Abstract
Rationale and Objectives
Renal perfusion measurements using non-invasive Arterial Spin Labeled (ASL) Magnetic Resonance Imaging (MRI) techniques are gaining interest. Currently, focus has been on perfusion in the context of renal transplant. Our objectives were to explore the use of ASL in patients with renal cancer, and to evaluate three-dimensional (3D) fast spin echo (FSE) acquisition, a robust volumetric imaging method for abdominal applications. We evaluate 3D ASL perfusion MRI in the kidneys compared to two-dimensional (2D) ASL in patients and healthy subjects.
Materials and Methods
Isotropic resolution (2.6×2.6×2.8 mm3) 3D ASL using segmented FSE was compared to 2D single-shot FSE. ASL used pseudo-continuous labeling, suppression of background signal, and synchronized breathing. Quantitative perfusion values and signal-to-noise-ratio (SNR) were compared between 3D and 2D ASL in four healthy volunteers and semi-quantitative assessments were made by four radiologists in four patients with known renal masses (primary renal cell carcinoma).
Results
Renal cortex perfusion in healthy subjects was 284 ± 21 mL/100g/min, with test-retest repeatability of 8.8 %. No significant differences were found between the quantitative perfusion value or SNR in volunteers between 3D and 2D ASL, or in 3D ASL with synchronized or free breathing. In patients, semi-quantitative assessment by radiologists showed no significant difference in image quality between 2D and 3D ASL. In one case, 2D ASL missed a high perfusion focus in a mass that was seen by 3D ASL.
Conclusions
3D ASL renal perfusion imaging provides isotropic-resolution images, with comparable quantitative perfusion values and image SNR in similar imaging time to single-slice 2D ASL.
Keywords: MRI, Arterial Spin Labeling, Perfusion, Blood flow, Renal Cell Carcinoma, Kidney disease
INTRODUCTION
Imaging the distribution and heterogeneity of tissue perfusion is an important component of clinical identification and characterization of primary and metastatic cancer. Quantitative perfusion measurements in tumours may be important for monitoring disease progression (1), in particular in response to antiangiogenic therapy (2–6), and may play a role in assessing the early changes of disease or in understanding normal physiology. There is increasing interest in perfusion measurements as a biomarker for assessing renal function and for characterizing renal masses. Quantitative perfusion is reduced in renal insufficiency and in hemodynamically significant renal artery stenosis (7–10). In renal cell carcinoma (RCC), perfusion has proven value because of the relationship between angiogenesis, prognosis, and response to different targeted therapies in these tumours (11–17).
Arterial Spin Labeling (ASL) is a well-established method for measuring tissue perfusion (18–20) that has been widely used in quantitative perfusion measurements of the brain with application to brain tumours (21–24), cerebrovascular disease and stroke, epilepsy and dementia (25). A major advantage of ASL is the relative ease with which ASL images can be converted to quantitative images of tissue perfusion. ASL employs external magnetic fields to label nuclear magnetization of endogenous water in arterial blood and then observes the effect on tissue signal after the water flows into and diffuses throughout the tissue. Freely diffusible endogenous water is an excellent tracer for perfusion that compares well with intra-venous-administered contrast material, because of its lower risk for renal patients and because signal is linear in concentration and independent of venous bolus dynamics and vessel permeability effects that complicate quantification of perfusion with intra-venous contrast agents.
ASL has been successfully applied to imaging perfusion in organs and lesions in the abdomen (8,10–13,15,16,26–40). Initial studies have focused on single-slice two-dimensional (2D) imaging. While these approaches have met with some success, their spatial coverage limits the ability to visualize the full extent of disease. Multi-slice 2D imaging with ASL is possible (41) but sequential imaging after ASL preparation causes time delays that complicate quantification across slices, and interfere with strategies for reducing motion errors by background suppression. Background suppression (BGS) (42) has previously been shown to reduce signal fluctuations from physiological motion (32), particularly for ASL in the abdomen (32,42,43). Since full BGS can be achieved for only a short time, it favors combination with 3D acquisitions where the entire volume can be excited at a single time-point. Though segmented 3D volumetric acquisition requires more time to acquire the entire image than 2D imaging, multiple averages of 2D acquisitions are usually required to achieve sufficient signal-to-noise ratio so scan times are often comparable. In addition, acquiring 3D images with isotropic resolution allows for reformatting of the image data to enhance lesion characterization as has been widely implemented for contrast enhanced MRI of abdominal pathology, including renal masses (44,45).
A 3D volumetric approach for renal ASL has recently been reported (34,40) and applied to the assessment of renal perfusion in healthy kidney donors (39), but these studies were limited to assessing whole-kidney perfusion in healthy volunteers. In this work we implement and evaluate a 3D volumetric, isotropic resolution ASL technique using a Fast Spin Echo (FSE) acquisition. After evaluation in healthy volunteers, we assess semi-quantitatively its use in patients with primary renal cell carcinoma (RCC). Here, the benefit of 3D reformatting is expected to be significant for evaluating renal masses. To the best of our knowledge, this is the first such demonstration of an isotropic, 3D ASL measurement in renal cancer patients.
MATERIALS AND METHODS
Four healthy volunteers (2 female, aged 23—53, average 31) and four patients referred for pre-surgical MRI evaluation of known renal masses (2 female, aged 39—67, average 53) were imaged in this study. Healthy volunteers had no contraindications to MRI and had no known recent health problems. Four patients with renal masses on one kidney undergoing MRI examination prior to nephrectomy consented to additional imaging sequences. In one volunteer, only one kidney was evaluated owing to prior surgery on the other kidney. No subjects were excluded.
MRI Imaging
This study was performed on a 1.5 T GE EXCITE HDx MRI scanner using the body-coil for transmission and the product 8-channel body-array-receiver (GE Healthcare, Waukesha, WI).
Arterial Spin Labeling
The pseudo-continuous labeling (pCASL) technique was used in all cases, and has been previously described in detail (46). Labeling is applied for 1.5 sec (average radiofrequency strength 1.4 uT, average/maximum labeling gradient strength 0.7 / 7 mT/m) followed by a 1.5-sec post-labeling delay (47) before image acquisition. The labeling plane was positioned slightly inferior to the diaphragm, not intersecting the heart, to label blood in the descending aorta, as shown in Fig. 1.
Figure 1.
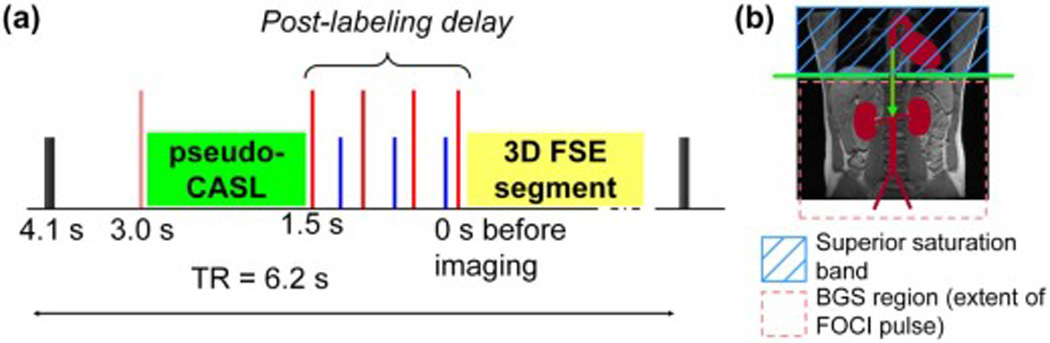
a) Pulse sequence diagram showing the arterial spin labeling preparation before the 3D FSE segment acquisition (yellow block): the 1.5-sec long pulsed-CASL labeling module is shown as a green block, saturation pulses (black) at 4.1 seconds prior to imaging, tall pulses are adiabatic inversion pulses, the inversion pulse before labeling is a frequency offset corrected pulse, shorter pulses during the 1.5-sec post-labeling delay are arterial saturation bands. A strong gradient killer pulse is played after the last inversion pulse before imaging (not shown for clarity). b) Labeling diagram showing pCASL labeling plane below the diaphragm labeling blood in the descending aorta, the extent of the background suppression (BGS) region, defined by the selective pre-saturation and FOCI pulses shown by the pink dashed-box, and the arterial saturation bands shown as a blue cross-hatched box.
