Abstract
Therapy resistance and poor outcome in prostate cancer is associated with increased expression of Cyclin D1. Androgens promote DNA double strand break repair to reduce DNA damage, and cyclin D1 was also shown to enhance DNA damage repair (DDR). In this study, we investigated the significance of cyclin D1 in androgen-induced DDR using established prostate cancer cells and prostate tissues from cyclinD1 knockout mice. We demonstrate that endogenous cyclin D1 further diminished the dihydrotestosterone (DHT)-dependent reduction of γH2AX foci in vitro. We also show that cyclin D1 was required for the androgen-dependent DNA damage response both in vitro and in vivo. Furthermore, cyclin D1 was required for androgen-enhanced DDR and radioresistance of prostate cancer cells. Moreover, microarray analysis of primary prostate epithelial cells from cyclin D1-deficient and wild-type mice demonstrated that most of the DHT-dependent gene expression changes are also cyclin D1-dependent. Collectively, our findings suggest that the hormone-mediated recruitment of cyclin D1 to sites of DDR may facilitate the resistance of prostate cancer cells to DNA damage therapies, and highlight the need to explore other therapeutic approaches in prostate cancer to prevent or overcome drug resistance.
Keywords: Cyclin D1, DNA damage, DNA repair, prostate epithelial cells
INTRODUCTION
The cyclin D1 gene governs diverse functions implicated in the onset and progression of tumorigenesis. As the regulatory subunit of a holoenzyme formed through heterodimerization with CDK4/CDK6, cyclin D1 governs G1/S phase progression of cultured cells through phosphorylation of the pRb protein. In addition cyclin D1-dependent kinases phosphorylate NRF-1 to regulate mitochondrial biogenesis (1,2). The expression of cyclin D1 is induced by growth factors, oncogenes, and cellular stress and the abundance of cyclin D1 is rate-limiting in the growth of a variety of human malignancies (3). Cyclin D1 overexpression promotes the growth of multiple tumor types. Cyclin D1 antisense abrogated the growth of breast tumors implanted in mice (4), and cyclin D1−/− mice were resistant to transgenic tumors induced by Ras in either the skin or the mammary gland (5,6). Furthermore, cyclin D1 heterozygote mice are resistant to gastrointestinal tumorigenesis induced by the APC gene (7). Cyclin D1 participates in several non-canonical functions, promoting angiogenesis (8), cellular invasion and migration (9–13).
Cyclin D1 regulates the activity of more than 30 transcription factors (TF) (14). Recently approximately 90% of estrogen receptor mediated gene expression in the mammary gland was shown to be determined by cyclin D1 (15). The mechanism by which cyclin D1 inhibits gene expression involves the recruitment of cyclin D1 to TF binding sites in the context of local chromatin, associated with the recruitment of histone deacetylase (HDAC) which contribute to the transcriptional repression of differentiation inducing genes (16,17). HDAC1, 2, 3 and 5 bind to cyclin D1 and HDACs are recruited in the context of the local chromatin to deacetylate histone 3 lysine 9 (H3K9) which occurs contemporaneous with the recruitment of HP1α and SUV39 (18). ChIP-ChIP studies using proximal promoter regions on tiled arrays demonstrated cyclin D1 occupied proximal promoter elements including both E2F and non-E2F sites. Genome wide ChIP-Seq identified cyclin D1 binding sites at both proximal and distal genomic sites with enrichment amongst genes regulating chromosomal stability. Cyclin D1 induces gene expression in vivo, in addition to reporter assays (1,19). Although cyclin D1 has been shown to regulate synthetic AR-responsive reporter genes in transformed cells, the role of cyclin D1 in regulating global gene expression induced by the AR in primary prostate epithelial cells was previously unknown.
In early studies down-regulation of cyclin D1 was shown to be necessary for PCNA relocation and repair of DNA in response to UV induced DNA damage (20). In subsequent studies cyclin D1 was shown to bind DNA damage repair proteins including BRCA1 (21). Using γH2AX as a marker of DNA damage and comet assays to assess repair of damaged DNA, endogenous cyclin D1 was shown to enhance DNA damage repair (22). Cyclin D1−/− MEFs showed enhanced apoptosis in response to UV irradiation (23). In addition, cyclin D1 induces the expression of, and binds, Rad51 and serves to recruit DNA repair factors to chromatin. The recruitment of DNA repair factors was shown to be enhanced by cyclin D1, but not the related cyclin D1b isoform (22). In contrast, breast cancer cells over-expressing cyclin D1 showed enhanced apoptosis in response to γ-irradiation, suggesting cell-type specific differences in response to DNA damage (22,24).
In order to determine the significance of endogenous cyclin D1 in androgen-induced DNA repair in vitro and in vivo, prostatic cancer cells and ventral prostates from cyclin D1−/− gene knockout animals were deployed. These studies identify a novel role for cyclin D1 in the DNA damage response in normal and malignant prostate epithelial cells. Microarray based gene expression analysis demonstrated that endogenous cyclin D1 restrains androgen dependent gene expression. In LNCaP cells, DHT treatment reduced the number of γH2AX foci and was contingent upon cyclin D1 abundance since shRNA to cyclin D1 ablated the DHT mediated reduction in γH2AX foci. Using a direct measure of DNA repair we demonstrate DHT induced DNA repair is cyclin D1 dependent. Finally, tethering cyclin D1 to local chromatin was sufficient to induce the DDR in the presence of AR.
