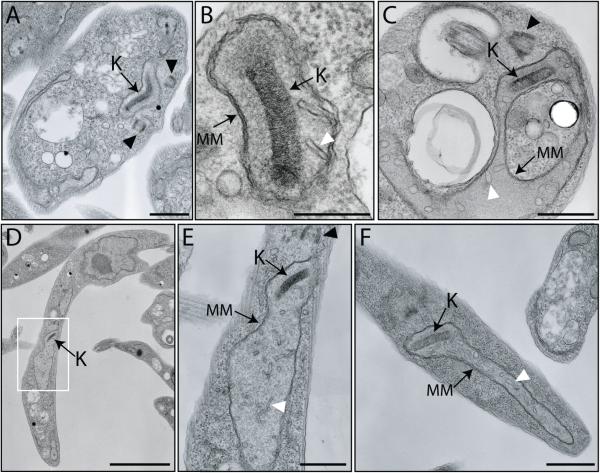
While the induction of TbLOK1 RNAi clearly disrupts mitochondrial structure, electron microscopy revealed that the internal architecture of the organelle was surprisingly unaffected (Fig. 6). After 4 days of RNAi, both the outer and inner membranes (Fig. 6CF, labeled MM) were indistinguishable from those in uninduced cells (Fig. 6A & B). Similarly, knockdown of TbLOK1 did not significantly disturb the number and organization of cristae, even though the average diameter of the mitochondrial tubule appeared greater in cells lacking the TbLOK1 protein. Only in a few cases did cristae appear slightly enlarged (white arrowhead in Fig. 6C, F). While RNAi eventually causes the loss of the kinetoplast, the kDNA disc and its connection to the basal body appeared normal in most cells at day 4. Our EM analyses support our immunofluorescence studies, indicating that the immediate consequence of the depletion of the TbLOK1 protein is the conversion of the mitochondrion into a thicker and less branched tubule.
Figure 6. Ultrastructure of the mitochondrion is normal in TbLOK1-depleted cells.
Uninduced cells (A, B) or those depleted of TbLOK1 for 4 days (C, D, F) were fixed, embedded in resin, and then thin sections were examined by EM. E. Enlargement of the area corresponding to the white box in D. K, kinetoplast; MM, mitochondrial membranes; black arrowheads, basal bodies; white arrowheads, cristae. Scale bar for A and F, 800 nm; for B, 300 nm; for C and E, 600 nm; for D, 3.0 μm
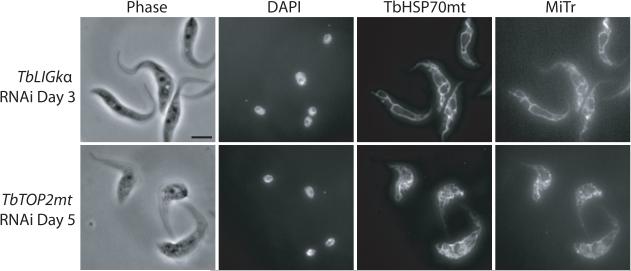
Although knockdown of TbLOK1 caused some cells to lose their kinetoplast, we found that the collapse of mitochondrial shape clearly preceded significant kDNA loss. Therefore, we reasoned that it was more likely that the change in mitochondrial structure promoted kDNA loss, rather than loss of the kinetoplast causing alterations in organelle shape. To test this idea, we forced cells to lose their kDNA by knocking down two different mitochondrial enzymes with known roles in kDNA replication—DNA ligase kα (TbLIGkα) (Downey et al., 2005) and topoisomerase II (TbTOP2mt) (Wang & Englund, 2001)—and examined the effect on mitochondrial morphology (Fig. 7). Although both enzymes are essential for cell viability, RNAi of TbTOP2mt stops cell division after 5 days, while TbLIGkα depletion stops growth in 2-3 days. Therefore, we evaluated kDNA loss and mitochondrial structure at the time of growth arrest in each of the lines. Confirming previous studies, DAPI staining showed that most cells knocked down for TbLIGkα and TbTOP2mt had lost their kinetoplast at the point when cells stopped dividing. Despite the absence of kDNA, we found that mitochondrial shape was undisturbed. Thus, the alteration of mitochondrial shape following TbLOK1 RNAi is not simply due to kDNA loss.
Figure 7. Disruption of kDNA replication or maxicircle transcription does not directly affect mitochondrial shape.
Following RNAi to either DNA ligase kα (TbLIGkα) or mitochondrial topoisomerase II (TbTOP2mt), trypanosomes were stained with DAPI, MitoTracker (MiTr), and antibodies recognizing mtHSP70 and then examined by phase or fluorescence microscopy.
Knockdown of the TbLOK1 protein leads to a loss of mitochondrial respiration
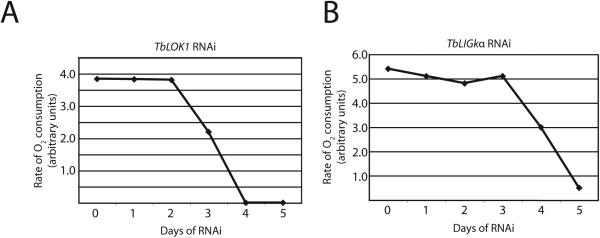
Along with the growth defect and loss of mitochondrial structure, TbLOK1 RNAi led to a reduction in mitochondrial staining with MitoTracker, a membrane potential-dependent dye (Poot et al., 1996). Remarkably, more than 60% of cells had little or no MitoTracker fluorescence after 4 days of RNAi. Since inner membrane potential is essential for mitochondrial function, we examined oxygen consumption in TbLOK1-depleted cells. After 3 days of RNAi, the rate of mitochondrial respiration was down ~ 50% and after 4 days it was undetectable (Fig. 8). These results suggested that the lack of MitoTracker staining was likely due to a defect in mitochondrial function, such as decreased inner membrane potential. Since more than half of the cells with reduced TbLOK1 protein levels still have their kinetoplast at day 4, it is unlikely that the respiration defect is strictly due to kDNA loss. Nonetheless, to further test this possibility we induced kDNA loss in cells by knocking down TbLIGkα. Although TbLIGkα RNAi caused at least 90% of cells to lose their kinetoplast by day 3, there was no immediate effect on oxygen consumption (Fig. 8) or MitoTracker staining (Fig. 7). A decrease in respiration occurred later, with a significant drop in O2 consumption occurring at days 4 and 5. Thus, the respiration and MitoTracker staining defects seen in TbLOK1 RNAi cells are not due to kDNA loss, suggesting that TbLOK1 plays a more immediate role in mitochondrial function.
Figure 8. TbLOK1 knockdown inhibits mitochondrial respiration.
TbLOK1 RNAi was induced for the indicated times and the rate of oxygen consumption measured. For comparison, cells knocked down for mitochondrial DNA ligase kα (TbLIGkα) were also evaluated.
