Abstract
Natural killer (NK) cell phenotype is partially mediated through binding of killer immunoglobulin-like receptors (KIR) with HLA class I ligands. The KIR gene family is highly polymorphic and not well captured by standard GWAS approaches. Here we tested the hypothesis that variations in KIR gene content combined with HLA class I ligand status is associated with keratinocyte skin cancers using a population-based study of basal cell carcinoma (BCC) and squamous cell carcinomas (SCC). We conduced an interaction analysis of KIR gene content variation and HLA-B (Bw4 vs Bw6) and HLA-C (C1 vs C2). KIR centromeric B haplotype was associated with significant risk of multiple BCC tumors (OR=2.39, 95% CI: 1.10–5.21), and there was a significant interaction between HLA-C and the activating gene KIR2DS3 for BCC (pinteraction = 0.005). Furthermore, there was significant interaction between HLA-B and telomeric KIR B haplotype (containing the activating genes KIR3DS1 and KIR2DS1) as well as HLA-B and the activating KIR gene 2DS5 (pinteraction 0.001 and 0.012, respectively). Similar but greatly attenuated associations were observed for SCC. Moreover, previous in vitro models demonstrated that p53 is required for upregulation of NK ligands, and accordingly, we observed there was a strong association between the KIR B haplotype and p53 alteration in BCC tumors, with a higher likelihood that KIR B carriers harbor abnormal p53 (p<0.004). Taken together, our data suggest functional interactions between KIR and HLA modify risks of BCC and SCC, and that KIR encoded by the B genes provide selective pressure for altered p53 in BCC tumors.
Introduction
Basal cell and squamous cell carcinoma of the skin are the most commonly diagnosed malignancies in the U.S. (1). Although these keratinocyte-derived tumors are treatable and rarely metastasize, they are associated with significant morbidity and considerable health care costs (2, 3). Exposure to ultraviolet radiation (UVR) is a major risk factor in the development of these skin cancers and acts as both a mutagen and an immune suppressing agent (4, 5). The effect of immunosuppression on skin cancer occurrence is highlighted in organ transplant recipients who are at high risk for the development of squamous cell carcinoma (SCC) and basal cell carcinoma (BCC) (rev in (6)). In addition, observations of spontaneous BCC regression underscore the importance of the immune response to tumor antigens in tumor progression (7, 8).
Natural killer (NK) cells are a major component of the innate immune system and serve as the first line of defense against virally infected and transformed cells (9). NK cell maturation involves “licensing”, a specific set of receptor-ligand interactions between NK receptors and MHC class I molecules (10). Inter-individual variation in these interactions is hypothesized to result in a spectrum of NK killing phenotypes. Once licensed, the cytolytic function of NK cells is orchestrated through a complex array of inhibitory and activating signals which is in part determined by the interaction of the killer-cell immunoglobulin-like receptors (KIR) with human leukocyte antigen (HLA) class I ligands, via a balance of inhibitory and activating KIR signals (11). Expression of cognate HLA class I molecules on target cells results in inhibition of NK cell mediated cytolysis via the inhibitory KIR, while the absence or aberrant presentation of these molecules leads to activation of the NK cell via activating receptors, including KIR (11).
The KIR locus is variable in both gene content and allelic polymorphisms (12–14). These structural complexities, as well as extensive gene homology, have resulted in poor coverage using GWAS methods, making KIR understudied as a cancer susceptibility trait. The KIR gene region on chromosome 19 is divided into two haplotype blocks, and haplotypes based on gene content have been established (15). Within each block are the ancestral haplotype A, and divergent B haplotypes with increasing diversity primarily of activating KIR genes (15). Novel KIR structural haplotypes arise from ongoing recombination and unequal crossover events (14, 16).
Most KIR-ligand pairs have not yet been identified, particularly for the activating KIR (11, 17). However, HLA-C and HLA-B are both ligands for inhibitory KIR receptors (11). Furthermore, there is an established dimorphism in both of these genes that results in differential binding of these ligands for KIR receptors, central to the NK licensing process. For example, KIR2DL2 and KIR2DL3 both bind HLA-C1 (defined by Asn80), while KIR2DL1 binds HLA-C2 (Lys80) (12, 18, 19). Similarly, KIR3DL1 binds specifically to the HLA-Bw4 ligand, but not HLA-Bw6 (defined by sequence at positions 77–83) (12, 19). Emerging evidence suggests it is necessary to investigate KIR*HLA interactions in order to observe true phenotypes (rev in (20)).
Gene content variability at the KIR locus has become central in hematopoietic transplant research (21) and has also been investigated as a susceptibility trait for a variety of infectious and autoimmune diseases, as well as cancer (rev in (20)) Moreover, interactions between the independently segregating KIR and HLA genes dictate biologic function of NK cells, yet their interaction in skin cancer susceptibility has only been described for melanoma (22–24). Finally, in vitro models demonstrate that functional p53 is necessary for the up-regulation of NK ligands (25). In this study we tested the hypothesis that variation in KIR gene content and HLA polymorphisms are associated with risk of keratinocyte-derived skin cancers and are a driver of p53 alteration in these common skin cancers.
Materials and Methods
Study population
The New Hampshire Health Study (NHHS) is a population-based case-control study of NMSC and has been previously described (26, 27). Briefly, a statewide incidence survey was used to identify incident BCC and SCC, and eligible cases were recruited into a case-control study. Cases were: 1) 25 –74 years of age, 2) had a listed telephone number, and 3) spoke English. Controls were frequency-matched on age and sex to the combined distribution of cases. Controls aged 25–64 years were identified from the New Hampshire State Department of Transportation records and those aged 65–74 years were obtained from enrollment lists of the Center for Medicaid and Medicare Services. The participants filled out a work history and residence questionnaire and were administered an interview including demographic factors, pigmentation characteristics, sun exposure and sensitivity, and other factors as previously described (27, 28). Participants provided a bio-specimen of blood or mouthwash sample for genotyping work. There were 3247 cases and controls (1139 controls, 1235 BCC, 873 SCC) with available DNA. For this study, we first selected all cases of multiple BCC within 30 days of initial diagnosis (≥2 BCC n=163, which included 44 subjects ≥3 BCC), followed by a random selection of single BCC (n=259), SCC (n=263), and controls (n=421).
