Abstract
Principal component analysis (PCA) is widely used in genome-wide association studies (GWAS), and the principal component axes often represent perpendicular gradients in geographic space. The explanation of PCA results is of major interest for geneticists to understand fundamental demographic parameters. Here, we provide an interpretation of PCA based on relatedness measures, which are described by the probability that sets of genes are identical-by-descent (IBD). An approximately linear transformation between ancestral proportions (AP) of individuals with multiple ancestries and their projections onto the principal components is found.
In addition, a new method of eigenanalysis “EIGMIX” is proposed to estimate individual ancestries. EIGMIX is a method of moments with computational efficiency suitable for millions of SNP data, and it is not subject to the assumption of linkage equilibrium. With the assumptions of multiple ancestries and their surrogate ancestral samples, EIGMIX is able to infer ancestral proportions (APs) of individuals. The methods were applied to the SNP data from the HapMap Phase 3 project and the Human Genome Diversity Panel. The APs of individuals inferred by EIGMIX are consistent with the findings of the program ADMIXTURE.
In conclusion, EIGMIX can be used to detect population structure and estimate genome-wide ancestral proportions with a relatively high accuracy.
Keywords: PCA, Relatedness, Coancestry, IBD, SNP, Admixture
Introduction
Principal component analysis was introduced for the study of genetic data almost thirty years ago by Menozzi et al. (1978), and has since become a standard tool. Population differentiation can be inferred from multivariate statistical methods such as PCA of allele frequencies (Menozzi et al., 1978; Cavalli-Sforza and Feldman, 2003). In a new approach, Patterson et al. (2006) applied PCA to SNP genotypic data for individuals rather than populations. Their method, implemented in a software package “EIGENSTRAT”, has been widely used to correct for population stratification in genome-wide association studies (GWAS) (Price et al., 2010). Although PCA is not based on a population genetics model, and may seem like a “black box” method, principal component axes often represent perpendicular gradients in geographic space (Cavalli-Sforza and Feldman, 2003; Price et al., 2006; Novembre et al., 2008). The relationship of PCA results to fundamental demographic parameters is of major interest to geneticists.
Novembre and Stephens (2008) showed that the gradient and wave patterns of principal components do not necessarily reflect migration events in history. From the perspective of coalescent theory, McVean (2009) provided a genealogical interpretation of PCA. He showed that the projection of samples onto the principal components could be obtained from the pairwise coalescence times between study individuals. Ma and Amos (2010) proposed a formulation of PCA based on the variance-covariance matrix of the sample allele frequencies.
We now provide an alternative interpretation of PCA based on relatedness measures: probabilities that sets of genes have descended from a single ancestral gene and so are identical by descent (ibd). The ibd concept is essential for genetic analyses such as linkage studies for mapping disease genes and forensic DNA profiling (Weir et al., 2006; Thompson, 2013). In population genetics, Weir and Hill (2002) extended the work of Weir and Cockerham (1984) by allowing different levels of coancestry for different populations, and by allowing non-zero coancestries between pairs of populations. Our further extension is to allow different coancestries between pairs of individuals and different inbreeding coefficients for individuals. The coancestry coefficient between two populations defined in the model of Weir & Hill is now replaced by the average kinship coefficient among pairs of study individuals from these two populations respectively, relative to a single ancestral population, so that the assumption of random rating can be relaxed. These individual-perspective measures of population structure can be used to explain the behavior of PCA.
Ancestral proportions (AP) of an individual refer to the fractions of the genome derived from specific ancestral populations (Pritchard et al., 2000; Falush et al., 2003; Tang et al., 2005; Alexander et al., 2009). The early approach for estimating AP can track back to Hanis et al. (1986), and the ancestral allele frequencies should be known to allow estimating allele admixture in this method. However, ancestral allele frequencies are usually estimated from surrogate ancestral samples in practice and later studies took into account in describing the uncertainty of estimated ancestral information.
A Bayesian approach, STRUCTURE, was developed to infer population substructure using unlinked genotypes (Pritchard et al., 2000). Later, it was extended to model linked markers (Falush et al., 2003) through admixture linkage disequilibrium (LD). STRUCTURE is computationally intensive and not likely to be suitable for large-scale studies, like GWAS, involved with thousands of individuals and hundreds of thousands of SNPs. SNP pruning has to be done before applying STRUCTURE, and this can introduce selection bias with respect to different SNP sets. A maximum-likelihood estimation method, frappe, has also been proposed to estimate AP with much less computation than STRUCTURE, but it assumes the markers are unlinked (Tang et al., 2005). The ADMIXTURE method was developed to analyze thousands of markers – it adopts the likelihood model embedded in STRUCTURE with an assumption of linkage equilibrium among the markers (Alexander et al., 2009).
Instead of estimating global ancestry via genome-wide markers, detection of local ancestry from chromosomal segments in admixed populations becomes of great interest. Recently, HAPMIX and MULTIMIX were proposed to infer local ancestry from dense SNP markers based on approximate coalescent models modeling linkage disequilibrium with two or more ancestries (Price et al., 2009; Churchhouse and Marchini, 2013). However, their methods require a fine genetic map.
The potential connection between ancestral proportions and principal components in the eigenanalysis has been investigated by the previous studies with a limited number of numerical simulations (Patterson et al., 2006; Engelhardt and Stephens, 2010). McVean (2009) indicated it is possible to identify relative admixture proportions from principal components. Ma and Amos (2012) showed how to estimate two-way admixture proportions with a proof under their framework of variance-covariance matrix. They also observed that an admixed population could divide the triangle of three parental populations in the PC plot into three small triangles with areas according to the three-way admixture proportions. However, none of these studies provided a sufficient proof for inferring admixture fractions from the principal components under their theoretical framework in the cases of more than two ancestral populations.