TbLOK1-depleted cells are defective in maxicircle transcription
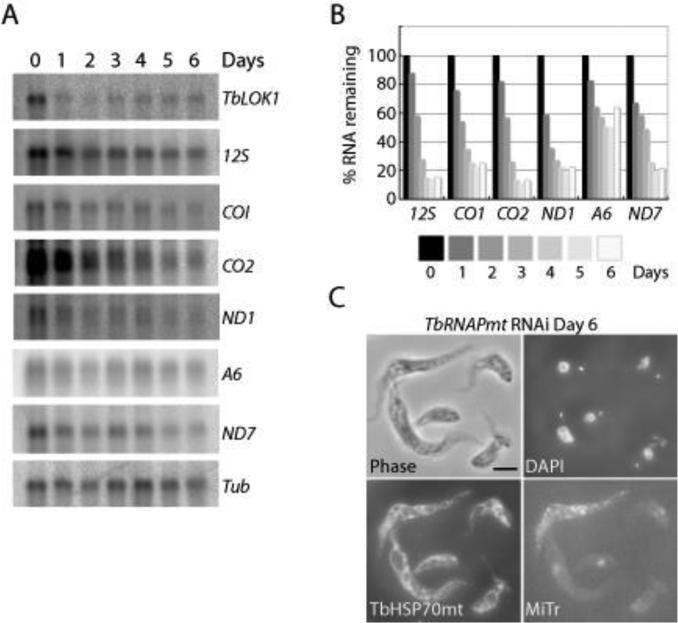
We found that knockdown of TbLOK1 caused a rapid reduction in the levels of many mitochondrial transcripts (Fig. 9A, B). Northern blots showed that following 3 days of TbLOK1 RNAi, the levels of the 12S ribosomal RNA, cytochrome oxidase subunits I and II, and NADH dehydrogenase subunit I fell below 40%, while the transcript for the F1-ATPase subunit 6 and NADH dehydrogenase subunit 7 were down more than 50%. To examine the connection between kDNA transcription and organelle shape, we depleted trypanosomes of the mitochondrial RNA polymerase (TbRNAPmt) (Grams et al., 2002). Although reduced levels of TbRNAPmt caused a more potent disruption in transcription than TbLOK1 RNAi, with most maxicircle transcripts disappearing within 3 days (Grams et al., 2002), we observed almost no change in mitochondrial structure. Unlike TbLOK1-depletion, normal branched mitochondria were seen in more than 80% of cells even following 6 days of RNAi to TbRNAPmt (Fig. 9C,). While there was no discernable effect on mitochondrial morphology, TbRNAPmt knockdown had a profound effect on inner membrane potential. Little or no MitoTracker staining was seen in TbRNAPmt RNAi cells at day 6 (Fig. 9C,). Thus, it is possible that the defect in maxicircle transcription seen in both TbLOK1 and TbRNAPmt-depleted cells causes the loss of membrane potential and respiration. However, decreased transcription does not account for the abnormal mitochondrial shape seen in TbLOK1 RNAi cells.
Figure 9. TbLOK1 RNAi causes a defect in maxicircle transcription.
A. After TbLOK1 depletion, mRNA was electrophoresed and northern blots probed for TbLOK1 (to show knockdown efficacy) and tubulin (a loading control), as well as the maxicircle-encoded 12S rRNA, cytochrome oxidase subunit I (CO1) and subunit II (CO2), NADH dehydrogenase subunit I (NDI) and subunit 7 (ND7), and the A6 subunit of the F1/Fo-ATPase. B. Quantitation of the blots shown in A, normalized to nuclear-encoded tubulin. The amount of each transcript present at day 0 of RNAi was arbitrarily set to 100. C. Following RNAi-mediated knockdown of TbRNAPmt, trypanosomes were stained with MitoTracker (MiTr), subjected to immunofluorescence with antibodies recognizing mtHSP70, stained with DAPI, and examined by phase and fluorescence microscopy. Although TbRNAPmt RNAi induces maxicircle loss, minicircle levels are not initially affected (Grams et al., 2002). Thus, DAPI staining of the kinetoplast is not significantly reduced following TbRNAPmt depletion.
TbLOK1 is a mitochondrial outer membrane protein with a punctate distribution along the organelle surface
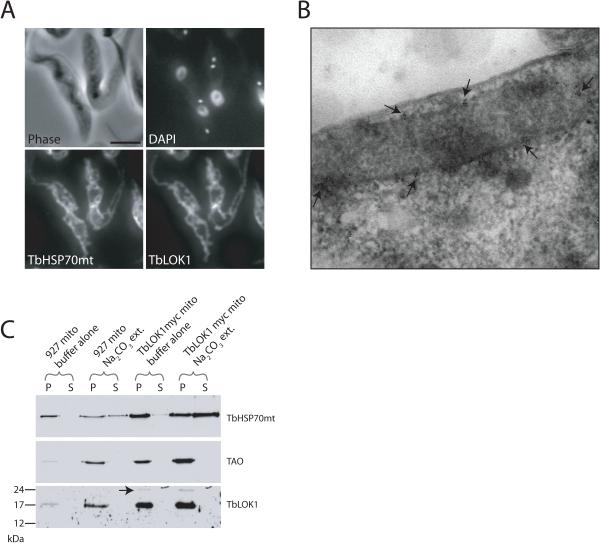
Using immunofluorescence to localize TbLOK1 in cells, we found that the staining of TbLOK1 was the same as that seen using antibodies to the matrix-localized TbHSP70mt protein (Guler et al., 2008), indicating that TbLOK1 is a mitochondrial protein (Fig. 10A). To further pinpoint its cellular location, we used immunogold electron microscopy of cells expressing TbLOK1::myc, an epitope-tagged version of the TbLOK1 protein. Micrographs of thin sections showed that the TbLOK1::myc fusion protein was at or very near the mitochondrial membranes (Fig. 10B, see arrows). Interestingly, the distribution of TbLOK1::myc was not uniform, but instead clusters of gold labeling were observed every few microns along the mitochondrial surface. Although this method could not resolve whether TbLOK1 was in the outer or inner membrane, its location on the mitochondrial surface was consistent with the prediction that TbLOK1 has two transmembrane segments.
Figure 10. TbLOK1 is a mitochondrial membrane protein.
A. Immunofluorescence studies show TbLOK1 is mitochondrial. Wild-type 927 cells were fixed and permeabilized, then decorated with antibodies to TbLOK1 and TbHSP70mt. Following DAPI staining, cells were examined by phase and fluorescence microscopy. B. Immunoelectron microscopy shows TbLOK1 at the mitochondrial membranes. Cryosections from trypanosomes where both alleles express the TbLOK1::myc fusion protein were fixed, and thin-sections decorated with antibodies to the myc epitope. Following incubation with gold-conjugated secondary antibodies, sections were osmium stained and examined by EM. Arrows indicate clusters of myc signal along the mitochondrial double membrane. C. TbLOK1 is an integral membrane protein. Mitochondria isolated from wild-type trypanosomes, or cells in which one allele expresses the TbLOK1::myc fusion, were treated with either buffer alone or with sodium carbonate, pH 11.5. Pellets (P) and supernatants (S) were analyzed by western blotting with antibodies to TbHSP70mt (a soluble protein), TAO (a membrane protein), or TbLOK1. Migration of TbLOK1::myc is indicated with an arrow.
Confirming that TbLOK1 is an integral membrane protein, we found it was resistant to carbonate extractions (Fig. 10C). Mitochondria from wild-type cells and from cells expressing the TbLOK1::myc fusion protein were treated with 0.1 M sodium carbonate, which extracts soluble proteins and those peripherally-associated with membranes (Fujiki et al., 1982). When carbonate extracted samples were centrifuged, western blotting showed that the soluble, matrix-localized TbHSP70mt protein was readily released into the supernatant, while the trypanosome alternative oxidase (TAO), a integral protein of the inner membrane, remained in the mitochondrial pellet. Since TbLOK1 was detected in the pellet fraction along with TAO, we conclude that TbLOK1 is also an integral membrane protein. Both TbLOK1::myc (Fig. 10C, arrow) and the untagged TbLOK1 protein behaved the same in our assays.