Background Suppression
Spatially selective pre-saturation pulses were played 4100 ms prior to imaging followed by a spatially selective “C-shaped” frequency-offset-corrected (FOCI) inversion pulse (48) (10.8-kHz bandwidth, 15.36-ms, β = 809 s−1, µ = 3.9), chosen for superior spatial selectivity, played 3000 ms prior to imaging. Four additional non-selective Silver-Hoult pulses 10-ms duration, 1.8-kHz bandwidth, β = 1242 s−1, µ = 4.5), were played at times: 1500, 680, 248, and 57 ms prior to imaging, as shown in the pulse sequence diagram in Fig. 1. These pulse timings were chosen to null signal from magnetization exhibiting a range of T1-recovery rates (49). Spatially selective BGS pre-saturation and inversion pulses were applied over the entire abdomen (380 mm) with the superior edge of the slab coinciding with the labeling plane. In this study focused on the kidneys, the imaging volume was centered on the renal arteries. The selective BGS pulses were extended inferiorly (100 mm below the image center) and asymmetrically about the imaging center to suppress signal in the veins flowing from foot-to-head that may flow into the imaging region during the preparation period prior to imaging. A strong killer gradient pulse was applied to de-phase residual transverse magnetization after the last BGS pulse.
Post-Labeling Arterial Saturation
Spatially selective saturation pulses (followed by a spoiling gradient) were applied to a 10-cm wide slab superior to the labeling plane to null post-labeling arterial inflow. Three pulses were played at times: 1037, 392, and 116 ms prior to imaging.
Perfusion Quantification
The perfusion difference image (dM) was calculated on the scanner as a complex subtraction in k-space between label and control images. For normal volunteers, this perfusion difference was divided by an M0 image acquired using the same acquisition sequence to match T2-weighted image contrast and resolution. To give M0 weighting, the same pre-saturation pulse was applied 4.1 sec prior to imaging and without any BGS inversion pulses or labeling pulses. For perfusion, one data set was acquired for each of the label and control images. For the M0 image, one data set was acquired, thus taking half the acquisition time of the perfusion data. The M0 image was not acquired in the renal cancer patients because of limited available time. Perfusion quantification used the expression given in Eq. 1 to calculate flow, f.
| [1] |
A constant T1,tissue of 970 ms, as previously measured for the renal cortex (50), was assumed across the entire image; for blood, T1,blood of 1300 sec was used (26). The arrival time of blood from the labeling location to the kidneys was taken to be δt = 750 ms (51); the labeling time was τ = 1500 ms, and the post-labeling delay was w = 1500 ms. The tissue/blood partition coefficient used was λ = 0.9 (52,53). The inversion efficiency of the pCASL labeling was 0.8 (46), with an additional inversion inefficiency from the background suppression inversion pulses of 0.75 (54), giving a total inversion efficiency of α = 0.6.
3D FSE acquisition
Continual spin-echo refocusing in the FSE approach offers robustness to increased off-resonance and non-uniform magnetic fields in the abdomen compared to gradient-echo based approaches. A segmented 3D FSE acquisition was used to limit the extent of T2 decay during the echo train. The k-space trajectory followed a radial sampling on a Cartesian grid pattern (55,56). Centric k-space ordering allowed a short echo time and reduced T2-contrast on the largest spatial scales. Subsequent echoes were ordered by radial distance in ky-kz space (56). To reduce the total number of phase-encodes required, sampling was limited to within an elliptical boundary in the ky-kz plane, in conjunction with 9/16 partial-Fourier acquisition in the ky dimension (56). The FSE acquisition employed a constant-flip-angle refocusing to limit sensitivity to physiological and residual respiratory motion. An echo train of 72 sinc-shaped refocusing pulses was used, each of flip angle 130°, with an echo spacing of 6.4 ms giving a 461-ms echo train. The echo train is long enough to expect T2-decay during the read-out, however, the radial sampling pattern imposes a symmetric modulation of k-space, and reasonably benign image blurring is expected as a result. Crusher gradients imposed a phase variation of 2 cycles per pixel in both the slice and read-out directions. One extra dummy refocusing pulse of flip angle 130° was used to stabilise the echo train.
Sagittal-orientated acquisition slabs were used to allow the smallest volume (i.e. slab thickness) and hence scan time to cover an entire single-kidney. The field-of-view (FOV) was 340 × 238 mm, acquired with a 128 × 92 matrix and a receiver bandwidth of ±19.63 kHz, with frequency-encoding in the head-foot direction. Images of a single kidney were acquired with slab-selective pulses; 30—34 slices with 2.8-mm thickness were acquired to cover the entire kidney, giving a near isotropic 2.6 × 2.6 × 2.8-mm resolution. Each echo-train was acquired after a single excitation following a spin-labeling preparation. The repetition time between echo-train acquisitions was 6.2 sec. Typically, 23 echo-trains (depending on the number of slices, 92 × 9/16 × 32 slices / 72 = 23 shots) were needed to acquire volumetric data, giving a scan time of ~2.5 minutes for each of the labelled and control images, and therefore a scan time of ~5 minutes for each perfusion difference image. Two extra repetitions at the beginning of the acquisition were acquired and discarded to establish a steady state of the background signal. The 3D M0 image took ~2.5 minutes to acquire. Only one image was acquired without signal averaging as volumetric phase-encoding provides noise reduction comparable to the 2D image averaging normally employed in ASL imaging. Subjects were coached so that respiration was synchronized to follow the 6.2-sec repetition time, as has been successfully demonstrated in previous studies (26,32,57). Each subject was instructed to begin breathing after the sound of the image acquisition, and cease breathing at end-expiration before the next acquisition. The trace from the respiratory abdominal bellows was monitored during the acquisition to ensure compliance during the acquisition.
2D Single-Shot-FSE (SSFSE) Acquisition
A coronal 2D single-shot FSE (SSFSE) acquisition was acquired covering both kidneys with the frequency-encoding in the superior-inferior direction. The FOV was 340 mm, as for the 3D acquisition, with a 128 × 128 image matrix, 2.6 × 2.6-mm pixel resolution and 10-mm slice thickness. The 2D SSFSE acquisition comprised a similar train of 130° sinc-shaped refocusing pulses with an echo spacing of 6.9 ms. 9/16 partial Fourier acquisition gave a 72-pulse echo train and a 497 ms read-out. Crusher gradients on slice- and frequency-encode dimensions were matched to the 3D acquisition, imparting 2 cycles per pixel. 16 label-control pairs were signal averaged. An M0 image with the pre-saturation pulse and no other BGS inversion pulses was also acquired as part of the imaging sequence. The repetition time between acquisitions was 6 sec, giving a scan time of 3.5 minutes. Synchronized breathing was used with 2D acquisitions. The same ASL labeling preparation and quantification was used with both the 3D FSE and 2D SSFSE acquisitions.
Quantitative Perfusion Imaging in Volunteers
In the four healthy volunteers, a 3D volumetric ASL perfusion image of a single kidney ("test") was acquired followed by a repeated perfusion image acquisition ("retest") as well as an M0 reference image. This block of images was obtained for the left and right kidneys separately. A coronal single-slice 2D ASL perfusion image was also acquired covering both kidneys. Each of the blocks: “3D left”, “3D right” and “2D” were acquired in a randomized order in each of the volunteers. Finally, a 3D perfusion image of each kidney was acquired during free breathing. The whole imaging session was approximately 1 hour.
Renal Masses
In four patients, each with a known renal mass involving one of their kidneys, 2D and 3D perfusion difference images were acquired prior to their clinical MRI imaging protocol. 3D perfusion imaging encompassed the entire kidney, whilst the 2D perfusion slice was placed through the middle of the lesion as identified on multi-slice 2D T2-weighted SSFSE imaging. To limit the additional time of the patients’ studies, no M0 reference image was acquired, and consequently, quantification of perfusion could not be performed. Instead, semi-quantitative assessment of perfusion difference image quality was performed by four radiologists.
