Abstract
Background
An increasing number of neuroscientific studies gain insights by focusing on differences in functional connectivity – between groups, individuals, temporal windows, or task conditions. We found using simulations that additional insights into such differences can be gained by forgoing variance normalization, a procedure used by most functional connectivity measures. Simulations indicated that these functional connectivity measures are sensitive to increases in independent fluctuations (unshared signal) in time series, consistently reducing functional connectivity estimates (e.g., correlations) even though such changes are unrelated to corresponding fluctuations (shared signal) between those time series. This is inconsistent with the common notion of functional connectivity as the amount of inter-region interaction.
New Method
Simulations revealed that a version of correlation without variance normalization – covariance – was able to isolate differences in shared signal, increasing interpretability of observed functional connectivity change. Simulations also revealed cases problematic for non-normalized methods, leading to a “covariance conjunction” method combining the benefits of both normalized and non-normalized approaches.
Results
We found that covariance and covariance conjunction methods can detect functional connectivity changes across a variety of tasks and rest in both clinical and non-clinical functional MRI datasets.
Comparison with Existing Method(s)
We verified using a variety of tasks and rest in both clinical and non-clinical functional MRI datasets that it matters in practice whether correlation, covariance, or covariance conjunction methods are used.
Conclusions
These results demonstrate the practical and theoretical utility of isolating changes in shared signal, improving the ability to interpret observed functional connectivity change.
Keywords: functional connectivity, functional MRI, task functional connectivity, resting-state functional connectivity, schizophrenia
INTRODUCTION
Extensive neuroscientific research has identified consistent patterns of brain activity associated with a variety of behavioral processes. In trying to understand the systems-level mechanisms underlying these activation patterns, researchers have increasingly relied on functional connectivity – the statistical dependence among brain activity time series. Functional connectivity has been used across a wide variety of systems and a wide variety of neuroscientific approaches, such as functional MRI (fMRI), electroencephalography (EEG), and multi-unit recording (Nolte et al. 2004; Smith et al. 2011b; Buschman et al. 2012). Much of this research has focused on identifying the basic systems-level architecture of the brain via the detection of functional connections during resting state (Biswal et al. 2010; Brookes et al. 2011; Power et al. 2011; Yeo et al. 2011; Craddock et al. 2013). In order to link functional connectivity to cognition and behavior, however, researchers are increasingly focusing on functional connectivity differences. Such differences can be between groups (e.g., patients vs. healthy controls), individuals (e.g., correlating with IQ), temporal windows (i.e., functional connectivity dynamics), or task conditions. We focus here on measuring and interpreting such functional connectivity differences.
Despite the common statistical definition of functional connectivity stated above, functional connectivity results are typically interpreted in terms of neural interactions. This is likely due to the distinction between what is of underlying theoretical interest – true neural interactions – and methodological reality. Therefore, we suggest that one can make progress here by reducing the gap between methods and the phenomena of theoretical interest. In other words, we suggest that any functional connectivity measure that more closely reflects true neural interactions is a better functional connectivity measure.
Here we developed a simulation framework to systematically characterize relationships between functional connectivity measures and ground truth interactions. We designed the framework 1) to involve signals (neurons/regions) influencing one another, and 2) to be as simple as possible to facilitate interpretation and to make as few assumptions about the true nature of brain region interactions as possible. Briefly, the framework involves simply summing Gaussian random time series consisting of shared signal (time series copied between source and target), unshared signal (time series that are not copied between source and target), and noise. The simulations allowed us to identify measures that better reflect neural interactions, highlighting the appropriateness of some functional connectivity measures over others when neural interaction changes are of primary interest.
The most common statistical measures used to estimate functional connectivity across a wide variety of neuroscientific approaches are Pearson correlation and related methods (e.g., coherence, partial correlation). These and many other common statistical measures utilize the concept of “percent variance explained” – dividing an estimate of shared variance by overall variance (i.e., variance normalization) – to produce standardized estimates of association. While these measures are frequently useful in other contexts, it was recently suggested that they are inappropriate for estimating functional connectivity differences (Friston 2011)1. If true, this would have major implications for the study of brain network function, as an increasing number of studies use Pearson correlation and related measures when studying functional connectivity differences across groups, individuals, or conditions (Zalesky et al. 2012a; 2012b).
As an illustration of a limitation of Pearson correlation, it has been shown that increased noise in neuronal recordings decreases correlations between neuronal time series, even when the underlying neuronal interactions are unchanged (Behseta et al. 2009). The sensitivity of correlations to unshared signal (rather than noise per se) may be especially problematic, however, as this would reduce the interpretability of any detected functional connectivity difference. For instance, a significant change in inter-region correlation could be driven solely by increased neural processing by only one of the two tested brain regions. Thus, we use the term “unshared signal” to emphasize that these effects could be driven by functionally important neural processes. The same conclusions also apply to the more general concept of “unshared variance”, which encompasses both signal and noise.
We used simulations to ground our systematic exploration of shared and unshared signal changes. These simulations revealed a functional connectivity method (covariance) immune to systematic bias from unshared signal. However, simulations also revealed that this method is sensitive to possible increases in overall variance/power that may be unrelated to true brain interaction change. We therefore developed a conjunctive method, in which a functional connectivity change is only considered significant if it is detected using both a variance normalized measure (e.g., correlation) and covariance. We then applied this method to empirical data, determining that it not only provides increased interpretability of results but also often provides results distinct from current methods in practice. These findings validate a new theoretical and methodological framework for characterizing functional connectivity differences, improving interpretability of brain network dynamics.
Due to the complex and potentially counterintuitive nature of the results, we encourage readers to run the simple simulations themselves, available here: https://github.com/ColeLab/simplesims/. Seeing and running the code may facilitate development of an improved intuition for the nature of these functional connectivity measures. Modifications of the code, including testing of other functional connectivity measures and different conditions, are encouraged as well.
MATERIALS AND METHODS
Functional connectivity estimation
Estimates of time series association were calculated using either MATLAB (version R2012a) or R (version 2.15.1). Covariance was the simplest measure we used, and was calculated as:
Where X and Y are brain activity time series, n is the number of time points, and X̄ and Ȳ are the time series means.
Pearson correlation was calculated as:
Where S is the time series standard deviation. Most analyses also involved the Fisher’s z-transform of the resulting Pearson correlation, which increases the dynamic range of correlation values to go beyond ±1.0. This is critical when investigating changes in functional connectivity, as forgoing the Fisher’s z-transform would result in artificial restrictions in dynamics. The Fisher’s z-transform:
Psycho-physiological interaction (PPI) was estimated using simple linear regression, which was calculated using the lm function in R, equivalent to:
Where var is the time series variance. The beta for each condition was estimated separately for each condition and subtracted, consistent with generalized PPI (McLaren et al. 2012). The additional step of including task regressors in the regression model was not included here because we did not simulate mean task activity amplitude changes (such that they were already effectively removed from the simulated time series).
Partial correlation was computed as the inverse covariance matrix (i.e., the inverse variance-covariance matrix). This is a standard procedure, which computes the correlations between pairs of time series after the variances from all other time series have been linearly removed. This procedure also normalizes the resulting statistic by the tested time series’ variance (the diagonal in the variance-covariance matrix), thus implementing variance-based normalization like standard Pearson correlation.
Basic simulations
Simulations were conducted using R (version 2.15.1) (R Development Core Team 2009). Two brain region time series (X and Y) were simulated as linear mixtures of shared signal (sharedXY; identical across regions), unshared signal (unsharedX and unsharedy), and unshared noise (noiseX and noisey):
Data for each variable were created using the function rnorm, which produced 200 normally distributed time points. The original amplitudes were modified only in the case of the noise variables, which were multiplied by 0.25 such that noise accounted for proportionally less of the variance than the signal variables. This was repeated 25 times, producing 25 distinct time series per simulated region.
Manipulations of shared and unshared signals consisted of scaling the relevant component (i.e., multiplying each time point by a constant) prior to mixing with the other component to produce the relevant time series. For instance, when increasing shared variance in both regions, these formulas apply:
Functional connectivity estimates were then applied to the manipulated time series, and compared to the estimates from the original time series. Statistical significance was assessed using t-tests (two-tailed, independent samples) on the functional connectivity estimates across simulated subjects. R and MATLAB code implementing these simulations can be found at: https://github.com/ColeLab/simplesims/
Phase locked value simulations
We carried out a standard PLV analysis using publically available software (available at the time of publication at: https://praneethnamburi.wordpress.com/2011/08/10/plv/). The software implements the standard PLV algorithm (Lachaux et al. 1999). The simulation data were generated in the same manner as for the other simulations, except that each simulated subject contributed 50 trials. This was necessary for the software, which required multiple trials to estimate PLV. Each trial consisted of 200 time points, and PLV was implemented as though they were collected at a 100 Hz sampling rate, with a filter order of 50 and frequency range of 10–20 Hz. The first 25 PLV time point estimates were discarded to exclude the initial transient (a common artifact of the PLV approach) and the remaining values were averaged for each trial. Note that the final z-normalization step often recommended for PLV was not applied, since this would more clearly necessitate that PLV results would look similar to Pearson correlation and related measures (due to dividing by the standard deviation of the time series, like Pearson correlation).
Biophysical computational model simulations
In addition to the simpler conceptual simulation we used a well-validated computational model of resting-state functional connectivity (Deco et al. 2013), extending a local circuit model (Wong and Wang 2006), to incorporate biologically plausible mechanisms for multiple interacting brain regions, with their activity translated to BOLD signal fluctuations. We recently applied this model to investigate specific synaptic parameters in relation to clinical effects. Here we explicitly studied the effects of shared versus unshared signal alterations in the simulated network. The network is composed of 66 nodes and is a dynamic mean-field model (Wong and Wang 2006), coupled through a large-scale anatomical connectivity matrix, which was derived from diffusion tractography in humans as reported in Hagmann et al. (Hagmann et al. 2008). For our simulations, we extracted the anatomical connectivity matrix from Figure 1 of Deco & Jirsa (Deco and Jirsa 2012), described in detail in our prior work (Yang et al. 2014). BOLD signals were simulated via the Balloon-Windkessel model, as done previously (Deco et al. 2013). All model parameter values were set to those used in our prior work (Yang et al. 2014). Specifically, default values were set to w=0.531, G=1.25, and sigma=0.0004 (amplitude of unshared noise or signal), with remaining values set to those of Deco et al.
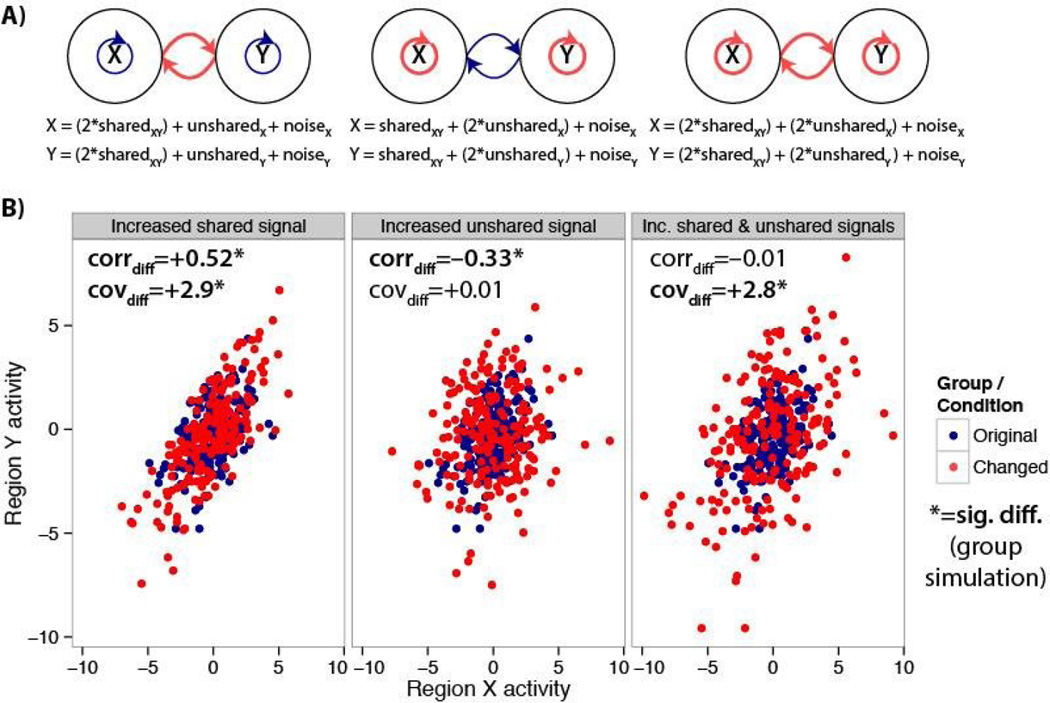
Figure 1. Differences between correlations and covariances for estimating functional connectivity differences.
A) Diagrams and equations illustrating simulated communication changes between brain regions (or neurons) X and Y. Left, only the portion of the time series shared across both regions is amplified relative to the unshared and noise portions. Center, only the unshared portion is amplified. Right, both the shared and unshared portions are amplified. B) A single subject’s simulated data are shown for illustration across the three conditions. Results of the group simulation are shown in the upper left of each panel. The correlation (corrdiff) and covariance (covdiff) results are in agreement when only shared signal is increased, but not for the other two cases.