In our study, an approximately linear transformation between ancestral proportions (AP) of individuals with multiple ancestries and their projections onto the principal components is revealed, and a proof is given under the framework of identity by descent. This linear transformation could explain the perpendicular gradients in geographic space, and it also justifies the observation that the ratios of triangle areas correspond to admixture fractions in the study of Ma and Amos (2012). We also propose a new method of eigenanalysis “EIGMIX” to estimate individual ancestries. EIGMIX uses method of moments estimation with computational efficiency suitable for millions of SNP data, and it is not subject to the assumption of linkage equilibrium. Ancestral proportions can be estimated by making assumptions of surrogate samples for ancestral populations, but inferring ancestral allele frequencies is not necessary. The calculation uses all study individuals simultaneously without projecting the remaining individuals onto the existing axes of surrogates.
We applied various methods to the SNP data of 1,198 founders from the HapMap Phase 3 project and 938 unrelated individuals from the Human Genome Diversity Project (HGDP). The ancestral proportions of individuals inferred by PCA and EIGMIX are consistent with the findings of the program ADMIXTURE. All eigenanalysis in the study are implemented in the R package “SNPRelate” (Zheng et al., 2012), allowing users to apply our method to their SNP data.
Methods
We develop our approach with a series of indicator variables xijkl for the kth allele, k = 1, 2, at the lth locus, l = 1, 2, …, L, in the jth individual sampled from the ith population, j = 1, 2, … ni; i = 1, 2 …, N. The total sample size is n = Σi ni. The variables take the value 1 for alleles of a specific type, e.g. the reference allele, at a locus, and the value 0 otherwise. Genotypes are indicate by gijl = xij1l + xij2l, and these take the values 0,1,2.
Population Coancestry Framework of Weir & Hill (2002)
Under the framework of Weir & Hill (2002), the expectations for first and second moments of the x’s are
Here expectation is over both repeated samples from the population and over evolutionary replicates of the populations. These expressions introduce the total inbreeding coefficient Fij, the within-population coancestries θi, and the between-population-pair coancestries θii′. The quantities pl are the overall, or ancestral, frequencies of the reference alleles if all study individuals can be traced back to a single reference population. This reference population could be common ancestors at a point in time of the past. The equal values for ℰ[xij1l xij2l] and ℰ[xijkl xij′k′l] require an assumption of random mating.
The coancestry coefficient θi refers to the ibd probability for a random pair of alleles in population i, and the pair of alleles can come from the same individual. The coancestry coefficient θii′ refers to the ibd probability for a random pair of alleles, one from population i and the other is from population i′. Note that we implicitly assume θi and θii′ are the same at each locus, and in practice θi and θii′ are actually the average inbreeding and coancestry coefficients over all L loci.
Now consider an individual perspective measures of population structure, i.e., a special case of Weir & Hill’s model where each population i has only one sampled individual (ni = 1) so j = 1 for each population. The assumption of random mating is relaxed, and the sample size n is also the number of populations r.
Therefore,
| (1) |
where , the average inbreeding coefficient among all study individuals, and , the average kinship coefficient among all study individuals. The individual perspective measures do not account for familial data and the relatedness of individuals is established from evolutionary history.
Each study individual is assigned to one population, thereby the genetic covariance matrix defined by Patterson et al. (2006) at the individual level can be expressed using an index i, :
| (2) |
The expected values of the numerator in Equation 2 is:
where (setting θii = θi).
When the number n of study individuals is large,
| (3) |
Eigen-decomposition in PCA
If we are interested in individual inbreeding coefficients (1 + θj)/2 (the coancestry of an individual with itself) and individual-pair coancestries θii′, the factors (1 − θT) and (θT − ψj − ψj′)/(1 − θT) in Equation 3 will confound the estimates when is used. This may explain why a large proportion of are negative, whereas the true θj and θjj′ are always between zero and one.
The Population Perspective
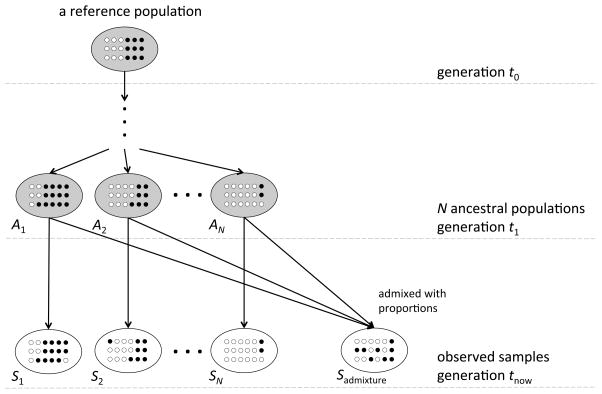
PCA conducts eigen-decomposition on the stochastic matrix 𝕄P, and it is possible to investigate the structural features of 𝕄P with its expectation. To illustrate what eigen-decomposition does, we introduce a genetic model consisting of populations at three points in time as shown in Figure 1. The alleles of all study individuals at tnow can be tracked to a single reference population at t0 through at least one of distinct ancestral populations at t1. The study samples S1, …, SN are directly inherited from the ancestral populations A1, .., AN without admixture, and the sample Sadmixture is admixed from N ancestral populations.
Figure 1.
A genetic model at a single locus for observed samples. The alleles of all study individuals at tnow can be tracked to a single reference population at t0, and there are N distinct ancestral populations at t1. The relationships among ancestral populations are described by a coancestry matrix ΘA.
What we can observe are the genomes of study individuals at tnow. It could be appropriate to assume there are N ancestral populations at t1 which is between t0 and tnow, and the samples S1, …, SN are good candidates (or pseudo-ancestors) to represent the ancestral populations. For example, in the initial phase of the HapMap Project, genetic data were gathered from four populations (CEU, YRI, CHB and JPT) with European, African and Asian ancestry respectively. Here, N = 3, S1 represents CEU individuals, S2 for YRI and S3 for CHB+JPT.