KIR genotyping
DNA was extracted from buffy coat using Qiagen genomic DNA extraction kits. Multiplex KIR genotyping was performed with a Luminex-based typing kit (Lifecodes Product #545110, Gen-Probe Transplant Diagnostics Inc.). This method captured the presence/absence of 14 KIR genes: 2DS1, 2DS2, 2DS3, 2DS4, 2DS5, 2DP1, 2DL1, 2DL2, 2DL3, 2DL4, 2DL5, 3DL1, 3DS1, and 3DL2. The samples were analyzed by the University of Minnesota Cytokine Reference Laboratory on a Bio-Plex instrument.
HLA-C genotyping
The HLA-C alleles are classified as group C1 ligands which contain serine (AGC) at codon 77 and asparagine (AAC) at codon 80, and group C2 ligands which contain asparagine (AAC) at codon 77 and lysine (AAA) at codon 80 (18). Genotyping of the HLA-C codon 77 polymorphism was performed using an allelic discrimination SNP genotyping assay (Life Technologies, Carlsbad, CA) on a LightCycler 480 instrument (Roche). The genotyping was performed according to Roche guidelines, using 10 ng of genomic DNA.
HLA-B genotyping
HLA-B genotyping was performed through two amplification steps, followed by pyrosequencing. The first amplification step (PCR-I) was used to reduce noise, using the method of Pozzi et.al (29). Briefly, 20ng of purified genomic DNA were used in a 25ul PCR amplification containing: 2.5 µl of 10× buffer, 5ul Q-solution (Qiagen), 0.5µl 10mM dNTPs, 0.5µl of 10µM HLA-B forward primer (Bx1), 0.5µl 10µM HLA-B reverse primer (BINT3) and 0.2ul HotStarTaq DNA polymerase (Qiagen). PCR conditions were: 1 cycle at 94°C for 10min, followed by 8 cycles at 94°C for 20 s, 65°C for 30 s, 72°C for 2 min, followed by 32cycles at 94°C for 20 s, 60°C for 30 s, 72°C for 2 min, followed by 72°C for 7 min, and 4°C hold. This HLA-B specific PCR-I product was then used for a second amplification step, as described by Yun et.al (19). PCR-II reagents: 1ul of PCR-I product, 2.5ul 10X buffer, 0.5ul 10mM dNTPs, 0.5µl of 10µM HLA-B-forward, 0.5µl of 10µM HLA-B-reverse (biotinylated), and 0.2ul HotStarTaq DNA polymerase (Qiagen), in a 25µl amplification. PCR-II cycling conditions: 95°C for 10 min, followed by 40 cycles at: 94°C for 30 s, 57°C for 30 s, 72°C for 1 min; followed by 72°C by 10 min, and 4°C Hold. 20 ul of PCR-II product were used for pyrosequencing. Sequencing was carried out using a Pyromark Q96 MD (Qiagen) pyrosequencing apparatus and HLA-B sequencing primer 5'- CACAGACTGACCGAGAG -3'. HLA-B genotype was determined by manual pyrogram reading by two readers. The assay consisted of 27 nucleotide dispensations that read the genotype of HLA-B codons 77–83 (Sequence to analyze: RRCCTGCGSAHC[CT][GC]GCKCSGCTACTACAACCAGAGCGAGGCCGGTG). The reads from codon 77, 79 and 80–83 were combined to determine HLA-B genotype. HLA-B genotype was undetermined for 14 (1.4%) study samples.
KIR haplotype determination
KIR haplotypes were assigned based on gene content and known linkage disequilibrium, as described by Pyo et.al. (16). The KIR 2DL5-2DS3/2DS5 genic block was not used for haplotype assignment due to ambiguity of location. The KIR region was considered as two independent haplotype blocks (centromeric and telomeric). Within each haplotype block an individual was assigned as A/A, A/B, or B/B, reflecting haplotypes inherited from each parent. The centromeric haplotypes were defined as follows: A/A (containing both 2DL3 and 2DP1/2DL1, with no D2L2 or 2DS2 present), A/B (containing 2DL3, 2DL2 and 2DS2), and B/B (containing both 2DS2 and 2DL2 with no 2DL3 present). Telomeric haplotypes were defined as: A/A (containing 3DL1 and 2DS4, with no 3DS1 or 2DS1 present), A/B (containing 3DL1, 2DS4, 2DS1 and 3DS1), and B/B (containing 2DS1 and 3DS1, with no 3DL1 or 2DS4 present). Using this definition of KIR, individuals have 4 KIR haplotypes; this definition that reflects the haplotype block structure (centromere and telomere) and the inheritance of a haplotype from each parent for each block. Missing KIR data were imputed using the established linkage disequilibrium among genes to categorize the participants into these most common KIR haplotypes. For 16 samples telomeric haplotype could not be determined due to 2DS4 control probe failure. 83 (7.5%) samples (23 Controls, 35 SCC, and 25 BCC) were excluded due to poor DNA sample quality.
P53
We requested the original paraffin-embedded tumor specimens for histopathology re-review. The study dermatopathologist performed a standard histopathology re-review that included classification of tumor morphology, grade, associated actinic keratoses, degree of histologic solar elastosis, and percent of tumor present in the tissue specimen. In a subset of tumors, immunohistochemical (IHC) analysis of p53 was performed using standard approaches (30) and were scored as “negative”, “trace” staining or the percent of tumor cells staining positively (negative, trace, 0–100%) for each intensity (1,2,3+). Tumors with a 3+ intensity score in 10% or more of tumorous cells were considered p53 IHC positive (as described in (31)). TP53 mutation for this study was previously described in Almquist et al (32). IHC and mutation data were combined into a single metric to indicate abnormal p53 status, and was available for 100 BCC and 100 SCC tumors.