MATERIALS AND METHODS
Cell culture and DNA transfection
Cell lines were authenticated by examination of morphology, growth profile and are mycoplasma free. LNCaP cells (ATCC; Passage=26) were maintained in RPMI containing 1% penicillin/streptomycin supplemented with 10% FBS (Life Technologies). NIH2/4 cells (A gift from Dr. T. Misteli) were cultured in DMEM containing 1% penicillin/streptomycin supplemented with 10% FBS. The NIH2/4 (parental, NIH3T3) stable cell line that 256 repeats of the lac operator sequence (lacO) stably integrated into chromosome 3. The NIH2/4 stable cell line were transfected using the Nucleofector kit for immortalized cell lines (Amaxa, Nucleofector R). All cell lines were cultured for less than 6 months. Mouse primary prostate epithelial cells (PEC) were prepared from the ventral prostate of FVB cyclin D1−/− and FVB cyclin D1+/+ mice (25,26). The ventral prostate was rinsed 3 times in PBS containing 1% penicillin/streptomycin and Gentamicin. The tissue was finely chopped in a 6cm cell culture plate with a sterile razor blade and suspended in 1.5ml of collagenase blend type L (0.5 mg/ml) and placed at 37°C in a 5% CO2 incubator for 16hrs. Cells were then transferred to a polypropylene tubes and washed 3 times with 5ml of PBS, centrifuging at 2000rpm for 5 min between washes. Resuspend pellet in F-12 medium supplemented with 10% FBS, Insulin 5ug/ml, EGF 10ng/ml, Hydrocortisone 1ug/ml, transferrin 5ng/ml, bovine pituitary extract 30ug/ml, 1% penicillin/streptomycin, and 1% Gentamicin (growth medium). Cells were plated in 10cm culture dishes and placed at 37°C in a 5% CO2 incubator for 4hrs, during this period fibroblasts will attach and supernatant containing epithelial cells withdrawn and plated onto 10cm poly-lysine coated dishes. Growth media changed every 48hrs.
Chemicals and reagents
Experimental procedures with transgenic mice were approved by the ethics committee of Thomas Jefferson University. Cyclin D1−/− mice (23) were in the FVB strain (23). Murine prostate epithelial cell culture were isolated from prostate glands and maintained as previously described and analyzed after 25 passages with at least three lines of each genotype (7,27). Fluorescence activated cell sorting, for Ki67 and BrdU positive cells (28), as previously described (29). The DHT was used at physiological concentration of 10nM (Sigma).
siRNA transfection
The siRNA to cyclin D1 (Santa Cruz, Biotechnology) and control siRNA were transfected using oligofectamine (Invitrogen). Cells were seeded at 60% density in 6 well plates, washed once in PBS followed by transfection. Control siRNA or cyclin D1 siRNA (80nM) was used to treat cells in phenol red free, serum and antibiotic free RPMI. Cells were arrested by starvation in 0.1% FBS or 5% charcoal stripped medium for 72 hrs. Cells were stimulated with either 10% serum or DHT at physiological concentrations for 24 hrs.
Statistical Analysis
Comparisons between groups were analyzed by two-sided t-test. A difference of P < 0.05 was considered to be statistically significant. Data are expressed as mean ± SEM.
RNA extraction from prostate epithelial cell samples
RNA was extracted from epithelial cells using RecoverAll™ Total Nucleic Acid Isolation Kit (Applied Biosystems, Foster City, CA) followed by RQ1 DNase I (Promega Inc, Madison, WI) mediated removal of contaminating DNA from RNA preparations followed by RNA clean-up using RNAEasy Kit (Qiagen Inc, Valencia, CA). Purified RNA was reverse transcribed into c-DNA using Iscript Reverse transcriptase kit (Bio-Rad, Hercules, CA). RNA was labeled for hybridization with affymetrix chip-420_2.0 as previously described (15).
Microarray analysis methods
We used quantile normalization of the chip intensities as our preferred method of normalization (30). The limma package (31) was utilized to perform the analysis of the microarray experiments with p-values for each gene measuring the degree of association between gene expression and phenotype. We used the Bonferonni correction, which would consider an association significant if the p-value is less than ±/m, where m is the number of genes tested and ± is the nominal Type I error level. The False Discovery Rate (FDR) which is the expected proportion of false positives (32) was also used (data files submitted to GEO-GSE57867). Pathway analysis of differentially regulated genes was conducted in DAVID using the PANTHER database resource (33,34).
Immunofluorescence
NIH2/4-AR cells stably expressing AR were serum deprived (0.05% serum) for 24 hrs prior to transfection (lipofectamine 200) with Cherry-LacR-NLS-MCD1, Cherry-LacR-NLS-Cyclin D1 or Cherry-LacR-NLS. Following 5 hrs after transfection media was changed to DMEM containing 10% normal serum plus and minus DHT (100nM) for 24 hrs. Slides were fixed with 10% formalin for 10 min at room temperature (RT). The slides were then treated with 0.2% Triton X-100 for 5 min at RT and blocked with 2% BSA overnight at 4°C. The primary antibodies used were mouse monoclonal anti-phospho-Histone H2AX (Ser139) (clone JBW301) (Millipore Corporation, Billerica, MA) (1/1,000) and AR (Santa Cruz Biotechnology, Santa Cruz, CA) (1/200).
DNA repair assays
DR-GFP was provided by Dr. Jeremy M. Stark (35). DR-GFP is used to quantify homology-directed repair (HDR) in transient transfection of LNCaP cells. LNCaP cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) containing penicillin and streptomycin (100 mg of each/liter) and supplemented with 10% fetal bovine serum (FBS). Cells were in fected with lentiviral particles shRNA vector or shRNA cyclin D1 (pTRIPZ system). Positive cells were selected using Puromycin (2 mg/ml) for 2 weeks. Puromycin resistance clones were pooled. To analyze the efficiency of HDR LNCaP (pTRIPZ shVector and shcyclin D1) cells were cultured in phenol red-free DMEM with 10% charcoal/dextran–treated FBS and 2 mM glutamine and 1μM doxycycline for 24 hrs. Then the cells were transiently co-transfected with DR-GFP and pCβA-Sce (provided by Dr. Jeremy M. Stark) using the Lipofectamine 2000 (Invitrogen, Carlsbad, CA). At the same time the cells were treated with DHT (10 nM, 100 nM) or vehicle (ethanol) control. 24 hrs and 48 hrs post transfection the cells were changed with fresh DHT or vehicle control. The cells were cultured for 3 days to allow completion of repair, then the percentage of GFP-positive (GFP+) cells were analyzed by FACS analysis. The GFP expression plasmid in the same backbone, pCAGGS-NZEGFP was used as a transfection efficiency control. The empty vector pCAGGS-BSKX was used as a negative control.
Clonogenic assay
LNCaP cells were stable transduced with either pTRIPZ lentiviral inducible vector short-hairpin RNA (shRNA) control (shRNA-Ctrl) or shRNA against CCND1 (shRNA-CCND1) and seeded 24 hours prior to induction of shRNA. Expression of shRNA-Ctrl and shRNA-CCND1 was induced by treating cell with doxycycline (1μg/ml) for 72 hours and then seeded in cell culture flasks (2.5 × 105). The following day, cells were irradiated with 0, 0.5, 1.0, 1.5, 2 or 3 grays. After 14 days, colonies were stained with crystal violet solution and counted. The clonogenic survival fraction (SF) was set to the cell plating efficiency.