A coancestry matrix ΘA is used to describe the relationships among N ancestral populations at t1 based on population perspective measures, where
| (4) |
That is, is the average IBD probability for a pair of alleles randomly sampled with replacement from the hth ancestral population, and is the coancestry coefficient for random pairs of individuals from the hth and h′th ancestral populations respectively. Since we track all individuals back to the reference population at t0, the sample allele frequencies at t1 are treated as random variables over a probability space, which starts from the reference population at t0 and arrives at t1 with the coancestry state ΘA.
Ancestral Proportions
In practice individuals may have recent ancestors in more than one population, and an admixture model is introduced in which each individual is assumed to have inherited some proportion of its ancestry from each population. For an individual j, let the ancestral proportions be a vector aj = (ai,1, …, ai,N)T, where and 0 ≤ ai,h ≤ 1. Let Ziklh = 1 when the kth allele of individual i at SNP l is inherited from the hth ancestral population at t1, and Ziklh = 0 otherwise. The vector Zikl = {Zikl1, …, ZiklN}T is modeled as a random variable with probabilities ai, i.e., ℰ[Ziklh] = ai,h. Further,
| (5) |
represent the genomic ancestral proportions. Note that Equation 5 still holds even if loci are correlated due to linkage disequilibrium. We assume that the two alleles in individual i at SNP l are independently derived from ancestral populations, since pairs of chromosomes of an individual are independently inherited from two parents respectively. Then the expected value of the inbreeding coefficient at SNP l for individual i is , the same for each SNP. The average inbreeding coefficient over L loci is , assuming the coancestry matrix of ancestral populations is identical at each locus.
For a pair of individuals i and i′, we assume that any pair of alleles, one from i and the other from i′ are independently derived from ancestral populations. Then the expected value of the kinship coefficient at SNP l is , and the average kinship coefficient over L loci is also . This assumption is appropriate to model relatedness in structured population with admixture, with as background relatedness due to evolutionary history. However, the validity of the assumption could be violated if individuals i and i′ are in a family, e.g., parent and offspring.
Matrix Decomposition
For a study sample, there are n unrelated individuals. Each individual i has AP ai with respect to N ancestral populations. Let A = [a1, a2, …, an]T be a n-by-N matrix with rows representing ancestral proportions of individuals. Then the coancestry matrix of study individuals ΘS can be expressed as
| (6) |
We rewrite Equation 3 in matrix notation for large n,
| (7) |
where Jn is a matrix of dimension n × n with entries equal to one, since
The diagonal is considered as a bias term in the PCA with respect to ancestral proportions.
Note that because we lose a dimension by forcing each column to sum to zero. The eigenvectors corresponding to the largest N − 1 eigenvalues of ΘM form a new coordinate with N − 1 dimensions while AP form an old N-dimensional coordinates. The mapping from the old coordinate to the new one is a linear transformation, and the proof is given in the appendix A1. In addition, this mapping is actually an affine transformation equivalent to a (N − 1)-dimensional linear transformation followed by a translation, and the affine transformation can be represented as an linear transformation on the higher dimensional space.
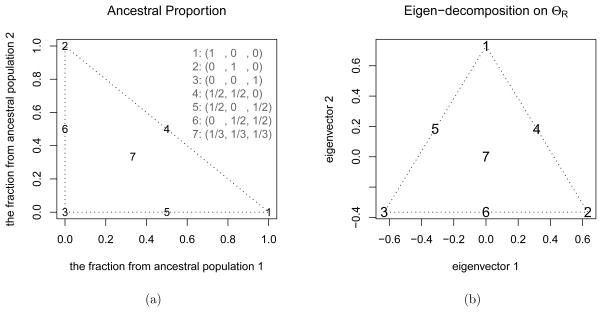
For example, assume there are three ancestral populations and seven individuals, in which individuals 1, 2, 3 are inherited from the ancestral populations without admixture, individuals 4, 5, 6 have two ancestral populations with equal contributions and individual 7 has three ancestral populations with equal contributions. The matrix A of ancestral proportions is
and ΘA is assumed to diag(0.05, 0.05, 0.05). The AP coordinates are shown in Figure 2a, and the new eigen-decomposition coordinates are shown in Figure 2b.
Figure 2.
The relationship between ancestral proportions and eigen-decomposition: a) seven admixture fractions from three ancestral populations are plotted in the figure; b) the first and second eigenvectors of matrix , where the ancestral coancestry matrix ΘA is assumed to diag(0.05, 0.05, 0.05), A is an n-by-N matrix with rows representing admixture proportions of individuals, n = 7 and N = 3. The mapping from the two-dimensional coordinate in (a) to that of (b) is a linear transformation followed by a translation.
EIGMIX – Inferring Ancestral Proportions
The mapping in Figure 2 suggests an approach to estimate ancestral proportions using the largest principal components. Let S1, …, SN be the observed surrogate samples for the ancestral populations, as shown in Figure 1. Now we look at the largest (N − 1) principal components, and identify each location of pseudoancestor i ∈ {1, …, N} in the eigen coordinates, by averaging the locations of the sample Si. So we have N positions in the eigen coordinates, which corresponds to N independent components in the AP coordinates. Then a linear transformation can be made to reverse the original mapping, i.e., the principal components of all study individuals are reversed to the AP coordinates by a linear transformation. In addition, the property of linear mapping makes the inferred ancestral proportions unique if N surrogate samples are specified and their locations in the eigen coordinates are distinct.
For example, the positions of individuals 1, 2 and 3 with ancestral proportions (1,0,0), (0,1,0) and (0,0,1) in the eigen coordinate of Figure 2(b) are denoted by es1, es2 and es3 respectively. Let T2×2 be a linear transformation and L be a translation operator. A transformation from the AP coordinates to the eigen coordinates is:
| (8) |
Therefore, L3×2 = [es3, es3, es3]′ (moving every point a constant distance) and T2×2 = [es1 − es3, es2 − es3]′.
The inverse transformation is:
| (9) |
where eadmix is an arbitrary point in the eigen coordinate.
