Abstract
The human intestinal microbiome plays a major role in human health and diseases, including colorectal cancer. Colorectal carcinogenesis represents a heterogeneous process with a differing set of somatic molecular alterations, influenced by diet, environmental and microbial exposures, and host immunity. Fusobacterium species are part of the human oral and intestinal microbiota. Metagenomic analyses have shown an enrichment of Fusobacterium nucleatum (F. nucleatum) in colorectal carcinoma tissue. Using 511 colorectal carcinomas from Japanese patients, we assessed the presence of F. nucleatum. Our results showed that the frequency of F. nucleatum positivity in the Japanese colorectal cancer was 8.6% (44/511), which was lower than that in United States cohort studies (13%). Similar to the United States studies, F. nucleatum positivity in Japanese colorectal cancers was significantly associated with microsatellite instability (MSI)-high status. Regarding the immune response in colorectal cancer, high levels of infiltrating T-cell subsets (i.e., CD3+, CD8+, CD45RO+, and FOXP3+ cells) have been associated with better patient prognosis. There is also evidence to indicate that molecular features of colorectal cancer, especially MSI, influence T-cell-mediated adaptive immunity. Concerning the association between the gut microbiome and immunity, F. nucleatum has been shown to expand myeloid-derived immune cells, which inhibit T-cell proliferation and induce T-cell apoptosis in colorectal cancer. This finding indicates that F. nucleatum possesses immunosuppressive activities by inhibiting human T-cell responses. Certain microRNAs are induced during the macrophage inflammatory response and have the ability to regulate host-cell responses to pathogens. MicroRNA-21 increases the levels of IL-10 and prostaglandin E2, which suppress antitumor T-cell-mediated adaptive immunity through the inhibition of the antigen-presenting capacities of dendritic cells and T-cell proliferation in colorectal cancer cells. Thus, emerging evidence may provide insights for strategies to target microbiota, immune cells and tumor molecular alterations for colorectal cancer prevention and treatment. Further investigation is needed to clarify the association of Fusobacterium with T-cells and microRNA expressions in colorectal cancer.
Keywords: BRAF, CpG island methylator phenotype, Colon neoplasia, Fusobacterium species, miR-21
Core tip: The human intestinal microbiome plays a major role in human health and diseases, including colorectal cancer. Metagenomic analyses have shown an enrichment of Fusobacterium nucleatum (F. nucleatum) in colorectal carcinoma tissue. Our results showed that the frequency of F. nucleatum positivity in Japanese colorectal cancer was 8.6%, which was lower than that in United States cohort studies (13%). F. nucleatum positivity was significantly associated with microsatellite instability-high status. Additionally, F. nucleatum possesses immunosuppressive activities by inhibiting T-cell responses. Thus, emerging evidence may provide insights for strategies to target microbiota, immune cells, and molecular alterations for colorectal cancer prevention and treatment.
INTRODUCTION
The human intestinal microbiome encompasses at least 100 trillion (1014) microorganisms and plays a major role in human health and diseases, including colorectal cancer[1-3]. Colorectal carcinogenesis represents a heterogeneous process with a differing set of somatic molecular alterations, influenced by diet, environmental and microbial exposures, and host immunity[4,5]. Fusobacterium species (a group of non-spore-forming, anaerobic gram-negative bacteria) are a part of the normal human oral and intestinal microbiota. The species of the Fusobacterium genera are highly heterogeneous, and some of them have been recognized as opportunistic pathogens implicated not only in periodontitis[6-8] but also in inflammatory bowel disease (IBD)[9-11], pancreatic abscess[12,13], and hepatic abscess[12-15]. Regarding gastrointestinal cancer, metagenomic analyses have shown an enrichment of Fusobacterium nucleatum (F. nucleatum) in colorectal carcinoma tissue, which has been confirmed by quantitative PCR for the 16S ribosomal RNA gene DNA sequence of F. nucleatum[16,17]. Studies have shown that a greater amount of F. nucleatum in colorectal carcinoma tissue is associated with high degrees of microsatellite instability (MSI-high) and CpG island methylator phenotype (CIMP)[18].