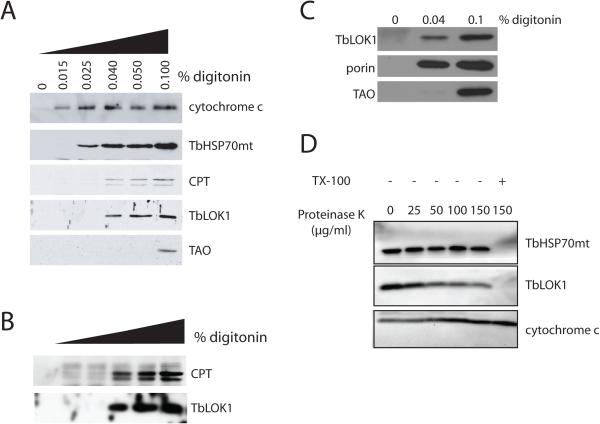
Studies with whole-cell detergent extractions suggest that the TbLOK1 protein resides in the mitochondrial outer membrane (Fig. 11A). Trypanosomes were treated with increasing concentrations of digitonin, a detergent known to preferentially solubilize the mitochondrial outer membrane (Levy et al., 1966, Morton et al., 1968, Schnaitman et al., 1967) and the plasma membrane (Lineberger et al., 1989, Plutner et al., 1992). Supernatant fractions were analyzed for TbLOK1 and marker proteins for the outer membrane (carnitine palmitoyl transferase or CPT; (Detke & Elsabrouty, 2008)), the intermembrane space (cytochrome c; (Esseiva et al., 2004)), the inner membrane (TAO; (Chaudhuri et al., 1998)), and the matrix (TbHSP70mt; (Guler et al., 2008)). As expected, in the absence of detergent, none of the mitochondrial proteins were released into the supernatant. With 0.015% digitonin, the plasma membrane and the mitochondrial outer membranes were permeabilized, causing release of cytochrome c from the intermembrane space. Under these conditions, the outer membrane was not fully solubilized and none of the outer membrane CPT enzyme was found in the supernatant fraction (see Fig. 11B for darker exposure). With 0.025% digitonin, the inner membrane was partially disrupted and the matrix TbHSP70mt protein was released to the supernatant. With 0.04% detergent, the outer membrane was completely solubilized and the integral CPT protein was now extracted, whereas the inner membrane TAO enzyme required 0.1% digitonin for its complete release. TbLOK1 behaved like the CPT protein and was largely extracted by 0.04% digitonin.
Figure 11. TbLOK1 is a mitochondrial outer membrane protein.
A. TbLOK1 is in the outer mitochondrial membrane. Wild-type cells were treated with either buffer, or with different concentrations of digitonin. Samples were centrifuged and the supernatants analyzed by western blotting with antibodies to TbLOK1 and the following marker proteins: cytochrome c (intermembrane space), TbHSP70mt (matrix), carnitine palmitoyl transferase (CPT, outer membrane), and trypanosome alternative oxidase (TAO, inner membrane). B. Darker exposure of the gel shown in ‘A’. C. TbLOK1 cofractionates with the outer membrane porin protein. Cells were treated with 0.04% digitonin, 0.1% digitonin, or with buffer alone, centrifuged and the supernatants blotted with antibodies to the TbLOK1, porin or TAO proteins. D. Protease digestions of isolated mitochondria. Mitochondria from 927 trypanosomes were treated with the indicated amounts of proteinase K and then Western blotted for TbLOK1, TbHSP70mt and cytochrome c. As indicated, some samples were treated with 1% Triton X-100 (TX-100) prior to the addition of protease.
Although CPT is reported to be a mitochondrial outer membrane protein in trypanosomes (Detke & Elsabrouty, 2008)(L. Simpson, unpublished observations), its location has not been definitively established. Therefore, we repeated our digitonin extractions using porin (Pusnik et al., 2009) as a marker for the outer membrane (Fig. 11C). We found that TbLOK1 and porin were similarly extracted by 0.04% digitonin, while TAO, the inner membrane marker, required more detergent (0.1% digitonin) for its release into the supernatant. Thus, our results with the porin protein complement those with CPT, both suggesting that TbLOK1 is in the outer membrane.
Two additional experiments support our digitonin extraction studies. First, we found that TbLOK1 was accessible to protease digestion in isolated mitochondria (Fig. 11D). Mitochondria purified from trypanosomes were treated with proteinase K and aliquots Western blotted for TbLOK1, the intermembrane space cytochrome c protein, and matrix-localized TbHSP70mt. While the levels of TbLOK1 were clearly reduced with increasing concentrations of protease, the amounts of TbHSP70mt remain unchanged. TbHSP70mt was only digested when the mitochondrial membranes were solubilized by treatment with Triton X-100. In an attempt to demonstrate that the mitochondrial outer membrane remained intact in our studies, we examined the protease sensitivity of intermembrane space-localized cytochrome c. Unfortunately, due to its tight folding we found that cytochrome c was resistant to digestion even in the presence of detergent. However, since cytochrome c is readily lost from mitochondrial preparations whose outer membrane is ruptured (Schneider et al., 2007b), its persistence in our fractions argues that our isolated mitochondria were largely intact. Also suggesting that TbLOK1 is in the outer membrane, we found that the TbLOK1 protein was accessible to antibodies when trypanosomes were perforated by very small amounts of detergent (0.015% digitonin; Fig. S3). In contrast, TbHSP70mt, residing within the organelle, could be decorated by antibodies only when the mitochondrial membranes were solubilized with 0.1% Triton X-100.
Discussion
Using a genomic RNAi library in T. brucei, we have identified TbLOK1, a novel mitochondrial outer membrane protein. TbLOK1 is an uncharacterized 19 kDa protein with extensive similarity to proteins in other kinetoplastids, but with no obvious counterparts in more distantly related eukaryotes. We find that TbLOK1 is an integral protein in the mitochondrial membranes, consistent with hydropathy analyses predicting two transmembrane segments. Depletion of the TbLOK1 protein by RNAi causes a strong growth defect, indicating that TbLOK1 is essential for trypanosome viability.
Three independent assays indicate that TbLOK1 is a mitochondrial outer membrane protein—digitonin extractions of whole cells, protease digestions with isolated mitochondria, and antibody accessibility studies. Nonetheless, since all of these assays can be technically troublesome, we are confident but not absolutely certain of this outer membrane localization. For example, although we found that the levels of TbLOK1 in isolated mitochondria decreased after exogenous proteinase K digestion, significant amounts of the protein remained even after treatment with high concentrations of the protease. TbLOK1 could be poorly susceptible to protease because it is tightly folded or resides very close to the membrane surface, like the outer membrane porin (Pusnik et al., 2009) or TbTOB55 (Sharma et al., 2010) proteins. On the other hand, the partial protease resistance of TbLOK1 could indicate a location different from typical outer membrane proteins. For example, the punctate distribution of TbLOK1 is consistent with it residing in contact sites, regions where the outer and inner membranes connect (Toulmay & Prinz, 2011)). Regardless, we are encouraged by the recent result showing TbLOK1 in the T. brucei mitochondrial outer membrane proteome (A. Schneider, communication prior to publication). Also consistent with our conclusions, we could find no obvious mitochondrial import signal in TbLOK1 using programs that detect N-terminal presequences such as PSORT (Nakai & Horton, 1999), TargetP (Emanuelsson et al., 2000), or MitoProt (Claros & Vincens, 1996). Since the targeting information for outer membrane proteins is not well characterized, but clearly differs from that of proteins imported into the matrix or inner membrane (Chacinska et al., 2009), this result may not be so surprising.
We identified TbLOK1 in an RNAi screen for trypanosomes that lose their kDNA, but subsequently found that the defect in kinetoplast maintenance after TbLOK1 knockdown is delayed and incomplete. We argue that that the TbLOK1 protein does not play a direct role in kDNA replication, and that kDNA loss instead follows the more immediate mitochondrial structural and functional defects caused by TbLOK1 depletion. After the induction of TbLOK1 RNAi, cells stop their growth after 3 or 4 days, but at this point more than 80% of cells still have a normal kinetoplast. At day six, about a third of the trypanosomes still contain normal kDNA and another third carry a small kinetoplast. Upon close examination, we do see subtle effects on kDNA homeostasis, including a small decrease in the total amount of both minicircles and maxicircles, and a modest decrease in minicircle replication intermediates. All of these effects are much less severe than those seen during RNAi-mediated depletion of known kDNA replication proteins, and most of the TbLOK1 RNAi effect on kDNA maintenance occurs well after the onset of growth arrest. By comparison, trypanosomes knocked down for DNA ligase kα (Downey et al., 2005) or mitochondrial topoisomerase II (Wang & Englund, 2001) lost more than 90% of their kDNA at the point when cells stopped dividing or often even before cell growth ceased. Moreover, the mitochondrial outer membrane location of the TbLOK1 protein argues against a direct role in kDNA replication, which takes place in the mitochondrial matrix.