Note that there is a bias term in the diagonal shown in Equation 7. A scheme for bias removal is to define a new genetic covariance matrix, the EIGMIX coancestry matrix , EIGMIX coancestry matrix:
| (10) |
Then ℰ[𝕄*] = ΘM/(1 − θT) without any bias when there are a large number of individuals. We have previously (Weir and Cockerham, 1984) suggested the simple modification of taking the ratios of the sums over loci of the numerators and denominators instead of averaging the ratios to reduce the variance, in part by reducing the impact of rare variants. Since the ratio of expected values is an approximation for the expected value of an ratio of two random variables, our modification tends to have an advantage of bias correction due to division compared to the original PCA.
In practice, the matrix 𝕄* of real data could have more than N − 1 significant eigenvalues when we assume the number of ancestral populations N to be a specific number (e.g., N = 3 for Europe, Asia and Africa). The largest N − 1 eigenvalues with their eigenvectors form a low-rank approximation of 𝕄* (a real symmetric matrix), which minimizes the Frobenius norm with respect to a n-by-n matrix M with rank(M) ≤ N − 1:
where 𝕄* − M = [mj,j′]n×n. The closest matrix to 𝕄* is , as measured in the Frobenius norm, where |λ1| ≥ |λ2| ≥ … ≥ |λn| are the eigenvalues of 𝕄* and ei is the eigenvector corresponding to λi, and
𝕄* is not necessarily a nonnegative definite matrix, i.e., its eigenvalues are not necessarily all nonnegative. Here “largest eigenvalues” refer to the absolute values of eigenvalues in descending order.
In addition, the estimates of EIGMIX given an arbitrary number of ancestral populations are not always bounded from 0 to 1, although we force the proportions to sum to one. If the inferred ancestral proportions lie much outside the range [0,1], signaling outliers, we could conclude that the assumption of N ancestral populations with their surrogates is not appropriate or that the SNP markers have no power to distinguish ancestral populations.
According to PCA, we might expect the eigen-decomposition of and ℰ[𝕄*] could result in similar eigenvectors corresponding to a few most significant eigenvalues when there are true structural feature in data, since the difference between and ℰ[𝕄*] depends only on the diagonal. The average difference per entry in the term of Frobenius norm becomes small when the total number of study individuals n is large:
A few largest eigenvalues and eigenvectors could capture the similar structure information of and ℰ[𝕄*]. Here, “similar” means similar relative positions in the eigen coordinates, since numerical calculation does not guarantee that the resulting eigenvectors will have the same absolute positions in the coordinate, e.g., if a vector v is an eigenvector then −v is also the eigenvector according to the same eigenvalue. A further numerical study is shown in the appendix A2.
Results
Materials
The Phase 3 HapMap data consist of SNP genotypes generated from 1,397 samples in total, collected using two platforms: the Illumina Human1M (by the Wellcome Trust Sanger Institute) and the Affymetrix SNP 6.0 (by the Broad Institute) (International HapMap 3 Consortium et al., 2010). Data from the two platforms have been merged for the release. The PLINK format of HapMap 3 data were downloaded from http://hapmap.ncbi.nlm.nih.gov/downloads/genotypes/hapmap3_r3/plink_format/. The consensus and polymorphic data set of 1198 founders were used in the study analyses, which include only SNPs that passed quality control in all populations, as shown in Table 1.
Table 1.
Summary of population samples in the eigenanalysis.
| Name | Population | # of samples |
|---|---|---|
| HapMap Phase III (1,198 founders): | ||
| ASW | African ancestry in Southwest USA | 53 |
| CEU | Utah residents with Northern and Western European ancestry from the CEPH collection | 112 |
| CHB | Han Chinese in Beijing, China | 137 |
| CHD | Chinese in Metropolitan Denver, Colorado | 109 |
| GIH | Gujarati Indians in Houston, Texas | 101 |
| JPT | Japanese in Tokyo, Japan | 113 |
| LWK | Luhya in Webuye, Kenya | 110 |
| MEX | Mexican ancestry in Los Angeles, California | 58 |
| MKK | Maasai in Kinyawa, Kenya | 156 |
| TSI | Toscani in Italia | 102 |
| YRI | Yoruba in Ibadan, Nigeria | 147 |
|
| ||
|
The Human Genome Diversity Panel (HGDP, 938 unrelated individuals):
| ||
| Africa | 101 | |
| Europe | 157 | |
| Middle East | 163 | |
| Central & South Asia | 199 | |
| East Asia | 228 | |
| Oceania | 26 | |
| America | 64 | |
The Human Genome Diversity Panel data consists of 1043 individuals from 51 populations over the world: sub-Saharan Africa, North Africa, Europe, the Middle East, Central & South Asia, East Asia, Oceania and the Americas (Cann et al., 2002). The study individuals were genotyped on the Illumina 650K platform, and the SNP data could be downloaded from http://www.hagsc.org/hgdp/files.html. The dataset contains a small number of relatives, and 938 individuals were remained in the analysis after filtering out first and second degree relatives of which were suggested by Rosenberg (2006).
To reduce potential effects of linkage disequilibrium, SNP pruning was conducted by randomly selecting autosomal SNPs for which each pair was at least as far apart as 200kb: 9,949 remaining SNPs for HapMap Phase 3 and 9,790 for HGDP. All analyses were performed on both of the pruned and full SNP sets, and the unbound estimates of ancestral proportion are reported. In the full sets, there are 1,423,833 and 644,258 autosomal SNPs for HapMap3 and HGDP respectively.