Next, we introduced a common input to all nodes – that is “shared” signal (amplitude=0.0005). The amplitude of unshared signal specific to each node was represented by sigma. We parametrically varied both shared and unshared signal parameters across the entire network. Finally, to compute a measure of model-derived connectivity we used a measure of connectivity across the entire network termed global brain connectivity (GBC) (Cole et al. 2010). To compute the GBC of each node, we first generated a correlation matrix of each node’s signal with signals from all other (65) nodes. Values in the correlation matrix were then converted using a Fisher’s r-to-z transform. Next, for each node, GBC was computed as the mean value of each of the 65 columns (corresponding to the other 65 nodes) in this transformed matrix. To compute the average GBC of the network, we took the mean GBC value across the 66 nodes. This effectively yielded a GBC index of the model across parameters. We also repeated the same calculation using covariance (rather than correlation) prior to calculating GBC, to examine the functional connectivity of the model as derived using covariance instead of correlation.
7-task fMRI data collection
The 7-task fMRI dataset was collected as part of the Washington University-Minnesota Consortium Human Connectome Project (Van Essen et al. 2013). Participants were recruited from Washington University (St. Louis, MO) and the surrounding area. All participants gave informed consent. The data used were from the first and second quarter releases, consisting of data from 139 participants. Data from 21 subjects were not used because one or more of the data runs was not collected for these subjects, such that data from 118 subjects were included in the final analyses. Whole-brain echo-planar imaging acquisitions were acquired with a 32 channel head coil on a modified 3T Siemens Skyra with TR = 720 ms, TE = 33.1 ms, flip angle = 52°, BW = 2290 Hz/Px, in-plane FOV = 208 × 180 mm, 72 slices, 2.0 mm isotropic voxels, with a multi-band acceleration factor of 8 (Ugurbil et al. 2013). Data were collected over two days. On each day 28 minutes of rest (eyes open with fixation) fMRI data across two runs were collected (56 minutes total), followed by 30 minutes of task fMRI data collection (60 minutes total). Each of the 7 tasks was completed over two consecutive fMRI runs. Resting-state data collection details for this dataset can be found elsewhere (Smith et al. 2013), as can task data details (Barch et al. 2013).
7-task fMRI dataset analysis
The 7-task dataset preprocessing consisted of standard functional connectivity preprocessing (typically performed with resting-state data), with several modifications given that analyses were also performed on task-state data. Resting-state and task-state data were preprocessed identically in order to facilitate comparisons between them. Spatial normalization to a template, motion correction, intensity normalization (normalized to a 4D whole brain mean of 10,000) were already implemented in a minimally processed version of the 7-task dataset described elsewhere (Glasser et al. 2013), so we began preprocessing with this version of the data. With the volume (rather than the surface) version of the minimally preprocessed data, we used AFNI (Cox 1996) to additionally remove nuisance time series (motion, ventricle, whole brain, and white matter signals, along with their derivatives) using linear regression, remove the linear trend for each run, and spatially smooth the data (4mm full width at half maximum). Note that the main results were broadly similar with and without whole brain (global) signal regression, though this question is outside the scope of the present study. This preprocessing step was included to reduce potential motion and other artifacts (Power et al. 2014). Unlike standard resting-state functional connectivity preprocessing a low-pass temporal filter was not applied. This was done due to the possible presence of task signals at higher frequencies than the relatively slow resting-state fluctuations. In order to make this dataset comparable to most other current fMRI datasets, however, the data were temporally down-sampled (as the last step of preprocessing) by averaging data from every three consecutive volumes (making a 2160 ms TR, close to the 2000 ms TR in most ‘legacy’ single-band fMRI datasets). This had an effect similar to a mild low-pass temporal filter on the data (removing frequencies above 0.46 Hz). We found that effects were similar with and without this down-sampling step, however.
Data were sampled from a set of 264 brain regions (rather than individual voxels) in order to make inferences at the region and systems level. We used an independently identified set of putative functional brain regions (Power et al. 2011) so as to reduce any potential circularity in analyses (Kriegeskorte et al. 2009). This particular set of regions was also used rather than anatomically defined sets of regions in order to reduce the chance of combining signal from multiple functional regions (Wig et al. 2011). These brain regions were identified using a combination of resting-state functional connectivity parcellation (Cohen et al. 2008) and task neuroimaging meta-analysis (Power et al. 2011). Data were summarized for each region by averaging signal in all voxels falling inside each region.
Preprocessing was carried out using Freesurfer, AFNI (Cox 1996), and custom code in MATLAB 2012b (Mathworks) for the 7-task dataset (using the minimally preprocessed version of the data (Glasser et al. 2013)). Further analysis was carried out with MATLAB and R.
We estimated functional connectivity using Pearson correlations and covariances between time series from all pairs of brain regions using MATLAB (version R2012a). For Pearson correlations, all computations used Fisher’s z-transformed values. Functional connectivity estimation was straightforward for resting-state data, as there were no additional steps after preprocessing prior to calculating these values.
For task data, we sought to suppress or remove influences of (across-trial mean) task-related activations on task-related changes in functional connectivity. Therefore, we ran a standard fMRI general linear model analysis, and calculated functional connectivity based on the residuals. Specifically, each region’s task time series was modeled using a standard general linear model with one regression coefficient per task. To improve removal of task-related activation variance, a separate regressor was included for each major 7-task dataset condition (e.g., face stimuli vs. tool stimuli in the N-back task; 24 regressors total). Note that regressing out task events using general linear modeling primarily removes the across-trial signal means, retaining trial-to-trial and sub-trial fluctuations in time series such that these sources of variability likely contribute the most to task FC estimates (Truccolo et al. 2002; Rissman et al. 2004). The residuals from this regression model were used for FC estimation, restricted to time points corresponding to the current task (including a standard hemodynamic lag). Note that results were similar with and without task activation regression.
Functional connectivity differences were assessed using two-way t-tests paired by subject. Multiple comparisons were corrected for using false discovery rate (Genovese et al. 2002).
Schizophrenia fMRI dataset analysis
To test the clinical relevance of shared signal analyses, we examined functional connectivity in a large sample of patients diagnosed with chronic schizophrenia (SCZ). Specifically, we studied 71 SCZ patients and 74 demographically matched HCS obtained from a publicly-distributed dataset provided by the Center for Biomedical Research Excellence (COBRE) (http://fcon_1000.projects.nitrc.org/indi/retro/cobre.html). All the processing and analyses procedures followed our recently published work (Yang et al. 2014). Briefly, SCZ patients were excluded if they had: i) history of neurological disorder, ii) history of mental retardation, ii) history of severe head trauma with more than 5 minutes loss of consciousness; iv) history of substance abuse or dependence within the last 12 months. Diagnostic decisions were reached using the SCID interview for the DSM-IV. SCZ patients (N=71) and their respective HCS (N=74) underwent data collection at Center for Biomedical Research Excellence using a Siemens Tim-Trio 3T scanner. Full acquisition details for the SCZ replication sample and HCS is detailed previously (Mayer et al. 2013). Briefly, BOLD signal was collected with 32 axial slices parallel to the AC-PC using a T2*-weighted gradient-echo, echo-planar sequence (TR/TE=2000/29ms, flip angle=75°, acquisition matrix=64×64, voxel size=3×3×4mm). The acquisition lasted 5 minutes and produced 150 volumetric images per subject. Structural images were acquired using a 6 minute T1-weighted, 3D MPRAGE sequence (TR/TE/TI=2530/[1.64, 3.5, 5.36, 7.22, 9.08]/900, flip angle=7°, voxel size [isotropic]=1mm, image size=256×256×176 voxels), with axial slices parallel to the AC-PC line. All the described parameters were provided via the publically-distributed website (http://fcon_1000.projects.nitrc.org/indi/retro/cobre.html).
The processing of this dataset was completed independently of the other fMRI dataset in order to test if the specific processing steps included in the other dataset were necessary for the reported results (distinct effects with correlation versus covariance). All BOLD data underwent the following processing steps: i) slice-time correction, ii) first 5 images removed from each run, iii) rigid body motion correction, iv) 12-parameter affine transform of the structural image to the Talairach coordinate system, and v) co-registration of volumes to the structural image with 3×3×3mm re-sampling, ensuring all BOLD images across both scanners were interpolated to the same resolution.
In addition, all BOLD images for the clinical analyses had to pass stringent quality assurance criteria to ensure that all functional data were of comparable and high quality: i) signal-to-noise ratios (SNR) > 100, computed by obtaining the mean signal and standard deviation for a given slice across the BOLD run, while excluding all non-brain voxels across all frames. Furthermore, all image frames with possible movement-induced artifactual fluctuations in intensity were identified via two criteria: First, frames in which sum of the displacement across all 6 rigid body movement correction parameters exceeded 0.5mm (assuming 50mm cortical sphere radius) were identified; Second, root mean square (RMS) of differences in intensity between the current and preceding frame was computed across all voxels divided by mean intensity and normalized to time series median. Frames in which normalized RMS exceeded the value of 3 were identified. The frames flagged by either criterion were marked for exclusion (logical or), as well as the one preceding and two frames following the flagged frame. Collectively, these quality assurances add confidence that typical neuroimaging confounds (i.e. SNR or movement) are not driving present effect. Lastly, to remove spurious signal in resting-state data we completed additional preprocessing steps, as is standard practice (Cordes et al. 2001): all BOLD time-series underwent high (>0.009 Hz) and low (<0.08 Hz) pass temporal filtering, removal of nuisance signal extracted from anatomically-defined ventricles, white matter, and the remaining brain voxels (i.e. global signal) (all identified via individual-specific FreeSurfer segmentations (Desikan et al. 2006)), as well as 6 rigid-body motion correction parameters, and their first derivatives using in-house MATLAB tools.
RESULTS
Simulating neural interactions with minimal assumptions
We conducted a series of simulations that modeled changes in brain region interactions. We designed these simulations to be as simple as possible so as to make as few assumptions about the true nature of brain region interactions as possible. Each brain region was modeled as a mixture of shared signal (sharedXY; identical across regions), unshared signal (unsharedX; distinct across regions), and unshared noise (noiseX; also distinct across regions). Each region’s time series was composed of 200 time points, with equal parts of shared and unshared signals and one-quarter part noise (see Methods for details). Changes in brain region activity consisted of differential scaling of each of these components (e.g., multiplying the shared signal by 2) (Figure 1A). Each simulation was run 25 times, each with two conditions. These simulations could be considered as simulating 25 subjects during two brain states each (e.g., a cognitive manipulation, or spontaneous changes across time), or differences between two groups of 25 subjects (e.g., patients versus healthy controls, during a resting-state experiment). Note that we focus primarily on brain region interactions but conclusions are likely identical for interactions between individual neurons as well as other forms of brain interaction, though (as we address with the spectral covariance approach) in many scenarios it will be necessary to account for temporal lag between time series.
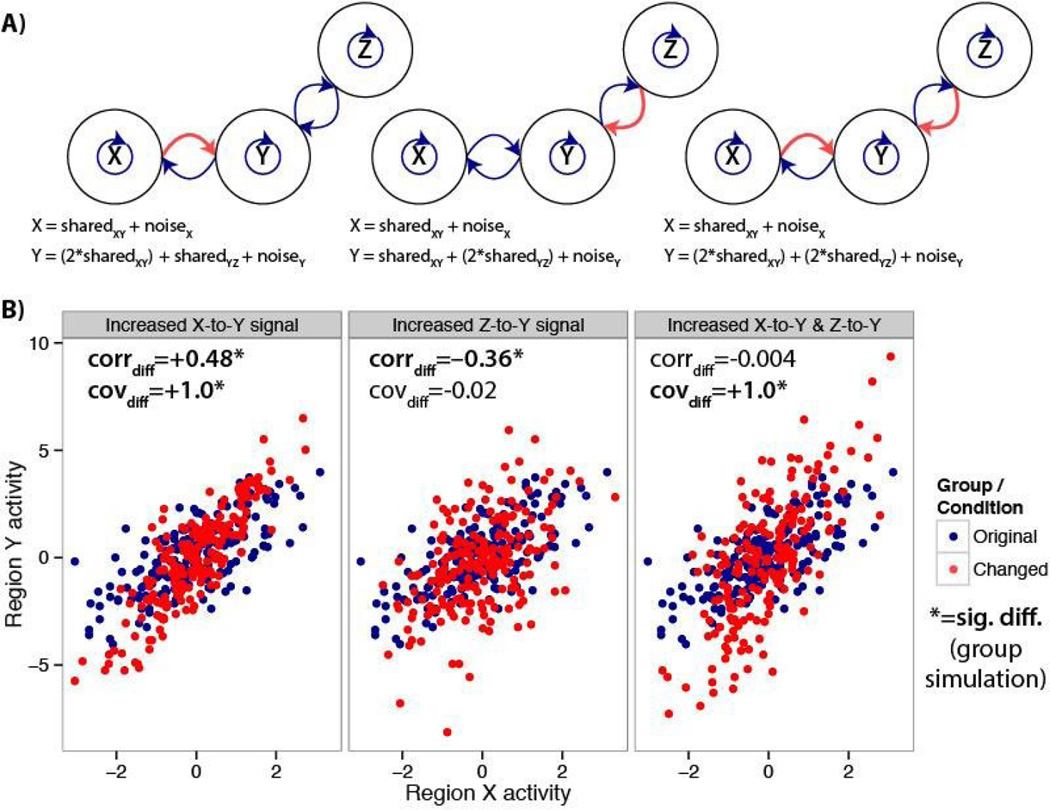
There are multiple possible underlying physiological changes that could result in the simulated functional connectivity changes (i.e. changes in coupling). For instance, the simulated increases in shared variance could result from increased synaptic strengths (e.g., due to short-term or long-term plasticity (Zucker and Regehr 2002; Yao et al. 2007)) or increased synchrony due to entrainment of neural populations to the same oscillations (Fries 2005). One mechanism for this type of change could be mediated via pre-synaptic glutamate release along with an action potential from the presynaptic neuron activating the N-methyl-D-aspartate glutamate receptor (NMDAR) (Krystal et al. 2003). Alternatively, a change in coupling may reflect elevated dopamine tone in the same cortical circuit (Vijayraghavan et al. 2007). Importantly, we remain agnostic with regard to these types of assumptions in our simple model, allowing generalizability of our conclusions across a spectrum of biological mechanisms. In contrast to shared variance, the simulated increases in unshared variance could result from increased neural activity unrelated to shared signals communicated to/from the regions of interest. For instance, there could be increased processing in one of the two tested regions, or more interaction between one of the tested regions and another unrelated region (e.g., increased communication between regions Y and another region Z, rather than between X and Y; Figure 2A). There are likely other scenarios involving changes in shared and/or unshared signals not mentioned here that these simulations nonetheless account for.