Statistical analysis
We used unconditional logistic regression to compute odds ratios (OR) and 95% confidence intervals (95% CI) for BCC and SCC. All models were adjusted for age at diagnosis (continuous), sex, and skin type. Skin type was defined as the reaction to 1 hour of sun exposure the first time in the summer; those who responded that they had a severe sunburn with blistering, or painful sunburn for a few days followed by peeling were classified as skin type “Burn”, and those who responded that they tanned without any sunburn, or had a mild burn followed by tanning, were classified as “Tan”. We conducted interaction testing for KIR*HLA and skin cancer risk using the likelihood ratio test comparing logistic regression models with and without the interaction term. Separate analyses were performed for SCC and BCC. Statistical analyses were performed in R v2.13.1 using the EpiCalc package.
Results
We conducted genotype analyses of KIR and HLA-C and B dimorphisms in 1023 participants (398 Controls, 387 BCC, 238 SCC). The demographic characteristics of study subjects are presented in Table 1. To investigate the association of KIR gene content and keratinocyte cancer risk, we conducted haplotype level analysis of the independently inherited centromeric and telomeric haplotype blocks of the KIR locus (Table 2). No overall associations were found for SCC or BCC in haplotype level analysis (Table 2). Of the 387 BCC there were 44 subjects with ≥ 3 tumors removed within 30 days of diagnosis. Centromeric B/B haplotypes were associated with significant elevation in risk of multiple (≥ 3) BCC (OR=2.39, 95% CI: 1.10 – 5.21) (Table 2). We similarly investigated the single gene effects of HLA-C and HLA-B dimorphism (Table 3). While no differences in risk of keratinocyte cancer were found with HLA-C genotype, the HLA-Bw4 homozygous genotype was associated with a significant reduction in risk of SCC (OR=0.58, 95% CI: 0.34–0.99) (Table 3).
Table 1.
Selected demographic characteristics of study participants
| Controls | BCC | Multiple BCCa | SCC | |
|---|---|---|---|---|
| N (%) | 398 (39) | 387 (38) | 44 (4) | 238 (23) |
| Age | ||||
| Mean (SD) | 58 (12) | 59 (11) | 60 (10) | 64 (8) |
| Sex | ||||
| Male (%) | 236 (59) | 219 (57) | 31 (70) | 163 (68) |
| Female (%) | 162 (41) | 168 (43) | 13 (30) | 75 (32) |
≥3 concomitant BCCs
Table 2.
The association between KIR haplotypes and keratinocyte skin cancers
| Controls (n=398) |
BCC (n=387) |
Multiple BCCa (n= 44) |
SCC (n=238) |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haplotypes | n | (%) | n | (%) | ORb | (95%CI) | n | (%) | ORb | (95%CI) | n | (%) | ORb | (95%CI) |
| Centromeric | ||||||||||||||
| A/A | 189 | (47) | 81 | (47) | 1.00 | ref. | 18 | (41) | 1.00 | ref. | 117 | (49) | 1.00 | ref. |
| A/B | 144 | (36) | 149 | (39) | 1.06 | (0.77–1.44) | 12 | (27) | 0.83 | (0.38–1.83) | 84 | (35) | 0.97 | (0.66–1.40) |
| B/B | 65 | (16) | 57 | (15) | 0.91 | (0.60–1.38) | 14 | (32) | 2.39 | (1.10–5.21) | 37 | (16) | 0.96 | (0.58–1.56) |
| Telomericc | ||||||||||||||
| A/A | 239 | (60) | 235 | (61) | 1.00 | ref. | 25 | (57) | 1.00 | ref. | 144 | (61) | 1.00 | ref. |
| A/B | 140 | (35) | 129 | (33) | 0.94 | (0.69–1.27) | 19 | (43) | 1.31 | (0.69–2.51) | 86 | (36) | 1.01 | (0.70–1.45) |
| B/B | 15 | (4) | 15 | (4) | 1.14 | (0.54–2.41) | - | - | - | - | 4 | (<2) | 0.49 | (0.15–1.55) |
≥3 concomitant BCCs
OR adjusted for age, sex and skin type
Telomeric genotype missing due to 2DS4 probe failure for 16 subjects
Table 3.
The association between HLA dimorphism in KIR ligand domains and keratinocyte cancers
| Controls | BCC | Multiple BCCa | SCC | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | (%) | n | (%) | ORb | (95%CI) | n | (%) | ORb | (95%CI) | n | (%) | ORb | (95%CI) | |
| HLA-Cc | ||||||||||||||
| C1 | 127 | (32) | 142 | (37) | 1.00 | ref. | 14 | (32) | 1.00 | ref. | 93 | (39) | 1.00 | ref. |
| C1C2 | 163 | (41) | 149 | (39) | 0.81 | (0.58–1.13) | 17 | (39) | 0.90 | (0.42–1.93) | 101 | (42) | 0.81 | (0.55–1.20) |
| C2 | 53 | (13) | 53 | (14) | 0.89 | (0.56–1.42) | 7 | (16) | 1.37 | (0.51–3.72) | 34 | (14) | 0.78 | (0.45–1.35) |
| HLA-Bd | ||||||||||||||
| Bw6 | 134 | (34) | 146 | (38) | 1.00 | ref. | 15 | (30) | 1.00 | ref. | 94 | (39) | 1.00 | ref. |
| Bw4/Bw6 | 188 | (47) | 169 | (44) | 0.84 | (0.61–1.16) | 17 | (40) | 0.95 | (0.45–2.01) | 112 | (47) | 0.92 | (0.63–1.34) |
| Bw4 | 66 | (17) | 68 | (18) | 0.93 | (0.62–1.42) | 9 | (20) | 1.25 | (0.51–3.08) | 32 | (13) | 0.58 | (0.34–0.99) |
≥3 concomitant BCCs
OR adjusted for age, sex and skin type
HLA-C genotype missing for 108 subjects
HLA-B genotype missing for 14 subjects
The biologic interaction between HLA class I ligand and inhibitory KIR determines NK cell licensing and subsequent potential to respond to activating signals. We therefore conducted an interaction analysis of KIR gene content variation and HLA-C (Table 4) and HLA-B (Table 5). There was a statistically significant interaction between HLA-C and KIR2DS3 for both BCC (p<0.005) and SCC (p=0.038), with those having both HLA-C1/C1 and the KIR2DS3 gene demonstrating elevated risk of both types of keratinocyte caner (Table 4). In addition, for BCC, there were statistically significant interactions between HLA-B and telomeric B haplotypes (containing KIR2DS1 and KIR3DS1, p<0.001) as well as HLA-B and KIR2DS5 (p=0.012) (Table 5). For both interactions those with the Bw4/Bw4 genotype and the activating KIR gene had reduced risk of BCC, where the effect was attenuated, or possibly reversed, among those carrying a Bw6 allele. Similar non-significant trends were observed for SCC.