Statistical Analysis
Statistical significance between the numbers of γH2AX foci in the prostate cancer cell line LNCaP stable transduced either with shRNA-Ctrl or shRNA-CCND1 in the presence of DHT or vehicle control were calculated using 2-way ANOVA followed by Bonferroni posthoc test. The % ratio of γH2AX positive cells in cyclin D1+/+ and cyclin D1−/− mice ventral prostates were analyzed by a 2-way ANOVA followed by Bonferroni posthoc test. The number of γH2AX foci in NIH2/4 cells co-expressing AR either with Cherry-lacR-NLS-CCND1 or control were analyzed by a unpaired t-student test. In order to compare the differences in CCND1 and γH2AX foci co-localization between the different experimental conditions a 2-way ANOVA followed by Bonferroni posthoc test were performed. For the DNA repair rate in LNCaP cells stable for either shRNA-Ctrl or shRNA-CCND1 the surviving fraction was analyzed by a 2-way ANOVA followed by Bonferroni posthoc test. All data are expressed as mean ± standard error of the mean (SEM).
RESULTS
Cyclin D1 promotes DHT-dependent DNA synthesis and is required for DHT-induced reduction of γH2AX foci in prostate cancer cell lines
DHT induced cell proliferation of LNCaP cells compared to vehicle control cells (Supplemental Fig. 1A), consistent with a previous report (36). In order to examine the requirement for cyclin D1 in prostate cancer cellular proliferation, LNCaP cells were treated with either cyclin D1 siRNA or control siRNA. The DNA synthetic phase was reduced 50% upon transduction with cyclin D1 siRNA (Supplemental Fig. 1B). The reduction in cyclin D1 was observed in DHT-treated or in the basal state. AR abundance was increased in DHT treated cells (Supplemental Fig. 1C), cyclin D1 siRNA reduced both basal and DHT-induced cellular proliferation (Supplemental Fig. 1D).
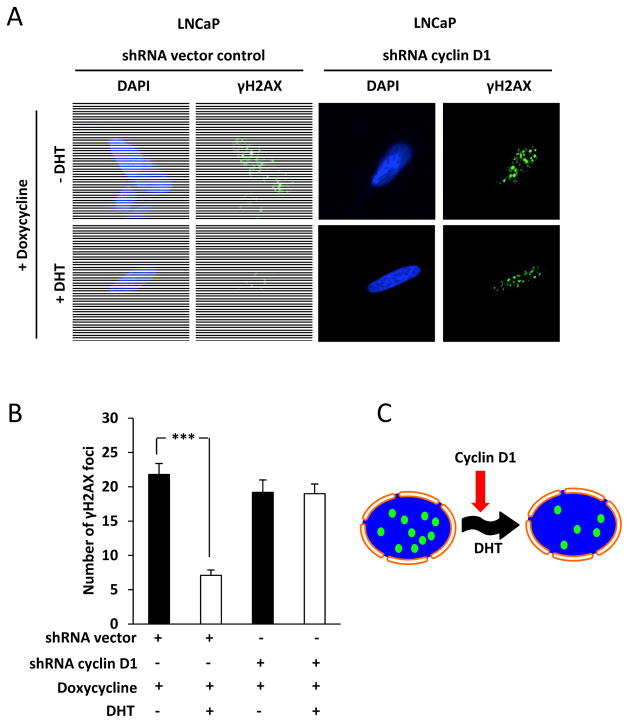
In view of the finding that liganded AR promotes Homologous DNA Repair and that cyclin D1 inhibits AR activation through binding N-terminal region, we determined the role for cyclin D1 in DHT signaling and the DNA damage response in LNCaP cells. Histone H2AX phosphorylation on Serine 139 producing γH2AX is a sensitive marker for DNA double strand breaks (37). Large foci of γH2AX are important for the accumulation and retention of DSB repair factors (38). We assessed the assemblage of nuclear repair foci containing γH2AX in LNCaP cells transduced with either inducible cyclin D1 shRNA or control shRNA vector and treated with DHT (10 nM) (Fig. 1A). γH2AX foci were reduced after 30 mins of DHT treatment (P< 0.001) (Fig. 1B). Cyclin D1 shRNA abrogated the DHT mediated reduction of γH2AX foci. Thus cyclin D1 is required for the androgen-dependent attenuation of the DNA damage response (Fig. 1C).
Figure 1. Cyclin D1 is required for DHT mediated reduction in γH2AX foci.
(A) Confocal microscopy of LNCaP cells with nuclear staining using 4,6-diamidino-2-phenylindole (DAPI) and quantitation of γH2AX foci following 30 min of DHT treatment of shRNA vector control cells and shRNA cyclin D1 cells. (B) Number of γH2AX foci were quantitated for N>25 cells for shRNA vector control and shRNA cyclin D1 cells. Data are mean ±SEM. (C) Schematic representation of cyclin D1 regulated DHT dependent reduction in γH2AX foci. All data are mean ± SEM, *** P< 0.001.
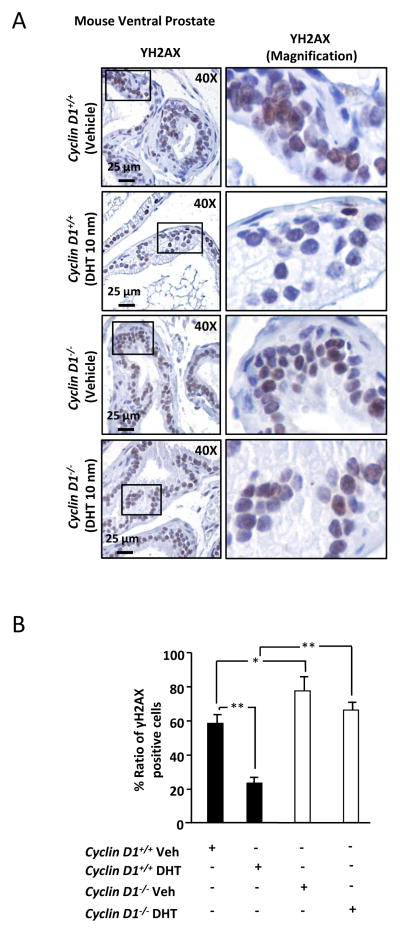
In view of the in vivo finding that endogenous cyclin D1 reduces γH2AX foci in vitro, we conducted immunohistochemical staining for evidence of cyclin D1 reduction of DNA damage in the prostate gland in vivo. As a well established marker of the DNA damage response, we conducted immunohistochemical staining for γH2AX assessing ventral prostate of cyclin D1−/− vs cyclin D1+/+ littermate controls after castration and DHT replacement (Fig. 2A and 2B). Basal level γH2AX was increased in cyclin D1−/− vs. cyclin D1+/+ mice (P< 0.05). DHT reduced γH2AX in vivo (P< 0.01) but the effect of DHT on the DDR was abrogated in the cyclin D1−/− mice. Thus endogenous cyclin D1 is required for DHT-mediated reduction of DNA damage in the prostate in vivo.