Figure 2. Differences between correlations and covariances for estimating functional connectivity differences, due to interaction with a third region.
Interaction between region Y and Z can stand in for “unshared signal” (in Figure 1) when testing for functional connectivity differences for regions Y and X. A) Diagrams and equations illustrating simulated communication changes between brain regions (or neurons) X, Y, and Z. Left, only the portion of the time series shared between regions X and Y is amplified in region Y. Center, only the portion shared between regions Z and Y is amplified in region Y. Right, the XY shared and ZY shared portions are both amplified in region Y. B) A single subject’s simulated data are shown for illustration across the three conditions. Results of the group simulation are shown in the upper left of each panel. The correlation (corrdiff) and covariance (covdiff) results are in agreement when only XY shared signal is increased, but not for the other two cases. This suggests correlation-like measures are sensitive to a wider variety of interactions irrelevant to the interactions between the two regions being tested.
We compared covariances (cov) and Fisher’s z-transformed Pearson correlations (corr) before and after manipulating the amount of shared and/or unshared signals. Note that we applied the Fisher’s z-transform so changes among high correlations were not restricted as they approached ±1.0, but conclusions were the same without this transform. We found that both correlations (mean corrdiff=+0.52, t(24)=28, p<0.00001) and covariances (mean covdiff=+2.9, t(24)=30, p<0.00001) increased when the shared signal was increased (Figure 1, left side), consistent with the mathematical formulation described below. This reflected a larger effect of region X’s activity on Y’s activity, and vice versa (a 2× increase in each direction). This result is consistent with common notions of functional connectivity differences as a change in the amount of inter-region communication.
We next sought to simulate circumstances in which correlation and covariance would differ. We found that an increase in unshared signal significantly decreased correlations (mean corrdiff=−0.33, t(24)=−15, p<0.00001), while covariances (mean covdiff=+0.01, t(24)=0.1, p=0.9) were unchanged (Figure 1, center). Critically, only covariance tracked the shared variance (which was left unchanged), whereas correlation was decreased by amplification of signal unassociated with inter-region communication.
Simulations also demonstrated that the results differed when both shared and unshared signals were increased (Figure 1, right side). With this manipulation correlation changes could be significantly positive, negative, or show no difference, depending on the amount of unshared signal change. We focused on the simple case in which there was a 2× increase in both shared and unshared signals: There was no difference between correlations (mean corrdiff=−0.01, t(24)=−0.4, p=0.7), but there was a difference between covariances (mean covdiff=+2.8, t(24)=29, p<0.00001).
Again, only covariance tracked changes in the shared signal, consistent with increased influence of the regions’ activities on each other. However, the correlation result could be considered correct if there was some confound affecting both shared and unshared variances in a similar manner. For example, there could be an increase in overall variance/power that would result in apparent increases in shared signal along with unshared signal. In the absence of a confound, however, the simulated scenario seems quite plausible, as increased inter-region communication could result in increased shared signal along with increased unshared signal due to greater within-region computation (e.g., information received in a region that needs to be processed but is unshared with the other region).
Taken together, these simple simulations suggest that covariance differences are associated with shared signal differences, reflecting true inter-region communication differences (in the absence of confounds). Given that overall variance/power confounds are possible with all neuroscientific methods, the conservative approach would be to use both correlation and covariance in conjunction: those changes in functional connectivity detected using both correlation and covariance are more likely to be true changes in functional connectivity.
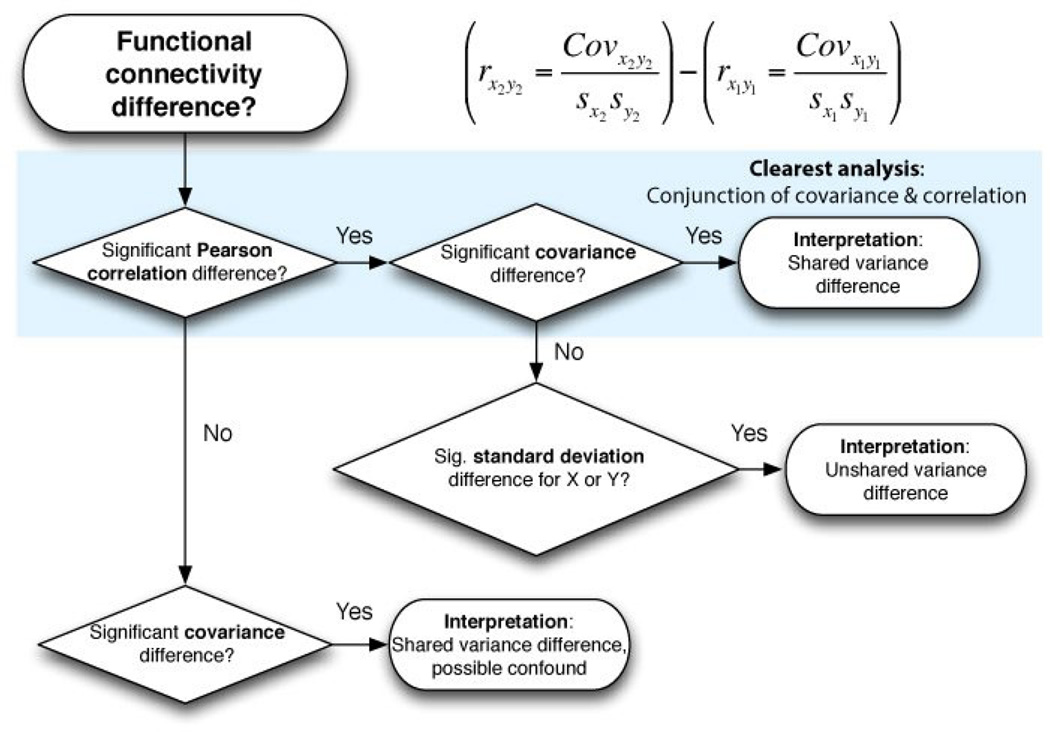
We constructed a flowchart to illustrate this logic (Figure 3). Simulation results of all possible combinations of shared and unshared variance changes are reported in Figure 4. We also found that interactions with a third region Z can produce similar effects as a change in unshared signal when investigating regions X and Y (Figure 2), suggesting correlation-like measures are also sensitive to a wider variety of interactions irrelevant to the interactions between the two regions being tested. This was not the case for partial correlations, due to linear removal of the third region’s variance. However, like Pearson correlations, partial correlations normalize by overall variance such that results were virtually identical to Pearson correlation in the simulations reported in Figure 1. This suggests partial correlations retain many of the limitations of standard Pearson correlations.
Figure 3.
A flowchart illustrating a “covariance conjunction” approach to interpreting functional connectivity differences. A similar line of reasoning would also work for most functional connectivity measures (not just correlation; e.g., PPI). Note that simply using covariance would result in a simpler line of reasoning: a significant covariance difference signifies a shared variance difference. However, as noted, a potential confound related to a change in overall variance could invalidate a result significant for covariance only. We suggest that the most conservative approach involves conducting both covariance and correlation analyses, assigning the most confidence to results that are consistent across both approaches (the upper-most route in the flowchart).
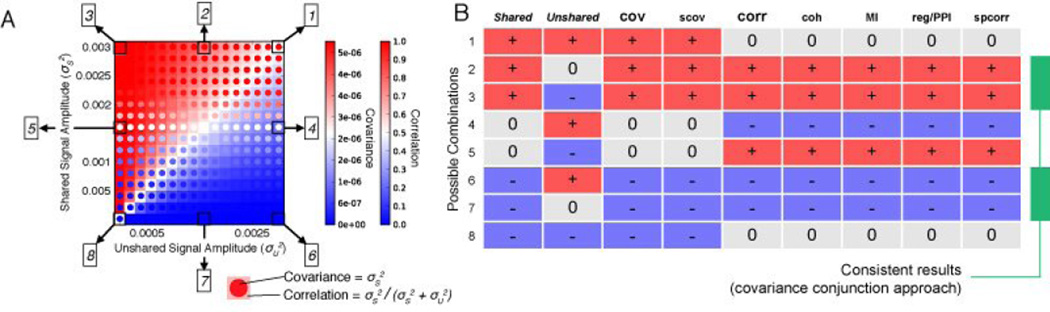
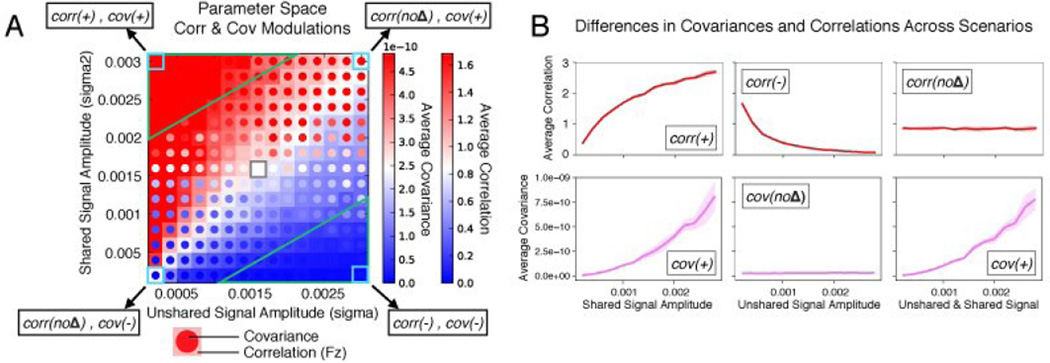
Figure 4. All possible shared and unshared variance change combinations.
A) The full parameter space is shown for changes in shared and unshared variance relative to a central point (in white). The correlation and covariance values were calculated using the simple mathematical formulation described in the Results section (not the simulations, though note the similarity to results with the realistic neural simulations presented in Figure 5). The boxed numbers refer to the combinations listed in part B. B) Group simulation results (using the same methods as Figure 1) are shown across all possible manipulations of shared variance and unshared variance (p<0.05). Figure 1 illustrates cases 2, 4, and 1. Note that correlation and covariance give different answers in 4 out of the 8 cases, and that covariance matches shared variance changes in all cases. The suggested covariance conjunction approach results are highlighted in green. A variety of other common functional connectivity measures are also included to illustrate how general these results are.
Code for these simulations can be found at: https://github.com/ColeLab/simplesims/ + increase, 0 no change, - decrease, cov=covariance, scov=spectral covariance, corr=Pearson correlation, coh=coherence, MI=mutual information, reg=regression, PPI=psycho-physiological interaction, spcorr=Spearman correlation.
Mathematical formulation
We next examined a simple mathematical formulation to verify and illustrate the problem that arises when assessing changes in functional connectivity based on the measure of correlation. We consider two time series, X and Y, which could be physiological signals from two brain areas (or neurons). Covariance (cov) provides a measure of how strongly X and Y change together: cov(X,Y) = ((X − (X))(Y − (Y))), where (…) is the average over time. Correlation (corr) is a rescaled measure that normalizes covariance by the variances of X and Y: corr(X,Y) = cov(X,Y)/sqrt(var(X)var(Y)). This normalization in correlation complicates interpretation of changes in functional connectivity, because a change in correlation can reflect a change in covariance or a change in variance.
We framed this problem by considering that changes in correlation-based estimates of functional connectivity can be driven by both shared and unshared brain signals. We considered the case where X and Y can each be decomposed into two components: a signal that is shared between X and Y with variance σs2, and a signal that is unshared between X and Y with variance σU2. Then cov(X,Y) = σs2, and corr(X,Y) = σs2/(σs2 + σU2). Thus covariance reflects the shared signal and is not systematically altered by the unshared signal. In contrast, correlation depends on both shared and unshared signals due to the normalization step. This formulation illustrates the difficulty in interpreting changes in connectivity based on correlation rather than covariance. For instance, a decrease in covariance purely reflects a decrease in shared signal. In contrast, a decrease in correlation could reflect a decrease in shared signal or an increase in unshared signals. We next turn to a numerical demonstration of this problem with simulated neural data.