Analyses of HapMap Phase 3 Data
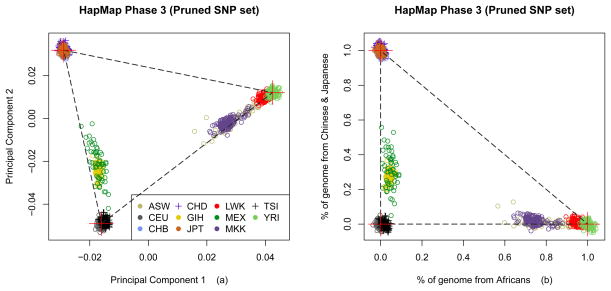
To avoid the confounding effect of relatives, 1,198 founders were selected for the PCA analysis by removing the offspring. The first two principal components are the focus, since more eigenvectors provide little additional information for inferring primary population structure. As shown in Figure 3a, the samples from CEU, YRI and CHB+JPT correspond to three vertices of a triangle, and the other populations tend to be admixtures from these three ancestries. Inferring ancestral proportions was conducted by a coordinate transformation, assuming three ancestral populations with surrogate samples: CEU, YRI and CHB+JPT. The X and Y axes in Figure 3b represent the proportions of genome from African and Asian ancestries respectively. Gujarati Indians in Houston (GIH, yellow) and Mexican ancestry in Los Angeles (MEX, green) appear to be admixtures between Europeans and Asians. ASW, MKK and LWK tend to be more related to African ancestry with some admixture, while CHD and TSI are quite close to the surrogate samples of Asia. The PCA plot with the largest two principal components generated by the full SNP set is shown in Supplemental Figure S1, which is similar to Figure 3.
Figure 3.
The principal component analysis on HapMap Phase 3 data, using a pruned set of 9,949 SNPs and 1,198 founders consisting of 11 populations: a) the first and second eigenvectors; b) a linear transformation of coordinate from a) followed by a translation, assuming three ancestral populations with surrogate samples: CEU, YRI and CHB+JPT. The average positions of three surrogate samples are masked by a red plus sign.
The population admixture proportions are estimated by averaging ancestral proportions of individuals using the full SNP set. African Americans (ASW) are a typically admixed sample, estimated with ~78% of genome from YRI and 21% from CEU, and approximately no genome from CHB+JPT. The result confirms the estimates of 78% African and 22% European ancestry shown in the supplementary materials of the HapMap Phase 3 report (International HapMap 3 Consortium et al., 2010). The HAPMIX algorithm (Price et al., 2009) was used in HapMap Phase 3 project, the optimal linear combination of 74% YRI and 26% CEU was observed for MKK, and a combination of 94% YRI and 6% CEU for LWK. In our analyses, the PCA-inferred combinations are 74% YRI + 24% CEU for MKK and 94% YRI + 5% CEU for LWK. Our results are consistent with the admixture proportions previously estimated.
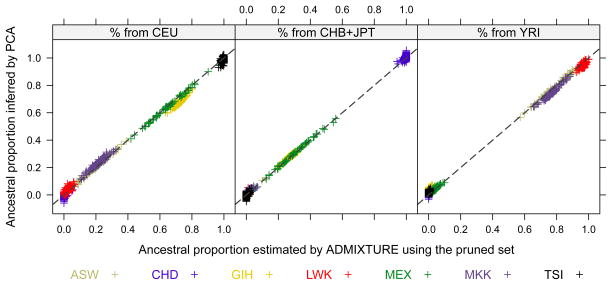
The supervised ADMIXTURE and EIGMIX methods were applied to the HapMap3 SNP data assuming three ancestral populations with surrogate samples CEU, YRI and CHB+JPT. ADMIXTURE is a model-based method with an assumption of markers in linkage equilibrium, therefore a pruned SNP set was used to avoid the strong influence of SNP clusters. The pseudo-ancestors (YRI, CHB+JPT and CEU) are specified in the analyses of ADMIXTURE according to the AP (1, 0, 0), (0, 1, 0) and (0, 0, 1). As shown in Figure 4, the AP inferred by PCA tend to be consistent with those estimated by ADMIXTURE using the same SNP set. However, the offsets are observed for admixed populations, such like GIH and MKK. The PCA-based proportions of genome from CEU are lower than ADMIXTURE for GIH, and those are higher for MKK. Actually, our inference on MKK was actually consistent with what HapMap Phase 3 has reported. Note that PCA is a dimension reduction technique and may lose information if we look only at the largest two principal components, and the assumption of pseudo-ancestors (CEU, YRI, CHB+JPT) might not truly represent the ancestors in human evolution.
Figure 4.
A comparison between PCA and supervised ADMIXTURE with respect to ancestral proportions for the HapMap Phase 3 data. A pruned set of 9,949 SNPs was used by both PCA and ADMIXTURE.
The EIGMIX coancestry matrix was used in the eigenanalysis instead of the PCA covariance matrix. As shown in Table 2, the differences of ancestral proportions at the individual level between ADMIXTURE and PCA/EIGMIX were calculated to evaluate the potential biases compared to the estimates of ADMIXTURE. The estimated proportions of EIGMIX tend to be less biased than PCA’s except Chinese in Metropolitan Denver (CHD), whereas the differences are relatively small overall for the HapMap3 data. The variances of EIGMIX are comparable to PCA if the ADMIXTURE estimates are assumed to be true values.
Table 2.
The differences on ancestral proportions of individuals between supervised ADMIXTURE and the eigenanalysis for the HapMap Phase 3 data.