Figure 2. Cyclin D1 reduces γH2AX foci in the prostate in vivo.
(A) Immunohistochemical staining for γH2AX in the ventral prostate from cyclin D1−/− and cyclin D1+/+ mice treated with vehicle or DHT (10nm). Right panels are magnification of the inset box shown in left panels. (B) Quantitation of γH2AX positive cells from cyclin D1−/− and cyclin D1+/+ mice treated with vehicle or DHT. All data are mean ± SEM, * P<0.05 and ** P<0.01.
Cyclin D1 promotes DNA repair and radioresistence in presence of DHT
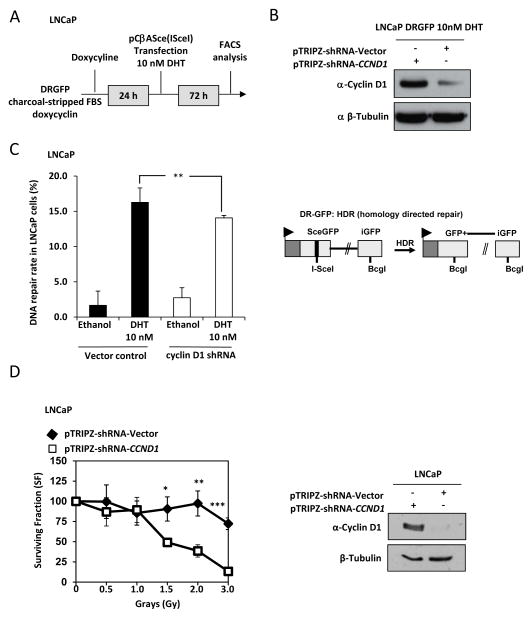
A DNA repair assays were deployed to directly test the role of cyclin D1 in homologous directed repair (HDR). The repair assay was conducted on LNCaP cells stably transduced with a doxycycline inducible cyclin D1 shRNA or a control shRNA under androgen-depleted conditions. After 24hrs cells were transfected with SceI expressing plasmid or control plasmid together with DG-GFP (homology directed repair). Cells were maintained in androgen deprivation condition together with doxycycline and treated with vehicle or DHT (100nM) (Fig. 3A). Western blot analysis was conducted to confirm the reduction in cyclin D1 abundance following induction of cyclin D1 shRNA (Fig. 3B). FACS analysis of the cells revealed that DHT mediated induction of HDR repair was dependent on cyclin D1 (P< 0.008) (Fig. 3C). We next investigated if cyclin D1 promoted radioresistence. LNCaP cells were transduced with doxycycline inducible cyclin D1 shRNA or a control shRNA under physiological levels of DHT (Fig. 3D, left panel). Irradiation of shcyclin D1 LNCaP cells resulted in a dose dependent precipitous decline in clonogenic survival from 1.5 Gray to 3 Grays (P= 0.023, P< 0.02 and P< 0.001, respectively). At 3 Grays cyclin D1 promoted clonogenic survival of LNCaP cells by ~6 fold. Western blot analysis was conducted to confirm the reduction in cyclin D1 abundance following induction of cyclin D1 shRNA (Fig. 3D, right panel).
Figure 3. Cyclin D1 has a distinct role in DNA repair pathways and promotes radioresistance.
(A) Schematic depiction of the protocol used to treat LNCaP cells. (B) Western blot analysis to verify cyclin D1 abundance is reduced following doxycycline stimulation of shRNA to cyclin D1 in DHT treated LNCaP cells. (C) Cyclin D1-dependent DHT regulated response to homology directed repair (DR-GFP, left panel) in LNCaP cells and schematic of reporter constructs (right panel). (D, left panel) LNCaP cells transduced with doxycycline inducible cyclin D1 shRNA or shRNA control vector were exposed to increasing doses of irradiation followed by clonogenic assays to determine cell survival. Western blotting confirmed the shRNA-CCND1 reduced cyclin D1 abundance (right panel). All data are mean ±SEM, * P<0.05, ** P<0.01 and *** P< 0.001.
Cyclin D1 governs DHT-dependent signaling in primary prostate epithelial cells
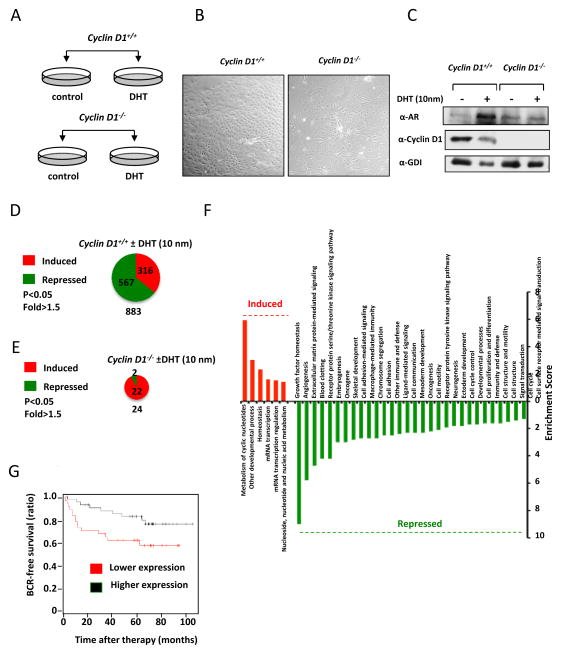
In order to understand the role of cyclin D1 in prostate epithelial cell-dependent gene expression we deployed primary murine prostate epithelial cell cultures that express wild type AR. The primary prostate epithelial cells (PEC) from either wild type or cyclin D1−/− mice were of similar morphology assessed by phase contrast microscopy and were treated either with vehicle or DHT (10 nm) (Fig. 4A, B). Western blot analysis demonstrated a reduction in the abundance of cyclin D1 in cyclin D1−/− cells (Fig. 4C). In the wild type cells, the addition of DHT induced AR abundance, however, the relative abundance of the AR was not induced by DHT in cyclin D1−/− cells (Supplemental Fig. 1C).