Accumulating evidence indicates that innate and adaptive immunity influences tumor evolution[19]. Attesting to an important role of T-cell-mediated adaptive immunity in inhibiting tumor progression, therapeutic antibodies against immune checkpoint molecules, including CTLA4, PDCD1 (programmed cell death 1; PD-1), and CD274 (programmed cell death 1 ligand 1; PD-L1), can effectively enhance antitumor T-cell activity in various malignancies[20,21]. Emerging evidence indicates that tumor genetic alterations and tumor-host interactions have complex roles in the effectiveness of T-cell-based immunotherapies[22-25]. Although these immunotherapies appeared to be less effective for colorectal cancer, high-level infiltrates of T-cells in colorectal cancer tissue have been associated with better patient survival[26-28], and a recent study has suggested a potential role for the immune checkpoint pathway in suppressing the antitumor immune response in a subset of colorectal cancers[29].
Regarding the association between the gut microbiome and immunity, a number of studies have shown that F. nucleatum has immunosuppressive activities via inhibiting human T-cell responses to mitogens and antigens[30-35]. Additionally, F. nucleatum inhibitory protein has been shown to arrest human T-cells in the G1 phase of the cell cycle[33]. Furthermore, F. nucleatum can induce apoptotic cell death in peripheral blood mononuclear cells and Jurkat T-cells[31]. This F. nucleatum-induced cell death is mediated through the aggregation of the immune cells, which might have important implications for the pathogenesis of this bacterial species[35]. These findings indicate that F. nucleatum suppressively modulates the tumor-immune microenvironment.
Thus, the results of these studies suggest a complex link between the gut microbiome, immunity, and molecular alterations in colorectal tumorigenesis. A better understanding of the relationship between microorganisms and immune cells in the tumor microenvironment is needed in order to effectively target the microbiota and immunity for colorectal cancer prevention and therapy.
AMOUNT OF F. NUCLEATUM IN COLORECTAL CARCINOMA TISSUE AND THE ASSOCIATION WITH MSI
Using quantitative PCR, Mima et al[36] have reported that F. nucleatum was detected in 76 (13%) of 598 colorectal carcinomas (stages I-IV) within the well-known United States cohort studies (the Nurses’ Health Study and the Health Professionals Follow-up Study) and in adjacent non-tumor tissue in 19 (3.4%) of 558 cases analyzed. In the 558 pairs of colorectal carcinoma and adjacent non-tumor tissues, the amount of F. nucleatum was higher in colorectal carcinoma tissue than in paired adjacent non-tumor tissue[36].
We also collected 511 colorectal carcinoma tissues (stages I-IV) from Japanese patients who underwent endoscopic resection or other surgical treatment and assessed the presence of F. nucleatum via gene expression analysis. Genomic DNA was extracted from formalin-fixed paraffin-embedded (FFPE) tissues. The amount of F. nucleatum in colorectal carcinoma tissue was measured by quantitative PCR assay as previously described[36]. Considering the influence of contaminating stromal cells, we performed microdissection only in cases with F. nucleatum positivity and conducted quantitative PCR again using the DNA extracted from the carcinoma component. Our current data demonstrated that F. nucleatum positivity was detected in 44 (8.6%) of the 511 Japanese patients with colorectal cancer (Table 1). The frequency of F. nucleatum positivity in the Japanese patients was significantly lower than that in the United States cohort study[36].
Table 1.