| Pop. | PCA – ADMIXTURE | EIGMIX – ADMIXTURE | ||||
|---|---|---|---|---|---|---|
| mean ± sd
|
mean ± sd
|
|||||
| % CEU | % CHB+JPT | % YRI | % CEU | % CHB+JPT | % YRI | |
| ASW | 0.30 ± 0.75 | −0.29 ± 1.11 | −0.01 ± 0.90 | 0.18 ± 1.09 | −0.27 ± 1.24 | 0.09 ± 0.73 |
| CHD | −1.10 ± 1.65 | 1.09 ± 1.66 | 0.01 ± 1.15 | −1.16 ± 1.82 | 1.11 ± 1.70 | 0.05 ± 1.34 |
| GIH | −4.15 ± 0.85 | 0.42 ± 0.57 | 3.74 ± 0.87 | −3.39 ± 0.84 | −0.60 ± 0.74 | 3.98 ± 1.12 |
| LWK | 1.50 ± 1.33 | 0.24 ± 1.22 | −1.74 ± 1.25 | 0.86 ± 1.42 | −0.02 ± 1.35 | −0.84 ± 1.03 |
| MEX | −0.62 ± 0.70 | 0.23 ± 0.57 | 0.40 ± 0.75 | −0.31 ± 0.91 | 0.02 ± 0.83 | 0.29 ± 1.01 |
| MKK | 1.60 ± 0.85 | 0.61 ± 1.10 | −2.21 ± 0.77 | 0.72 ± 1.07 | 0.33 ± 1.03 | −1.05 ± 0.66 |
| TSI | −1.07 ± 1.74 | −0.78 ± 1.69 | 1.84 ± 1.12 | −0.85 ± 1.73 | −1.14 ± 1.80 | 1.99 ± 1.32 |
Analyses of HGDP
As suggested by Rosenberg (2006), a standardized subset of HGDP data consisting of 938 unrelated individuals was employed in the admixture analyses with a pruned set of 9,790 SNPs. The number of ancestral populations is suggested by geographic regions, the worldwide human relationship inference (Rosenberg et al., 2002; Li et al., 2008) and the plots of eigenvectors (shown in Supplementary Figure S2), and we used six ancestries in our primary analyses. The surrogate samples are suggested by the previous inferred regional ancestry (Li et al., 2008) and relative positions in the plots of eigenvectors: Sardinian for Europe (n = 28), Chinese Han for East Asia (n = 44), Kalash for Central & South Asia (n = 22), Pygmy for Africa (n = 34), Karitiana for America (n = 14) and Papuan for Oceania (n = 16).
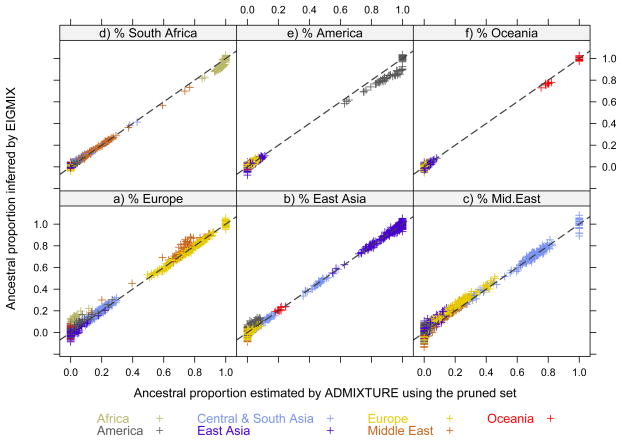
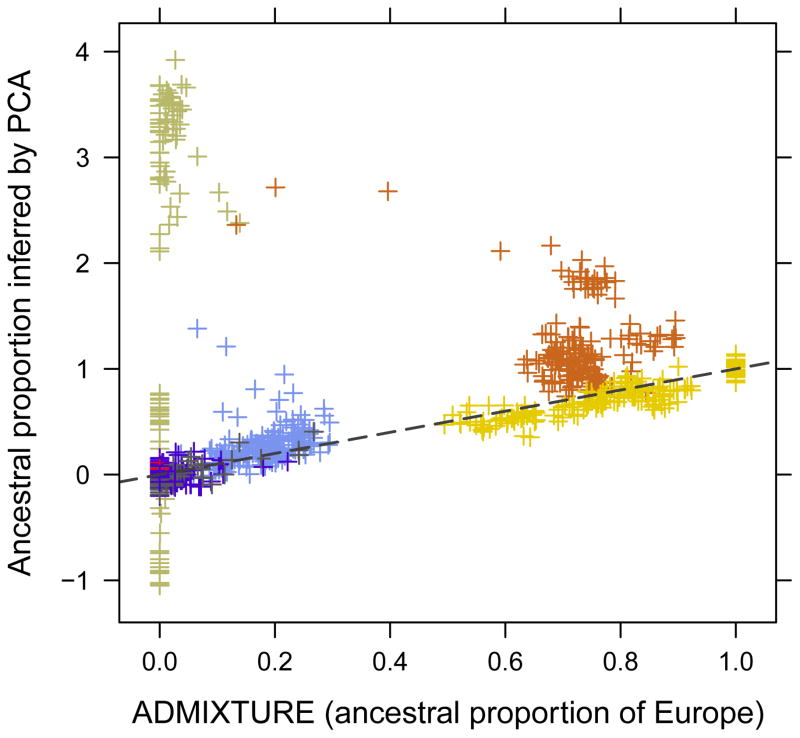
The supervised ADMIXTURE and EIGMIX methods were both applied to the HGDP SNP data with six ancestral populations. The estimated ancestral proportions of individuals are shown in Figure 5. Overall the estimates of EIGMIX are consistent with what ADMIXTURE does, however a difference of 10% admixture proportion is observed for samples from Africa and Middle East when the percents of Europe are inferred. In Figure 5e, the samples of America are also observed to be off the diagonal line. PCA was applied to the same study individuals and SNP set: the PCA-inferred admixed ancestries are shown in Figure 6 and Supplementary Figure S3. The PCA method is observed to have higher variance than EIGMIX, especially for the samples from Africa and Middle East. The variance reduction in EIGMIX is primarily due to the modification of taking the ratios of the sums over loci, rather than diagonal bias removal.
Figure 5.
A comparison of ancestral proportions between EIGMIX and supervised ADMIXTURE with 6 ancestral populations for the HGDP data. A pruned set of 9,790 SNPs was used by both EIGMIX and ADMIXTURE.
Figure 6.
A comparison of ancestral proportions between PCA and supervised ADMIXTURE with 6 ancestral populations with a pruned set of 9,790 SNPs. The color legend is as the same as Figure 5, and EIGMIX is more robust than PCA when inferring admixture fractions.