Figure 4. Cyclin D1 determines DHT-signaling in primary prostate epithelial cells.
(A) Schematic representation of protocol in which cells were treated with DHT for 24 hours post-dissection. (B) Phase-contrast microscopy of prostate epithelial cells (PEC) from cyclin D1+/+ and cyclin D1−/− mice. (C) Western blot for cyclin D1 and AR. Chart display the number of genes that were altered in expression by DHT in (D) cyclin D1+/+ PECs, (E) DHT dependent genes in cyclin D1−/− PECs. (F) Functional pathway analysis (PANTHER) of DHT regulated genes in cyclin D1+/+ PECs. (G) Kaplan Meier Analysis of cyclin D1 down-regulated gene expression in PECs.
In order to determine if cyclin D1 promoted DHT dependent DNA repair through a transcriptional program a genome wide analysis of the genes regulated by endogenous cyclin D1 in primary prostate epithelial cells was conducted. An analysis of the genes regulated by DHT in cyclin D1+/+ (883 genes) and cyclin D1−/− PECs (24 genes) reveals that 93% of the genes regulated by DHT are dependent upon cyclin D1 (Fig. 4D, E and Supplemental Data Set 1). In cyclin D1+/+ PECs most of the genes (64%) were down regulated in response to DHT. Using PANTHER functional annotation identified the pathways regulated by DHT in a cyclin D1 dependent manner (Fig. 4F). Many of the pathways inhibited by DHT in a cyclin D1 dependent manner are associated with cell growth and proliferation, cell cycle, development, signal transduction and growth factor signaling. We next asked if there is any clinical relevance of the cyclin D1 dependent gene signature. Signatures were tested for prognostic value as described by (39). We used K-means clustering to define two groups of tumors on the basis of the up-regulated and down-regulated signatures and measured the association with risk of BCR (biochemical recurrence) by Kaplan-Meier analysis. The up-regulated signature was not associated with BCR (hazard ratio = 1.72, log-rank test, P = 0.15). The down-regulated signature was significant and associated with biochemical recurrence (hazard ratio = 2.12, log-rank test, P = 0.045) (Fig. 4G)
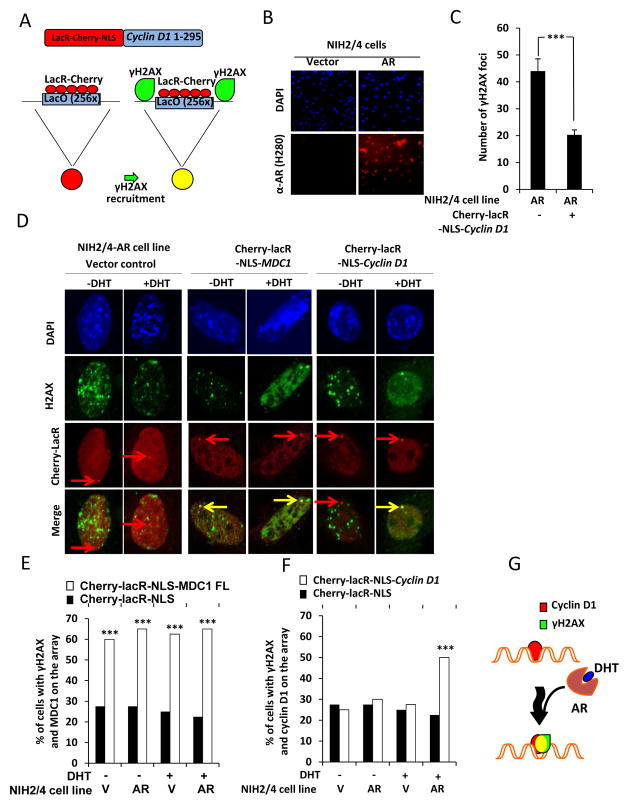
Cyclin D1 inhibits DHT-dependent recruitment of repair factors to chromatin
Previous studies had shown that stable association of DDR factors with chromatin amplified the DDR signal via an ATM and DNA-PK-mechanism (40). Cyclin D1 is tethered to chromatin in a similar manner (22). In order to examine the role of DHT in the recruitment of DDR factors to chromatin, we used a LacR system. The cDNAs encoding DNA repair factors or cyclin D1 were fused to the Escherichia coli lac-repressor (LacR) and tagged with Cherry-red fluorescent protein. These expression plasmids were examined in an NIH3T3 cell line that contains 256 repeats of the LacO stably integrated into chromosome 3 (NIH 2/4) (Fig. 5A). The NIH 2/4 cells were transduced by retrovirus encoding the AR to produce the androgen-responsive cell line NIH 2/4-AR (Fig. 5B). Recruitment of this chimeric DNA repair factor fusion protein Cherry-lacR-NLS-Cyclin D1 compared to Cherry-LacR-NLS control vector demonstrated that the total number of γH2AX foci was reduced by expression of either the AR or cyclin D1 alone. In the presence of both cyclin D1 and the AR there was further reduction in the number of γH2AX foci (P< 0.001) (Fig. 5C). Transfection with the positive control expression plasmid encoding Cherry-lacR-NLS-MDC1 was sufficient to activate the DDR as demonstrated by γH2AX positive foci at the lacO site (Fig. 5D and E). Phosphorylation of H2AX at the LacO site was substantially enhanced in the presence of cyclin D1 and DHT (P< 0.001) (Fig. 5D and F).
Figure 5. Cyclin D1 is recuited to γH2AX foci by DHT.