Statistical Analysis
For perfusion analysis in the volunteers, an average quantitative perfusion value was found for a region of interest (ROI) encompassing the whole cortex, excluding the renal medulla, of each kidney and an average over all kidneys in all four subjects was found. For 3D images, ROIs were drawn on sagittal slices formed from four consecutive perfusion difference image-slices averaged together to match slice thickness to the 2D acquisition and improve depiction of the renal anatomy. The bright perfusion signal on difference images was used to identify the renal cortex ROI, and then the ROI was transferred to the quantitative perfusion maps for regional averaging. ROIs in all slices were used to find an average perfusion value from the whole of the imaged renal cortex. For free-breathing studies, where the cortex perfusion signal was blurred by respiratory motion, drawing cortex ROIs occasionally required estimation of edges by comparison to the images acquired with synchronized breathing. For 2D images, ROIs were drawn on the perfusion difference images in the native coronal plane to encompass the whole of the imaged renal cortex. A two-sided paired t-test was used to compare the average perfusion values from the 3D FSE and 2D SSFSE acquisitions. Average perfusion at test and retest was compared by a two-sided paired t-test across all 4 volunteers (one kidney in one subject was not imaged); and similarly for respiratory synchronized and free breathing. For perfusion quantification t-tests, the significance threshold was set at p=0.05. Test-retest repeatability (R) was taken to be the standard deviation of differences of flow at test and retest, divided by the average flow of all kidneys.
Perfusion difference signal in the cortex was measured in one 2.8-mm slice from the 3D set ("thin-slice"), an additional higher SNR slice formed from 4 consecutive 3D partitions averaged together, giving an 11.2-mm slice ("thick-slice"), as well as on 2D images, which had a slice thickness of 10 mm. Image reconstruction for partial Fourier data resulted in phase-corrected real images from which contiguous slices were averaged. Coil sensitivities estimated from the center of k-space (58) used in partial Fourier reconstruction were used in the coil-weighted image combination (59). A region of noise outside the body was selected and noise standard deviation estimated. Without parallel imaging and with coil-sensitivities estimated from the image itself, there was no spatially dependent noise amplification resulting in a fair SNR comparison between 3D and 2D sequences (60). SNR values were averaged over all kidneys in all subjects. In addition, normalized SNR (SNRnorm) was also calculated according to Eq. 2, where the SNR was normalized to the slice thickness (s2D,3D) and square-root-of-imaging-time of the 2D acquisition. Imaging time was taken as the number of effective signal averages, 23 echo trains (NET) for 3D FSE and 16 label control pairs (Nlcp) for 2D SSFSE, according to Eq. 2. Average perfusion difference SNR values were compared between 2D acquisitions and 3D acquisitions with thick slices, as well as each of thin slices and thick slices when normalized. In addition, comparison was made between 3D-acquired, normalized thick-slice SNR for respiratory synchronized and free breathing acquisition. In each case a two-sided paired t-test was used with the significance threshold set at p=0.05.
| Eq. [2] |
For semi-quantitative assessment in patients, orthogonal coronal and axial images were obtained by reformatting the 3D dataset. Four radiologists independently rated side-by-side perfusion difference images from 2D and 3D ASL datasets. A reformatted coronal slice from the 3D data-set matching the location of the 2D image was chosen. In each case, reviewers, who were blinded to the MRI acquisition technique used, indicated whether the 2D or 3D ASL showed sharper delineation of the perfusion features. For each reader, the probability of finding their particular set of preferred images, calculated from the binomial distribution given that it was equally likely to prefer either 2D or 3D, was found. This probability was averaged across all four readers. The cohort of four subjects supports a significance threshold of p=0.1 to indicate whether one sequence was preferred over another, given that a unanimous read in favor of one sequence would have a probability of p=0.0625. In addition, overall correspondence of the 2D and 3D perfusion images was assessed using a five-point scale. 1: no correspondence between perfusion features is seen; 2: some features are consistent between images; 3: most features are consistent between images; 4: features are consistent with some noticeable difference in SNR or blurring; 5: images are entirely consistent and extremely similar in appearance.
This study was conducted with Institutional Review Board approval and HIPAA-compliance. Written informed consent was obtained from all subjects.
RESULTS
Qualitative perfusion difference images, reformatted in orthogonal planes, and quantitative perfusion images from a volunteer with 3D FSE and 2D SSFSE acquisitions are shown in Fig. 2. For quantitative perfusion measurements (Table 1), there was no significant difference between perfusion values measured by 3D FSE and 2D SSFSE (p=0.9). There was no significant difference between perfusion values at test and retest (p=0.6), the test-retest repeatability of whole-cortex perfusion measurements for 3D FSE was R = 8.8%. There was no significant difference between perfusion values measured by 3D FSE with synchronized breathing and free breathing (p=0.7). For SNR measurements (Table 2), there was no significant difference in the SNR of perfusion difference signal in thick-slice 3D and 2D images (p=0.6) but it was lower for 3D (without statistical significance) when normalized for slice thickness and imaging time (p=0.06). SNR was higher for 3D compared to 2D (without statistical significance) for normalized thin-slice 3D images (p=0.07). There was no significant difference in the SNR of the perfusion difference 3D-images acquired with synchronized breathing or free breathing (p=0.2).
Figure 2.
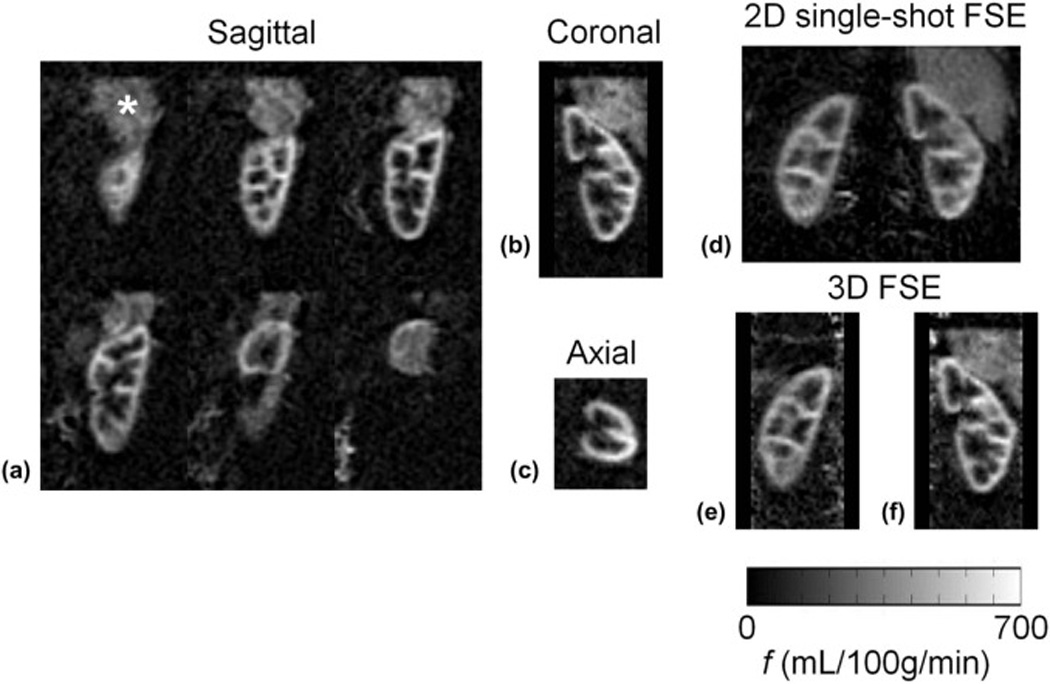
Perfusion difference images from one 5-minute 3D FSE sagittal acquisition of the left kidney (a) shown with reformatted orthogonal planes (b,c) covering the whole left kidney in a volunteer. Data was acquired with near-isotropic 2.6×2.6×2.8 mm resolution; images are displayed with 4 consecutive slices averaged giving 11.2 mm slice thickness to improve SNR. Excellent depiction of the renal cortex is seen; the bright signal above the kidney (*) is perfusion in the spleen. Quantitative perfusion images are shown for one volunteer from d) 2D single-shot FSE data, and (e–f) coronal reformats of 3D FSE data of the left and right kidneys. Agreement of quantification between acquisition methods is good.
Table 1.
Quantitative Perfusion Measurements (mL/100g/min)
| Sequence | 3D FSE | 2D single- shot FSE |
||
|---|---|---|---|---|
| Test | Test | Retest | Free Breathing |
Test |
| Kidney 1 | 239 | 236 | 227 | 231 |
| Kidney 2 | 222 | 227 | 201 | 194 |
| Kidney 3 | 318 | 290 | 285 | 272 |
| Kidney 4 | 254 | 206 | 348 | 280 |
| Kidney 5 | 377 | 375 | 298 | 347 |
| Kidney 6 | 319 | 337 | 345 | 333 |
| Kidney 7 | 261 | 282 | 236 | 316 |
| Mean ± SE | 284 ± 21 | 279 ± 23 | 277 ± 22 | 282 ± 21 |
Quantitative renal cortex perfusion measurements are given in mL/100g/min for each of the tests and sequences. Kidneys 1–7 are left and right from normal volunteers 1–4. One kidney in one volunteer was not imaged. Mean ± standard error over 7 kidneys is also given. 2D/3D = two/three-dimensional; FSE = fast spin echo.