Other regression-based methods such as psycho-physiological interaction are similar to correlations
In the original review suggesting correlations may be problematic when testing for functional connectivity differences, it was suggested that the psycho-physiological interaction (PPI) method does not suffer from the hypothesized issues with correlation (Friston 2011). PPI is essentially the regression of one time series on another (with simultaneously fit nuisance variables) (McLaren et al. 2012), such that changes in which time series is the “source” and “target” can give different estimates. Mathematically, the regression beta estimates are equivalent to covariance divided by the source time series variance (see Methods). Thus, we predicted that manipulations to the source time series would appear similar to correlations, whereas manipulations to the target time series would appear similar to covariances. Consistent with this, we found that increasing region X’s unshared signal (the source) decreased the beta estimate (mean betadiff=−0.29, p<0.00001), while increasing region Y’s unshared signal (the target) did not change the beta estimate (mean betadiff=+0.01, p=0.7). Thus, PPI and related regression approaches are similar to correlation with regard to the source time series, but reflect covariance with regard to the target time series. When manipulating both time series (as in Figure 1), the regression/PPI approach results were the same as correlations. Note that some of the sensitivity to unshared variance may be reduced by including task timing estimates as nuisance regressors (as typically done with PPI, and as done with the empirical analyses below), yet this would only account for across-trial mean activity such that much of the unshared variance (e.g., of moment-to-moment and trial-to-trial signals) would remain.
Overall, these results are mostly inconsistent with the previous claim regarding PPI (Friston 2011). Specifically, contrary to the original claim, there can be a change in a PPI estimate even when shared signal does not change: when unshared signal changes in both (or just the target) time series. We found that a variety of other common functional connectivity measures also showed similar results to Pearson correlation, such as coherence – a commonly used method for investigating functional connectivity using electrophysiological signals (see results below, and Figure 4B).
Preliminary extension of approach to lag-invariant methods
Correlation is the most common functional connectivity approach with fMRI, likely because its low temporal resolution results in only minimal lags between time series. In contrast, methods such as electroencephalography (EEG) and intracranial recording obtain data at high temporal resolution, resulting in lagged correspondence between time series (e.g., 10 to 100 ms delays in inter-region signal propagation). Therefore, one of the most common functional connectivity measures with these methods has been coherence, which is robust to lags. This is possible because coherence measures the correspondence between each time series’ power across frequencies (i.e., the spectral density distribution). Importantly, coherence is similar to Pearson correlation in that it is normalized by overall variance. We therefore hypothesized that coherence would be sensitive to changes in unshared signal, just like correlation.
We tested this hypothesis using identical simulations as used in Figure 1, but tested using coherence and with a 5 time point lag between the time series. Confirming our hypothesis, we found that coherence showed the same pattern of results as correlation (in contrast to covariance). Specifically, there was an increase in coherence when shared signal was increased (mean cohdiff = 0.40, t(24)=49, p<0.00001), a decrease when unshared signal was increased (mean cohdiff = −0.07, t(24)=−7, p<0.00001), and no change when both shared and unshared signals were increased (mean cohdiff = 0.007, t(24)=0.7, p=0.46). Note that there were no significant changes detected using correlations or covariance with the 5 time point lag.
We next developed a new spectral measure based on covariance, which we hypothesized would be unchanged by temporal lags or differences in unshared signal. We call this measure “spectral covariance” (scov). This measure is computed by estimating the spectral density distribution (i.e., a periodogram) for each time series, followed by measuring the covariance between these distributions. Intuitively, this is the same covariance approach used above, but now on the pattern of power across frequencies rather than the raw time series. Consistent with our hypothesis, we found that this measure matched covariance even with a temporal lag between the time series. Specifically, there was an increase in spectral covariance when shared signal was increased (mean scovdiff = 1.4, t(24)=28, p<0.00001), no change when unshared signal was increased (mean scovdiff = 0.003, t(24)=0.03, p=0.98), and an increase when both shared and unshared signals were increased (mean scovdiff = 0.71, t(24)=5, p=0.00002). It therefore appears possible to implement a covariance conjunction approach that is lag invariant, combining results from coherence and spectral covariance to increase confidence in a detected functional connectivity change.
We next tested if this method would work when only a small subset of frequencies are altered, rather than all frequencies. Spectral covariance was not robust to unshared signal changes in this case. Specifically, there was a decrease in spectral covariance when unshared signal was increased in a single frequency (mean scovdiff = −0.9, t(24)=−10, p<0.00001). This suggests this new approach is limited to cases in which broad sets of frequencies are altered. It will be important for future research to investigate ways to overcome this limitation in spectral covariance. One possibility may be the use of lagged covariance (testing covariance at various lags between time series), though this could result in overfitting data due to multiple comparison testing across many lags. Overall, these results demonstrate a proof of principle for a way to estimate lag-invariant functional connectivity change that is robust to changes in unshared signal. More generally, these results suggest it may be possible to modify a variety of other functional connectivity measures to be robust to changes in unshared signal.
Extension to phase locked value
Some characterizations of functional connectivity have focused on phase synchronization of oscillations (Lachaux et al. 1999; Engel et al. 2001; Aydore et al. 2013). One prominent method for isolating phase synchronization is phase locked value (PLV) (Lachaux et al. 1999). PLV characterizes time series in terms of oscillations at a particular frequency range, quantifying how close the phase is between two time series. Unlike correlation, coherence, and the other measures, PLV removes fluctuation amplitudes to focus exclusively on the timing of the fluctuations. One might therefore assume that PLV would be immune to the changes in signal amplitude implemented by our simulations. We carried out a standard PLV analysis using publically available software (see Methods) to test this possibility.
Surprisingly, we found that PLV acted very similarly to Pearson correlations and related measures. We found that PLV significantly increased when shared signal was amplified (mean PLVdiff=+0.29, t(24)=64, p<0.00001), significantly decreased when unshared signal was amplified (mean PLVdiff=−0.21, t(24)=−42, p<0.00001), and showed no significant change when both shared and unshared signals were increased (mean PLVdiff=0.01, t(24)=1.85, p=0.07). Note the marginally significant effect for the last simulation, hinting at a possible difference from Pearson correlations and related measures (but not as initially expected).
It is beyond the scope of the present study to fully characterize why these simulations resulted in PLV changes. However, one possibility is that PLV can be conceptualized as counting the number of identified shared fluctuations in two time series, with added unshared signal reducing the number of identified shared fluctuations. This may be due to greater corruption of the perfectly in-phase oscillations present in the shared signal by larger unshared oscillations. In particular, cases in which a shared oscillation would be nearly canceled out by an anti-phasic unshared oscillation may be completely canceled out with greater unshared signal amplitude. This is all despite there actually being true phase synchronization underlying the signals, given the presence of the shared signal across both time series. It will be important for future research to explore this and other possibilities, as well as developing alternative phase locking estimates that are not systematically biased by changes in unshared signal.
Biologically realistic simulations illustrate relationships between brain network dynamics and functional connectivity measures
We next utilized a previously developed biophysically based computational model (Deco et al. 2013; Yang et al. 2014) to (1) test if the effects identified above are present in a model that captures neurobiologically realistic neuronal dynamics in a larger network, and (2) to explore the effect of many possible brain region activity changes on functional connectivity measures using plausible neuronal dynamics. Population spiking activity in 66 nodes was simulated by a dynamical mean-field model (Wong and Wang 2006), coupled through structured long-range projections derived from diffusion-weighted imaging in humans (Hagmann et al. 2008). Simulated electrophysiological signals were then converted to simulated fMRI blood-oxygen level dependent (BOLD) signals using the Balloon-Windkessel hemodynamic model (Friston et al. 2003) to mimic empirical BOLD connectivity data presented below. We quantified effects across the entire simulated network by utilizing a simple graph theoretical measure known as global brain connectivity (GBC) (Cole et al. 2010). GBC involves averaging of a given region’s functional connectivity estimates with the rest of the brain (i.e., connectivity with all other regions). In this case we compared GBC using correlation versus covariance. We examined each measure in response to systematically manipulating the amount of unshared and shared signal between all nodes in the model (Figure 5).
Figure 5. Neurobiologically realistic simulations reveal the relationship between network changes and functional connectivity measures.
A) Shared and unshared neural signals were systematically manipulated across 66 simulated brain regions. The two-dimensional parameter space illustrates the effects of these manipulations for correlations (corr, squares, far right color bar) and covariances (cov, circles in each square, the adjacent color bar), averaged across all connections for parsimony (i.e., global connectivity across the entire set of simulated regions). The color scales indicate increases (red) and decreases (blue) relative to the central point in the parameter space (white, marked with gray border). The approximate portion of the parameter space in which both correlation and covariance gave the same results (i.e., the conjunction) is highlighted by green triangles in the upper left and lower right corners. Note that these large-scale neural network dynamics are nearly isomorphic to the pure mathematical solution (see Figure 4A), supporting the theoretical formulation. B) The same simulations for each variable in one dimension, indicating that simulations of a neurobiologically realistic network are consistent with the simpler simulations in Figure 1. Note that correlation here (as throughout this article) is the Fisher’s Z-transformed Pearson correlation, which can exceed 1.
The modeling simulations converged with the simpler conceptual illustration: only covariance matched the changes in shared signal (Figure 5A, diagonal). Specifically, covariance was unchanged from the central “baseline” point in Figure 5A as unshared signal was changed (horizontally). However, covariance was highly sensitive to changes in shared signal (vertically). In contrast, correlations interacted with both shared and unshared signals, remaining unchanged when shared and unshared signals changed equally (diagonally). However, correlations increased (upper left) or decreased (lower right) depending on the relative dominance of shared versus unshared signal changes. Note that these manipulations were implemented at the neural level, and fMRI related BOLD signals were simulated from the resulting neural activity prior to functional connectivity estimation. Thus, these results support the possibility that covariance analysis of fMRI data may more accurately reflect changes in shared signal across brain regions as opposed to artifacts of non-shared signal changes.
One potential concern with using covariances (rather than correlations) for functional connectivity is the possibility that the entire signal (shared and unshared) could be amplified artificially. Correlations are unaffected by such scaling due to normalization by the time series’ standard deviations. It is unclear how this could occur in neural populations, however. This could reflect a change of scale in the data recording equipment (e.g., scale shifts with fMRI), suggesting a potential advantage of correlation over covariance in practice. Further along this line of reasoning, it remains unclear how often correlation and covariance changes diverge in practice. We evaluate the feasibility of using covariance to measure functional connectivity further below, both in terms of providing reasonable estimates given the possibility of scale shifts and also in terms of whether it actually matters which method is used in practice.
Empirically validating covariance as a functional connectivity measure
We next sought to test for the general feasibility of using covariance as a functional connectivity measure based on empirical data. We used the publicly available WU-Minn Human Connectome Project fMRI dataset (118 subjects) (Van Essen et al. 2013). One way correlation has been empirically validated as a functional connectivity measure is via its consistency with known neural systems. For instance, regions in the visual system are especially correlated with each other relative to other brain systems during resting state, and the same is true of other known systems as well (Power et al. 2011; Yeo et al. 2011). We used this approach with covariance, with the expectation that covariance would also be higher within than between neural systems, validating covariance as a functional connectivity measure. Importantly, we observed correlation and covariance effects relative to zero (i.e., their ability to detect the presence of functional connectivity) rather than differences between brain states or individuals, such that both methods should provide similar results.
We used a set of 264 brain regions (Figure 6A) that were previously identified using fMRI meta-analysis and an approach for identifying areas of locally homogeneous functional connectivity (Power et al. 2011). These regions were used because they were identified in a distinct dataset – reducing potential statistical biases in the present results (Kriegeskorte et al. 2009) – and because these regions have an associated regional community partition (Power et al. 2011) that is consistent with known brain systems.