(A) Schematic depiction of the control LacR-Cherry-NLS and the chimeric fusion protein LacR-Cherry-NLS-cyclin D1. (B) Immunofluorescent imaging of NIH2/4 cells stably expressing AR or vector control stained for DAPI and anti-AR. (C) Confocal immunofluorescence microscopy of NIH2/4 cells stably expressing with AR and transfected with LacR-Cherry-NLS-cyclin D1 compared to appropriate vector control cell. The number of γH2AX foci from 10 cells was quantitated for each condition. Cells were cultured in FBS without DHT (D) Confocal immunofluorescence microscopy of NIH2/4 cells transiently transfected with cyclin D1 fused to Cherry-lacR-NLS (red). Phosphorylation of γH2AX (green). Quantitation of co-localization of γH2AX and (E) MDC1 and (F) cyclin D1 (yellow) for N=40 cells and depicted as percentage of γH2AX coincident with MDC1 or cyclin D1 at the LacO array. (G) Schematic representation of cyclin D1 occupancy at LacO array. In presence of AR and DHT, LacO array demonstrates γH2AX and cyclin D1 positivity (yellow). All data are mean ±SEM, ** P< 0.01 and *** P< 0.001.
DISCUSSION
The current studies demonstrate that DHT induced recruitment of cyclin D1 to sites of DNA damage associated with a reduction in the formation of γH2AX repair foci. The recruitment of the DNA repair protein MDC1 was unaffected by DHT. Cyclin D1 reduced the number of γH2AX repair foci both in primary prostate epithelial cells in vivo and in transformed LNCaP cell. Consistent with these findings cyclin D1 promoted DNA repair mediated via homology directed repair in a DHT dependent manner. The contemporaneous induction of DNA synthesis and cellular division with the induction of DNA repair by cyclin D1 may contribute to therapy DNA damage therapy resistance in prostate cancer cells.
In human prostate cancer cells the abundance of cyclin D1 is induced by growth factors and siRNA to cyclin D1 reduced ErbB2-mediated DNA synthesis in LNCaP cells (41). Forced expression of cyclin D1 enhanced cellular proliferation in two studies (42,43) and inhibited proliferation in another study (44). Cyclin D1 physically associates with the AR in IP-Western blot analysis and forced overexpression of cyclin D1 is capable of inhibiting AR reporter gene activity (45). The difference between our results in which endogenous cyclin D1 enhanced cell proliferation and DNA synthesis contrasts with findings of cyclin D1 cDNA overexpression in LNCaP cells (44). These differences therefore may relate to the experimental approach in which we have examined the role of endogenous cyclin D1 rather than assessing the effect of cyclin D1 overexpression. In the current studies DHT promoted the DNA damage signaling assessed by γH2AX foci in the prostate in vivo and in LNCaP cells in tissue culture. Recently, Polkinghorn et al. (46) demonstrated that AR signaling promoted expression of genes that enhance DNA repair. Using γH2AX foci and a comet assay as a marker of DNA damage androgen depletion decreased DNA repair in LNCaP cells. The decreased DNA repair of irradiated LNCaP cells treated with anti-androgens was associated with reduced colony survival by clonogenic assay. Finally, in assays to directly measure non-homologous end joining (NHEJ) and homologous repair (HR) there was significantly decreased NHEJ in the anti-androgen treated LNCaP cells however there was no change in HR. More recently, Goodwin et al. (47) demonstrate that AR signaling promoted DNA repair in a castrate resistance prostate cancer model (C4-2 cells). When C4-2 cells were challenged with DNA damage, AR was activated, promoted AR recruitment to genes that regulate DNA repair leading to repair of double stranded DNA breaks. However in cyclin D1+/+ PECs we did not observe changes in gene expression by microarray anslysis consistent with DHT induction of DNA repair. We note that most of the genes in the PEC microarray of DHT dependent gene expression changes are cyclin D1-dependent. This observation in PECs is similar to the 17β-estradiol stimulated cyclin D1-dependent gene expression changes in the mouse mammary gland (48). Genome-wide expression profiling conducted of 17β-estradiol-treated castrated virgin mice deleted of the ccnd1 gene demonstrated that cyclin D1 determines estrogen-dependent gene expression for 88% of estrogen-responsive genes in vivo. In the case of PECs we observed that 97% of the genes are dependent on cyclin D1 (Supplemental Table 1). Over the last 2 decades, a substantial body of evidence has suggested cyclin D1 plays a direct role in transcriptional regulation. Cyclin D1 physically associates with, and regulates the transcriptional activity of more than 30 transcription factors (TFs), including neuroD, myoD, CEBPβ, PPARγ and estrogen receptor. Previous studies examined transcriptional regulation of the cyclin D1 target genes, LPL, aP2 (49) and miR17/20 (50). ChIP-ChIP demonstrated cyclin D1 and p300 together occupied genes in close proximity to the transcriptional start site (51), and whole genome ChIP-Seq demonstrated enrichment of cyclin D1 at genes that regulate mitosis and chromosomal instability (CIN) in close proximity to the transcriptional start site (52). Together, these studies are consistent with a model in which cyclin D1 functions to regulate transcription in a cdk-independent manner.
DNA damage induces a step wise change in chromatin structure, which participates in maintaining genomic integrity (53) and chromatin topology impacts the genotoxic stress responses. Hyper-condensed chromatin is associated with lower efficiency of DNA repair and more compact chromatin leads to DNA-damage resistance (54). In the current studies, endogenous cyclin D1 enhanced DNA synthesis and cell proliferation. Cyclin D1 reduced the number of DNA lesion assessed by γH2AX repair foci in vitro and in vivo in a DHT dependent fashion. Cyclin D1 is recruited to sites of DNA damage in a DHT dependent manner. These findings are consistent with a role for cyclin D1 in promoting DNA repair.
Supplementary Material
Acknowledgments
This work was supported in part by awards from R01CA070896, R01CA075503, R01CA086072, R01CA137494, and R01CA132115 (R.G.P.). The Kimmel Cancer Center is supported by the NIH Cancer Center Support Grant (1P30CA56036-08) (R.G.P.). This project is funded in part from the Dr. Ralph and Marian C. Falk Medical Research Trust and a grant from Pennsylvania Department of Health (R.G.P.). Supported in part by an American-Italian Cancer Foundation Post-Doctoral Research Fellowship [G.D.]. The Department specifically disclaims responsibility for an analysis, interpretations or conclusions. There are no conflicts of interest associated with this manuscript. Michael P. Lisanti and his laboratory were supported via the resources of Thomas Jefferson University. We thank David Strahan for preparation of this manuscript.