In contrast to the modest effect on kDNA replication, knockdown of TbLOK1 has a more pronounced effect on kDNA segregation. Normally, replication produces a double-size network that undergoes scission to form kinetoplasts that are equal in size to each other and to the parent. We found that TbLOK1 RNAi often caused the network to divide asymmetrically, yielding one daughter kDNA which is larger than normal and another which is correspondingly smaller. Using FISH probes for mini- and maxicircle DNA, we found TbLOK1-depletion also caused an accumulation in the number of long ‘threads’ of maxicircles connecting the two dividing minicircle networks. The separation of maxicircles was recently shown to mark the final stages in kinetoplast segregation (Gluenz et al., 2011). Therefore, knockdown of TbLOK1 leads to a defect in the distribution of both mini- and maxicircles during cytokinesis. Although the increase in asymmetric kDNA division and the accumulation of maxicircle threads are indicative of a defect in kinetoplast segregation, there are a surprising variety of mutant phenotypes that also demonstrate these defects, including RNAi-mediated knockdown of TbTOP2mt (involved in kDNA replication)(Wang et al., 2002), subunits of the mitochondrial proteasome (controls the levels of mini- and maxicircle replication) (Li et al., 2008) and the acyl carrier protein (involved in mitochondrial fatty acid synthesis)(Clayton et al., 2011) as well as overexpression of the maxicircle-specific replication factor TbPIF2 (controls maxicircle replication) (Liu et al., 2009). Thus, the reason that knockdown of TbLOK1 causes a defect in kDNA segregation remains obscure. Nonetheless, we speculate on one possibility below.
We found that depletion of the TbLOK1 protein causes an immediate and substantial effect on mitochondrial shape and function. The single mitochondrion in PCF cells contains numerous branches and interconnections, and the organelle has a fishnet-like appearance. Coincidental with the growth arrest during TbLOK1 RNAi, the complex mitochondrial structure seen in normal cells was replaced by a simple, thickened tubule that lacks branches. Paralleling the growth defect and loss of mitochondrial shape, TbLOK1 RNAi caused a marked defect in overall mitochondrial function, such as the inability to stain mitochondria with the potential-dependent dye, MitoTracker, and a large drop in mitochondrial O2 consumption. Both defects point to problems with electron transport from the cytochrome-mediated pathway. The collapse of mitochondrial shape and function seen in TbLOK1-depleted cells clearly preceded significant loss of kDNA. Also arguing that kinetoplast loss does not explain the phenotypes seen in TbLOK1 RNAi, we showed that while knockdowns of mitochondrial topoisomerase II and DNA ligase kα both caused an almost complete loss of kDNA (Downey et al., 2005, Wang & Englund, 2001), they had little or no immediate effect on organelle structure, MitoTracker staining, or respiration.
Depletion of TbLOK1 also led to a defect in maxicircle transcription, and the reduction of maxicircle-encoded subunits of the oxidative phosphorylation complexes could explain the lack of MitoTracker staining and respiration seen in TbLOK1 RNAi cells. Supporting this idea, we found that knockdown of the mitochondrial RNA polymerase, TbRNAPmt, which completely blocks maxicircle transcription (Grams et al., 2002), similarly disrupts MitoTracker staining. However, unlike TbLOK1 RNAi, knockdown of TbRNAPmt did not significantly perturb mitochondrial morphology. Thus, all of the problems seen in TbLOK1-depleted cells cannot be explained simply by kDNA loss or a defect in maxicircle expression. We speculate that TbLOK1 plays a broader role in mitochondrial architecture and activity.
Defects in several different cellular processes have been found to impact both mitochondrial shape and mitochondrial function, offering several tantalizing possibilities for the role of TbLOK1. These processes include mitochondrial fusion and division (Otera & Mihara, 2011), phospholipid homeostasis (Osman et al., 2011), protein import (Abelovska, 2011), mitochondrial-cytoskeletal attachment (Jensen, 2005), and energy production (Ishihara et al., 2003). Although the precise molecular role of TbLOK1 is not known, the unique properties of this protein make it reasonable to briefly consider its role in a few of these diverse possibilities. For example, the interplay between division and fusion is thought to be a critical regulator of both the number and shape of mitochondria, and defects in either process can often affect organelle function (Westermann, 2008). TbDLP, a dynamin-like protein related to a fission component in yeast and mammals, has been shown to be, like TbLOK1, essential for cell growth (Chanez et al., 2006, Morgan et al., 2004). In one study (Morgan et al., 2004), knockdown of TbDLP produced a thickened, unbranched mitochondrial tubule much like that seen in TbLOK1 RNAi cells. Although TbLOK1 does not show similarity to any known fusion or division proteins, we note that counterparts to many of the yeast and mammalian proteins are not apparent in the T. brucei genome, suggesting the machinery for trypanosome fusion and fission may be divergent. Regardless, in yeast and mammalian cells, some of the division machinery shows a punctate distribution along mitochondria (Youngman et al., 2004, Hobbs et al., 2001, Boldogh et al., 2003, Pitts et al., 1999), similar to the discrete clusters of TbLOK1 on the mitochondrial surface seen in our immunoelectron microscopy images. Moreover, a defect in yeast mitochondrial fusion leads to a subsequent loss of mitochondrial DNA (mtDNA) (Berger et al., 1997, Dimmer et al., 2005, Hobbs et al., 2001, Sogo & Yaffe, 1994, Youngman et al., 2004), resembling the delayed kDNA loss seen in TbLOK1 RNAi cells.
Changes in the lipid composition of the mitochondrial membranes are known to affect organelle shape and function, providing an alternative possibility for the function of TbLOK1. For example, the yeast ERMES (ER-mitochondrial encounter structure) mediates the attachment between the mitochondria and the endoplasmic reticulum, and mutations in one or more of the ERMES subunits result in impaired phospholipid traffic between the organelles, as well as altered mitochondria lipid composition (Kornmann & Walter, 2010, Kornmann et al., 2011). Like knockdown of TbLOK1, loss of ERMES function leads to a collapse of mitochondrial shape, abnormal mitochondrial function and a loss of mitochondrial DNA. Resembling the distribution of TbLOK1, many of the ERMES proteins are located in dot-like structures in the mitochondrial outer membrane.
Defects in trypanosome lipid synthesis have also been found to affect mitochondrial shape and function (Fridberg et al., 2008, Gibellini et al., 2009, Guler et al., 2008, Perez-Moreno et al., 2012, Signorell et al., 2009). For example, inhibiting fatty acid synthesis by knocking down the acyl carrier protein (TbACP) results in aberrant mitochondrial morphology, defective kDNA segregation, reduced cytochrome-mediated respiration, and decreased levels of several phospholipids in the mitochondrial membranes (Clayton et al., 2011, Guler et al., 2008, Stephens et al., 2007). It was hypothesized that the altered membrane composition disrupted the attachment of the kinetoplast to segregation machinery, i.e., the tripartate attachment complex (TAC). Although the ultrastructure of the mitochondria in cells depleted for TbACP differs from that in TbLOK1 RNAi cells, both knockdowns lead to similar increases in asymmetric kDNA segregation and kDNA loss. Regardless, these studies clearly highlight the importance of maintaining normal phospholipid levels for both shape and function of mitochondria.
In PCFs, knockdown of TbLOK1 did not simply cause the complete loss of mitochondrial shape, but instead led to a specific loss of branching and interconnections between tubules. The unbranched mitochondrial tubule seen following RNAi-mediated depletion of TbLOK1 in PCF cells is strikingly reminiscent of the mitochondrion seen in BSF trypanosomes. It is tempting to speculate that TbLOK1 plays a crucial role in the mitochondrial morphogenesis that occurs during the conversion of one life cycle stage to another. Consistent with this idea, we found that the level of TbLOK1 protein was reduced in BSFs compared to PCFs. Another study found that TbLOK1 is downregulated in BSF cells at the transcript level (Siegel et al., 2010). Further studies are clearly needed to pinpoint the exact role that this novel and essential outer membrane protein plays in the shape and function of trypanosome mitochondria.