Table 2.
Quantitative Perfusion Difference Image SNR
| Sequence | 3D FSE | 2D single-shot FSE |
||
|---|---|---|---|---|
| Slice thickness | 11.2 mm | 2.8 mm | 11.2 mm | 10 mm |
| Synchronized Breathing | Yes | Yes | No | Yes |
| Kidney 1 | 5.85 | 3.0 | 4.9 | 7.8 |
| Kidney 2 | 5.82 | 3.2 | 4.1 | 6.0 |
| Kidney 3 | 8.07 | 4.6 | 5.7 | 8.9 |
| Kidney 4 | 6.37 | 4.2 | 9.3 | 8.8 |
| Kidney 5 | 14.2 | 8.3 | 5.2 | 14.1 |
| Kidney 6 | 11.1 | 6.2 | 8.3 | 12.5 |
| Kidney 7 | 5.9 | 3.4 | 4.3 | 6.0 |
| Average SNR (± SD) | 8.2 ± 3.3 | 4.7 ± 1.9 | 6.0 ± 2.0 | 9.2 ± 3.1 |
| Average SNRnorm (± SD) | 6.1 ± 2.1 | 14.1 ± 4.7 | 4.4 ± 1.3 | 9.2 ± 3.1 |
SNR values are calculated from average ROI signal on perfusion difference images on native 2.8-mm thick slices in the 3D sequence, or from the 11.2-mm thick average of 4 contiguous slices. The 2D slice was 10-mm thick. Kidneys 1–7 are left and right from normal volunteers 1–4. One kidney in one volunteer was not imaged. The average SNR ± standard deviation over 7 kidneys is also given, and finally the SNR value normalized for slice thickness and square-root-imaging time (SNRnorm). Also shown in one column are results from measurements made during free breathing compared to those with synchronized breathing. 2D/3D = two/three-dimensional; FSE = fast spin echo; ROI = region of interest; SNR = signal to noise ratio.
In all four patients, highly perfused masses and sub-regions within masses were identified that correlated with their appearance on 2D perfusion images and with anatomical T2-weighted imaging. Lesions in all patients were later identified as renal cell carcinoma, clear cell type by histology. In Fig. 3, a heterogeneously hyper-perfused lesion is assessed in reformatted orthogonal planes; images from a second patient are shown in Fig 4. The utility of 3D imaging for clinical applications is demonstrated in Fig. 5 where the 2D slice acquisition of this lesion misses the higher perfusion focus demonstrated posteriorly within the mass in the 3D dataset. Here, 3D coverage is obtained in approximately 5 minutes compared to 3.5 minutes for the single slice 2D acquisition. Fig. 6 shows the comparison of 2D images and reformatted 3D perfusion images for each of the four patients. Semi-quantitative assessment by radiologists indicated that there was no significant difference between 3D and 2D sequences when radiologists were asked to choose which image showed preferred sharpness (Table 3). The average probability of finding the radiologists' preferences between sequences was p=0.28. The average score of all subjects from all readers was 3.6 indicating modest agreement between 3D and 2D images (Table 4). All subjects showed similar agreement when averaged over readers, scoring between 3 and 3.75. Readers differed in their assessment of the agreement between 3D and 2D images, with averages over subjects between 2.75 and 4.25.
Figure 3.
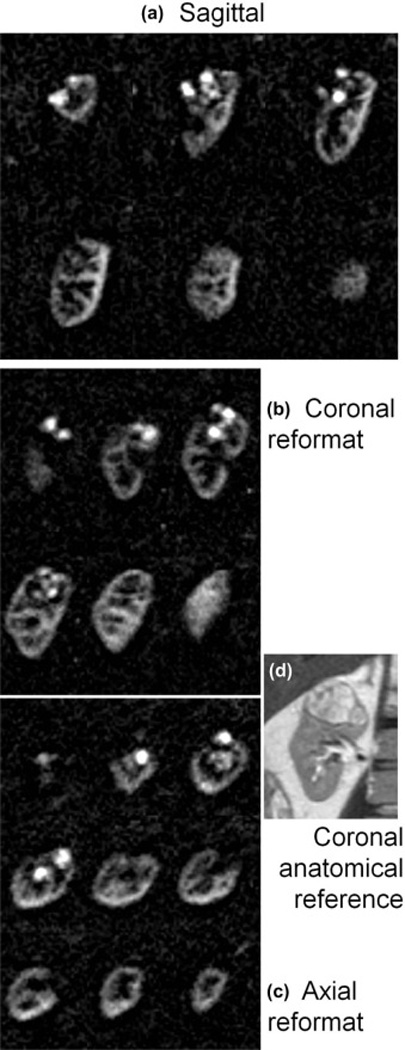
Three dimensional perfusion difference images in Patient 2 initially acquired in the sagittal plane with near-isotropic resolution 2.6×2.6×2.8 mm (a) allowed for reformatted images in b) coronal, and c) axial orientations, displayed with 11.2 mm slice thickness. Perfusion is clearly high compared to surrounding parenchyma and of a heterogeneous nature, which correlates well with the anatomical appearance of the lesion (2D multi-slice single-shot FSE) shown in d). Scan time for this 3D image data was ~5 min.
Figure 4.
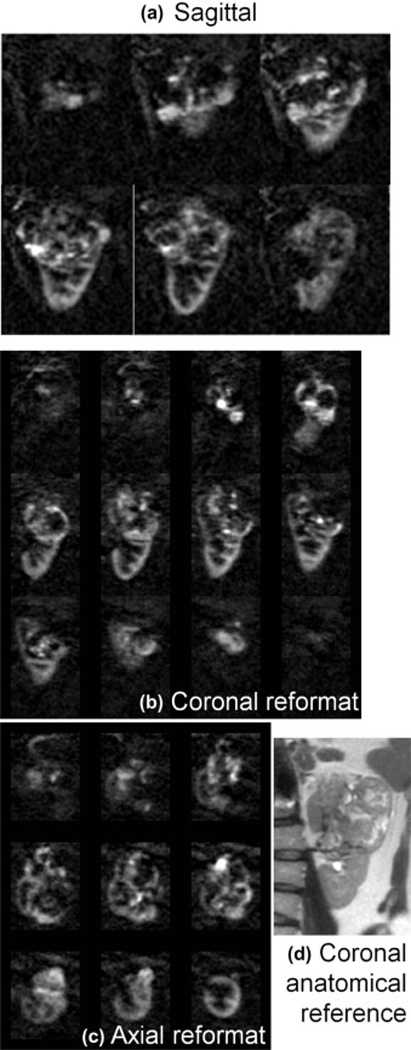
Three dimensional perfusion difference images in Patient 1 initially acquired in the sagittal plane with near-isotropic resolution 2.6×2.6×2.8 mm (a) allowed for reformatted images in b) coronal, and c) axial orientations, displayed with 11.2 mm slice thickness. Perfusion is clearly high compared to surrounding parenchyma and of a heterogeneous nature, which correlates well with the anatomical appearance of the lesion (2D multi-slice single-shot FSE) shown in d). The lesion is very large and complex, extending beyond the borders of the kidney. Scan time for this 3D image data was ~5 min.
Figure 5.