Footnotes
Conflicts of Interest:
The authors have no potential conflicts of interest.
References
- 1.Wang C, Li Z, Lu Y, Du R, Katiyar S, Yang J, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci U S A. 2006;103(31):11567–72. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakamaki T, Casimiro MC, Ju X, Quong AA, Katiyar S, Liu M, et al. Cyclin D1 determines mitochondrial function in vivo. Mol Cell Biol. 2006;26(14):5449–69. doi: 10.1128/MCB.02074-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pestell RG. New roles of cyclin D1. Am J Pathol. 2013;183(1):3–9. doi: 10.1016/j.ajpath.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee RJ, Albanese C, Fu M, D’Amico M, Lin B, Watanabe G, et al. Cyclin D1 is required for transformation by activated Neu and is induced through an E2F-dependent signaling pathway. Molecular and Cellular Biology. 2000;20(2):672–83. doi: 10.1128/mcb.20.2.672-683.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robles AI, Rodriguez-Puebla ML, Glick AB, Trempus C, Hansen L, Sicinski P, et al. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 1998;12(16):2469–74. doi: 10.1101/gad.12.16.2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411(28 June):1017–21. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 7.Hulit J, Wang C, Li Z, Albanese C, Rao M, Di Vizio D, et al. Cyclin D1 genetic heterozygosity regulates colonic epithelial cell differentiation and tumor number in ApcMin mice. Molecular and Cellular Biology. 2004;24(17):7598–611. doi: 10.1128/MCB.24.17.7598-7611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hulit J, Lee RJ, Li Z, Wang C, Katiyar S, Yang J, et al. p27Kip1 repression of ErbB2-induced mammary tumor growth in transgenic mice involves Skp2 and Wnt/beta-catenin signaling. Cancer Res. 2006;66(17):8529–41. doi: 10.1158/0008-5472.CAN-06-0149. [DOI] [PubMed] [Google Scholar]
- 9.Li Z, Wang C, Prendergast GC, Pestell RG. Cyclin D1 functions in cell migration. Cell cycle. 2006;5(21):2440–2. doi: 10.4161/cc.5.21.3428. [DOI] [PubMed] [Google Scholar]
- 10.Neumeister P, Pixley FJ, Xiong Y, Xie H, Wu K, Ashton A, et al. Cyclin D1 governs adhesion and motility of macrophages. Molecular Biology of the Cell. 2003;14(5):2005–15. doi: 10.1091/mbc.02-07-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Z, Wang C, Jiao X, Lu Y, Fu M, Quong AA, et al. Cyclin D1 regulates cellular migration through the inhibition of thrombospondin 1 and ROCK signaling. Molecular and Cell Biology. 2006;26(11):4240–56. doi: 10.1128/MCB.02124-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Jiao X, Wang C, Ju X, Lu Y, Yuan L, et al. Cyclin D1 induction of cellular migration requires p27(KIP1) Cancer Res. 2006;66(20):9986–94. doi: 10.1158/0008-5472.CAN-06-1596. [DOI] [PubMed] [Google Scholar]
- 13.Li Z, Wang C, Jiao X, Katiyar S, Casimiro MC, Prendergast GC, et al. Alternate cyclin D1 mRNA splicing modulates p27KIP1 binding and cell migration. J Biol Chem. 2008;283(11):7007–15. doi: 10.1074/jbc.M706992200. [DOI] [PubMed] [Google Scholar]
- 14.Fu M, Wang C, Li Z, Sakamaki T, Pestell RG. Minireview: Cyclin D1: Normal and Abnormal Functions. Endocrinology. 2004;145(12):5439–47. doi: 10.1210/en.2004-0959. [DOI] [PubMed] [Google Scholar]
- 15.Casimiro MC, Wang C, Li Z, Di Sante G, Willmart NE, Addya S, et al. Cyclin D1 determines estrogen signaling in the mammary gland in vivo. Mol Endocrinol. 2013;27(9):1415–28. doi: 10.1210/me.2013-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Pattabiraman N, Zhou JN, Fu M, Sakamaki T, Albanese C, et al. Cyclin D1 repression of peroxisome proliferator-activated receptor gamma expression and transactivation. Mol Cell Biol. 2003;23(17):6159–73. doi: 10.1128/MCB.23.17.6159-6173.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu M, Wang C, Rao M, Wu X, Bouras T, Zhang X, et al. Cyclin D1 represses p300 transactivation through a cyclin-dependent kinase-independent mechanism. J Biol Chem. 2005;280(33):29728–42. doi: 10.1074/jbc.M503188200. [DOI] [PubMed] [Google Scholar]
- 18.Fu M, Rao M, Bouras T, Wang C, Wu K, Zhang X, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. J Biol Chem. 2005;280(17):16934–41. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 19.Watanabe G, Albanese C, Lee RJ, Reutens A, Vairo G, Henglein B, et al. Inhibition of cyclin D1 kinase activity is associated with E2F-mediated inhibition of cyclin D1 promoter activity through E2F and Sp1. Mol Cell Biol. 1998;18(6):3212–22. doi: 10.1128/mcb.18.6.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pagano M, Theodoras AM, Tam SW, Draetta GF. Cyclin D1-mediated inhibition of repair and replicative DNA synthesis in human fibroblasts. Genes Dev. 1994;8(14):1627–39. doi: 10.1101/gad.8.14.1627. [DOI] [PubMed] [Google Scholar]
- 21.Wang C, Fan S, Li Z, Fu M, Rao M, Ma Y, et al. Cyclin D1 antagonizes BRCA1 repression of estrogen receptor alpha activity. Cancer Res. 2005;65(15):6557–67. doi: 10.1158/0008-5472.CAN-05-0486. [DOI] [PubMed] [Google Scholar]
- 22.Li Z, Jiao X, Wang C, Shirley LA, Elsaleh H, Dahl O, et al. Alternative cyclin d1 splice forms differentially regulate the DNA damage response. Cancer Res. 2010;70(21):8802–11. doi: 10.1158/0008-5472.CAN-10-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Albanese C, D’Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274(48):34186–95. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Q, Fukushima P, DeGraff W, Mitchell JB, Stetler Stevenson M, Ashkenazi A, et al. Radiation and the Apo2L/TRAIL apoptotic pathway preferentially inhibit the colonization of premalignant human breast cells overexpressing cyclin D1. Cancer Res. 2000;60(10):2611–5. [PubMed] [Google Scholar]