Clinical and molecular features in Japanese patients with colorectal cancer according to Fusobacterium nucleatum status n (%)
| Clinical and molecular features | Total (n) |
F. nucleatum expression |
||
| Negative | Positive | P-value | ||
| All cases | 511 | 467 (91) | 44 (8.6) | |
| Gender | ||||
| Male | 286 (56) | 267 (57) | 19 (43) | 0.075 |
| Female | 225 (44) | 200 (43) | 25 (57) | |
| Age (mean ± SD) | 67.1 ± 11.8 | 67.3 ± 11.7 | 65.0 ± 12.1 | 0.220 |
| Tumor size (mm) (mean ± SD) | 49.8 ± 24.1 | 49.2 ± 24.5 | 56.4 ± 19.6 | 0.063 |
| Tumor location | ||||
| Rectum | 207 (41) | 194 (42) | 13 (30) | |
| Distal colon | 133 (26) | 121 (26) | 12 (27) | 0.240 |
| (Sigmoid colon to splenic flexure) | ||||
| Proximal colon | 171 (33) | 152 (33) | 19 (43) | |
| (Transverse colon to cecum) | ||||
| Disease stage | ||||
| I | 56 (11) | 53 (11) | 3 (6.8) | 0.470 |
| II | 160 (31) | 142 (30) | 18 (41) | |
| III | 235 (46) | 216 (46) | 19 (43) | |
| IV | 60 (12) | 56 (12) | 4 (9.0) | |
| KRAS mutation (codon 12/13/61/146) | ||||
| Wild-type | 354 (69) | 324 (69) | 30 (68) | 0.870 |
| Mutant | 157 (31) | 143 (31) | 14 (32) | |
| BRAF mutation (codon 600) | ||||
| Wild-type | 483 (95) | 445 (95) | 38 (86) | 0.031 |
| Mutant | 28 (5.5) | 22 (4.7) | 6 (14) | |
| PIK3CA mutation (exon 9/20) | ||||
| Wild-type | 451 (88) | 414 (89) | 37 (84) | 0.390 |
| Mutant | 60 (12) | 53 (11) | 7 (16) | |
| MSI status | ||||
| MSS/MSI-low | 470 (92) | 435 (93) | 35 (80) | 0.0059 |
| MSI-high | 41 (8.0) | 32 (6.9) | 9 (20) | |
Percentage (%) indicates the proportion of cases with a specific clinical or molecular feature within a given category of F. nucleatum detection by quantitative PCR. The P-values were calculated using t-test for age and tumor size and by means of the χ2 test or Fisher’s exact test for all other variables. F. nucleatum: Fusobacterium nucleatum; MSI: Microsatellite instability; MSS: Microsatellite stable.
Some cohort studies observed associations of highly enriched Fusobacterium in colorectal cancer tissues with CIMP-high, MSI-high, and MLH1 methylation in patients with colorectal cancer[18,36,37]. Consistent with these reports, our current data using Japanese populations showed that high expression of F. nucleatum in colorectal cancers was significantly associated with MSI-high status (Table 1). We also examined the relationship between the amount of F. nucleatum and patient mortality; however F. nucleatum status in colorectal cancers was not associated with cancer-specific survival. The role of F. nucleatum in colorectal carcinogenesis remains uncertain. Recent studies showed that F. nucleatum increases the production of reactive oxygen species (ROS) and inflammatory cytokines (e.g., IL-6 and TNF) in colorectal cancer[38]. Inflammation and ROS can reduce the enzymatic activity of mismatch repair (MMR) proteins and cause epigenetic silencing of the mismatch repair protein MLH1 leading to MSI[39].
ASSOCIATION BETWEEN IMMUNE CELL AND CLINICAL OUTCOME IN COLORECTAL CANCER
The abundance of tumor-infiltrating T-cells has been associated with improved clinical outcomes in colorectal cancer patients[28,40]. Although the exact mechanism remains uncertain, the adaptive immune system may play an important role in suppressing tumor progression[27,41]. Tumor-infiltrating T-cells may be an indicator of a host immune response to tumors and are attractive targets for immunotherapy[42-45]. Tumor-infiltrating lymphocytes may also reflect specific molecular alterations associated with indolent tumor behavior. Previous studies have shown that lymphocytic infiltration is associated with MSI in colorectal cancer[40,46-48]. Truncated peptides produced by frameshift mutations due to MSI may be immunogenic and contribute to host immune response[41,43,49]. However, little is known about the relationship between tumor-infiltrating T-cells and other tumor molecular features, including the CIMP status, and KRAS, BRAF and PIK3CA mutations.