Experimental procedures
Trypanosome strains, media, transfection
Wild-type T. brucei PCF strain 927 (from E. Ullu; Yale University, New Haven) and BSF strain 427 (from G. Cross; Rockefeller University, New York) were grown as described (Hirumi & Hirumi, 1989, Brun & Schonenberger, 1979). The RNAi library (Englund et al., 2005, Morris et al., 2002) and other constructs in pZJM (Wang et al., 2000) were transfected into 29-13 procyclic cells (from G. Cross), which carries the tet repressor and tetracycline-inducible T7 RNA polymerase (Wirtz and Clayton, 1995; Wirtz 1998). Cells with stable integrations of the RNAi library constructs were selected and maintained in SDM-79 medium with 15 μg/ml G418 (Invitrogen), 50 μg/ml hygromycin (Roche) and 2.5 μg/ml phleomycin (Sigma).
RNAi and library screening
~300 clones of PCF strain 29-13, each carrying a different ~ 500 bp genomic DNA segment in pZJM, were screened for cells that lose kDNA following the procedure of Morris et al. ((Morris et al., 2002)). Briefly, cell lines were cloned by limiting dilution (0.5 cells/well in a 96-well plate) in SDM-79 medium (supplemented with 15% FBS and the appropriate antibiotics). RNAi to the genomic DNA insert in pZJM was induced by adding 1 μg/ml tetracycline (Sigma) to the medium and then cells were assayed for kDNA loss by fluorescence microscopy after staining with the DNA dye, Hoechst 33342 (Fisher Scientific). Since kDNA loss is lethal to PCFs, the relevant gene in each candidate was identified by PCR of the pZJM insert in a clone of the same cells that had not been induced for RNAi. kDNA loss was evaluated after 5 and 8 days of RNAi. Five clones showed significant kDNA loss, and two had identical inserts corresponding to TbLOK1 (XP_827340, GeneDB: Tb09.211.1940) with orthologs in the kinetoplastids Leishmania major (XP_843534), Leishmania infantum (XP_001469236), and Trypanosoma cruzi (XP_821663). The other three clones, with an insert homologous to the RHS (retrotransposon hot spot) family of genes (Bringaud et al., 2002), were not studied further.
Since the library insert contained sequences from the 5′ and 3′ untranslated regions of TbLOK1, we created an RNAi construct specific for TbLOK1 by PCR amplifying an ~ 500 bp portion of the open reading frame and inserting it into XhoI-HindIII cut pZJM. After transfection of the NotI-digested plasmid into 29-13 cells, two cell lines were isolated and cloned by limiting dilution. Both clones (and the two original library isolates) had similar growth and loss of kDNA phenotypes. One clone (1.C2) was used for all subsequent studies. For growth assays, cells were counted using a Model Z1 Coulter Counter. As observed previously with other genes, the trypanosomes eventually recovered from the TbLOK1 RNAi, with the transcript returning to near normal levels at day 6, despite the continued production of dsRNA corresponding to the TbLOK1 sequence (not shown).
RNAi strains with stem-loop RNAi constructs for the mitochondrial topoisomerase II or RNA polymerase have been described (Wang & Englund, 2001). To knockdown mitochondrial DNA ligase kα, pZJM-LIGB1 was constructed by PCR amplifying a 554 bp fragment of the coding sequence and cloning it into XhoI-HindIII cut pZJM. Transfection into 29-13 and cloning of cells resulted in RNAi strain LigB1.B2.
Construction of the TbLOK1::myc fusion protein
The TbLOK1::myc fusion, which carries 3 tandem copies of the myc epitope tag inserted at the C-terminus of TbLOK1 was constructed by amplifying TbLOK1 from genomic DNA (list of primers used for this and other constructs are in Table S1) and inserting the PCR product into pXS3XmycNEO (Zhao et al., 2008). The entire cassette was PCR amplified, transfected into 29-13 cells, and neomycin resistant clones were selected. A strain in which both alleles of TbLOK1 are myc tagged was constructed by PCR amplifying TbLOK1 and inserting it into pXS3XmycBLAST(Zhao et al., 2008). The cassette was PCR amplified, transfected into the TbLOK1::myc-expressing strain described above, and neomycin/blasticidin resistant clones were selected. Although stable clones were obtained, PCR analysis of genomic DNA from the transfected cells indicated that the wild-type TbLOK1 locus had duplicated, raising the possibility that the myc-tagged version of TbLOK1 is not fully functional. Consistent with this idea, western blots showed reduced levels of the TbLOK1::myc fusion protein compared to the wild-type TbLOK1 in cells, possibly due to degradation. Nonetheless, the myc tag does not appear to interfere with localization, since the distribution of TbLOK1::myc was indistinguishable from that of the untagged protein by immunofluorescence and subcellular fractionation.
Antibodies to TbLOK1
The complete TbLOK1 open reading frame was PCR amplified, cloned into pET14b (Novagen), and transformed into E. coli strain BL21 Rosetta (DE3) pLysS (Novagen). Two liters of cells (OD600 of 0.6) were induced with 1 mM IPTG for 3 h at 37°C, and the his6-tagged TbLOK1 protein was purified under denaturing conditions using the Quick Start Protein Expression Kit (Qiagen). Purified protein was used to raise antibodies in rats (Cocalico Biologicals, Reamstown, PA). All bleeds recognized a single ~ 19 kDa band on western blots and showed a mitochondrial distribution by immunofluorescence and cell fractionation.
Western blotting antibodies
Lysates were prepared by pelleting trypanosomes at 3,000 rpm in a microfuge for 5 min, washing pellets once with phosphate buffered saline (PBS), resuspending the cell pellets in SDS-sample buffer, and heating them at 95°C for 5 min. After centrifugation for 1 min, aliquots (~106 cell equivalents/lane) were run on 12% or 15% SDS-polyacrylamide gels and blotted onto PVDF membranes (Millipore) using standard methods. Membranes were probed with antibodies to TbLOK1 (1:1000 dilution), TbHSP70mt (Guler et al., 2008), (1:2000 dilution), enolase (Bakshi & Shapiro, 2004) (1:200 dilution), cytochrome c (Esseiva et al., 2004) and porin (Pusnik et al., 2009)(from A. Schneider, University of Bern, Switzerland) (1:100 dilution), trypanosome alternate oxidase (TAO; from M. Chaudhuri, Meharry Medical College, Nashville, TN) (1:200 dilution), Rieske iron-sulfur protein (ISP, from T. Ochsenreiter and S. Hajduk, University of Georgia, Athens, GA) (1:500 dilution), and carnitine O-palmitoly transferase (CPT, a gift from L. Simpson, UCLA (1:200 dilution). Antibodies to CPT were raised against L. tarentole CPT1 protein ((Detke & Elsabrouty, 2008) and L. Simpson, personal communication) and were found to cross-react with two T. brucei proteins corresponding to Tb927.8.590 and Tb927.7.2250 (both predicted to be ~ 75 kDa) and Tb927.3.3900 (predicted to be ~ 67 kDa). Immune complexes were visualized with species-specific HRP-conjugated secondary antibody (Invitrogen), chemiluminescence (HyoGLO, Denville Scientific) and a Versadoc imaging system with Quantity One software (Bio-Rad, Hercules, CA), or with X-ray film.
Immunofluorescence microscopy
Indirect immunofluorescence of mid-log phase cells (~ 6 × 106 cells/ml) adhered to poly-L-lysine coated slides has been described (Wang & Englund, 2001). Antibodies to TbLOK1 (1:500 dilution) or TbHSP70mt (1:200 dilution) were used, followed by anti-rat Alexa 488 and anti-rabbit Alexa 594 secondary antibodies (Invitrogen, 1:500 dilution. Slides were washed, stained with 2 μg/ml DAPI (4′,6-diamidino-2-phenylindole; Sigma) and mounted in Vectashield (Vector Laboratories). In some experiments, live trypanosomes were stained in growth medium with 1 μM MitoTracker Red (Invitrogen) before fixing and immunofluorescence. Images were captured using a Zeiss Axioskop microscope, a Retiga Exi CCD camera and IPLab software (Biovision). At least 140 cells were counted at each timepoint.