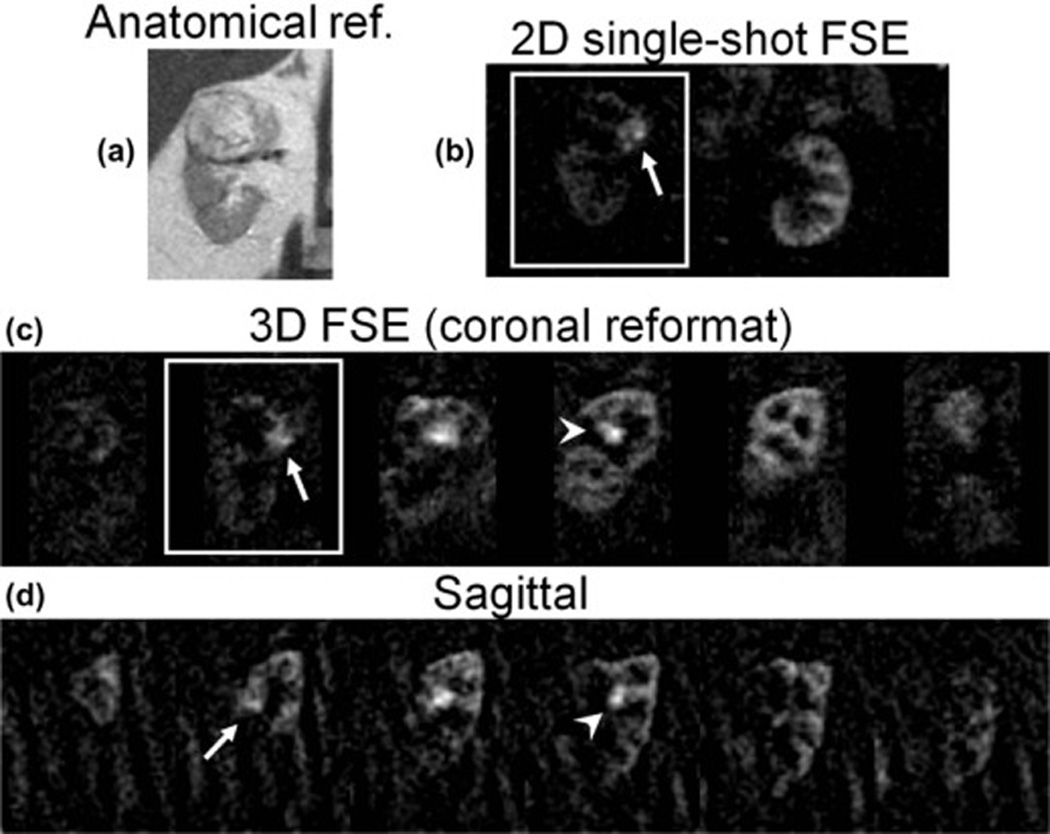
Utility of 3D coverage in clinical applications is demonstrated in images of a renal mass in Patient 3, shown in a 2D single-shot FSE anatomical reference image (a). Good agreement of perfusion difference images in the same slice (solid box) is seen between b) 2D single-shot FSE, and c) 3D FSE coronal reconstructed images, showing a highly perfused nodule (arrow); low perfusion is seen in the surrounding parenchyma. In the 3D perfusion image data (coronal, (c) and sagittal (d)), additional highly perfused foci are seen more anteriorly in the kidney (arrowhead), as well as perfusion of the parenchyma in posterior portions of the kidney.
Figure 6.
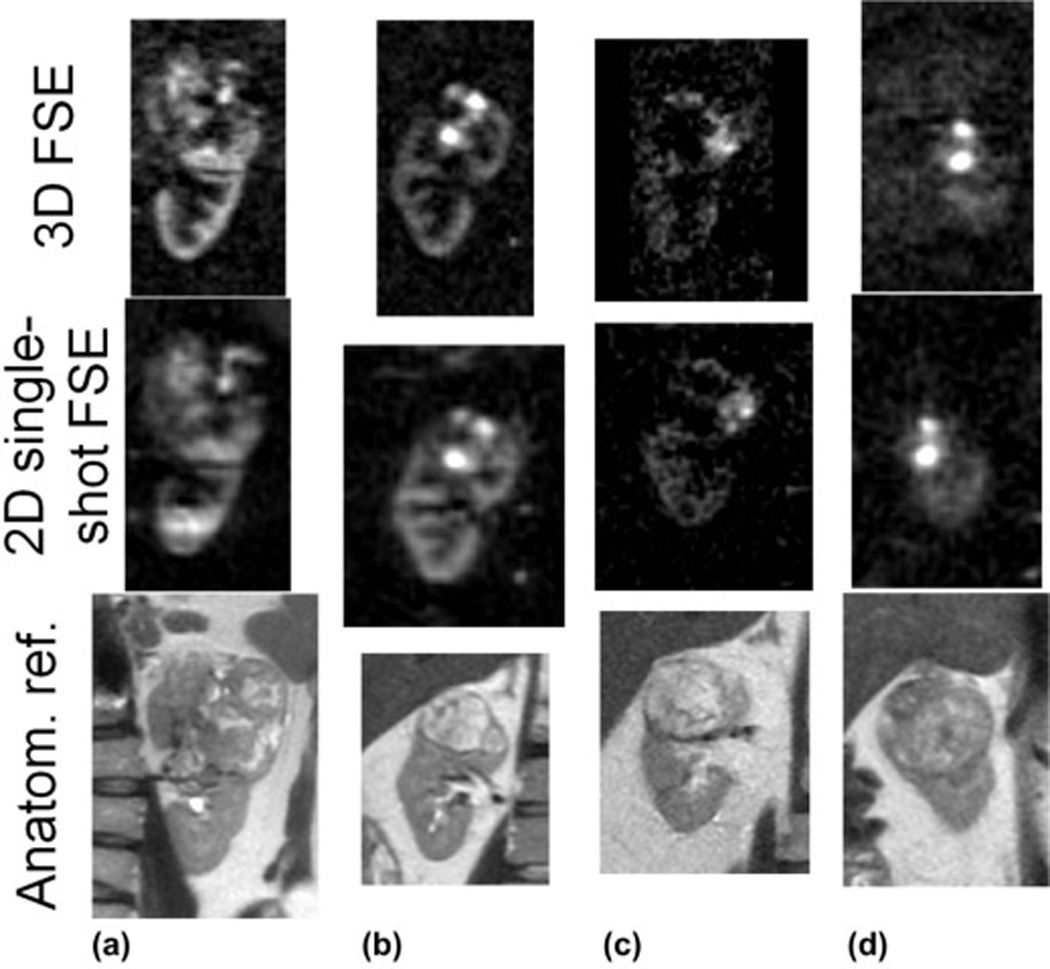
Comparison of perfusion difference images of renal masses in all patients. Single coronal slices reformatted from 3D FSE sagittal datasets (top) and coronal 2D single-shot FSE acquisitions (middle) are shown. Anatomical reference images (multi-slice single-shot FSE) are also shown (bottom). Perfusion image scan-time for 3D scans was ~5 min and for 2D single-shot FSE was 3.5 min.
Table 3.
Semi-quantitative assessment by radiologists: Rating of image sharpness by acquisition sequence.
| Reader | Patient | Totals | ||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | n2D | n3D | |
| 1 | 3D | 3D | 2D | 3D | 1 | 3 |
| 2 | 2D | 3D | 2D | 2D | 3 | 1 |
| 3 | 3D | 3D | 2D | 2D | 2 | 2 |
| 4 | 3D | 3D | 2D | 3D | 1 | 3 |
The sequence preferred for image sharpness is given for each patient and each reader. The data are summarized per reader on the right, with n2D and n3D being the number of patients for which 2D and 3D imaging was preferred. 2D/3D = two/three-dimensional.
Table 4.
Semi-quantitative assessment by radiologists: Scores for similarity of perfusion features between sequences.
| Reader | Patient | Average | |||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | ||
| 1 | 4 | 4 | 4 | 5 | 4.25 |
| 2 | 4 | 3 | 2 | 2 | 2.75 |
| 3 | 4 | 4 | 2 | 3 | 3.25 |
| 4 | 3 | 4 | 4 | 5 | 4 |
| Average | 3.75 | 3.75 | 3 | 3.75 | 3.56 |
Scores for the similarity in perfusion features between 2D and 3D sequences for each patient given by each reader are given, as well as averages by patient and reader.
DISCUSSION
Our study has confirmed that 3D segmented acquisition for renal ASL is feasible despite the initial concern that inter-segment motion might degrade image quality. In this study we sought to assess quantitatively the effect of segmented acquisition in a 3D FSE ASL sequence by establishing the test-retest repeatability of ASL measurements, and in addition by comparison of quantitative perfusion and SNR values with a single-shot 2D sequence. We investigated both a structured scenario with synchronized breathing where inter-segment differences of physiological and breath displacements dominate as potential sources of image degradation, and a free breathing scenario where averaged, gross respiratory motion dominates as potential sources of image degradation, In both cases, we found no significant difference in the quantitative perfusion values or image SNR between segmented 3D and single-shot 2D ASL. These results indicate that this segmented 3D scheme with FSE read-out, radial sampling, pCASL labeling with post-labeling delay, and background suppression is applicable to a patient population where control of breathing and involuntary motions may be poor, although larger studies will be needed to support this suggestion. In all of our patient examinations, perfusion images were of sufficient quality to assess perfusion of the lesion. However, in the volunteer study, it was observed that free breathing can introduce residual image blurring and artefact that can degrade image quality.