Discussion
In this study, we provide an interpretation of principal components analysis (PCA) based on relatedness measures, i.e., the probability that sets of genes are identical-by-descent. The expected values of pairwise estimates in the genetic covariance matrix of PCA are relative kinship coefficients with an additional term with respect to a single reference population in the past. An approximately linear transformation between ancestral proportions of individuals with multiple ancestries and their projections onto the principal components is revealed. A new method “EIGMIX” is proposed to estimate ancestries, allowing both linked and unlinked genetic markers regardless of linkage disequilibrium. The ancestral proportions can be estimated by making assumptions of surrogate ancestral samples. EIGMIX is a method of moments with high computational efficiency compared to existing MLE and Bayesian methods such like ADMIXTURE and STRUCTURE, and it is suitable to large-scale GWAS data with thousands of individuals and millions of SNPs. We applied the PCA, EIGMIX and supervised ADMIXTURE methods to the real SNP data from the HapMap Phase 3 project and the Human Genome Diversity Panel. The ancestral proportions inferred by PCA and EIGMIX are consistent with the findings of ADMIXTURE, but EIGMIX proportions are observed to be less biased and more robust than PCA.
Novembre et al. (2008) showed that SNP profiles of individuals within Europe can be used to infer their geographic origin with relatively high accuracy by PCA. The reason why the PC axes often represent perpendicular gradients in geographic space can be explained by ancestral proportions with two or more ancestries. In our genetic model (see Figure 1), the time t0 of single reference population is not specified explicitly, and it could be many generations ago – even the time before modern humans’ ancestors migrated out of Africa. The repeated migration in the history of Europe could create gene frequency clines as suggested by isolation-by-distance models (Wright, 1943). Starting from the single reference population at t0, such as the population at the time before humans migrated out of Africa, it would be possible to treat the observed alleles and the hidden pattern of ibd in the current generation as a sample from the probability space of a long-term evolutionary process. However, this strategy could be confounded by the unknown allele frequencies in the reference population. To avoid this problem, the derivation of the formulas in PCA and EIGMIX have removed explicit use of the allele frequencies.
Ma and Amos (2012) observed that a three-way admixed population could divide the triangle of parental populations in the PC plot into three small triangles with areas according to their admixture proportions. They also tried to extend this observation to the general case of more than three parental populations. A closed-form estimator of ancestral proportion is difficult to find so they solved the eigenequation numerically to confirm the observation. Our mathematical derivation of the linear transformation between ancestral proportions and eigenvectors can be used to confirm the observation of Ma and Amos (2012). Here, we adopt an three-way admixed example with four populations (P1, P2, P3 and P4) shown in Figure 5 of the paper of Ma and Amos (2012), where P4 is an admixed population. It is shown in Supplementary Figure S4. The mapping from the two-dimensional coordinate in Figure S4 (a) to that of (b) is an affine transformation. Sets of parallel lines remain parallel after an affine transformation, and it also preserves ratios of distances between points lying on a straight line. Therefore, the ratio of heights in the triangles remain the same. Ma and Amos’s observation can be confirmed theoretically under their framework with our linear transformation proof.
It is important to realize the potential limitations and our findings should be interpreted with caution. The assumption of ancestral populations used in inferring admixture fractions from the largest principal components could be confounded by the fact that human evolution is complex and has involved repeated migration and admixture from and out of Africa (Cavalli-Sforza and Feldman, 2003; Abi-Rached et al., 2011). Therefore, the selection of surrogate samples could be biased due to lack of historical knowledge or true unknown ancestries. For example, it is known that Mexicans have mainly Native Americans and European ancestry, with a small African contribution (Price et al., 2007). The ancestral proportions of MEX in HapMap Phase 3 data are confounded by an unknown link between Amerindians and CHB+JPT, although Amerindian seems closely related to Asian rather than European and African in genetics. Also, CHB+JPT and Native Americans represent two evolution branches from their common ancestors, and it may not be appropriate to assume a simple linear combination to reflect genetic difference in Native Americans.
The number of ancestral populations N is another important issue when we infer admixture proportions. A statistical test for how many significant eigenvalues in SNP data has been proposed, which is based on the approximate Tracy–Widom distribution (Patterson et al., 2006). The potential impacts on this test include linkage disequilibrium and categorical genetic data, since the Tracy–Widom distribution was originally developed for the case of independent Gaussian matrix entries. The MLE method for selecting N based on AIC (Akaike information criterion) and BIC (Bayesian information criterion) statistics was also introduced with ADMIXTURE (Alexander et al., 2009). However, we suggest that the choice of N should rely on the knowledge of the history of a population, with limited advice from statistical significance.
In summary, we provide a genetic interpretation of PCA, and propose EIGMIX to infer ancestral proportions with relatively high accuracy. EIGMIX could help us better understand population structure for isolated and admixed populations.
Supplementary Material
Acknowledgments
We appreciate the input from W.G. Hill. This work was supported in part by NIH grants GM 075091 and GM 099568.
Appendix
A1 Proof of Eigen-decomposition
Here, we perform eigen-decomposition on in Equation 7, and the mapping from A to the eigenvectors of ΘM is a linear transformation, where A is a n-by-N matrix with rows representing ancestral proportions of individuals and ΘA is a N-by-N coancestry matrix. Let , where In is an identity matrix and Jn is a matrix n × n with entries equal to one, then ΘM = Y ΘAYT.
Proof
Note that ΘM and ΘA are not necessarily non-negative definite matrices, and some of the eigenvalues could be negative. To avoid a complex matrix, we perform eigen-decomposition on , since and ΘM have the same eigenvectors and the square of eigenvalues of ΘM correspond to the eigenvalues of .