Table 4.
Regression models for the joint effects of KIR and HLA-C dimorphism
| KIR | HLA-C | Controls | BCC | ORa (95%CI) | SCC | ORa (95%CI) |
|---|---|---|---|---|---|---|
| Centromere | ||||||
| A/A | C1C1 | 69 | 68 | Ref. | 54 | Ref. |
| Any B | 58 | 74 | 1.21 (0.74–1.97) | 39 | 0.88 (0.49–1.56) | |
| A/A | Any C2 allele | 96 | 95 | 0.95 (0.61–1.49) | 58 | 0.70 (0.42–1.17) |
| Any B | 120 | 107 | 0.88 (0.57–1.35) | 77 | 0.81 (0.49–1.31) | |
| Pinteraction | 0.393 | 0.465 | ||||
| Telomere | ||||||
| A/A | C1C1 | 74 | 83 | Ref. | 54 | Ref. |
| Any B | 53 | 59 | 0.95 (0.58–1.57) | 39 | 0.95 (0.53–1.69) | |
| A/A | Any C2 allele | 136 | 127 | 0.81 (0.54–1.21) | 86 | 0.78 (0.48–1.25) |
| Any B | 80 | 75 | 0.83 (0.53–1.30) | 49 | 0.80 (0.47–1.36) | |
| Pinteraction | 0.821 | 0.834 | ||||
| 2DS3 absent | C1C1 | 108 | 100 | Ref. | 68 | Ref. |
| 2DS3 present | 19 | 42 | 2.29 (1.24–4.26) | 25 | 1.89 (0.92–3.87) | |
| 2DS3 absent | Any C2 allele | 148 | 148 | 1.06 (0.74–1.52) | 103 | 0.99 (0.65–1.51) |
| 2DS3 present | 68 | 54 | 0.84 (0.53–1.33) | 32 | 0.74 (0.43–1.27) | |
| Pinteraction | 0.005 | 0.038 | ||||
| 2DS5 absent | C1C1 | 79 | 94 | Ref. | 61 | Ref. |
| 2DS5 present | 48 | 48 | 0.83 (0.50–1.38) | 32 | 0.88 (0.48–1.59) | |
| 2DS5 absent | Any C2 allele | 151 | 149 | 0.81 (0.55–1.19) | 94 | 0.75 (0.48–1.18) |
| 2DS5 present | 65 | 53 | 0.69 (0.42–1.11) | 41 | 0.79 (0.46–1.37) | |
| Pinteraction | 0.959 | 0.651 | ||||
OR adjusted for age, sex, and skin type
2DS3 and 2DS5 genes are present on B haplotypes and can occur in either the centromere or telomere block
Table 5.
Regression models for the joint effects of KIR and HLA-B dimorphism
| KIR | HLA-B | Controls | BCC | ORa (95%CI) | SCC | ORa (95%CI) |
|---|---|---|---|---|---|---|
| Centromere | ||||||
| A/A | Bw4/Bw4 | 27 | 30 | Ref. | 13 | Ref. |
| Any B | 39 | 38 | 0.85 (0.42–1.71) | 19 | 1.00 (0.40–2.47) | |
| A/A | Any Bw6 allele | 155 | 149 | 0.88 (0.49–1.57) | 104 | 1.67 (0.80–3.56) |
| Any B | 167 | 166 | 0.90 (0.51–1.59) | 102 | 1.61 (0.76–3.39) | |
| Pinteraction | 0.645 | 0.924 | ||||
| Telomere | ||||||
| A/A | Bw4/Bw4 | 30 | 48 | Ref. | 20 | Ref. |
| Any B | 36 | 20 | 0.35 (0.17–0.73) | 12 | 0.56 (0.22–1.39) | |
| A/A | Any Bw6 allele | 207 | 184 | 0.57 (0.35–0.95) | 126 | 1.20 (0.63–2.31) |
| Any B | 115 | 131 | 0.75 (0.44–1.26) | 80 | 1.36 (0.69–2.68) | |
| Pinteraction | 0.001 | 0.16 | ||||
| 2DS3 absent | Bw4/Bw4 | 40 | 49 | Ref. | 22 | Ref. |
| 2DS3 present | 26 | 19 | 0.60 (0.29–1.26) | 10 | 0.88 (0.34–2.26) | |
| 2DS3 absent | Any Bw6 allele | 247 | 227 | 0.77 (0.49–1.23) | 156 | 1.56 (0.86–2.83) |
| 2DS3 present | 75 | 88 | 0.98 (0.58–1.67) | 50 | 1.62 (0.82–3.18) | |
| Pinteraction | 0.074 | 0.749 | ||||
| 2DS5 absent | Bw4/Bw4 | 40 | 55 | Ref. | 23 | Ref. |
| 2DS5 present | 26 | 13 | 0.37 (0.17–0.81) | 9 | 0.65 (0.25–1.73) | |
| 2DS5 absent | Any Bw6 allele | 221 | 213 | 0.72 (0.45–1.13) | 139 | 1.40 (0.78–2.54) |
| 2DS5 present | 101 | 102 | 0.77 (0.47–1.27) | 67 | 1.49 (0.79–2.83) | |
| Pinteraction | 0.012 | 0.359 | ||||
OR adjusted for age, sex, and skin type
2DS3 and 2DS5 genesare present on B haplotypes and can occur in either the centromere or telomere block
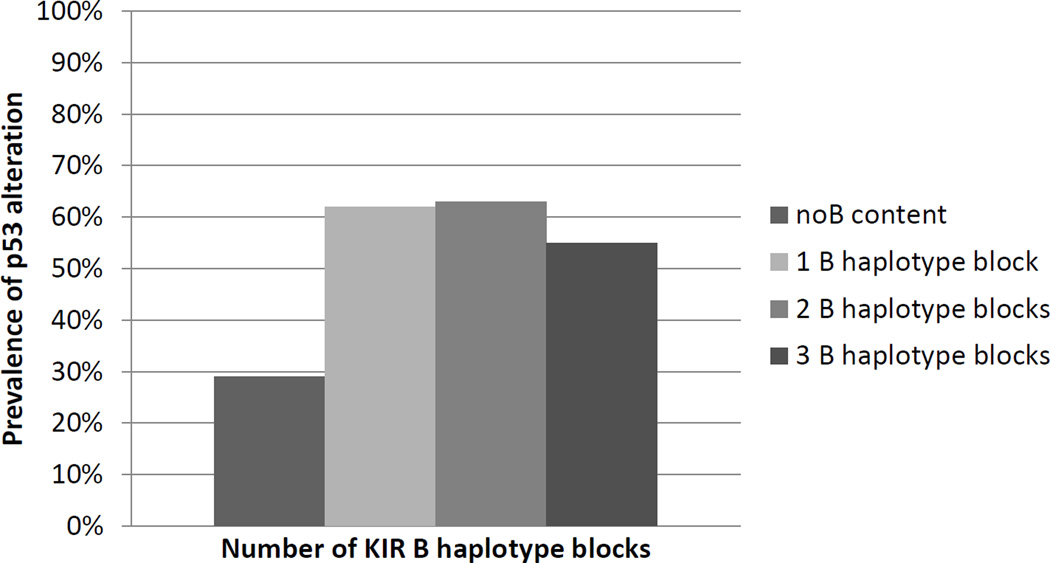
Finally, we tested the hypothesis that KIR B haplotype blocks were associated with alteration at p53 (defined as either mutation or altered IHC) (Figure 1). Among 100 BCC tumors, there was a strong association of altered p53 and KIR B haplotype blocks, with 62% (40/65) of B carriers having altered p53 and only 29% (10/35) of those with no B haplotype blocks having altered p53 (p<0.004). For the centromere haplotypes the prevalence of p53 alterations was 35% (17/48) in the cenA/A group and 63%(33/52) in the cenB group. For the telomere haplotypes the prevalence of mutation was 45% (28/62) in the telA/A group and 57%(20/35) in the telB group. There were no associations with p53 status in 100 SCC tumors (data not shown).
Figure 1.