The repeatability of ASL in the abdomen has already been investigated by many authors, with which we find comparable values of cortical perfusion and repeatability, in the range of 5–10% (30,32,34).
Assessment by four radiologists indicated a trend for preferring the image quality of 3D segmented perfusion images over the 2D single-shot technique. For subjects 1 and 3 where there was higher perfusion in both the tumor and parenchyma, all readers rated 3D preferable for subject 3, and 3 of 4 rated 3D preferable for subject 1. We attribute this to the absence of partial volume degradation, a result of the thinner intrinsic slice thickness and wider spatial coverage in the 3D acquisition. An additional possibility is that the slice profile of the single-shot FSE acquisition may also be degraded by the short, weakly selective RF refocusing pulses employed. Further studies with larger cohorts of patients might elucidate whether statistically significant differences between the acquisition strategies exist.
We were able to acquire an image of an entire kidney at an isotropic resolution of approximately 2.6 mm in a reasonable scan time of 5 minutes. This resolution and scan time were appropriate for our application of single kidney lesion assessment. For studies of bilateral disease or cortical thinning, a different resolution or scan time may be better. It is worth noting that this acquisition did not yet employ techniques of parallel imaging for dual slab acquisition (61) or phase encode acceleration, both of which could provide options for faster scanning and greater coverage. Image acceleration could also be especially helpful for accelerating the reference image used for quantification, since this image is not SNR challenged.
Moving to higher field-strength, such as 3 tesla, might also be beneficial because of higher intrinsic SNR and longer blood and tissue T1, but higher field acquisitions will need to consider the power deposition and systematic errors from higher magnetic and RF transmit field non-uniformities.
The 3D acquisition presented here differs in a number of respects from one presented previously (34). First, we employ pCASL labeling at a single labeling duration compared to the multi-TI labeling used by Cutajar et al. (34). Multi-TI labeling may offer more accurate perfusion modeling, however, pCASL, which typically has higher labeling sensitivity, together with a single long labeling time as used here could be more appropriate for imaging renal masses that may exhibit a range of perfusion and arrival time values. Second, the FSE read-out should be more resistant to phase errors and distortions due to susceptibility and should prove more resilient to off-resonance effects over the broader anatomic coverage typically needed for abdominal studies compared with the gradient and spin echo (GRASE) sequence used in Ref. (34). The technique used here obtains a higher spatial resolution and an isotropic pixel dimension, offering the opportunity for utilizing meaningful multi-planar reformations during clinical interpretation (44). The radial sampling on a Cartesian grid used here may allow more flexibility in choosing the sampling pattern as compared to the distribution of the gradient and spin echoes in the ky and kz dimensions as required with the GRASE technique.
This study indicates that 3D ASL imaging of kidney perfusion merits assessment as a diagnostic tool for evaluating renal masses and other pathologies. Our limited sample was targeted at assessing technical feasibility, but studies of diagnostic value in larger patient cohorts with renal masses are necessary and in progress. Such studies should include a wider range of pathological subtypes with varying perfusion characteristics (e.g. papillary renal cell carcinoma that is typically hypoperfused (15). Further work on comparing ASL with gold standard perfusion measures in renal pathology is also needed, though the absence of definitive high resolution gold standards for human studies of perfusion remains a challenge.
In conclusion, this study has demonstrated successful implementation of volumetric 3D near-isotropic resolution (2.6×2.6×2.8-mm) perfusion imaging in the kidneys with ASL. ASL with segmented 3D FSE image-acquisition provides robust quantitative measurements of renal perfusion comparable to 2D single-shot acquisition techniques. In patients, this 3D technique allows volumetric assessment and is thus amenable to demonstrating intra-lesion heterogeneity of renal masses. This 3D approach provides near-isotropic data sets that can be reformatted in multiple planes, and with comparable SNR and in similar scan times to the single-slice approach. In at least one case, 3D acquisition permitted identification of higher perfusion lesions not appreciated on single slice 2D imaging. However, further clinical evaluation of 3D ASL perfusion imaging is warranted in order to more completely assess efficacy. It is hoped that 3D ASL perfusion imaging will have an important impact on the assessment of organs or diseases with heterogeneous perfusion and in quantitatively monitoring perfusion changes in longitudinal studies, particularly in the response of tumors to anti-angiogenic therapy.
Acknowledgements
This work was supported by NIH grants: R21 CA121570-01A1 and P50 CA101942-07. DCA is an inventor on patents related to pseudo-continuous ASL methods and receives royalties from GE Healthcare and Philips Healthcare through his employer.
Abbreviations
- 2D
Two-dimensional
- 3D
Three-dimensional
- ASL
Arterial Spin Labeled
- BGS
Background Suppression
- FOCI
Frequency-Offset Corrected Inversion
- FSE
Fast Spin Echo
- GRASE
Gradient and Spin Echo
- MRI
Magnetic Resonance Imaging
- pCASL
pulsed Continuous Arterial Spin Labeling
- RCC
Renal Cell Carcinoma
- ROI
Region of Interest
- SNR
Signal-to-noise-ratio
- SSFSE
Single-shot Fast Spin Echo
- T1
T1 relaxation constant
- T2
T2 relaxation constant
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Zanelli GD, Fowler JF. The measurement of blood perfusion in experimental tumors by uptake of 86Rb. Cancer Res. 1974;34(6):1451–1456. [PubMed] [Google Scholar]
- 2.Murata R, Overgaard J, Horsman MR. Comparative effects of combretastatin A-4 disodium phosphate and 5,6-dimethylxanthenone-4-acetic acid on blood perfusion in a murine tumour and normal tissues. Int J Radiat Biol. 2001;77(2):195–204. doi: 10.1080/09553000010007695. [DOI] [PubMed] [Google Scholar]
- 3.Anderson H, Yap JT, Wells P, Miller MP, Propper D, Price P, Harris AL. Measurement of renal tumour and normal tissue perfusion using positron emission tomography in a phase II clinical trial of razoxane. Br J Cancer. 2003;89(2):262–267. doi: 10.1038/sj.bjc.6601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Faria SC, Ng CS, Hess KR, Phongkitkarun S, Szejnfeld J, Daliani D, Charnsangavej C. CT quantification of effects of thalidomide in patients with metastatic renal cell carcinoma. AJR Am J Roentgenol. 2007;189(2):378–385. doi: 10.2214/AJR.07.2164. [DOI] [PubMed] [Google Scholar]
- 5.Sabir A, Schor-Bardach R, Wilcox CJ, Rahmanuddin S, Atkins MB, Kruskal JB, Signoretti S, Raptopoulos VD, Goldberg SN. Perfusion MDCT enables early detection of therapeutic response to antiangiogenic therapy. AJR Am J Roentgenol. 2008;191(1):133–139. doi: 10.2214/AJR.07.2848. [DOI] [PubMed] [Google Scholar]
- 6.Yang HF, Du Y, Ni JX, Zhou XP, Li JD, Zhang Q, Xu XX, Li Y. Perfusion computed tomography evaluation of angiogenesis in liver cancer. Eur Radiol. 20(6):1424–1430. doi: 10.1007/s00330-009-1693-y. [DOI] [PubMed] [Google Scholar]