We previously utilized a database of clinically and molecularly annotated colorectal carcinoma cases (n = 768; stages I-IV) in the United States cohort studies[28]. Using tissue microarray and automated Ariol image analysis system, we quantified densities of CD3+, CD8+, CD45RO+, and FOXP3+ T-cells within neoplastic epithelial areas. Our data demonstrated that tumor-infiltrating CD45RO+ T-cell density is significantly associated with longer survival of colorectal cancer patients, independent of clinical, pathological, and molecular features (i.e., MSI, CIMP, and KRAS, BRAF and PIK3CA mutations). In addition, MSI-high is an independent predictor of CD45RO+ T-cell density. The strong association between MSI and CD45RO+ T-cell density supports the hypothesis that truncated peptides produced by MSI and frameshift mutations may elicit a host immune response and recruit CD45RO+ T-cells[41,49].
In most studies, MSI in colon cancer has been associated with improved survival[27,41,50,51], although the mechanism underlying this association is largely unknown. Similar to these reports from United States and Western countries[27,41,50,51], our current Japanese population-based study showed a significantly lower mortality rate (log-rank test: P = 0.048) in the MSI-high group than in the MSS/MSI-low group using the Kaplan-Meier method (data not shown). These results suggest one explanation that a host immune is stimulated in response to MSI-high colorectal cancer.
ASSOCIATION BETWEEN F. NUCLEATUM AND IMMUNE RESPONSE IN COLORECTAL CANCER
Myeloid-derived immune cells can inhibit T-cell proliferation and induce T-cell apoptosis[52]. Recently, Kostic et al[38] reported that F. nucleatum selectively expands myeloid-derived immune cells in colorectal cancer. In particular, myeloid-derived immune cells were enriched in F. nucleatum-fed mice vs controls. Myeloid-derived immune cells have been proposed to be myeloid cells present in the bone marrow, spleen, or tumor microenvironment that are able to suppress T-cell responses[53]. During tumor progression, reactive myeloid cells might mediate immunosuppression either by the self-limiting mechanism of T helper type (Th)1 inflammation resolution, such as ROS and IL-10 production, or by switching to a wound repair and angiogenic protumor Th2 inflammation with the expression of arginase, TGF-b, and IL-10[54]. These results indicate that F. nucleatum suppressively modulates the tumor-immune microenvironment because T-cell-mediated adaptive immunity plays an important role in preventing the development of tumors and inhibiting tumor progression[55]. Thus, immunosuppression by F. nucleatum may affect patient mortality in colorectal cancer. Additionally, the data in the United States cohort studies along with these lines of experimental evidence revealed that the amount of tissue F. nucleatum is inversely associated with CD3+ T-cell density in colorectal carcinoma tissue[36].
ASSOCIATION BETWEEN MICRORNA EXPRESSION AND IMMUNITY IN COLORECTAL CANCER
MicroRNAs constitute a class of small non-coding RNA molecules that function as post-transcriptional gene regulators and have been increasingly recognized as biomarkers of various human cancers[56-71]. Regarding colorectal cancer, we recently discovered that microRNA-31 (miR-31) expression is significantly up-regulated in BRAF-mutated cancers compared with that in wild-type cancers using microRNA array analysis[67]. Moreover, associations were identified between miR-31 expression and poor prognosis for colorectal cancers.
Certain microRNAs are induced during the macrophage inflammatory response and have the ability to regulate host-cell responses to pathogens[72]. In addition, pathogens themselves may regulate microRNA expression[73]. MicroRNAs influence networks that control innate and adaptive immunity and apoptosis by regulating signalling pathways[71,72]. Among the various microRNAs, microRNA-21 (miR-21) has been shown to play roles in immunity and colorectal carcinogenesis[74-76]. In fact, high-level miR-21 expression in colorectal cancer tissue has been associated with worse clinical outcome, suggesting that miR-21 could act as a prognostic tumor biomarker[77,78]. Studies have shown that miR-21 increases the levels of IL-10 and prostaglandin E2 (PGE2) in colorectal cancer cells[78-82]. IL-10 and PGE2 have been shown to suppress antitumor T-cell-mediated adaptive immunity through the inhibition of the antigen-presenting capacities of dendritic cells and T-cell proliferation and through the recruitment of myeloid-derived suppressor cells into the tumor microenvironment[83-86].