P53 alteration in BCC tumors as a function of increasing B haplotype blocks. The prevalence of p53 alteration (mutation or abnormal IHC) was calculated among individuals with varying numbers of KIR B haplotype blocks (summed across centromeric and telomeric blocks of the KIR locus). Those with p53 alterations were four times more likely to have KIR B then those with normal p53 (p<0.004); there was no evidence of a gene-dosage effect.
Discussion
Using a population-based case-control study of BCC and SCC we tested the hypothesis that KIR gene content and HLA class I dimorphisms play a role in the development of keratinocyte cancer, and result in selective pressure for p53 alteration. There was limited evidence for single gene effects. Not unexpectedly, since these gene products physically interact to alter NK cell phenotype, it was necessary to look at interactions between the genes to reveal disease associations. In particular, the presence of the activating KIR2DS3 gene with HLA-C1 was associated with an approximate doubling of risk for BCC and SCC. In contrast, telomeric B (containing KIR2DS1 and KIR3DS1) and 2DS5 were inversely associated with BCC among HLA-Bw4/Bw4 carriers. These findings suggest a complex interplay of KIR/HLA in the etiology of keratinocyte-derived cancers.
To date, there has been limited investigation of NK cell genetics and somatic mutations. One might expect that genetic variation in immune surveillance influences what somatic mutations go “unseen” and can develop into tumors. In vitro work has demonstrated that p53 is necessary for the up-regulation of stress ligands that would activate NK receptors (25). Extending this model to human populations would suggest that p53 must be inactivated among those with a robust NK response (those with B haplotypes). Our data are very consistent with this hypothesis.
Previous studies have investigated whether individual KIR genes act as risk traits for solid tumors. In breast and lung cancer, genes contained in B haplotypes (KIR 2DS1, 3DS1, and 2DS5) were associated with increased cancer risk (33). Also, increased numbers of activating KIR genes were associated with nasopharyngeal carcinoma (34). These studies suggest that an environment of active NK cells may contribute to carcinogenesis. Similarly, our finding that centromeric B/B haplotypes are associated with over 2 fold risk of multiple BCC (≥3), may also result from inappropriate or chronic activation of NK cells. In contrast to these studies, melanoma risk and progression were associated with inhibitory KIR and inhibitory KIR/HLA combinations (22–24).
Our approach focused on the joint effect of KIR and their licensing partners HLA-C and B. Increased BCC and SCC risk with KIR2DS3 in those homozygous for HLA-C1 was unexpected as there is no reported biologic interaction between KIR2DS3 and HLA-C, and KIR2DS3 has low cell-surface expression (35). Hierarchy of affinity in inhibitory KIR/HLA interactions has been suggested to play an important role in tuning NK cell response (9, 20). Presence of the activating KIR may further attenuate this process and enhance NK cell activation (36). Thus, it is possible that when interacting with homozygous C1 ligands NK cells have a lower activation threshold in the presence of activating KIR2DS3, resulting in NK cell hyperactivity. While such a response may be protective in infection control (37), it may be deleterious for cancer development as increased NK cell activation may result in prolonged inflammation and tissue destruction. Another explanation for our observed interaction between HLA-C and KIR2DS3 is rooted in the known genetic linkage of this region. KIR2DS3 occurs on B haplotypes (both centromeric and telomeric) and is therefore in partial linkage with centromeric genes known to interact with HLA-C. As the field progresses to more sophisticated KIR typing methods it will be possible to refine our definitions of haplotypes and directly address this question.