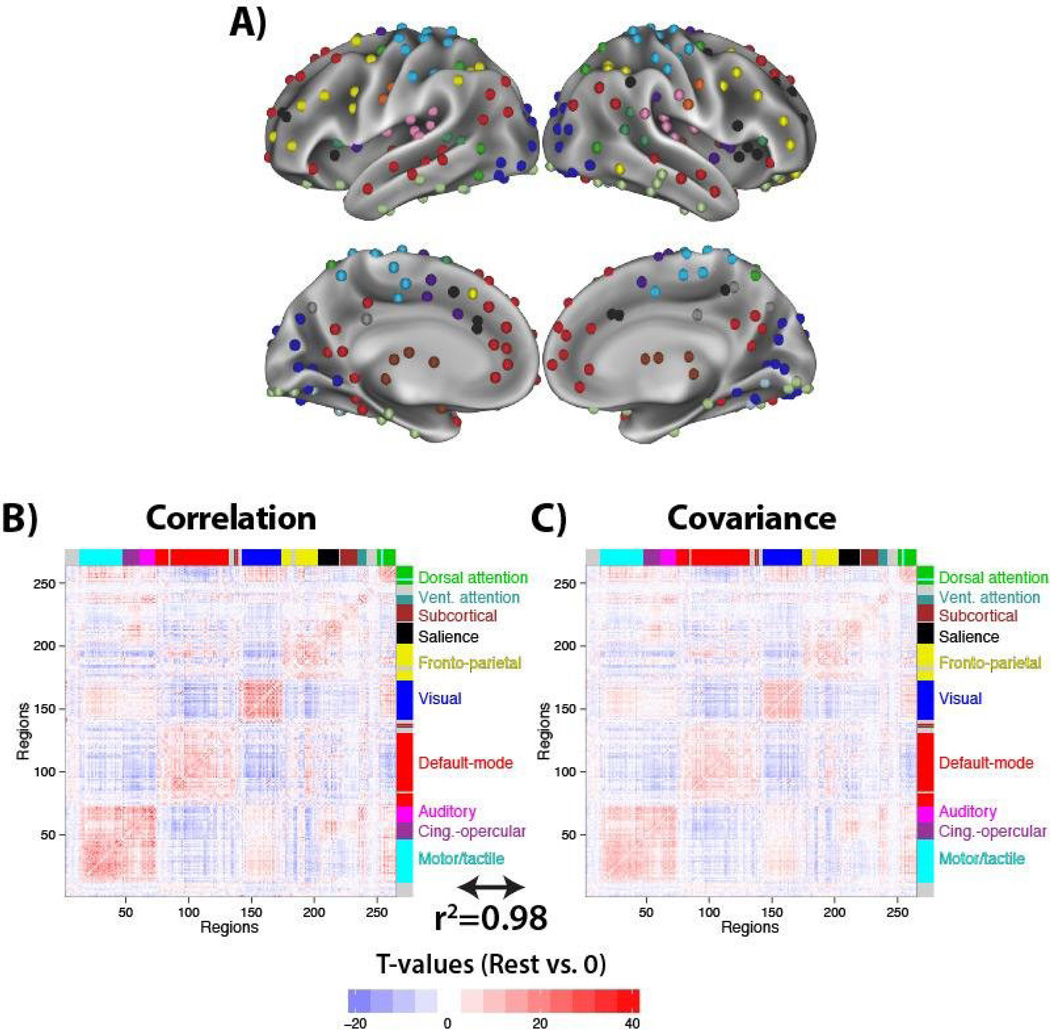
Figure 6. Validating covariance by estimating functional connectivity relative to zero.
A) A set of 264 previously identified regions were used because of an associated partition consistent with known neural systems (e.g., visual, auditory, default-mode). B) Standard resting-state functional connectivity estimation with fMRI was carried out with 118 subjects using Pearson correlation. Group t-tests versus 0 are reported for each connection, placing correlation results on the same scale as covariances. Labels are indicated on the right for the putative systems that the regions group into based on functional connectivity (Power et al. 2011). C) The analysis was repeated using covariance, resulting in a virtually identical whole-brain functional connectivity pattern (r2=0.98, p<0.00001 between the correlation and covariance t-value matrices). Results were similar for raw correlation and covariance matrices, and without global signal regression. These results validate covariance as a functional connectivity measure, while the following results focusing on functional connectivity change demonstrate distinctions between the measures.
We computed all pairwise correlations (Figure 6B) and covariances (Figure 6C) across the 264 regions. The regions were ordered based on previous community partition results (Power et al. 2011), such that connectivity clusters are apparent by visualizing the functional connectivity matrices (i.e., blocks of red along the diagonal in Figure 6). T-tests relative to zero were used to put the two functional connectivity measures on the same scale. Note that other measures of effect size could have been used as well (e.g., z values, Cohen’s d). We found that covariances revealed a large-scale brain network organization consistent with known systems, validating covariance as a functional connectivity measure. See the “Details regarding empirically validation of covariance as a functional connectivity measure” section below for more details.
These results suggest that while covariance and correlation give quite similar functional connectivity patterns generally, correlations may be better for detecting the absolute presence of functional connectivity. This is primarily due to generally higher t-values for correlations (mean absolute value t=5.4 for correlation, t=4.4 for covariance), associated with 10999 significant (P<0.05, FDR corrected for multiple comparisons) connections with correlation and 10120 with covariance – an advantage of 879 connections with correlation. We used simulations to confirm this advantage of correlations for detecting functional connections relative to 0 (though this was only true at low levels of noise; see the “Simulating the advantage of Pearson correlations for detecting shared signal relative to 0” section below). In contrast, the above simulations predict that covariance will be better than correlation when testing for functional connectivity change. We test this possibility next.
Does it matter in practice?: Testing for functional connectivity changes using empirical data
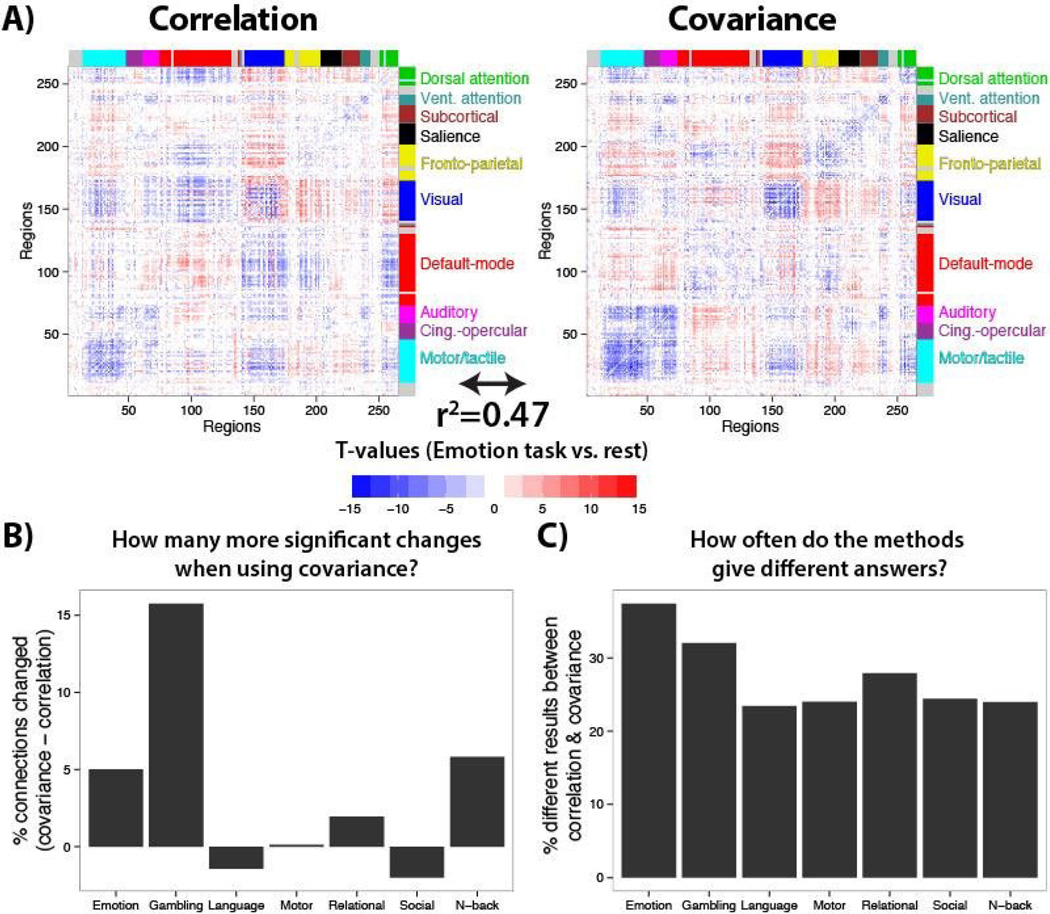
We next sought to test if correlation and covariance estimates of functional connectivity change differ in a meaningful way across a large set of tasks across a large set of brain regions. We estimated functional connectivity using both correlation and covariance during each of seven task brain states collected as part of the Human Connectome Project (Barch et al. 2013). These were the same subjects as in the resting-state results above (Figure 6), so we tested for task-driven changes in functional connectivity from the results above. Focusing on one of the tasks as a test case – the “Emotion” task versus rest (Figure 7A) – we found that there were 4% more significant functional connectivity differences using covariance than correlation, suggesting changes in unshared signal can often cancel out changes in shared signal with correlation (consistent with the white diagonal in Figure 5A). Further, we found that 37% of the results differed between the methods (e.g., a connection that was significantly decreased with covariance but significantly increased with correlation). This result strongly suggests that it matters in practice which method is used to estimate functional connectivity differences.
Figure 7. Correlation versus covariance across major brain systems and diverse cognitive domains.
A) T-tests compared all 34716 connections for an example task (the Emotion task) versus rest (p<0.05, corrected for multiple comparisons), separately using correlation and covariance. There was a rough similarity in the pattern of results, but also noticeable differences. Generally, there were many differences across methods that would alter interpretation of functional connectivity effects. Further, the above simulations suggest any observed difference with covariance has a clearer interpretation (i.e., results are unlikely to be driven by unshared signal changes). B) The percentage of connections significantly changed (each task versus rest) was computed when using covariance and correlation, then subtracted. Results from all seven tasks are shown. C) The percentage of the time that covariance and correlation gave different answers. For each task, the total number of differences in results (e.g., a connection that was significantly increased with covariance but significantly decreased with correlation) divided by the total number of significant results across both covariance and correlation approaches.
Focusing further on the whole-brain “Emotion” task versus rest results (Figure 7A), there was general similarity between the results using both methods. For instance, there was a general tendency toward reduced functional connectivity within brain systems (i.e., blue along the diagonal) and increased functional connectivity across brain systems (i.e., red off the diagonal). Notably, however, many increases within the default-mode system with correlations were not present with covariance, suggesting reductions in unshared signals drove these correlation results (as opposed to an actual increase in shared signals). Further, there were increases with covariance between the default-mode and visual systems that appeared as significant reductions with correlation. Thus, conclusions regarding these large systems-level interaction changes differ across the methods in meaningful ways.
We next quantified these patterns comprehensively across the seven task brain states. We found that covariance identified more functional connectivity changes for five of the seven tasks (Figure 7B). We used simulations to confirm the generally greater sensitivity of covariances (relative to correlations) for detecting shared signal differences (see the “Simulating the advantage of covariance for detecting shared signal differences” section below). Further, greater than 20% of results differed across the methods for every task (Figure 7C). These results again suggest – in a more comprehensive manner – that it matters in practice whether correlation or covariance is used when estimating functional connectivity differences between brain states.
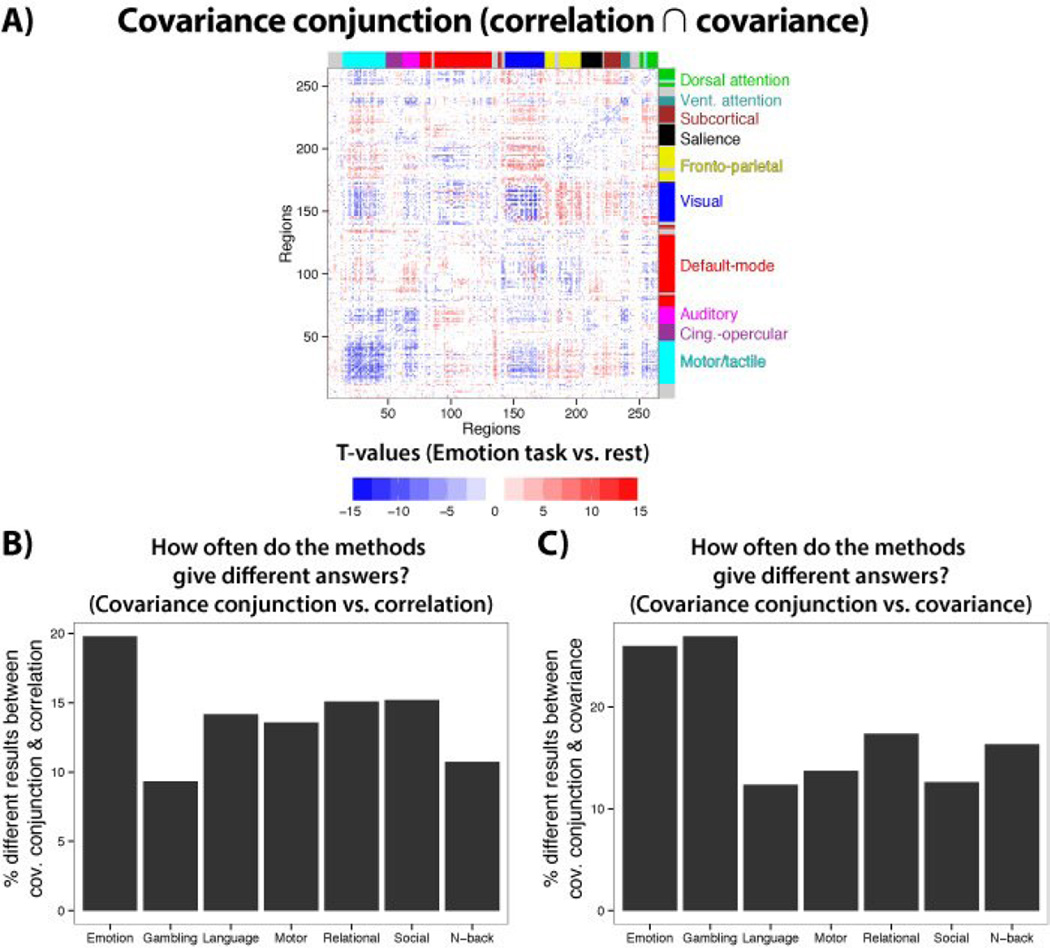
We next applied the covariance conjunction approach (see Figure 3), in which results are only considered to be statistically significant if they agree across both Pearson correlation and covariance. As expected, results were both similar and distinct from correlation and covariance (Figure 8). The Emotion task differed the most between correlation and covariance conjunction at 20% of the results being distinct, while the Gambling task differed the most between covariance and covariance conjunction at 27% of the results being distinct. While results differed more with covariance, both approaches involved a similar order of magnitude difference with covariance conjunction. These results suggest there may have been false positives in both correlations (likely from spurious changes in unshared signals) and covariances (possibly from scale/variance changes), which the conjunction approach controlled for.