CONCLUSION
The association between highly enriched F. nucleatum in colorectal carcinoma tissues and MSI-high status was observed in both the United States cohort studies and Japanese population-based study. Previous studies have reported that the frequency of colorectal cancers with MSI-high status in Japan (less than 10%)[63,67,87] tend to be lower than those in the United States and Western countries (approximately 15%)[28,88-91]. Therefore, the low rate of MSI-high colorectal cancer in Japan might be due to the amount of F. nucleatum in carcinoma tissues because our current data showed that the rate of F. nucleatum positivity in Japanese patients was significantly lower than that in the United States cohorts. MSI-high status in colorectal cancer has been associated with high levels of infiltrating T-cells, as mismatch repair defects in MSI-high tumors cause numerous frameshift mutations and truncated proteins, which elicit antitumor T-cell-mediated adaptive immunity[40,46,49,92]. However, MSI status is not the sole determinant of the immune response to colorectal cancer because the amounts of tumor-infiltrating T-cells considerably overlap between MSI-high and microsatellite stable (MSS) colorectal tumors[28,29,40]. Hence, there must be other factors that influence the antitumor immune response to colorectal cancer.
T-cell-mediated adaptive immunity plays an important role in regulating tumor evolution and in inhibiting tumor progression[55]. The immunity includes multiple steps involving the clonal selection of antigen-specific cells, their activation and proliferation in secondary lymphoid tissues, and their recruitment into the tumor microenvironment[93]. In a mouse model, F. nucleatum recruits myeloid-derived suppressor cells into the tumor microenvironment[38]. Myeloid-derived suppressor cells can inhibit T-cell proliferation and induce T-cell apoptosis[52]. Virulence factors derived from F. nucleatum also inhibit T-cell proliferation[33,94]. The experimental evidence may be consistent with a recent finding that a higher abundance of F. nucleatum in colorectal carcinoma tissue was associated with a lower density of T-cells, as measured by CD3 in the tumor microenvironment[36]. These findings support a role of F. nucleatum in down-regulating antitumor T-cell-mediated adaptive immunity.
Both tumor molecular and immunity analyses are increasingly important in cancer research and clinical practice. MicroRNAs play roles in carcinogenesis and immunity and can be potential biomarkers or therapeutic targets. MicroRNA-targeting therapies for human disease, including cancer, are currently being investigated[69,95,96]. Accumulating evidence suggest miR-21 increases the levels of IL-10 and PGE2 in the tumor microenvironment, which can lead to the suppression of antitumor T-cell-mediated adaptive immunity[84-86]. In light of these findings, it would be intriguing for future research to explore a potential strategy for inhibiting miR-21 and its immunosuppressive effect in immunotherapy and immunoprevention for colorectal cancer. In contrast, no study has reported whether F. nucleatum regulates microRNA expressions, including miR-21. Therefore, functional analysis and/or human population-based study are expected to identify the association between F. nucleatum and miR-21 expression in colorectal cancer.
We have summarized the hypothesis of the potential mechanism underlying the association of F. nucleatum in colorectal cancer with immune cells and molecular alterations in Figure 1. F. nucleatum increases the production of ROS and inflammatory cytokines in colorectal cancer. Inflammation and ROS can cause epigenetic silencing of the mismatch repair protein MLH1 leading to MSI. F. nucleatum possesses immunosuppressive activities by inhibiting human T-cell responses and modulates tumor-immune microenvironment suppressively. miR-21 increases the levels of IL-10 and PGE2, which suppress antitumor T-cell-mediated adaptive immunity in the tumor microenvironment.
Figure 1.