Antibody accessibility studies
Cells were allowed to settle and adhere to poly-L-lysine coated slides, then fixed with 4% paraformaldehyde in PBS, and treated with 0.1M glycine/PBS as described (Wang & Englund, 2001). Cells were then incubated with PBS, 0.015% digitonin (Roche) in PBS, or 0.1% Triton X-100 (Sigma) in PBS for 10 min. After washing twice for 5 min with PBS and blocking with 1% BSA/PBS for 60 min, trypanosomes were decorated with antibodies and imaged as described above.
Northern and Southern blotting
RNAi inductions were staggered so that all samples were collected on the same day. DNA was isolated as described (Wang & Englund, 2001). For free minicircle analysis, undigested DNA was run on a gel. For analysis of total minicircle and maxicircle content, the DNA was digested with Hind III/XbaI prior to agarose gel electrophoresis and Southern blotting (Wang & Englund, 2001). RNA was isolated using an RNA Isolation Kit (Stratagene) and Northern blotted as described (Zhao et al., 2008). Probes were made using the Random Primers DNA Labeling System (Invitrogen) and [α-32P]-dATP (50 μCi, 3000 Ci/mmol, New England Nuclear). For the minicircle probe, a DNA fragment from pJN6 (Ntambi & Englund, 1985) was used. The maxicircle probe was a gel-purified 1.8 kbp DNA fragment from XbaI-cut kDNA (Wang & Englund, 2001). The loading control was a portion of the trypanosome hexose transporter (THT) gene isolated by PCR (Wang & Englund, 2001). Oligos for amplifying probe fragments corresponding to TbLOK1 and different mitochondrially-encoded genes are in Table S1. Note that the oligos used to detect mitochondrial genes whose transcripts undergo editing will amplify the pre-edited mRNAs. Southern and Northern blots were analyzed and quantitated by a Fuji phosphorimager.
Thin-section and immunogold electron microscopy
Uninduced control cells and cells induced for RNAi to TbLOK1 for four days were fixed in 2.5% glutaraldehyde, post-fixed in osmium tetroxide and embedded in Epon as described (Baines & Gull, 2008). Thin sections were examined by an FEI Tecnai 12 transmission electron microscope. Immunogold electron microscopy was described previously (Rieder et al., 1996) and performed on ultrathin cryosections of cells where both alleles express the TbLOK1::myc fusion protein (see caveat for this cell line above).
Digitonin extraction assays
Wild-type 927 trypanosomes were permeabilized with digitonin (Schneider et al., 2007a). Briefly, 108 cells per sample were centrifuged, washed with 20 mM sodium phosphate, pH 7.9, 20 mM glucose, 0.15 M NaCl, and then resuspended in 500 μl SoTE buffer (20 mM Tris-HCl, pH 7.5, 0.6 M sorbitol, 2 mM EDTA). Next, 500 μl of SoTE buffer with different amounts of digitonin was added to each tube so that the final concentration of detergent was 0%, 0.015%, 0.025%, 0.04%, 0.05%, or 0.1%. Samples were incubated for 5 min on ice followed by centrifugation (3 min, 5,000 g, 4°C). Pellets and supernatants were resuspended in SDS-sample buffer, boiled for 5 min, and then analyzed by SDS-PAGE and western blotting.
Oxygen electrode measurements
As described (Guler et al., 2008), 9 × 107 cells were washed (150 mM NaCl, 5 mM glucose and 20 mM sodium phosphate, pH 7.9) and then suspended in 2 ml assay buffer (125 mM sucrose, 65 mM KCl, 5 mM HEPES, pH 7.2, 1 mM MgCl2, 2.5 mM potassium phosphate, 1 mM EDTA). Measurements of intact cells were performed at 28°C in a 2 ml Clark-type oxygen electrode (Yellow Springs Instruments). More than 95% of the oxygen consumed by the trypanosomes was inhibited by the addition of 100 μM oligomycin (Sigma; data not shown), indicating that our measured respiration rates were predominantly due to mitochondrial enzymes.
Mitochondria isolation, carbonate extractions and protease digestions
Mitochondria were isolated (Schneider et al., 2007b) from 2 liters (1 × 107 cells/ml) of wild-type 927 trypanosomes or from cells expressing TbLOK1::myc. Cells were pelleted (6000 g, 15 min, room temperature), washed twice with PBS, and then washed with SoTE buffer (0.6 M sorbitol, 2 mM EDTA, 10 mM MOPS/KOH, pH 7.2). Breaking buffer [SoTE buffer plus protease inhibitors (1:100, Roche) and 0.5% BSA was added to the cell pellets (1 × 109 cells/ml) and the cells were then resuspended with several strokes with a Dounce homogenizer. Cells were then lysed by two passages through a high-pressure (20,000 psi) emulsifier (EmulsiFlex-C-3, Avestin) and then centrifuged at 10,000 g for 20 min. The pellet was resuspended in breaking buffer and treated with 0.1 mg/ml DNase (Roche) for 15 min at room temperature. Intact cells and nuclei were removed by centrifugation at 490 g for 10 min, and the supernatant was centrifuged at 10,000 g for 15 min to collect the crude mitochondrial fraction. Mitochondria were washed once in SoTE by centrifugation at 10,000 g for 15 min. Mitochondrial pellets were resuspended in SoTE with 50% glycerol plus protease inhibitors at ~ 10 mg/ml (assuming an A280 of 0.21 = 10 mg/ml), aliquoted into ~ 1 mg samples, flash frozen in liquid nitrogen, and then stored at −80 °C.
For carbonate extractions, isolated mitochondria (containing 100 μg protein for each condition) were thawed on ice, pelleted in a Beckman Airfuge at 40 psi for 20 min, and washed once in 1X STE (250 mM sucrose, 20 mM Tris-HCl pH 8.0, 2 mM EDTA). Pellets were resuspended in 200 μl of either STE (buffer) or 0.1 M sodium carbonate, pH 11.5 and incubated on ice for 45 min, with occasionally mixing with a pipette. Samples were then spun in a Beckmann airfuge at 30 psi (100,000 g) for 1 h at room temperature. Pellets were resuspended in 25 μl 2X SDS-PAGE sample buffer (with ß-mercaptoethanol) and boiled for 5 min. 10 μl StrataClean resin (Stratagene) was added to supernatants and incubated at 4°C for 5 min with rotation. After centrifugation, the proteins were eluted from the resin with 25 μl 2X SDS-sample buffer. Equal amounts of pellet and supernatant were analyzed by SDS-PAGE and western blotting.
For protease digestions, frozen mitochondria were washed with 9 volumes of SEM buffer (0.25 M sucrose, 2 mM EDTA, 10 mM MOPS/KOH, pH 7.2) to remove glycerol. Mitochondria (100 μg for each condition) in 100 μl SEM were treated with proteinase K (0-150 μg/ml; Invitrogen Life Sciences) for 30 min on ice, and the reaction stopped by adding 4 mM PMSF. Mitochondria were re-isolated by centrifugation at 10,000 g for 15 min, pellets suspended in SDS-PAGE sample buffer and the proteins analyzed by Western blotting.
Supplementary Material
Acknowledgements
We thank all present and many past lab members for valuable discussions. We especially thank Andre Schneider for advice on protease digestions of isolated mitochondria and for communicating his results on the T. brucei mitochondrial OM proteome prior to publication. M.L.P. especially thanks Keith Gull for the opportunity to spend several months in his lab at Oxford University. We also thank John Burg and Peter Espenshade for help with trypanosome lysis, Joanne Hullihen and Pete Pedersen for their assistance with O2 measurements, and Michael McCaffery (and his Johns Hopkins Integrated Imaging Center) for wonderful expertise with thin-section EM and cryo-immuno EM. We are also thankful for antibodies to cytochrome c and porin (A. Schneider), trypanosome alternate oxidase (M. Chaudhuri), Rieske iron-sulfur protein (T. Ochsenreiter and S. Hajduk), and carnitine O-palmitoyl transferase (L. Simpson). This work was supported by NIH grant AI058613 to R.E.J. and P.T.E, and a Wellcome Trust principal fellowship and a Wellcome Trust program grant to K.G.