Note that rank(Y) ≤ N − 1, then rank(ΘM) ≤ N − 1. Let the eigenvalues of ΘM be |v1| ≥ |v2| ≥ … ≥ |vN−1| ≥ |vN| = … = |vn| = 0, and Q(M),i be the ith eigenvector with respect to vi. [Q(M),1, …, Q(M),n] forms an orthogonal matrix.
| (11) |
We perform singular value decomposition on Y,
Since rank(Y) ≤ N − 1, at least one of the singular values of Y is ZERO. Replace Y in Equation 11 by :
where forms an N × N diagonal matrix.
Let , where . SVD on . Again, at least one of the singular values of Z is ZERO.
Since
UZ is the eigenvector matrix of , i.e., [Q(M),1, …, Q(M),n] = UZ and the eigenvalue |vi| is the singular value of ZY (non-negative).
Note that
or,
| (12) |
The left hand side of Equation 12 is an n×N matrix where the last column is ZERO since vN = 0, where as the right hand side is the AP matrix times ( ) and . Note that this transformation matrix is a function of A. Given an AP matrix A, the transform matrix is determined, so each data point (ancestral proportion) in A maps to a new coordinate by a linear transformation.
A2 Numerical Evaluation of Diagonal Bias in PCA
To demonstrate the similarity of relative positions in the eigen coordinates of and ℰ[𝕄*], two pseudo-ancestor populations (N = 2) and three admixed populations (admixture fractions 25%, 50%, 75%) with equal sample sizes were utilized here. As shown in Table A1, as the sample size of each population grows, the bias for estimating the true admixture fraction 25% and 75% declines from 0.0424 to 0.0004. Another example is a spatially continuous admixed population, i.e., individuals with ancestral proportions uniformly distributed from 0 to 1. E.g, if n = 11 is the total number of study individuals, there are 11 individuals with admixture fractions of 0%, 10%, 20%, …, 90% and 100%. The maximum bias of the estimated ancestral proportions is shown in Table A2, and it decreases from 0.02270 to 0.00057 as the total number of individuals n increases.
Table A1.
The bias of estimating population admixture proportions in the example of two ancestral populations and three admixed populations with equal sample size npop.
| True ancestral proportion | 0 | 0.25 | 0.5 | 0.75 | 1 |
|---|---|---|---|---|---|
| Inferred population ancestral proportion from 1: | |||||
| npop = 1 | 0 | 0.20758 | 0.50000 | 0.79242 | 1 |
| npop = 25 | 0 | 0.24849 | 0.50000 | 0.75151 | 1 |
| npop = 50 | 0 | 0.24925 | 0.50000 | 0.75075 | 1 |
| npop = 100 | 0 | 0.24962 | 0.50000 | 0.75038 | 1 |
calculated by averaging admixture proportion of individuals.
Table A2.
The bias of estimating ancestral proportions in the example of a spatially continuous admixed population with n individuals in total1.
| # of individuals n | 11 | 51 | 101 | 251 | 501 |
|---|---|---|---|---|---|
| The maximum bias of inferred ancestral proportions of individuals from | 0.02270 | 0.00548 | 0.00281 | 0.00114 | 0.00057 |
ancestral proportions are uniformly distributed from 0 to 1 derived from two ancestral populations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, et al. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 2011;334:89–94. doi: 10.1126/science.1209202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, Legrand MF, Morel V, et al. A human genome diversity cell line panel. Science (New York, NY) 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L, Feldman M. The application of molecular genetic approaches to the study of human evolution. Nature Genetics. 2003;33:266–275. doi: 10.1038/ng1113. [DOI] [PubMed] [Google Scholar]
- Churchhouse C, Marchini J. Multiway admixture deconvolution using phased or unphased ancestral panels. Genetic epidemiology. 2013;37:1–12. doi: 10.1002/gepi.21692. [DOI] [PubMed] [Google Scholar]
- Engelhardt BE, Stephens M. Analysis of population structure: a unifying framework and novel methods based on sparse factor analysis. PLoS genetics. 2010;6:e1001117. doi: 10.1371/journal.pgen.1001117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates: disease associations and individual risk of diabetes and gallbladder disease among Mexican-Americans in Starr County, Texas. Am J Phys Anthropol. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- International HapMap 3 Consortium. Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science (New York, NY) 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Ma J, Amos CI. Theoretical formulation of principal components analysis to detect and correct for population stratification. PloS one. 2010;5 doi: 10.1371/journal.pone.0012510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Amos CI. Principal components analysis of population admixture. PloS one. 2012;7:e40115. doi: 10.1371/journal.pone.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McVean G. A genealogical interpretation of principal components analysis. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menozzi P, Piazza A, Cavalli-Sforza L. Synthetic maps of human gene frequencies in Europeans. Science. 1978;201:786–792. doi: 10.1126/science.356262. [DOI] [PubMed] [Google Scholar]
- Novembre J, Johnson T, Bryc K, Kutalik Z, Boyko AR, et al. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J, Stephens M. Interpreting principal component analyses of spatial population genetic variation. Nat Genet. 2008;40:646–649. doi: 10.1038/ng.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson N, Yu F, Cox DR, Waliszewska A, et al. A genome-wide admixture map for Latino populations. Am J Hum Genet. 2007;80:1024–1036. doi: 10.1086/518313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Rev Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the hgdp-ceph human genome diversity cell line panel, accounting for atypical and duplicated samples and pairs of close relatives. Annals of human genetics. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: analytical and study design considerations. Genet Epidemiol. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- Thompson EA. Identity by descent: variation in meiosis, across genomes, and in populations. Genetics. 2013;194:301–326. doi: 10.1534/genetics.112.148825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS, Anderson AD, Hepler AB. Genetic relatedness analysis: modern data and new challenges. Nat Rev Genet. 2006;7:771–780. doi: 10.1038/nrg1960. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–750. doi: 10.1146/annurev.genet.36.050802.093940. [DOI] [PubMed] [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;2:114–38. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, et al. A high-performance computing toolset for relatedness and principal component analysis of snp data. Bioinformatics (Oxford, England) 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.