In contrast to our observations of increased risk associated with centromeric activating KIR, reduced risk was observed with activating genes (KIR2DS1, KIR3DS1 and 2DS5) among HLA-Bw4 homozygous individuals. This pattern of risk reduction is consistent with expected enhancement of NK cell effector function, in the presence of activating receptors, when NK cells have been licensed through Bw4 (20, 38). Only Bw4, but not Bw6 epitopes bind to the inhibitory telomeric KIR3DL1 (which was present in >95% of our study population) and participates in licensing of NK cells (12). These results may be particularly robust for BCC as these tumors have low or absent HLA class 1 expression (39–41) making them particularly susceptible to NK killing.
HLA-Bw4 has been associated with increased NK cell responsiveness to tumor stimulation in vitro. NK cells from individuals with 3DL1+/Bw4/4, compared to 3DL1+/Bw4/6 or Bw6/6, reported greater response to tumor cells and increased IFN-γ production (42). Presence of activating KIR have also been shown to enhance IFN-γ secretion in cell culture (43). In addition, presence of KIR3DS1 was associated with increased IFN-γ levels and greater NK cell activity in early HIV-I infection (44). Our results suggest that Bw4 licensing combined with activating signals may be important in immune editing for BCC and SCC. IFN-γ is important in anti-tumor responses (45) and skin cancer surveillance (46–48). More efficient NK cell activation with increased IFN-γ production may contribute to the reduced risk in skin cancers associated with the activating telomeric KIR and HLA-Bw4 genotype.
While the KIR literature primarily focuses on NK cells which are the predominant KIR expressing cells, subsets of T cells also express KIR, including γδ T cells which are more abundant in the skin than NK cells (49, 50). The γδ T cells can respond to stress signals via T cell receptor adaptive and innate mechanisms, including the activating receptor NKG2D. Furthermore, γδ T cells can enhance innate immunity through up-regulation of T helper 1 cytokines including IFN-γ, to stimulate NK cell responses (50). Experiments in mice revealed that γδ T cells are able to inhibit early carcinogenesis and the progression of SCC and melanoma (50). Thus, the effects observed in this study may not only be a result of NK cell functionality, but may also reflect activity of the γδ T cells. Further research is needed to understand the role of KIR in γδ T cells, and whether they also undergo licensing through KIR-HLA interactions.
This study had several limitations, including multiplexed KIR genotyping which was prone to allelic dropout. To address this problem, we used published linkage disequilibrium information to impute missing data for KIR genes. Analyses restricted to those subjects without imputation did not change the study results. The KIR 2DL5-2DS3/2DS5 genic block could not be unambiguously assigned to centromeric and/or telomeric segments due to the limits of the genotyping methodology. Our HLA-Bw4 typing method was specific to HLA-B; there are a few HLA-A alleles that carry Bw4 epitopes (HLA-A23, 24, 25, and 32), which would be misclassified using our method (12). Approximately 7.5% of our study population was excluded from statistical analysis due to insufficient DNA sample quality for complex genotyping. Other limitations included lack of replication in an independent population and multiple statistical comparisons. In addition, we were underpowered to detect differences in risk based on Bw4 position 80 (isoleucine vs. threonine), which impacts KIR3DL1 binding (38), to test further gene*gene interactions, or conduct refined analyses of the multiple BCC phenotype, an important clinical problem which may have an underlying genetic component.
In conclusion, our findings suggest differential BCC and SCC risk with centromeric and telomeric activating KIR, and selective pressure from this inter-individual variability drives p53 alteration in skin tumors. Our study underscores the importance of investigating KIR in combination with HLA ligands as has been done for bone marrow transplant and melanoma (21–24). Future research should include high resolution typing of KIR and HLA, and further investigate the multiple BCC phenotype.
Acknowledgments
The authors wish to acknowledge Caitlin Canton and Kristen Evenson for technical assistance, the UMN Cytokine Reference Lab for Luminex measurements of KIR, Gong Yun and Dr. Jeffrey S. Miller for assistance in developing HLA typing assays, and M. Scot Zens of the New Hampshire Health Study.
Funding: R01CA82354, R01CA057494
Footnotes
Conflicts of interest: None to report.