- 25.Terracio L, Douglas HJ. Primary culture of rat ventral prostate epithelail cells. TCA manual/Tissue Culture Association. 1980;5(4):1169–71. [Google Scholar]
- 26.Ilio KY, Nemeth JA, Lang S, Lee C. The primary culture of rat prostate basal cells. Journal of andrology. 1998;19(6):718–24. [PubMed] [Google Scholar]
- 27.Ju X, Ertel A, Casimiro MC, Yu Z, Meng H, McCue PA, et al. Novel oncogene-induced metastatic prostate cancer cell lines define human prostate cancer progression signatures. Cancer Res. 2013;73(2):978–89. doi: 10.1158/0008-5472.CAN-12-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275(27):20853–60. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- 29.Wu K, Li A, Rao M, Liu M, Dailey V, Yang Y, et al. DACH1 is a cell fate determination factor that inhibits Cyclin D1 and breast tumor growth. Mol Cell Biol. 2006;26(19):7116–29. doi: 10.1128/MCB.00268-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 31.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 32.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B (Methodological) 1995;57(1):289–300. [Google Scholar]
- 33.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 34.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gunn A, Stark JM. I-SceI-based assays to examine distinct repair outcomes of mammalian chromosomal double strand breaks. Methods in molecular biology. 2012;920:379–91. doi: 10.1007/978-1-61779-998-3_27. [DOI] [PubMed] [Google Scholar]
- 36.Horoszewicz JS, Leong SS, Kawinski E, Karr JP, Rosenthal H, Chu TM, et al. LNCaP model of human prostatic carcinoma. Cancer Res. 1983;43(4):1809–18. [PubMed] [Google Scholar]
- 37.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–68. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 38.van Attikum H, Gasser SM. Crosstalk between histone modifications during the DNA damage response. Trends in cell biology. 2009;19(5):207–17. doi: 10.1016/j.tcb.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 39.Ding Z, Wu CJ, Chu GC, Xiao Y, Ho D, Zhang J, et al. SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature. 2011;470(7333):269–73. doi: 10.1038/nature09677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Soutoglou E, Misteli T. Activation of the cellular DNA damage response in the absence of DNA lesions. Science. 2008;320(5882):1507–10. doi: 10.1126/science.1159051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casimiro M, Rodriguez O, Pootrakul L, Aventian M, Lushina N, Cromelin C, et al. ErbB-2 induces the cyclin D1 gene in prostate epithelial cells in vitro and in vivo. Cancer Res. 2007;67(9):4364–72. doi: 10.1158/0008-5472.CAN-06-1898. [DOI] [PubMed] [Google Scholar]
- 42.Perry JE, Grossmann ME, Tindall DJ. Epidermal growth factor induces cyclin D1 in a human prostate cancer cell line. The Prostate. 1998;35(2):117–24. doi: 10.1002/(sici)1097-0045(19980501)35:2<117::aid-pros5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 43.Han AC, Hovenden S, Rosenblum NG, Salazar H. Adenocarcinoma arising in extragonadal endometriosis: an immunohistochemical study. Cancer. 1998;83(6):1163–9. [PubMed] [Google Scholar]
- 44.Petre CE, Wetherill YB, Danielsen M, Knudsen KE. Cyclin D1: mechanism and consequence of androgen receptor co-repressor activity. J Biol Chem. 2002;277(3):2207–15. doi: 10.1074/jbc.M106399200. [DOI] [PubMed] [Google Scholar]
- 45.Reutens AT, Fu M, Wang C, Albanese C, McPhaul MJ, Sun Z, et al. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol Endocrinol. 2001;15(5):797–811. doi: 10.1210/mend.15.5.0641. [DOI] [PubMed] [Google Scholar]
- 46.Polkinghorn WR, Parker JS, Lee MX, Kass EM, Spratt DE, Iaquinta PJ, et al. Androgen Receptor Signaling Regulates DNA Repair in Prostate Cancers. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goodwin JF, Schiewer MJ, Dean JL, Schrecengost RS, de Leeuw R, Han S, et al. A Hormone-DNA Repair Circuit Governs the Response to Genotoxic Insult. Cancer Discov. 2013 doi: 10.1158/2159-8290.CD-13-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Casimiro MC, Wang C, Li Z, Disante G, Willmart NE, Addya S, et al. Cyclin D1 Determines Estrogen Signaling in the Mammary Gland in Vivo. Mol Endocrinol. 2013 doi: 10.1210/me.2013-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fu M, Rao M, Bouras T, Wang C, Wu K, Zhang X, et al. Cyclin D1 inhibits peroxisome proliferator-activated receptor gamma-mediated adipogenesis through histone deacetylase recruitment. The Journal of Biological Chemistry. 2005;280(17):16934–41. doi: 10.1074/jbc.M500403200. [DOI] [PubMed] [Google Scholar]
- 50.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, et al. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. The Journal of Cell Biology. 2008;182(3):509–17. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bienvenu F, Jirawatnotai S, Elias JE, Meyer CA, Mizeracka K, Marson A, et al. Transcriptional role of cyclin D1 in development revealed by a genetic-proteomic screen. Nature. 2010;463(7279):374–8. doi: 10.1038/nature08684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Casimiro MC, Crosariol M, Loro E, Ertel A, Yu Z, Dampier W, et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. The Journal of Clinical Investigation. 2012;122(3):833–43. doi: 10.1172/JCI60256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lukas J, Lukas C, Bartek J. More than just a focus: The chromatin response to DNA damage and its role in genome integrity maintenance. Nature cell biology. 2011;13(10):1161–9. doi: 10.1038/ncb2344. [DOI] [PubMed] [Google Scholar]
- 54.Murga M, Jaco I, Fan Y, Soria R, Martinez-Pastor B, Cuadrado M, et al. Global chromatin compaction limits the strength of the DNA damage response. The Journal of cell biology. 2007;178(7):1101–8. doi: 10.1083/jcb.200704140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.