Figure 8. The covariance conjunction approach.
A) The statistically significant correlation (p<0.05, FDR corrected) and statistically significant covariance (p<0.05, FDR corrected) results from Figure 7A were combined via conjunction to implement the “covariance conjunction” approach. Conjunctions were calculated separately for increases and decreases from 0. B) The percentage of different results between covariance conjunction and correlation are shown. C) The percentage of different results between covariance conjunction and covariance are shown.
Beyond purely practical implications for which method is used, the above simulations suggest the observed effects with correlation have a clearer interpretation when combined with covariance (i.e., covariance conjunction). This logic applies not only to the effects that differ across the methods, but also to effects that are similar across the methods, since only covariance is diagnostic of whether shared signal (rather than unshared signal alone) changed in any given comparison.
Explaining empirical differences between correlation and covariance: changes in unshared brain activity variance
A key assumption of the mathematical formulation and computational models is that unshared variance can change across groups or time, such as when brain processing increases or decreases in a neuron (e.g., a change in spike frequency) or a brain region (e.g., an increase in fMRI activity amplitude variance). As a first pass at empirically testing this assumption, we assessed changes in overall time series variance between each of the 7 tasks and rest. We found that the variance of the following percentages of the 264 regions were significantly changed from rest for each of the 7 tasks (t-tests paired by subject, p<0.05, Bonferroni corrected for multiple comparisons): 97.3%, 98.9%, 73.1%, 56.8%, 79.6%, 74.2%, 97.7%. Note that because this analysis involved overall variance it did not isolate unshared variance changes, but rather indicates a combination of both shared and unshared variance changes. We next better isolated unshared variance by regressing out all other regions’ time series prior to estimating the variance for each region. This revealed the following percentages of regions (analyzed identically to the previous analysis other than the additional regressions): 82.2%, 92.0%, 73.5%, 78.4%, 74.6%, 75.4%, 93.9%. Thus, there were significant changes in variance unshared between the 264 regions investigated in the above analyses, which likely drove the differential results observed between correlation and covariance measures reported above. Note that all of these analyses were conducted after removing task regressor variance, such that trial-averaged mean amplitude effects are unlikely to explain the observed changes in unshared variance. This suggests moment-to-moment and/or trial-to-trial fMRI signal variability changes between rest and task performance – a proof of principle for unshared variance changes due to brain activity changes in other contexts (e.g., between groups, individuals, temporal windows).
Details regarding empirically validation of covariance as a functional connectivity measure
The distribution of functional connectivity measures across subjects must be approximately normally distributed in order to utilize standard parametric tests to test hypotheses at the second (group) level. Fisher’s z-transform is used to allow Pearson correlation to have this property. We verified this empirically using a standard test of normality, the Kolmogorov-Smirnov test. None of the 34716 correlations in Figure 6B significantly deviated from a normal distribution (p<0.05, Bonferroni corrected for multiple comparisons). Only 0.3% of the 34716 covariance in Figure 6C significantly deviated from a normal distribution (p<0.05, Bonferroni corrected for multiple comparisons). This suggests that it is likely appropriate to use second-level parametric tests (e.g., t-tests) with covariances.
The whole-brain pattern of covariances appeared similar to those of correlations. However, there were some large deviations in the raw value covariance matrix that were not present in the correlation matrix, which were controlled for using group-level t-tests (Figure 6C). These deviations reflect the fact that unlike correlations (and group-level t-values) each raw covariance value is in units dependent on the regions being tested (region X variance * region Y variance units). Thus, if some regions have substantially different activity variance amplitudes than most regions they will appear as large deviations in covariance. The identified deviations represented only a small fraction of the total number of connections: 36 covariances above a value of 2000, representing 0.1% of connections. Most of these deviations were covariances among subcortical regions. Note that this particular dataset is known to have lower subcortical signal-to-noise than most fMRI datasets, given that a 32-channel head coil was used (Van Essen et al. 2013). Correlations do not show these deviations because they standardize their values by dividing by the time series standard deviations – a solution to this problem but the cause of the problems identified in the simulations. These deviations were eliminated by standardizing covariances at the group level using t-tests, which involves dividing each connection’s across-subject mean value by its across-subject standard deviation (in contrast to dividing by the standard deviations of the time series with correlations).
Simulating the advantage of Pearson correlations for detecting shared signal relative to 0
We found that t-values tended to be higher for correlations than covariances when detecting connections (relative to 0) in the empirical data (Figure 6). We next sought to test if this effect was present in the simulations. Identifying this effect in the simulations would help generalize the result beyond the particular empirical tests we conducted.
We used the same simple simulation setup as used in Figure 1. We found that with small amounts of time series noise (0.25; the same as in Figure 1) group analysis t-values (testing vs. 0) were consistently much larger with correlations than covariances. To illustrate this we ran 100 simulations, finding that the mean correlation-based t-value was 126, while the mean covariance-based t-value was 52. We ran a t-test comparing the distributions of correlation-based and covariance-based t-values, to establish the consistency of this result: t(99)=29, p<0.00001. Importantly, however, this effect went away with high amounts of time series noise (4× the amplitude of the shared signal). In this scenario mean correlation-based t-value was 4.19, while the mean covariance-based t-value was 4.18 (t(99)=0.16, p=0.9). This suggests that correlations only have an advantage over covariances at low levels of noise. Generally, we found – using both empirical data and simulations – that correlations are more sensitive than covariances for detecting shared signal relative to 0.
Simulating the advantage of covariance for detecting shared signal differences
We found with most brain state comparisons that there were more significant differences in connectivity when using covariances than correlations (Figure 7) – the opposite of the result found when testing for connections relative to 0. We next sought to test if this effect was present in the simulations. Identifying this effect in the simulations would help generalize the result beyond the particular empirical tests we conducted.
We found that covariances showed consistently larger t-values when comparing large to small amounts of shared signal (the same test as in Figure 1, left side). To illustrate this consistency we ran 100 simulations: Mean covariance t-value: 38.83, mean correlation t-value: 34.82 (difference t(99)=5.05, p<0.00001). Unlike the advantage of correlation for detecting effects vs. 0, this result was stable at high levels of noise (4× the amplitude of the shared signal). Mean covariance t-value at high noise: 8.23, mean correlation t-value at high noise: 7.35 (difference t(99)=5.93, p<0.00001).
In summary, we found – using both empirical data and simulations – that correlations are more sensitive for detecting shared signal relative to 0, while covariances are more sensitive for detecting differences in shared signal. In most cases, however, we recommend using the covariance conjunction approach, which will only be as sensitive as the least sensitive measure (in this case correlation). We next examine a case in which one might choose to forgo this recommendation in order to take advantage of increased sensitivity of covariance to detect functional connectivity change.
Measuring functional connectivity differences across groups: Application to schizophrenia
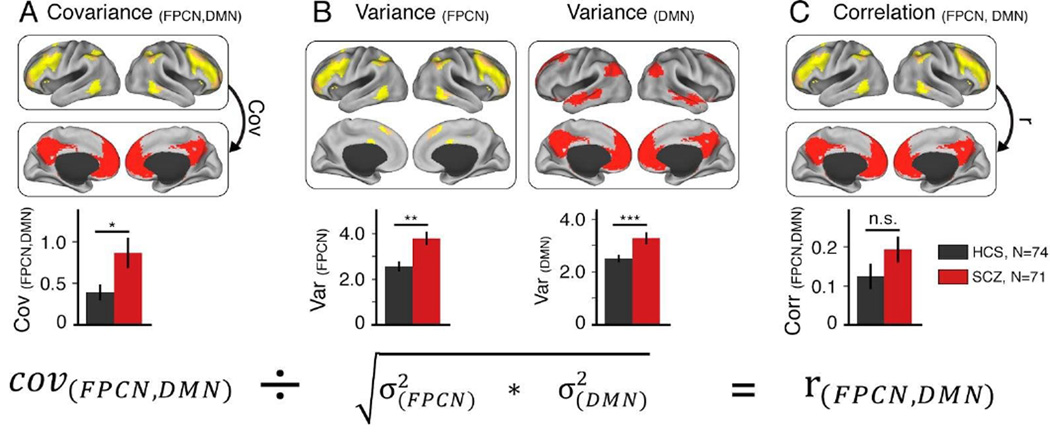
Above we demonstrated the impact of using covariance (versus correlation-like measures) as a functional connectivity measure with both simulated and empirically-derived data in healthy adults. We next examined if using covariance can have an impact on clinical between-group connectivity analyses where it is expected that one group would differ in connectivity patterns. To test this hypothesis, we analyzed a large sample of patients with schizophrenia (N=71) relative to a group of healthy matched controls (N=74). We focused on the statistical relationship between two large-scale neural systems that have been repeatedly implicated in schizophrenia – the default-mode network (DMN) and the frontoparietal control network (FPCN) (Baker et al. 2014). The networks were defined using a voxelwise partition previously identified in healthy adults (Power et al. 2011). We found that patients exhibited significantly increased covariance between the DMN and FPCN (Figure 9A). Interestingly, the effect was attenuated and no longer significant when using correlations (Figure 9C). This discrepancy occurred because of elevated variance within both DMN and FPCN for patients relative to controls (Figure 9B). Illustrating the reason for this effect, we present the full correlation equation in relation to these data (Figure 9, bottom panel). This illustrates that dividing the covariance by a relatively larger variance for patients will by definition result in a reduction of the correlation (Yang et al. 2014). Collectively, these clinical effects show how use of correlations can obscure a possible clinically-relevant difference in connectivity due to alterations in unshared signals. In contrast, covariance remained sensitive to the connectivity difference between the DMN and FPCN, a hypothesis suggested by recent work (Baker et al. 2014). Note, however, that even if there were no differences between covariance and correlation results, the above simulations demonstrate that we would gain additional insight into these effects by using covariance (e.g., confidence that results were not driven by unshared variance differences between patients and healthy controls). Further, it should be noted that this effect could plausibly (but not necessarily) be caused by an overall increase in variance/power, such that the increase in functional connectivity is apparent rather than actual. We recommend the covariance conjunction approach (Figure 3) in order to help rule out erroneous conclusions.
Figure 9. Detected disruptions across functional networks in schizophrenia differ between covariance and correlation.
A) Here we show altered covariance structure between large-scale associative networks in schizophrenia (SCZ), similar to recent findings (Baker et al. 2014) [t(143)=2.37, p=0.019, Cohen’s d=0.4]. B) We recently discovered elevated variance across the entire brain in chronic SCZ, which was particularly evident for associative networks (Yang et al. 2014). C) Based on this elevated non-shared variance, it follows that the difference in correlations between SCZ and healthy control subjects (HCS) across the two networks will be attenuated and no longer reveal a significant clinical effect [t(143)=1.48, p=0.14, Cohen’s d=0.25]. The equation on the bottom is presented for illustrative purposes, to highlight the importance of carefully decomposing the final correlation into variance and covariance components (Figure 3). FPCN, fronto-parietal control network; DMN, defaultmode network.
DISCUSSION
Despite decades of neuroscience research our basic understanding of what constitutes functional connectivity change (and how to measure it) is still evolving. Attention has recently been drawn to the issue of increased noise in one condition or group producing reductions in correlations (and related measures) (Behseta et al. 2009; Friston 2011), potentially resulting in false positives and false negatives across a range of studies. We postulated that the broader concept of “unshared signal” change is even more problematic for correlation-like measures. We reasoned that changes in independent neural processing would likely alter correlations despite no change in interactions among the tested regions/neurons. We verified this concern using a simple and generalizable simulation (Figure 1), mathematical theory, as well as a more complex biologically plausible simulation of large-scale neural dynamics (Figure 5). Across these analyses, we demonstrate that correlation changes were difficult to interpret due to their sensitivity to changes in unshared signal. In contrast, we demonstrate that covariance was sensitive to shared signal alone, increasing the interpretability of observed functional connectivity change. This was especially true for cases in which both covariance and correlation agree: the covariance conjunction approach. Following these simulations, we applied this method to empirical datasets, finding that it mattered in practice which functional connectivity measure was used, and that covariance provided robust results for both within-subject and across-group functional connectivity changes. Even if results had been less robust with covariance, however, we would still recommend its use as our comprehensive characterization of the relevant parameter space (Figure 4) indicates this measure – especially when combined with more standard measures – yields increased interpretability of functional connectivity effects generally.
Our findings suggest results reported by most previous studies of functional connectivity change (even most that did not use correlations, such as those using PPI) are difficult to interpret because of ambiguity concerning shared versus unshared signal contributions. This includes any studies involving a difference in functional connectivity estimates, such as across groups (e.g., clinical studies), tasks, individuals (e.g., individual difference correlations), or dynamics (e.g., resting-state dynamics across temporal windows). Note that correlation-based studies seeking to simply identify any difference (e.g., between groups) are likely valid in their identification of differences. However, due to the ambiguity of correlation-like measures, such studies may have misinterpreted results in terms of brain interaction change. The ability to accurately interpret observed differences will be important moving forward, given the ultimate neuroscientific goal of increasing mechanistic understanding of brain processes (rather than, e.g., simply identifying ambiguous group differences).