- 7.Berr SS, Mai VM. Extraslice spin tagging (EST) magnetic resonance imaging for the determination of perfusion. J Magn Reson Imaging. 1999;9(1):146–150. doi: 10.1002/(sici)1522-2586(199901)9:1<146::aid-jmri19>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 8.Fenchel M, Martirosian P, Langanke J, Giersch J, Miller S, Stauder NI, Kramer U, Claussen CD, Schick F. Perfusion MR imaging with FAIR true FISP spin labeling in patients with and without renal artery stenosis: initial experience. Radiology. 2006;238(3):1013–1021. doi: 10.1148/radiol.2382041623. [DOI] [PubMed] [Google Scholar]
- 9.Schoenberg SO, Rieger JR, Michaely HJ, Rupprecht H, Samtleben W, Reiser MF. Functional magnetic resonance imaging in renal artery stenosis. Abdom Imaging. 2006;31(2):200–212. doi: 10.1007/s00261-005-0388-3. [DOI] [PubMed] [Google Scholar]
- 10.Dong J, Yang L, Su T, Yang X, Chen B, Zhang J, Wang X, Jiang X. Quantitative assessment of acute kidney injury by noninvasive arterial spin labeling perfusion MRI: a pilot study. Sci China Life Sci. 56(8):745–750. doi: 10.1007/s11427-013-4503-3. [DOI] [PubMed] [Google Scholar]
- 11.De Bazelaire C, Rofsky NM, Duhamel G, Michaelson MD, George D, Alsop DC. Arterial spin labeling blood flow magnetic resonance imaging for the characterization of metastatic renal cell carcinoma(1) Acad Radiol. 2005;12(3):347–357. doi: 10.1016/j.acra.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 12.de Bazelaire C, Alsop DC, George D, Pedrosa I, Wang Y, Michaelson MD, Rofsky NM. Magnetic resonance imaging-measured blood flow change after antiangiogenic therapy with PTK787/ZK 222584 correlates with clinical outcome in metastatic renal cell carcinoma. Clin Cancer Res. 2008;14(17):5548–5554. doi: 10.1158/1078-0432.CCR-08-0417. [DOI] [PubMed] [Google Scholar]
- 13.Schor-Bardach R, Alsop DC, Pedrosa I, Solazzo SA, Wang X, Marquis RP, Atkins MB, Regan M, Signoretti S, Lenkinski RE, Goldberg SN. Does arterial spin-labeling MR imaging-measured tumor perfusion correlate with renal cell cancer response to antiangiogenic therapy in a mouse model? Radiology. 2009;251(3):731–742. doi: 10.1148/radiol.2521081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedrosa I, Alsop DC, Rofsky NM. Magnetic resonance imaging as a biomarker in renal cell carcinoma. Cancer. 2009;115(10 Suppl):2334–2345. doi: 10.1002/cncr.24237. [DOI] [PubMed] [Google Scholar]
- 15.Lanzman RS, Robson PM, Sun MR, Patel AD, Mentore K, Wagner AA, Genega EM, Rofsky NM, Alsop DC, Pedrosa I. Arterial spin-labeling MR imaging of renal masses: correlation with histopathologic findings. Radiology. 2012;265(3):799–808. doi: 10.1148/radiol.12112260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedrosa I, Rafatzand K, Robson P, Wagner AA, Atkins MB, Rofsky NM, Alsop DC. Arterial spin labeling MR imaging for characterisation of renal masses in patients with impaired renal function: initial experience. Eur Radiol. 2012;22(2):484–492. doi: 10.1007/s00330-011-2250-z. [DOI] [PubMed] [Google Scholar]
- 17.Rossi C, Artunc F, Martirosian P, Schlemmer HP, Schick F, Boss A. Histogram analysis of renal arterial spin labeling perfusion data reveals differences between volunteers and patients with mild chronic kidney disease. Invest Radiol. 47(8):490–496. doi: 10.1097/RLI.0b013e318257063a. [DOI] [PubMed] [Google Scholar]
- 18.Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. 1992;23(1):37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- 19.Williams DS, Detre JA, Leigh JS, Koretsky AP. Magnetic resonance imaging of perfusion using spin inversion of arterial water. Proc Natl Acad Sci U S A. 1992;89(1):212–216. doi: 10.1073/pnas.89.1.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang W, Williams DS, Detre JA, Koretsky AP. Measurement of brain perfusion by volume-localized NMR spectroscopy using inversion of arterial water spins: accounting for transit time and cross-relaxation. Magn Reson Med. 1992;25(2):362–371. doi: 10.1002/mrm.1910250216. [DOI] [PubMed] [Google Scholar]
- 21.Silva AC, Kim SG, Garwood M. Imaging blood flow in brain tumors using arterial spin labeling. Magn Reson Med. 2000;44(2):169–173. doi: 10.1002/1522-2594(200008)44:2<169::aid-mrm1>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 22.Warmuth C, Gunther M, Zimmer C. Quantification of blood flow in brain tumors: comparison of arterial spin labeling and dynamic susceptibility-weighted contrast-enhanced MR imaging. Radiology. 2003;228(2):523–532. doi: 10.1148/radiol.2282020409. [DOI] [PubMed] [Google Scholar]
- 23.Wolf RL, Wang J, Wang S, Melhem ER, O'Rourke DM, Judy KD, Detre JA. Grading of CNS neoplasms using continuous arterial spin labeled perfusion MR imaging at 3 Tesla. J Magn Reson Imaging. 2005;22(4):475–482. doi: 10.1002/jmri.20415. [DOI] [PubMed] [Google Scholar]
- 24.Kimura H, Takeuchi H, Koshimoto Y, Arishima H, Uematsu H, Kawamura Y, Kubota T, Itoh H. Perfusion imaging of meningioma by using continuous arterial spin-labeling: comparison with dynamic susceptibility-weighted contrast-enhanced MR images and histopathologic features. AJNR Am J Neuroradiol. 2006;27(1):85–93. [PMC free article] [PubMed] [Google Scholar]
- 25.Detre JA, Alsop DC, Vives LR, Maccotta L, Teener JW, Raps EC. Noninvasive MRI evaluation of cerebral blood flow in cerebrovascular disease. Neurology. 1998;50(3):633–641. doi: 10.1212/wnl.50.3.633. [DOI] [PubMed] [Google Scholar]
- 26.Martirosian P, Klose U, Mader I, Schick F. FAIR true-FISP perfusion imaging of the kidneys. Magn Reson Med. 2004;51(2):353–361. doi: 10.1002/mrm.10709. [DOI] [PubMed] [Google Scholar]
- 27.Boss A, Martirosian P, Schraml C, Clasen S, Fenchel M, Anastasiadis A, Claussen CD, Pereira PL, Schick F. Morphological, contrast-enhanced and spin labeling perfusion imaging for monitoring of relapse after RF ablation of renal cell carcinomas. Eur Radiol. 2006;16(6):1226–1236. doi: 10.1007/s00330-005-0098-9. [DOI] [PubMed] [Google Scholar]
- 28.Lanzman RS, Wittsack HJ, Martirosian P, Zgoura P, Bilk P, Kropil P, Schick F, Voiculescu A, Blondin D. Quantification of renal allograft perfusion using arterial spin labeling MRI: initial results. Eur Radiol. 20(6):1485–1491. doi: 10.1007/s00330-009-1675-0. [DOI] [PubMed] [Google Scholar]
- 29.Kiefer C, Schroth G, Gralla J, Diehm N, Baumgartner I, Husmann M. A feasibility study on model-based evaluation of kidney perfusion measured by means of FAIR prepared true-FISP arterial spin labeling (ASL) on a 3-T MR scanner. Acad Radiol. 2009;16(1):79–87. doi: 10.1016/j.acra.2008.04.024. [DOI] [PubMed] [Google Scholar]
- 30.Artz NS, Sadowski EA, Wentland AL, Djamali A, Grist TM, Seo S, Fain SB. Reproducibility of renal perfusion MR imaging in native and transplanted kidneys using non-contrast arterial spin labeling. J Magn Reson Imaging. 2011;33(6):1414–1421. doi: 10.1002/jmri.22552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Artz NS, Sadowski EA, Wentland AL, Grist TM, Seo S, Djamali A, Fain SB. Arterial spin labeling MRI for assessment of perfusion in native and transplanted kidneys. Magn Reson Imaging. 2011;29(1):74–82. doi: 10.1016/j.mri.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robson PM, Madhuranthakam AJ, Dai W, Pedrosa I, Rofsky NM, Alsop DC. Strategies for reducing respiratory motion artifacts in renal perfusion imaging with arterial spin labeling. Magn Reson Med. 2009;61(6):1374–1387. doi: 10.1002/mrm.21960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gardener AG, Francis ST. Multislice perfusion of the kidneys using parallel imaging: image acquisition and analysis strategies. Magn Reson Med. 2010;63(6):1627–1636. doi: 10.1002/mrm.22387. [DOI] [PubMed] [Google Scholar]