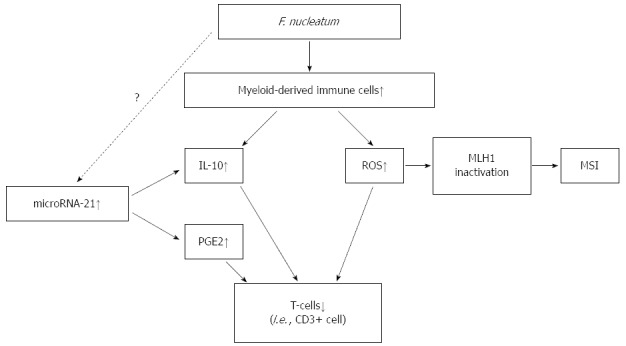
Hypothesis of potential mechanism about the association of Fusobacterium nucleatum in colorectal cancer with immune cells and molecular alterations. F. nucleatum increases the production of ROS and inflammatory cytokines in colorectal cancer. Inflammation and ROS can cause epigenetic silencing of the mismatch repair protein MLH1 leading to MSI. F. nucleatum possesses immunosuppressive activities by inhibiting human T-cell responses and modulates tumor-immune microenvironment suppressively. MicroRNA-21 increases the levels of IL-10 and PGE2, which suppress antitumor T-cell-mediated adaptive immunity in the tumor microenvironment. F. nucleatum: Fusobacterium nucleatum; MSI: Microsatellite instability; PGE2: Prostaglandin E2; ROS: Reactive oxygen species.
Thus, emerging evidence may provide insights for strategies to target microbiota, immune cells, and tumor molecular alterations for colorectal cancer prevention and treatment. Further investigation is needed to clarify the association of Fusobacterium with T-cells and microRNA expressions in colorectal cancer.
Footnotes
Supported by Japanese Society of Gastroenterology Research Foundation (to Nosho K); Pancreas Research Foundation of Japan (to Nosho K); Medical Research Encouragement Prize of The Japan Medical Association (to Nosho K); The Japan Society for the Promotion of Science Challenging Exploratory Research, grant No. 25670371 (to Shinomura Y); and Ono Cancer Research Foundation (to Ito M).
Conflict-of-interest statement: Authors declare no conflict of interests for this article.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: July 4, 2015
First decision: September 11, 2015
Article in press: November 13, 2015
P- Reviewer: Soucek P, Tsimogiannis K, Wang JY S- Editor: Ma YJ L- Editor: A E- Editor: Liu XM
References
- 1.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13:800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dickson RP, Martinez FJ, Huffnagle GB. The role of the microbiome in exacerbations of chronic lung diseases. Lancet. 2014;384:691–702. doi: 10.1016/S0140-6736(14)61136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kahn SE, Cooper ME, Del Prato S. Pathophysiology and treatment of type 2 diabetes: perspectives on the past, present, and future. Lancet. 2014;383:1068–1083. doi: 10.1016/S0140-6736(13)62154-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology--analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tjalsma H, Boleij A, Marchesi JR, Dutilh BE. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol. 2012;10:575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 6.Signat B, Roques C, Poulet P, Duffaut D. Fusobacterium nucleatum in periodontal health and disease. Curr Issues Mol Biol. 2011;13:25–36. [PubMed] [Google Scholar]
- 7.Michaud DS, Izard J, Wilhelm-Benartzi CS, You DH, Grote VA, Tjønneland A, Dahm CC, Overvad K, Jenab M, Fedirko V, et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut. 2013;62:1764–1770. doi: 10.1136/gutjnl-2012-303006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaud DS. Role of bacterial infections in pancreatic cancer. Carcinogenesis. 2013;34:2193–2197. doi: 10.1093/carcin/bgt249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohkusa T, Okayasu I, Ogihara T, Morita K, Ogawa M, Sato N. Induction of experimental ulcerative colitis by Fusobacterium varium isolated from colonic mucosa of patients with ulcerative colitis. Gut. 2003;52:79–83. doi: 10.1136/gut.52.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Minami M, Ando T, Okamoto A, Sasaki N, Ohkura T, Torii K, Hasegawa T, Ohta M, Goto H. Seroprevalence of Fusobacterium varium in ulcerative colitis patients in Japan. FEMS Immunol Med Microbiol. 2009;56:67–72. doi: 10.1111/j.1574-695X.2009.00550.x. [DOI] [PubMed] [Google Scholar]