References
- 1.Rogers HW, Weinstock MA, Harris AR, et al. Incidence estimate of nonmelanoma skin cancer in the united states, 2006. Arch Dermatol. 2010;146(3):283–287. doi: 10.1001/archdermatol.2010.19. [DOI] [PubMed] [Google Scholar]
- 2.Neville JA, Welch E, Leffell DJ. Management of nonmelanoma skin cancer in 2007. Nat Clin Pract Oncol. 2007;4(8):462–469. doi: 10.1038/ncponc0883. [DOI] [PubMed] [Google Scholar]
- 3.Housman TS, Feldman SR, Williford PM, et al. Skin cancer is among the most costly of all cancers to treat for the medicare population. J Am Acad Dermatol. 2003;48(3):425–429. doi: 10.1067/mjd.2003.186. [DOI] [PubMed] [Google Scholar]
- 4.IARC. IARC monographs on the evaluation of carcinogenic risks to humans. solar and ultraviolet radiation. 1992;55:1–316. [PMC free article] [PubMed] [Google Scholar]
- 5.Norval M, McLoone P, Lesiak A, Narbutt J. The effect of chronic ultraviolet radiation on the human immune system. Photochem Photobiol. 2008;84(1):19–28. doi: 10.1111/j.1751-1097.2007.00239.x. [DOI] [PubMed] [Google Scholar]
- 6.Athar M, Walsh SB, Kopelovich L, Elmets CA. Pathogenesis of nonmelanoma skin cancers in organ transplant recipients. Arch Biochem Biophys. 2011;508(2):159–163. doi: 10.1016/j.abb.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong DA, Bishop GA, Lowes MA, Cooke B, Barnetson RS, Halliday GM. Cytokine profiles in spontaneously regressing basal cell carcinomas. Br J Dermatol. 2000;143(1):91–98. doi: 10.1046/j.1365-2133.2000.03596.x. [DOI] [PubMed] [Google Scholar]
- 8.Barnetson RS, Halliday GM. Regression in skin tumours: A common phenomenon. Australas J Dermatol. 1997;38(Suppl 1):S63–S65. doi: 10.1111/j.1440-0960.1997.tb01013.x. [DOI] [PubMed] [Google Scholar]
- 9.Purdy AK, Campbell KS. Natural killer cells and cancer: Regulation by the killer cell ig-like receptors (KIR) Cancer Biol Ther. 2009;8(23):2211–2220. doi: 10.4161/cbt.8.23.10455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim S, Poursine-Laurent J, Truscott SM, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436(7051):709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 11.Long EO, Kim HS, Liu D, Peterson ME, Rajagopalan S. Controlling natural killer cell responses: Integration of signals for activation and inhibition. Annu Rev Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vilches C, Parham P. KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- 13.Shilling HG, Guethlein LA, Cheng NW, et al. Allelic polymorphism synergizes with variable gene content to individualize human KIR genotype. J Immunol. 2002;168(5):2307–2315. doi: 10.4049/jimmunol.168.5.2307. [DOI] [PubMed] [Google Scholar]
- 14.Traherne JA, Martin M, Ward R, et al. Mechanisms of copy number variation and hybrid gene formation in the KIR immune gene complex. Hum Mol Genet. 2010;19(5):737–751. doi: 10.1093/hmg/ddp538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Middleton D, Gonzelez F. The extensive polymorphism of KIR genes. Immunology. 2010;129(1):8–19. doi: 10.1111/j.1365-2567.2009.03208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pyo CW, Wang R, Vu Q, et al. Recombinant structures expand and contract inter and intragenic diversification at the KIR locus. BMC Genomics. 2013;14:89. doi: 10.1186/1471-2164-14-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: Tuning the genome for defense. Annu Rev Genomics Hum Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 18.Hollenbach JA, Ladner MB, Saeteurn K, et al. Susceptibility to crohn's disease is mediated by KIR2DL2/KIR2DL3 heterozygosity and the HLA-C ligand. Immunogenetics. 2009;61(10):663–671. doi: 10.1007/s00251-009-0396-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yun G, Tolar J, Yerich AK, et al. A novel method for KIR-ligand typing by pyrosequencing to predict NK cell alloreactivity. Clin Immunol. 2007;123(3):272–280. doi: 10.1016/j.clim.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat Rev Immunol. 2005;5(3):201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 21.Cooley S, Weisdorf DJ, Guethlein LA, et al. Donor selection for natural killer cell receptor genes leads to superior survival after unrelated transplantation for acute myelogenous leukemia. Blood. 2010;116(14):2411–2419. doi: 10.1182/blood-2010-05-283051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naumova E, Mihaylova A, Stoitchkov K, Ivanova M, Quin L, Toneva M. Genetic polymorphism of NK receptors and their ligands in melanoma patients: Prevalence of inhibitory over activating signals. Cancer Immunol Immunother. 2005;54(2):172–178. doi: 10.1007/s00262-004-0575-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naumova E, Mihaylova A, Ivanova M, Mihailova S. Impact of KIR/HLA ligand combinations on immune responses in malignant melanoma. Cancer Immunol Immunother. 2007;56(1):95–100. doi: 10.1007/s00262-006-0151-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campillo JA, Legaz I, Lopez-Alvarez MR, et al. KIR gene variability in cutaneous malignant melanoma: Influence of KIR2D/HLA-C pairings on disease susceptibility and prognosis. Immunogenetics. 2013;65(5):333–343. doi: 10.1007/s00251-013-0682-0. [DOI] [PubMed] [Google Scholar]
- 25.Textor S, Fiegler N, Arnold A, Porgador A, Hofmann TG, Cerwenka A. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 2011;71(18):5998–6009. doi: 10.1158/0008-5472.CAN-10-3211. [DOI] [PubMed] [Google Scholar]
- 26.Karagas MR, Tosteson TD, Blum J, Morris JS, Baron JA, Klaue B. Design of an epidemiologic study of drinking water arsenic exposure and skin and bladder cancer risk in a U.S. population. Environ Health Perspect. 1998;106(Suppl 4):1047–1050. doi: 10.1289/ehp.98106s41047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karagas MR, Nelson HH, Sehr P, et al. Human papillomavirus infection and incidence of squamous cell and basal cell carcinomas of the skin. J Natl Cancer Inst. 2006;98(6):389–395. doi: 10.1093/jnci/djj092. [DOI] [PubMed] [Google Scholar]