References
- Abelovska L. Mitochondria as protean organelles: membrane processes that influence mitochondrial shape in yeast. Gen. Physiol. Biophys. 2011;30 doi: 10.4149/gpb_2011_SI1_13. Spec No: S13-24. [DOI] [PubMed] [Google Scholar]
- Baines A, Gull K. WCB is a C2 domain protein defining the plasma membrane - sub-pellicular microtubule corset of kinetoplastid parasites. Protist. 2008;159:115–125. doi: 10.1016/j.protis.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Bakshi RP, Shapiro TA. RNA interference of Trypanosoma brucei topoisomerase IB: both subunits are essential. Mol. Biochem. Parasitol. 2004;136:249–255. doi: 10.1016/j.molbiopara.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Berger KH, Sogo LF, Yaffe MP. Mdm12p, a component required for mitochondrial inheritance that is conserved between budding and fission yeast. J. Cell Biol. 1997;136:545–553. doi: 10.1083/jcb.136.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldogh IR, Nowakowski DW, Yang HC, Chung H, Karmon S, Royes P, Pon LA. A protein complex containing Mdm10p, Mdm12p, and Mmm1p links mitochondrial membranes and DNA to the cytoskeleton-based segregation machinery. Mol. Biol. Cell. 2003;14:4618–4627. doi: 10.1091/mbc.E03-04-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F, Biteau N, Melville SE, Hez S, El-Sayed NM, Leech V, Berriman M, Hall N, Donelson JE, Baltz T. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot Cell. 2002;1:137–151. doi: 10.1128/EC.1.1.137-151.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun R, Schonenberger Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Short communication. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N. Importing mitochondrial proteins: machineries and mechanisms. Cell. 2009;138:628–644. doi: 10.1016/j.cell.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanez AL, Hehl AB, Engstler M, Schneider A. Ablation of the single dynamin of T. brucei blocks mitochondrial fission and endocytosis and leads to a precise cytokinesis arrest. J. Cell Sci. 2006;119:2968–2974. doi: 10.1242/jcs.03023. [DOI] [PubMed] [Google Scholar]
- Chaudhuri M, Ajayi W, Hill GC. Biochemical and molecular properties of the Trypanosoma brucei alternative oxidase. Mol. Biochem. Parasitol. 1998;95:53–68. doi: 10.1016/s0166-6851(98)00091-7. [DOI] [PubMed] [Google Scholar]
- Claros MG, Vincens P. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- Clayton AM, Guler JL, Povelones ML, Gluenz E, Gull K, Smith TK, Jensen RE, Englund PT. Depletion of mitochondrial acyl carrier protein in bloodstream-form Trypanosoma brucei causes a kinetoplast segregation defect. Eukaryot Cell. 2011;10:286–292. doi: 10.1128/EC.00290-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detke S, Elsabrouty R. Identification of a mitochondrial ATP synthase-adenine nucleotide translocator complex in Leishmania. Acta Trop. 2008;105:16–20. doi: 10.1016/j.actatropica.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Dimmer KS, Jakobs S, Vogel F, Altmann K, Westermann B. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast. J. Cell Biol. 2005;168:103–115. doi: 10.1083/jcb.200410030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downey N, Hines JC, Sinha KM, Ray DS. Mitochondrial DNA ligases of Trypanosoma brucei. Eukaryot Cell. 2005;4:765–774. doi: 10.1128/EC.4.4.765-774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Effron PN, Torri AF, Engman DM, Donelson JE, Englund PT. A mitochondrial heat shock protein from Crithidia fasciculata. Mol. Biochem. Parasitol. 1993;59:191–200. doi: 10.1016/0166-6851(93)90217-l. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Englund PT, Agbo EE, Lindsay ME, Liu B, Liu Y, Motyka SA, Yildirir G, Zhao Z. RNAi libraries and kinetoplast DNA. Biochem. Soc. Trans. 2005;33:1409–1412. doi: 10.1042/BST0331409. [DOI] [PubMed] [Google Scholar]
- Esseiva AC, Chanez AL, Bochud-Allemann N, Martinou JC, Hemphill A, Schneider A. Temporal dissection of Bax-induced events leading to fission of the single mitochondrion in Trypanosoma brucei. EMBO Rep. 2004;5:268–273. doi: 10.1038/sj.embor.7400095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridberg A, Olson CL, Nakayasu ES, Tyler KM, Almeida IC, Engman DM. Sphingolipid synthesis is necessary for kinetoplast segregation and cytokinesis in Trypanosoma brucei. J. Cell Sci. 2008;121:522–535. doi: 10.1242/jcs.016741. [DOI] [PubMed] [Google Scholar]
- Fujiki Y, Fowler S, Shio H, Hubbard AL, Lazarow PB. Polypeptide and phospholipid composition of the membrane of rat liver peroxisomes: comparison with endoplasmic reticulum and mitochondrial membranes. J. Cell Biol. 1982;93:103–110. doi: 10.1083/jcb.93.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibellini F, Hunter WN, Smith TK. The ethanolamine branch of the Kennedy pathway is essential in the bloodstream form of Trypanosoma brucei. Mol. Microbiol. 2009;73:826–843. doi: 10.1111/j.1365-2958.2009.06764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluenz E, Povelones ML, Englund PT, Gull K. The kinetoplast duplication cycle in Trypanosoma brucei is orchestrated by cytoskeleton-mediated cell morphogenesis. Mol. Cell. Biol. 2011;31:1012–1021. doi: 10.1128/MCB.01176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluenz E, Shaw MK, Gull K. Structural asymmetry and discrete nucleic acid subdomains in the Trypanosoma brucei kinetoplast. Mol. Microbiol. 2007;64:1529–1539. doi: 10.1111/j.1365-2958.2007.05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grams J, Morris JC, Drew ME, Wang Z, Englund PT, Hajduk SL. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 2002;277:16952–16959. doi: 10.1074/jbc.M200662200. [DOI] [PubMed] [Google Scholar]
- Guler JL, Kriegova E, Smith TK, Lukes J, Englund PT. Mitochondrial fatty acid synthesis is required for normal mitochondrial morphology and function in Trypanosoma brucei. Mol. Microbiol. 2008;67:1125–1142. doi: 10.1111/j.1365-2958.2008.06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Hobbs AE, Srinivasan M, McCaffery JM, Jensen RE. Mmm1p, a mitochondrial outer membrane protein, is connected to mitochondrial DNA (mtDNA) nucleoids and required for mtDNA stability. J. Cell Biol. 2001;152:401–410. doi: 10.1083/jcb.152.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Jofuku A, Eura Y, Mihara K. Regulation of mitochondrial morphology by membrane potential, and DRP1-dependent division and FZO1-dependent fusion reaction in mammalian cells. Biochem. Biophys. Res. Commun. 2003;301:891–898. doi: 10.1016/s0006-291x(03)00050-0. [DOI] [PubMed] [Google Scholar]
- Jensen RE. Control of mitochondrial shape. Curr. Opin. Cell Biol. 2005;17:384–388. doi: 10.1016/j.ceb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Osman C, Walter P. The conserved GTPase Gem1 regulates endoplasmic reticulum-mitochondria connections. Proc. Natl. Acad. Sci. U. S. A. 2011;108:14151–14156. doi: 10.1073/pnas.1111314108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornmann B, Walter P. ERMES-mediated ER-mitochondria contacts: molecular hubs for the regulation of mitochondrial biology. J. Cell Sci. 2010;123:1389–1393. doi: 10.1242/jcs.058636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Levy M, Toury R, Andre J. [Purification and enzymatic characterization of the external membrane of mitochondria]. C. R. Acad. Sci. Hebd. Seances Acad. Sci. D. 1966;263:1766–1769. [PubMed] [Google Scholar]
- Li Z, Lindsay ME, Motyka SA, Englund PT, Wang CC. Identification of a bacterial-like HslVU protease in the mitochondria of Trypanosoma brucei and its role in mitochondrial DNA replication. PLoS Pathog. 2008;4:e1000048. doi: 10.1371/journal.ppat.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lineberger B, Dawicki DD, Agarwal KC, Kessimian N, Steiner M. Permeabilization of platelets: an investigation of biochemical, ultrastructural and functional aspects. Biochim. Biophys. Acta. 1989;1012:36–45. doi: 10.1016/0167-4889(89)90008-6. [DOI] [PubMed] [Google Scholar]