- 34.Cutajar M, Thomas DL, Banks T, Clark CA, Golay X, Gordon I. Repeatability of renal arterial spin labelling MRI in healthy subjects. MAGMA. 2012;25(2):145–153. doi: 10.1007/s10334-011-0300-9. [DOI] [PubMed] [Google Scholar]
- 35.Gillis KA, McComb C, Foster JE, Taylor AH, Patel RK, Morris ST, Jardine AG, Schneider MP, Roditi GH, Delles C, Mark PB. Inter-study reproducibility of arterial spin labelling magnetic resonance imaging for measurement of renal perfusion in healthy volunteers at 3 Tesla. BMC Nephrol. 15(1):23. doi: 10.1186/1471-2369-15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heusch P, Wittsack HJ, Blondin D, Ljimani A, Nguyen-Quang M, Martirosian P, Zenginli H, Bilk P, Kropil P, Heusner TA, Antoch G, Lanzman RS. Functional evaluation of transplanted kidneys using arterial spin labeling MRI. J Magn Reson Imaging. doi: 10.1002/jmri.24336. [DOI] [PubMed] [Google Scholar]
- 37.Heusch P, Wittsack HJ, Pentang G, Buchbender C, Miese F, Schek J, Kropil P, Antoch G, Lanzman RS. Biexponential analysis of diffusion-weighted imaging: comparison of three different calculation methods in transplanted kidneys. Acta Radiol. 54(10):1210–1217. doi: 10.1177/0284185113491090. [DOI] [PubMed] [Google Scholar]
- 38.Park SH, Wang DJ, Duong TQ. Balanced steady state free precession for arterial spin labeling MRI: Initial experience for blood flow mapping in human brain, retina, and kidney. Magn Reson Imaging. 31(7):1044–1050. doi: 10.1016/j.mri.2013.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cutajar M, Hilton R, Olsburgh J, Marks SD, Thomas DL, Banks T, Clark CA, Gordon I. Renal blood flow using arterial spin labelling MRI and calculated filtration fraction in healthy adult kidney donors Pre-nephrectomy and post-nephrectomy. Eur Radiol. doi: 10.1007/s00330-015-3594-6. [DOI] [PubMed] [Google Scholar]
- 40.Cutajar M, Thomas DL, Hales PW, Banks T, Clark CA, Gordon I. Comparison of ASL and DCE MRI for the non-invasive measurement of renal blood flow: quantification and reproducibility. Eur Radiol. 24(6):1300–1308. doi: 10.1007/s00330-014-3130-0. [DOI] [PubMed] [Google Scholar]
- 41.Gardener AG, Francis ST. Multislice perfusion of the kidneys using parallel imaging: image acquisition and analysis strategies. Magn Reson Med. 63(6):1627–1636. doi: 10.1002/mrm.22387. [DOI] [PubMed] [Google Scholar]
- 42.Dixon WT, Sardashti M, Castillo M, Stomp GP. Multiple inversion recovery reduces static tissue signal in angiograms. Magn Reson Med. 1991;18(2):257–268. doi: 10.1002/mrm.1910180202. [DOI] [PubMed] [Google Scholar]
- 43.Ye FQ, Frank JA, Weinberger DR, McLaughlin AC. Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST) Magn Reson Med. 2000;44(1):92–100. doi: 10.1002/1522-2594(200007)44:1<92::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 44.Rofsky NM, Lee VS, Laub G, Pollack MA, Krinsky GA, Thomasson D, Ambrosino MM, Weinreb JC. Abdominal MR imaging with a volumetric interpolated breath-hold examination. Radiology. 1999;212(3):876–884. doi: 10.1148/radiology.212.3.r99se34876. [DOI] [PubMed] [Google Scholar]
- 45.Sun MR, Ngo L, Genega EM, Atkins MB, Finn ME, Rofsky NM, Pedrosa I. Renal cell carcinoma: dynamic contrast-enhanced MR imaging for differentiation of tumor subtypes--correlation with pathologic findings. Radiology. 2009;250(3):793–802. doi: 10.1148/radiol.2503080995. [DOI] [PubMed] [Google Scholar]
- 46.Dai W, Garcia D, de Bazelaire C, Alsop DC. Continuous flow-driven inversion for arterial spin labeling using pulsed radio frequency and gradient fields. Magn Reson Med. 2008;60(6):1488–1497. doi: 10.1002/mrm.21790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alsop DC, Detre JA. Reduced transit-time sensitivity in noninvasive magnetic resonance imaging of human cerebral blood flow. J Cereb Blood Flow Metab. 1996;16(6):1236–1249. doi: 10.1097/00004647-199611000-00019. [DOI] [PubMed] [Google Scholar]
- 48.Ordidge RJ, Wylezinska M, Hugg JW, Butterworth E, Franconi F. Frequency offset corrected inversion (FOCI) pulses for use in localized spectroscopy. Magn Reson Med. 1996;36(4):562–566. doi: 10.1002/mrm.1910360410. [DOI] [PubMed] [Google Scholar]
- 49.Maleki N, Dai W, Alsop D. A Systematic Approach to Optimizing Background Suppression for Arterial Spin Labeling Perfusion Imaging. Proc ISMRM. 2008:1929. [Google Scholar]
- 50.de Bazelaire CM, Duhamel GD, Rofsky NM, Alsop DC. MR imaging relaxation times of abdominal and pelvic tissues measured in vivo at 3.0 T: preliminary results. Radiology. 2004;230(3):652–659. doi: 10.1148/radiol.2303021331. [DOI] [PubMed] [Google Scholar]
- 51.Roberts DA, Detre JA, Bolinger L, Insko EK, Lenkinski RE, Pentecost MJ, Leigh JS., Jr Renal perfusion in humans: MR imaging with spin tagging of arterial water. Radiology. 1995;196(1):281–286. doi: 10.1148/radiology.196.1.7784582. [DOI] [PubMed] [Google Scholar]
- 52.Herscovitch P, Raichle ME. What is the correct value for the brain--blood partition coefficient for water? J Cereb Blood Flow Metab. 1985;5(1):65–69. doi: 10.1038/jcbfm.1985.9. [DOI] [PubMed] [Google Scholar]
- 53.Bjornerud A, Johansson LO, Briley-Saebo K, Ahlstrom HK. Assessment of T1 and T2* effects in vivo and ex vivo using iron oxide nanoparticles in steady state--dependence on blood volume and water exchange. Magn Reson Med. 2002;47(3):461–471. doi: 10.1002/mrm.10066. [DOI] [PubMed] [Google Scholar]
- 54.Garcia DM, Duhamel G, Alsop DC. Efficiency of inversion pulses for background suppressed arterial spin labeling. Magn Reson Med. 2005;54(2):366–372. doi: 10.1002/mrm.20556. [DOI] [PubMed] [Google Scholar]
- 55.Busse RF, Hariharan H, Vu A, Brittain JH. Fast spin echo sequences with very long echo trains: design of variable refocusing flip angle schedules and generation of clinical T2 contrast. Magn Reson Med. 2006;55(5):1030–1037. doi: 10.1002/mrm.20863. [DOI] [PubMed] [Google Scholar]
- 56.Busse RF, Brau AC, Vu A, Michelich CR, Bayram E, Kijowski R, Reeder SB, Rowley HA. Effects of refocusing flip angle modulation and view ordering in 3D fast spin echo. Magn Reson Med. 2008;60(3):640–649. doi: 10.1002/mrm.21680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martirosian P, Boss A, Fenchel M, Deimling M, Schafer J, Claussen CD, Schick F. Quantitative lung perfusion mapping at 0.2 T using FAIR True-FISP MRI. Magn Reson Med. 2006;55(5):1065–1074. doi: 10.1002/mrm.20871. [DOI] [PubMed] [Google Scholar]
- 58.McKenzie CA, Yeh EN, Ohliger MA, Price MD, Sodickson DK. Self-calibrating parallel imaging with automatic coil sensitivity extraction. Magn Reson Med. 2002;47(3):529–538. doi: 10.1002/mrm.10087. [DOI] [PubMed] [Google Scholar]
- 59.Roemer PB, Edelstein WA, Hayes CE, Souza SP, Mueller OM. The NMR phased array. Magn Reson Med. 1990;16(2):192–225. doi: 10.1002/mrm.1910160203. [DOI] [PubMed] [Google Scholar]
- 60.Robson PM, Grant AK, Madhuranthakam AJ, Lattanzi R, Sodickson DK, McKenzie CA. Comprehensive quantification of signal-to-noise ratio and g-factor for image-based and k-space-based parallel imaging reconstructions. Magn Reson Med. 2008;60(4):895–907. doi: 10.1002/mrm.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Han M, Beatty PJ, Daniel BL, Hargreaves BA. Independent slab-phase modulation combined with parallel imaging in bilateral breast MRI. Magn Reson Med. 2009;62(5):1221–1231. doi: 10.1002/mrm.22115. [DOI] [PMC free article] [PubMed] [Google Scholar]