To further illustrate the difficulty of interpreting changes in correlation-like measures, consider the possible range of distinct interpretations (as established by the simulations above) of an observed increase in correlation between two regions’ time series: (1) decreased independent activity in region X and/or Y, (2) decreased interaction between Y and another region Z resulting in less unshared signal in region Y (Figure 2), (3) increased interaction between X and Y. Covariance is also problematic, though less so, as an increase in covariance would have only two viable interpretations: (1) increased interaction between X and Y, or (2) increased overall variance/power in either X and/or Y. In contrast, an increase in the covariance conjunction measure (combining correlation and covariance) would have only a single viable interpretation: increased interaction between X and Y.
Correlations have been historically favored over covariance in many scientific applications because correlations are normalized by variance, making them insensitive to changes in scale and facilitating comparison across studies. These constitute major advantages in some cases, but we found that normalization by variance has unintended consequences when estimating functional connectivity differences. Rather than simply estimating change in an abstract measure of association, differences in correlation can be driven by changes in the unshared variance component, such that the very aspect we are trying to “control for” (the overall variance) actually drives the measured effect. This is not an issue only for correlation, but any measure that normalizes by some form of variance (or entropy), such as coherence, regression, and mutual information (Figure 4). We found that simply removing variance normalization from correlation (i.e., using covariance) circumvented these issues. We suggest that removing variance normalization from other measures may help solve this problem in other cases when estimating connectivity change (see Results for preliminary evidence with coherence). Such measures without variance normalization could then be combined with the original measures to allow implementation of the covariance conjunction approach – ruling out spurious changes in functional connectivity due to either changes in unshared variance or overall variance/power.
Limitations
As outlined above, there are several issues to consider when using covariance. First, since covariance is sensitive to changes in scale, care should be taken to ensure no scale shifts have occurred across conditions/groups/individuals that are being compared. Note, however, that this same issue is often present when comparing brain activity magnitudes across conditions/groups/individuals, such that this issue may be no worse here than in most existing neuroscientific studies of brain function (which have tended to test for activation magnitude changes rather than functional connectivity changes). Our recommendation is to sidestep this issue using the conservative “covariance conjunction” approach (Figure 3).
Another potential issue with covariance is its non-standard units: each covariance estimate is in units of region X activity * region Y activity. This reflects the non-normalized nature of covariance relative to correlation. This is not an issue in the case of functional connectivity change for a given pair of regions since the compared conditions/groups/individuals always have the same units (i.e., region X activity * region Y activity). This is problematic for performing across-connection comparisons, however, just as across-region activity comparisons are problematic with some methods (e.g., fMRI (Handwerker et al. 2004)) due to potential differences in activation scale across regions. However, we found that covariances could be normalized at the group level (using inter-subject variance) with t-tests, which eliminated scale differences across covariances (Figure 6). Note that having distinct units across connections may be an issue for some graph theoretical (Bullmore and Bassett 2011) analyses (especially at the single-subject level) that focus on network topology (e.g., community detection) but potentially not for others (e.g., degree centrality).
Using covariance does not eliminate all issues present when using correlation-like measures to estimate functional connectivity change. For instance, like correlations (Smith et al. 2011b), covariances do not estimate directionality of functional connectivity changes. It will be important to determine which of the existing directional/effective connectivity methods (Friston et al. 2003; Roebroeck et al. 2005; Nolte et al. 2008; Ramsey et al. 2011; Smith et al. 2011b) involve variance normalization, and if all of them do, then it will be important to develop new approaches that do not involve this analysis step for studies examining effective connectivity change. Importantly, there is already evidence of advantages when using an unnormalized version of a popular form of effective connectivity, Granger causality (Angelini et al. 2010; Stramaglia et al. 2015).
Another limitation is that, like correlations and most other functional connectivity measures (Smith et al. 2011b), covariances do not indicate if two regions are interacting directly or indirectly via a third (or fourth, fifth, etc.) region(s). This affects interpretation, but is often not problematic as long as it is taken into account. It may be possible, however, to estimate direct functional connectivity change using some form of partial correlation (or multiple regression) (Marrelec et al. 2006; Liang et al. 2011; Smith et al. 2011b; Ryali et al. 2012). It is important to note that partial correlation involves variance normalization and therefore faces limitations similar to standard correlations with respect to changes in unshared variance. Also note that, despite its name, inverse covariance also involves variance normalization. In the future it may be useful to identify partial correlation-like approaches that are adapted to not include variance-based normalization. One promising possibility is to use multivariate Granger causality without variance-based normalization (Angelini et al. 2010; Stramaglia et al. 2015), which estimates all time series simultaneously to achieve the main benefits of partial correlation in the context of directional connectivity. Note, however, that potential issues with using Granger causality with fMRI have been identified (Smith et al. 2011a; 2011b), such that this approach may be best applied to other modalities such as EEG. The inability to infer whether two regions are interacting directly notwithstanding, the present results suggest using covariances provides increased confidence that a change in connectivity between two regions reflects their shared signal change (irrespective of whether this occurred via a 3rd region; see Figure 2).
We recommend the use of covariance conjunction, in which the results of correlation and covariance analyses are combined, in order to remain conservative. This combines the benefits of both correlation (insensitivity to overall variance scaling) and covariance (insensitivity to changes in unshared variance) in terms of reducing Type I errors (false positives). However, as illustrated in Figures 4 and 5, this comes with the possibility of increased Type II errors (false negatives). For instance, a real increase in interaction may be accompanied by increases in independent processing in each tested region (unshared signal), resulting in an increase in covariance but not correlation. This would lead to a false negative when using the conjunction approach. We nonetheless recommend this approach given the possibility that an increase in both shared and unshared signal may also reflect an overall variance increase. It will be important for future work to look for ways to maintain all the benefits of covariance without its potential drawbacks.
We found that, in contrast to functional connectivity changes, correlations may be better for detecting the absolute presence of functional connections relative to zero. This supports the use of correlations to detect the absolute presence of functional connectivity, as performed by many resting-state functional connectivity studies (Biswal et al. 1995; Power et al. 2011; Yeo et al. 2011). Importantly, however, we reached the opposite conclusion in the case of detecting functional connectivity change.
Interpreting covariance differences in the empirical datasets
We specifically focused on functional connectivity differences between an emotion task and rest (Figure 7A), identifying numerous differences between the measures. There were similarities between the methods as well, however. For instance, there were within-network decreases for motor, auditory, and visual systems across both methods, consistent with recent findings (Cole et al. 2014). Notably, these decreases were more robust when using covariance, even extending the within-network decreases to other systems. This suggests that such within-network decreases in functional connectivity are either more widespread than indicated by correlations or, alternatively, that these decreases are largely due to decreases in overall variance/power. It will be important for future research to investigate the possible mechanisms underlying such widespread within-network decreased covariance. One possible interpretation is that task-focused attention (“cognitive set”) (Duncan 2013) requires reduced interactions within most brain systems to facilitate selection of task-relevant regional interactions, possibly including primarily inter-system interactions (e.g., visual-to-motor system interactions in a visual-motor task). This possibility is consistent with our recent study demonstrating extensive intersystem interaction changes across a variety of tasks (Cole et al. 2013). Note that in addition to using correlation and PPI we also found those effects using covariance differences (see that paper’s supplementary results) (Cole et al. 2013).
There is growing interest in establishing functional connectivity differences across different groups and clinical states to characterize dysfunctional neural dynamics. Functional connectivity has become a particularly powerful and widely used approach to characterize large-scale neural dynamics in severe neuropsychiatric illnesses such as schizophrenia (Anticevic et al. 2013; 2014). Use of correlation in such cases could be problematic for the same reasons articulated above: correlation differences can be driven by changes in unshared signal in one group relative to another, resulting in false positives or false negatives. To provide evidence for this, we examined functional connectivity differences between patients diagnosed with chronic schizophrenia and healthy controls. We focused on two well-characterized systems with known disruptions in schizophrenia (Baker et al. 2014): DMN and FPCN. As predicted, we found that covariance revealed a connectivity alteration in patients relative to controls that was not evident when using correlations – consistent with our network simulations and demonstrating that covariance can reveal a distinct set of functional connectivity differences from correlation in a clinical context.
Practical recommendations for the use of covariance as a functional connectivity measure
We have demonstrated the complexity of interpreting changes in correlation (and a variety of other measures), and the relatively improved clarity of interpreting changes in covariance. This suggests covariance may be preferred when testing for brain interaction changes. Despite this, caution suggests the use of more typical functional connectivity measures in addition to covariance. As outlined above (Figure 3 and Figure 8), one possibility would be to conduct both correlation and covariance analyses, assigning the most confidence to results that are consistent across both approaches. In addition, there may be cases in which correlation is more sensitive than covariance, such as when each subject’s data are scaled differently. In such cases there would be additional irrelevant inter-subject variance that would reduce statistical confidence in effects of interest. In cases where within-subject manipulations are used, subtracting covariances prior to the group analysis (as in a paired t-test) can reduce such inter-subject variance concerns. Alternatively, after subtracting covariances at the single subject level the resulting difference can then be divided by the standard deviation (aggregate across both conditions) to rescale the result prior to group analysis. Finally, it may be possible in some cases to rescale time series based on the time series mean – a percent signal change normalization approach often used with fMRI that is unlikely to be biased by changes in unshared variance.
Conclusion
We used mathematical and biologically realistic simulations to arrive at a theoretically important conclusion: variance normalization (as performed by most commonly used measures) can obscure estimates of functional connectivity change. This applies primarily to cases that involve unshared signal alterations, though even cases without such alterations are obscured for the investigator due to uncertainty whether such unshared signal alteration occurred (e.g., in Figure 7A decreased correlations were only interpretable due to similar decreases in covariance). This theoretical insight, corroborated by empirical evidence, has implications for a wide variety of previous and future studies, as estimating functional connectivity change is central to understanding the functional relevance of brain connections (by associating them with task conditions, individual differences, and group differences) and for characterizing brain connectivity dynamics. Removing variance normalization from other measures may similarly improve clarity in other context as well (e.g., lag-invariant functional connectivity using coherence). Generally, these findings suggest a need to reconceptualize functional connectivity change in terms of shared signal differences, rather than in terms of abstract measures of association that may obscure effects of interest.
Highlights.
Interpretability limits of functional connectivity measures identified with modeling
Most connectivity measures can change with no brain region interaction change
Decomposition of correlation reveals covariance as an important check on results
Empirical tests demonstrate that covariance and correlation often differ in practice
Even when results are identical between methods covariance provides an important check
Acknowledgements
We would like to thank Jeremy Reynolds, Kai Hwang, Zhen Yang, Joset Etzel, Robert Kass, Todd Braver, Deanna Barch, and members of the Petersen and Schlaggar Lab for helpful conversations during preparation of this manuscript. Data were provided in part by the Human Connectome Project, WU-Minn Consortium (Principal Investigators: David Van Essen and Kamil Ugurbil; 1U54MH091657) funded by the 16 NIH Institutes and Centers that support the NIH Blueprint for Neuroscience Research; and by the McDonnell Center for Systems Neuroscience at Washington University. Data were also provided in part by the Mind Research Network and the University of New Mexico funded by a National Institute of Health Center of Biomedical Research Excellence (COBRE) grant 1P20RR021938-01A2. Our work was supported by the US National Institutes of Health under awards K99-R00 MH096801 (M.W.C.), and T32GM 007205 (G.J.Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Friston emphasized the inadequacy of Pearson correlations in terms of estimating indirect influences, undirected influences, and their tendency for changing due to changes in noise. We focus here on the last criticism, and touch upon the other criticisms in the Discussion.