- Liu B, Liu Y, Motyka SA, Agbo EE, Englund PT. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 2005;21:363–369. doi: 10.1016/j.pt.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Liu B, Wang J, Yaffe N, Lindsay ME, Zhao Z, Zick A, Shlomai J, Englund PT. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle replication. Mol. Cell. 2009;35:490–501. doi: 10.1016/j.molcel.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukes J, Hashimi H, Zikova A. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 2005;48:277–299. doi: 10.1007/s00294-005-0027-0. [DOI] [PubMed] [Google Scholar]
- Morgan GW, Goulding D, Field MC. The single dynamin-like protein of Trypanosoma brucei regulates mitochondrial division and is not required for endocytosis. J. Biol. Chem. 2004;279:10692–10701. doi: 10.1074/jbc.M312178200. [DOI] [PubMed] [Google Scholar]
- Morris JC, Wang Z, Drew ME, Englund PT. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 2002;21:4429–4438. doi: 10.1093/emboj/cdf474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morton DJ, Hoppel C, Cooper C. The action of digitonin on rat liver mitochondria. Electron microscopy. Biochem. J. 1968;107:377–380. doi: 10.1042/bj1070377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motyka SA, Drew ME, Yildirir G, Englund PT. Overexpression of a cytochrome b5 reductase-like protein causes kinetoplast DNA loss in Trypanosoma brucei. J. Biol. Chem. 2006;281:18499–18506. doi: 10.1074/jbc.M602880200. [DOI] [PubMed] [Google Scholar]
- Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- Ntambi JM, Englund PT. A gap at a unique location in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 1985;260:5574–5579. [PubMed] [Google Scholar]
- Ogbadoyi EO, Robinson DR, Gull K. A high-order trans-membrane structural linkage is responsible for mitochondrial genome positioning and segregation by flagellar basal bodies in trypanosomes. Mol. Biol. Cell. 2003;14:1769–1779. doi: 10.1091/mbc.E02-08-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Voelker DR, Langer T. Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 2011;192:7–16. doi: 10.1083/jcb.201006159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Mihara K. Molecular mechanisms and physiologic functions of mitochondrial dynamics. J Biochem. 2011;149:241–251. doi: 10.1093/jb/mvr002. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno G, Sealey-Cardona M, Rodrigues-Poveda C, Gelb MH, Ruiz-Perez LM, Castillo-Acosta V, Urbina JA, Gonzalez-Pacanowska D. Endogenous sterol biosynthesis is important for mitochondrial function and cell morphology in procyclic forms of Trypanosoma brucei. Int. J. Parasitol. 2012 doi: 10.1016/j.ijpara.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Pitts KR, Yoon Y, Krueger EW, McNiven MA. The dynamin-like protein DLP1 is essential for normal distribution and morphology of the endoplasmic reticulum and mitochondria in mammalian cells. Mol. Biol. Cell. 1999;10:4403–4417. doi: 10.1091/mbc.10.12.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plutner H, Davidson HW, Saraste J, Balch WE. Morphological analysis of protein transport from the ER to Golgi membranes in digitonin-permeabilized cells: role of the P58 containing compartment. J. Cell Biol. 1992;119:1097–1116. doi: 10.1083/jcb.119.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot M, Zhang YZ, Kramer JA, Wells KS, Jones LJ, Hanzel DK, Lugade AG, Singer VL, Haugland RP. Analysis of mitochondrial morphology and function with novel fixable fluorescent stains. J. Histochem. Cytochem. 1996;44:1363–1372. doi: 10.1177/44.12.8985128. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J. Bioenerg. Biomembr. 1994;26:179–191. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- Pusnik M, Charriere F, Maser P, Waller RF, Dagley MJ, Lithgow T, Schneider A. The single mitochondrial porin of Trypanosoma brucei is the main metabolite transporter in the outer mitochondrial membrane. Mol. Biol. Evol. 2009;26:671–680. doi: 10.1093/molbev/msn288. [DOI] [PubMed] [Google Scholar]
- Rieder SE, Banta LM, Kohrer K, McCaffery JM, Emr SD. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 1996;7:985–999. doi: 10.1091/mbc.7.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnaitman C, Erwin VG, Greenawalt JW. The submitochondrial localization of monoamine oxidase. An enzymatic marker for the outer membrane of rat liver mitochondria. J. Cell Biol. 1967;32:719–735. doi: 10.1083/jcb.32.3.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Bouzaidi-Tiali N, Chanez AL, Bulliard L. ATP production in isolated mitochondria of procyclic Trypanosoma brucei. Methods Mol. Biol. 2007a;372:379–387. doi: 10.1007/978-1-59745-365-3_27. [DOI] [PubMed] [Google Scholar]
- Schneider A, Charriere F, Pusnik M, Horn EK. Isolation of mitochondria from procyclic Trypanosoma brucei. Methods Mol. Biol. 2007b;372:67–80. doi: 10.1007/978-1-59745-365-3_5. [DOI] [PubMed] [Google Scholar]
- Sharma S, Singha UK, Chaudhuri M. Role of Tob55 on mitochondrial protein biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010;174:89–100. doi: 10.1016/j.molbiopara.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlomai J. The structure and replication of kinetoplast DNA. Curr Mol Med. 2004;4:623–647. doi: 10.2174/1566524043360096. [DOI] [PubMed] [Google Scholar]
- Siegel TN, Hekstra DR, Wang X, Dewell S, Cross GA. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010;38:4946–4957. doi: 10.1093/nar/gkq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorell A, Gluenz E, Rettig J, Schneider A, Shaw MK, Gull K, Butikofer P. Perturbation of phosphatidylethanolamine synthesis affects mitochondrial morphology and cell-cycle progression in procyclic-form Trypanosoma brucei. Mol. Microbiol. 2009;72:1068–1079. doi: 10.1111/j.1365-2958.2009.06713.x. [DOI] [PubMed] [Google Scholar]
- Sogo LF, Yaffe MP. Regulation of mitochondrial morphology and inheritance by Mdm10p, a protein of the mitochondrial outer membrane. J. Cell Biol. 1994;126:1361–1373. doi: 10.1083/jcb.126.6.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens JL, Lee SH, Paul KS, Englund PT. Mitochondrial fatty acid synthesis in Trypanosoma brucei. J. Biol. Chem. 2007;282:4427–4436. doi: 10.1074/jbc.M609037200. [DOI] [PubMed] [Google Scholar]
- Stuart KD, Schnaufer A, Ernst NL, Panigrahi AK. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Toulmay A, Prinz WA. Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 2011;23:458–463. doi: 10.1016/j.ceb.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vickerman K. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 1985;41:105–114. doi: 10.1093/oxfordjournals.bmb.a072036. [DOI] [PubMed] [Google Scholar]
- Wang Z, Drew ME, Morris JC, Englund PT. Asymmetrical division of the kinetoplast DNA network of the trypanosome. EMBO J. 2002;21:4998–5005. doi: 10.1093/emboj/cdf482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Englund PT. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 2001;20:4674–4683. doi: 10.1093/emboj/20.17.4674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Morris JC, Drew ME, Englund PT. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 2000;275:40174–40179. doi: 10.1074/jbc.M008405200. [DOI] [PubMed] [Google Scholar]
- Westermann B. Molecular machinery of mitochondrial fusion and fission. J. Biol. Chem. 2008 doi: 10.1074/jbc.R800011200. [DOI] [PubMed] [Google Scholar]
- Youngman MJ, Hobbs AE, Burgess SM, Srinivasan M, Jensen RE. Mmm2p, a mitochondrial outer membrane protein required for yeast mitochondrial shape and maintenance of mtDNA nucleoids. J. Cell Biol. 2004;164:677–688. doi: 10.1083/jcb.200308012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Lindsay ME, Roy Chowdhury A, Robinson DR, Englund PT. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 2008;27:143–154. doi: 10.1038/sj.emboj.7601956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.