- 28.Miller KL, Karagas MR, Kraft P, et al. XPA, haplotypes, and risk of basal and squamous cell carcinoma. Carcinogenesis. 2006;27(8):1670–1675. doi: 10.1093/carcin/bgi376. [DOI] [PubMed] [Google Scholar]
- 29.Pozzi S, Longo A, Ferrara GB. HLA-B locus sequence-based typing. Tissue Antigens. 1999;53(3):275–281. doi: 10.1034/j.1399-0039.1999.530308.x. [DOI] [PubMed] [Google Scholar]
- 30.Kelsey KT, Hirao T, Hirao S, et al. TP53 alterations and patterns of carcinogen exposure in a U.S. population-based study of bladder cancer. Int J Cancer. 2005;117(3):370–375. doi: 10.1002/ijc.21195. [DOI] [PubMed] [Google Scholar]
- 31.Torti DC, Christensen BC, Storm CA, et al. Analgesic and nonsteroidal anti-inflammatory use in relation to nonmelanoma skin cancer: A population-based case-control study. J Am Acad Dermatol. 2011;65(2):304–312. doi: 10.1016/j.jaad.2010.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almquist LM, Karagas MR, Christensen BC, et al. The role of TP53 and MDM2 polymorphisms in TP53 mutagenesis and risk of non-melanoma skin cancer. Carcinogenesis. 2011;32(3):327–330. doi: 10.1093/carcin/bgq256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ozturk OG, Gun FD, Polat G. Killer cell immunoglobulin-like receptor genes in patients with breast cancer. Med Oncol. 2012;29(2):511–515. doi: 10.1007/s12032-011-9932-x. [DOI] [PubMed] [Google Scholar]
- 34.Butsch Kovacic M, Martin M, Gao X, et al. Variation of the killer cell immunoglobulin-like receptors and HLA-C genes in nasopharyngeal carcinoma. Cancer Epidemiol Biomarkers Prev. 2005;14(11 Pt 1):2673–2677. doi: 10.1158/1055-9965.EPI-05-0229. [DOI] [PubMed] [Google Scholar]
- 35.Vanden Bussche CJ, Mulrooney TJ, Frazier WR, Dakshanamurthy S, Hurley CK. Dramatically reduced surface expression of NK cell receptor KIR2DS3 is attributed to multiple residues throughout the molecule. Genes Immun. 2009;10(2):162–173. doi: 10.1038/gene.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart CA, Laugier-Anfossi F, Vely F, et al. Recognition of peptide-MHC class I complexes by activating killer immunoglobulin-like receptors. Proc Natl Acad Sci U S A. 2005;102(37):13224–13229. doi: 10.1073/pnas.0503594102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopez-Vazquez A, Rodrigo L, Martinez-Borra J, et al. Protective effect of the HLA-Bw4I80 epitope and the killer cell immunoglobulin-like receptor 3DS1 gene against the development of hepatocellular carcinoma in patients with hepatitis C virus infection. J Infect Dis. 2005;192(1):162–165. doi: 10.1086/430351. [DOI] [PubMed] [Google Scholar]
- 38.Foley BA, De Santis D, Van Beelen E, Lathbury LJ, Christiansen FT, Witt CS. The reactivity of Bw4+ HLA-B and HLA-A alleles with KIR3DL1: Implications for patient and donor suitability for haploidentical stem cell transplantations. Blood. 2008;112(2):435–443. doi: 10.1182/blood-2008-01-132902. [DOI] [PubMed] [Google Scholar]
- 39.Walter A, Barysch MJ, Behnke S, et al. Cancer-testis antigens and immunosurveillance in human cutaneous squamous cell and basal cell carcinomas. Clin Cancer Res. 2010;16(14):3562–3570. doi: 10.1158/1078-0432.CCR-09-3136. [DOI] [PubMed] [Google Scholar]
- 40.Cabrera T, Garrido V, Concha A, et al. HLA molecules in basal cell carcinoma of the skin. Immunobiology. 1992;185(5):440–452. doi: 10.1016/s0171-2985(11)80086-0. [DOI] [PubMed] [Google Scholar]
- 41.Kageshita T, Ono T, Hirai S, et al. Ganglioside, adhesion molecule, and HLA antigen expression in basal cell carcinoma lesions. Cancer Res. 1992;52(11):3201–3207. [PubMed] [Google Scholar]
- 42.Kim S, Sunwoo JB, Yang L, et al. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105(8):3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mandelboim O, Kent S, Davis DM, et al. Natural killer activating receptors trigger interferon gamma secretion from T cells and natural killer cells. Proc Natl Acad Sci U S A. 1998;95(7):3798–3803. doi: 10.1073/pnas.95.7.3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Long BR, Ndhlovu LC, Oksenberg JR, et al. Conferral of enhanced natural killer cell function by KIR3DS1 in early human immunodeficiency virus type 1 infection. J Virol. 2008;82(10):4785–4792. doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alshaker HA, Matalka KZ. IFN-gamma, IL-17 and TGF-beta involvement in shaping the tumor microenvironment: The significance of modulating such cytokines in treating malignant solid tumors. Cancer Cell Int. 2011;11:33. doi: 10.1186/1475-2867-11-33. 2867-11-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedrich M, Holzmann R, Sterry W, et al. Ultraviolet B radiation-mediated inhibition of interferon-gamma-induced keratinocyte activation is independent of interleukin-10 and other soluble mediators but associated with enhanced intracellular suppressors of cytokine-signaling expression. J Invest Dermatol. 2003;121(4):845–852. doi: 10.1046/j.1523-1747.2003.12482.x. [DOI] [PubMed] [Google Scholar]
- 47.Huang SJ, Hijnen D, Murphy GF, et al. Imiquimod enhances IFN-gamma production and effector function of T cells infiltrating human squamous cell carcinomas of the skin. J Invest Dermatol. 2009;129(11):2676–2685. doi: 10.1038/jid.2009.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wenzel J, Uerlich M, Haller O, Bieber T, Tueting T. Enhanced type I interferon signaling and recruitment of chemokine receptor CXCR3-expressing lymphocytes into the skin following treatment with the TLR7-agonist imiquimod. J Cutan Pathol. 2005;32(4):257–262. doi: 10.1111/j.0303-6987.2005.00297.x. [DOI] [PubMed] [Google Scholar]
- 49.Ebert LM, Meuter S, Moser B. Homing and function of human skin gammadelta T cells and NK cells: Relevance for tumor surveillance. J Immunol. 2006;176(7):4331–4336. doi: 10.4049/jimmunol.176.7.4331. [DOI] [PubMed] [Google Scholar]
- 50.Girardi M. Immunosurveillance and immunoregulation by gammadelta T cells. J Invest Dermatol. 2006;126(1):25–31. doi: 10.1038/sj.jid.5700003. [DOI] [PubMed] [Google Scholar]