References
- Angelini L, de Tommaso M, Marinazzo D, Nitti L, Pellicoro M, Stramaglia S. Redundant variables and Granger causality. Phys Rev E. 2010 Mar 5;81(3):037201–037204. doi: 10.1103/PhysRevE.81.037201. [DOI] [PubMed] [Google Scholar]
- Anticevic A, Cole MW, Repovs G, Murray JD, Brumbaugh MS, Winkler AM, et al. Characterizing thalamo-cortical disturbances in schizophrenia and bipolar illness. Cerebral Cortex. 2014 Dec;24(12):3116–3130. doi: 10.1093/cercor/bht165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anticevic A, Cole MW, Repovs G, Savic A. Connectivity, pharmacology, and computation: toward a mechanistic understanding of neural system dysfunction in schizophrenia. Frontiers in …․ 2013. doi: 10.3389/fpsyt.2013.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydore S, Pantazis D, Leahy RM. A note on the phase locking value and its properties. NeuroImage. 2013 Jul 1;74:231–244. doi: 10.1016/j.neuroimage.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JT, Holmes AJ, Masters GA, Yeo BTT, Krienen F, Buckner RL, et al. Disruption of Cortical Association Networks in Schizophrenia and Psychotic Bipolar Disorder. JAMA Psychiatry. 2014 Feb 1;71(2):109. doi: 10.1001/jamapsychiatry.2013.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barch DM, Burgess GC, Harms MP, Petersen SE, Schlaggar BL, Corbetta M, et al. NeuroImage. C. Vol. 80. Elsevier B.V; 2013. May 22, Function in the human connectome: Task-fMRI and individual differences in behavior; pp. 169–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behseta S, Berdyyeva T, Olson CR, Kass RE. Bayesian Correction for Attenuation of Correlation in Multi-Trial Spike Count Data. J Neurophysiol. 2009 Feb 11;101(4):2186–2193. doi: 10.1152/jn.90727.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995 Oct;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Biswal BB, Mennes M, Zuo X-N, Gohel S, Kelly C, Smith SM, et al. Toward discovery science of human brain function. Proceedings of the National Academy of Sciences. 2010 Feb 22;:1–6. doi: 10.1073/pnas.0911855107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes MJ, Woolrich M, Luckhoo H, Price D, Hale JR, Stephenson MC, et al. Proc Natl Acad Sci USA. 40. Vol. 108. National Acad Sciences; 2011. Investigating the electrophysiological basis of resting state networks using magnetoencephalography; pp. 16783–16788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore ET, Bassett DS. Brain Graphs: Graphical Models of the Human Brain Connectome. Annual review of clinical psychology. 2011 Apr 27;7(1):113–140. doi: 10.1146/annurev-clinpsy-040510-143934. [DOI] [PubMed] [Google Scholar]
- Buschman TJ, Denovellis EL, Diogo C, Bullock D, Miller EK. Neuron. 4. Vol. 76. Elsevier Inc; 2012. Nov 21, Synchronous Oscillatory Neural Ensembles for Rules in the Prefrontal Cortex; pp. 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AL, Fair DA, Dosenbach NUF, Miezin FM, Dierker D, Van Essen DC, et al. Defining functional areas in individual human brains using resting functional connectivity MRI. NeuroImage. 2008 May;41(1):45–57. doi: 10.1016/j.neuroimage.2008.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014 Jul 2;83(1):238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Pathak S, Schneider W. Identifying the brain's most globally connected regions. NeuroImage. 2010 Feb 15;49(4):3132–3148. doi: 10.1016/j.neuroimage.2009.11.001. [DOI] [PubMed] [Google Scholar]
- Cole MW, Reynolds JR, Power JD, Repovs G, Anticevic A, Braver TS. Multi-task connectivity reveals flexible hubs for adaptive task control. Nat Neurosci. 2013 Sep;16(9):1348–1355. doi: 10.1038/nn.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes D, Haughton V, Arfanakis K, Carew J, Turski P, Moritz C, et al. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. AJNR Am J Neuroradiol. 2001 Jul 31;22(7):1326–1333. [PMC free article] [PubMed] [Google Scholar]
- Cox RW. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput Biomed Res. 1996 May 31;29(3):162–173. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Craddock RC, Jbabdi S, Yan C-G, Vogelstein JT, Castellanos FX, Di Martino A, et al. Imaging human connectomes at the macroscale. Nature Methods. 2013 May 30;10(6):524–539. doi: 10.1038/nmeth.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Jirsa VK. Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. The Journal of neuroscience. Soc Neuroscience. 2012;32(10):3366–3375. doi: 10.1523/JNEUROSCI.2523-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deco G, Ponce-Alvarez A, Mantini D, Romani GL, Hagmann P, Corbetta M. Resting-state functional connectivity emerges from structurally and dynamically shaped slow linear fluctuations. Journal of Neuroscience. Society for Neuroscience. 2013 Jul 3;33(27):11239–11252. doi: 10.1523/JNEUROSCI.1091-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage. 2006 Jun 30;31(3):968–980. doi: 10.1016/j.neuroimage.2006.01.021. [DOI] [PubMed] [Google Scholar]
- Duncan J. Neuron. 1. Vol. 80. Elsevier Inc; 2013. Oct 1, The Structure of Cognition:Attentional Episodes in Mind and Brain; pp. 35–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel AK, Fries P, Singer W. Dynamic predictions: oscillations and synchrony in top-down processing. Nat Rev Neurosci. 2001;2(10):704–716. doi: 10.1038/35094565. [DOI] [PubMed] [Google Scholar]
- Fries P. A mechanism for cognitive dynamics: neuronal communication through neuronal coherence. Trends Cogn Sci (Regul Ed) 2005 Oct;9(10):474–480. doi: 10.1016/j.tics.2005.08.011. [DOI] [PubMed] [Google Scholar]
- Friston KJ. Functional and Effective Connectivity: A Review. Brain Connectivity. 2011 Jan;1(1):13–36. doi: 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Harrison L, Penny W. NeuroImage. 4. Vol. 19. Elsevier; 2003. Dynamic causal modelling; pp. 1273–1302. [DOI] [PubMed] [Google Scholar]
- Genovese C, Lazar N, Nichols T. Thresholding of statistical maps in functional neuroimaging using the false discovery rate. NeuroImage. 2002 Mar 31;15(4):870–878. doi: 10.1006/nimg.2001.1037. [DOI] [PubMed] [Google Scholar]
- Glasser MF, Sotiropoulos SN, Wilson JA, Coalson TS, Fischl B, Andersson JL, et al. The minimal preprocessing pipelines for the Human Connectome Project. NeuroImage. 2013 Oct 15;80:105–124. doi: 10.1016/j.neuroimage.2013.04.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, et al. Mapping the Structural Core of Human Cerebral Cortex. Plos Biol. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handwerker D, Ollinger J, D'Esposito M. Variation of BOLD hemodynamic responses across subjects and brain regions and their effects on statistical analyses. NeuroImage. 2004 Mar 31;21(4):1639–1651. doi: 10.1016/j.neuroimage.2003.11.029. [DOI] [PubMed] [Google Scholar]
- Kriegeskorte N, Simmons WK, Bellgowan PSF, Baker CI. Circular analysis in systems neuroscience: the dangers of double dipping. Nat Neurosci. 2009 May 1;12(5):535–540. doi: 10.1038/nn.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krystal JH, D'Souza DC, Mathalon D, Perry E, Belger A, Hoffman R. Psychopharmacology. 3–4. Vol. 169. Springer; 2003. NMDA receptor antagonist effects, cortical glutamatergic function, and schizophrenia: toward a paradigm shift in medication development; pp. 215–233. [DOI] [PubMed] [Google Scholar]
- Lachaux JP, Rodriguez E, Martinerie J, Varela FJ. Measuring phase synchrony in brain signals. Hum Brain Mapp. 1999;8(4):194–208. doi: 10.1002/(SICI)1097-0193(1999)8:4<194::AID-HBM4>3.0.CO;2-C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N. Uncovering Intrinsic Connectional Architecture of Functional Networks in Awake Rat Brain. Journal of Neuroscience. 2011 Mar 9;31(10):3776–3783. doi: 10.1523/JNEUROSCI.4557-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrelec G, Krainik A, Duffau H, Pélégrini-Issac M, Lehéricy S, Doyon J, et al. Partial correlation for functional brain interactivity investigation in functional MRI. NeuroImage. 2006 Aug;32(1):228–237. doi: 10.1016/j.neuroimage.2005.12.057. [DOI] [PubMed] [Google Scholar]
- Mayer AR, Ruhl D, Merideth F, Ling J, Hanlon FM, Bustillo J, et al. Human brain mapping. 9. Vol. 34. Wiley Online Library; 2013. Functional imaging of the hemodynamic sensory gating response in schizophrenia; pp. 2302–2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren DG, Ries ML, Xu G, Johnson SC. NeuroImage. 4. Vol. 61. Elsevier B.V; 2012. Jul 16, A generalized form of context-dependent psychophysiological interactions (gPPI): a comparison to standard approaches; pp. 1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolte G, Bai O, Wheaton L, Mari Z, Vorbach S, Hallett M. Identifying true brain interaction from EEG data using the imaginary part of coherency. Clin Neurophysiol. 2004 Sep 30;115(10):2292–2307. doi: 10.1016/j.clinph.2004.04.029. [DOI] [PubMed] [Google Scholar]
- Nolte G, Ziehe A, Nikulin VV, Schlögl A, Krämer N, Brismar T, et al. Robustly Estimating the Flow Direction of Information in Complex Physical Systems. Phys Rev Lett. 2008 Jun 1;100(23):4. doi: 10.1103/PhysRevLett.100.234101. [DOI] [PubMed] [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional network organization of the human brain. Neuron. 2011 Nov 17;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. Methods to detect, characterize, and remove motion artifact in resting state fMRI. NeuroImage. 2014 Jan 1;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. 2009 [Google Scholar]
- Ramsey JD, Hanson SJ, Glymour C. NeuroImage. Elsevier Inc; 2011. Jul 19, Multi-subject search correctly identifies causal connections and most causal directions in the DCM models of the Smith et al. simulation study; pp. 1–11. [DOI] [PubMed] [Google Scholar]
- Rissman J, Gazzaley A, D'Esposito M. Measuring functional connectivity during distinct stages of a cognitive task. NeuroImage. 2004 Sep 30;23(2):752–763. doi: 10.1016/j.neuroimage.2004.06.035. [DOI] [PubMed] [Google Scholar]
- Roebroeck A, Formisano E, Goebel R. Mapping directed influence over the brain using Granger causality and fMRI. NeuroImage. 2005 doi: 10.1016/j.neuroimage.2004.11.017. [DOI] [PubMed] [Google Scholar]
- Ryali S, Chen T, Supekar K, Menon V. NeuroImage. 4. Vol. 59. Elsevier Inc; 2012. Feb 15, Estimation of functional connectivity in fMRI data using stability selection-based sparse partial correlation with elastic net penalty; pp. 3852–3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Bandettini PA, Miller KL, Behrens TEJ, Friston KJ, David O, et al. NeuroImage. Elsevier Inc; 2011a. Aug 20, The danger of systematic bias in group-level FMRI-lag-based causality estimation; pp. 1–2. [DOI] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, et al. NeuroImage. 2. Vol. 54. Elsevier; 2011b. Network modelling methods for FMRI; pp. 875–891. [DOI] [PubMed] [Google Scholar]
- Smith SM, Vidaurre D, Beckmann CF, Glasser MF, Jenkinson M, Miller KL, et al. Functional connectomics from resting-state fMRI. Trends Cogn Sci (Regul Ed) 2013 Dec;17(12):666–682. doi: 10.1016/j.tics.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stramaglia S, Angelini L, Cortés JM. Synergy, redundancy and unnormalized Granger causality. arXiv. 2015 doi: 10.1109/EMBC.2015.7319280. [DOI] [PubMed] [Google Scholar]
- Truccolo WA, Ding M, Knuth KH, Nakamura R, Bressler SL. Trial-to-trial variability of cortical evoked responses: implications for the analysis of functional connectivity. Clin Neurophysiol. 2002 Feb;113(2):206–226. doi: 10.1016/s1388-2457(01)00739-8. [DOI] [PubMed] [Google Scholar]
- Ugurbil K, Xu J, Auerbach EJ, Moeller S, Vu A, Duarte-Carvajalino JM, et al. NeuroImage. Elsevier; 2013. Pushing spatial and temporal resolution for functional and diffusion MRI in the Human Connectome Project. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TEJ, Yacoub E, Ugurbil K, et al. NeuroImage. Elsevier B.V; 2013. May 21, The WU-Minn Human Connectome Project: An overview; pp. 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan S, Wang M, Birnbaum SG, Williams GV, Arnsten AF. Nat Neurosci. 3. Vol. 10. Nature Publishing Group; 2007. Inverted-U dopamine D1 receptor actions on prefrontal neurons engaged in working memory; pp. 376–384. [DOI] [PubMed] [Google Scholar]
- Wig GS, Schlaggar BL, Petersen SE. Concepts and principles in the analysis of brain networks. Ann N Y Acad Sci. 2011 Apr 12;1224(1):126–146. doi: 10.1111/j.1749-6632.2010.05947.x. [DOI] [PubMed] [Google Scholar]
- Wong K-F, Wang X-J. A recurrent network mechanism of time integration in perceptual decisions. The Journal of neuroscience. Soc Neuroscience. 2006;26(4):1314–1328. doi: 10.1523/JNEUROSCI.3733-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang GJ, Murray JD, Repovs G, Cole MW, Savic A, Glasser MF, et al. Altered global brain signal in schizophrenia. Proceedings of the National Academy of Sciences. 2014 May 20;111(20):7438–7443. doi: 10.1073/pnas.1405289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H, Shi L, Han F, Gao H, Dan Y. Rapid learning in cortical coding of visual scenes. Nat Neurosci. 2007 Apr 29;10(6):772–778. doi: 10.1038/nn1895. [DOI] [PubMed] [Google Scholar]
- Yeo BTT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011 Sep;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Cocchi L, Fornito A, Murray MM, Bullmore E. NeuroImage. 2. Vol. 60. Elsevier Inc; 2012a. Apr 2, Connectivity differences in brain networks; pp. 1055–1062. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore E. On the use of correlation as a measure of network connectivity. NeuroImage. 2012b May 1;60(4):2096–2106. doi: 10.1016/j.neuroimage.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Zucker RS, Regehr WG. Short-term synaptic plasticity. Annu Rev Physiol. 2002;64:355–405. doi: 10.1146/annurev.physiol.64.092501.114547. [DOI] [PubMed] [Google Scholar]