- 11.Strauss J, Kaplan GG, Beck PL, Rioux K, Panaccione R, Devinney R, Lynch T, Allen-Vercoe E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 12.Shahani L, Khardori N. Fusobacterium necrophorum--beyond Lemierres syndrome. BMJ Case Rep. 2011;2011 doi: 10.1136/bcr.07.2011.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brook I, Frazier EH. Microbiological analysis of pancreatic abscess. Clin Infect Dis. 1996;22:384–385. doi: 10.1093/clinids/22.2.384. [DOI] [PubMed] [Google Scholar]
- 14.Yoneda M, Kato S, Mawatari H, Kirikoshi H, Imajo K, Fujita K, Endo H, Takahashi H, Inamori M, Kobayashi N, et al. Liver abscess caused by periodontal bacterial infection with Fusobacterium necrophorum. Hepatol Res. 2011;41:194–196. doi: 10.1111/j.1872-034X.2010.00748.x. [DOI] [PubMed] [Google Scholar]
- 15.Athavale NV, Leitch DG, Cowling P. Liver abscesses due to Fusobacterium spp that mimick malignant metastatic liver disease. Eur J Clin Microbiol Infect Dis. 2002;21:884–886. doi: 10.1007/s10096-002-0844-8. [DOI] [PubMed] [Google Scholar]
- 16.Kostic AD, Gevers D, Pedamallu CS, Michaud M, Duke F, Earl AM, Ojesina AI, Jung J, Bass AJ, Tabernero J, et al. Genomic analysis identifies association of Fusobacterium with colorectal carcinoma. Genome Res. 2012;22:292–298. doi: 10.1101/gr.126573.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M, Strauss J, Barnes R, Watson P, Allen-Vercoe E, Moore RA, et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012;22:299–306. doi: 10.1101/gr.126516.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tahara T, Yamamoto E, Suzuki H, Maruyama R, Chung W, Garriga J, Jelinek J, Yamano HO, Sugai T, An B, et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res. 2014;74:1311–1318. doi: 10.1158/0008-5472.CAN-13-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 20.Wolchok JD, Kluger H, Callahan MK, Postow MA, Rizvi NA, Lesokhin AM, Segal NH, Ariyan CE, Gordon RA, Reed K, et al. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369:122–133. doi: 10.1056/NEJMoa1302369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goede V, Fischer K, Busch R, Engelke A, Eichhorst B, Wendtner CM, Chagorova T, de la Serna J, Dilhuydy MS, Illmer T, et al. Obinutuzumab plus chlorambucil in patients with CLL and coexisting conditions. N Engl J Med. 2014;370:1101–1110. doi: 10.1056/NEJMoa1313984. [DOI] [PubMed] [Google Scholar]
- 22.Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Powles T, Eder JP, Fine GD, Braiteh FS, Loriot Y, Cruz C, Bellmunt J, Burris HA, Petrylak DP, Teng SL, et al. MPDL3280A (anti-PD-L1) treatment leads to clinical activity in metastatic bladder cancer. Nature. 2014;515:558–562. doi: 10.1038/nature13904. [DOI] [PubMed] [Google Scholar]
- 25.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagès F, Berger A, Camus M, Sanchez-Cabo F, Costes A, Molidor R, Mlecnik B, Kirilovsky A, Nilsson M, Damotte D, et al. Effector memory T cells, early metastasis, and survival in colorectal cancer. N Engl J Med. 2005;353:2654–2666. doi: 10.1056/NEJMoa051424. [DOI] [PubMed] [Google Scholar]
- 27.Salama P, Phillips M, Grieu F, Morris M, Zeps N, Joseph D, Platell C, Iacopetta B. Tumor-infiltrating FOXP3+ T regulatory cells show strong prognostic significance in colorectal cancer. J Clin Oncol. 2009;27:186–192. doi: 10.1200/JCO.2008.18.7229. [DOI] [PubMed] [Google Scholar]
- 28.Nosho K, Baba Y, Tanaka N, Shima K, Hayashi M, Meyerhardt JA, Giovannucci E, Dranoff G, Fuchs CS, Ogino S. Tumour-infiltrating T-cell subsets, molecular changes in colorectal cancer, and prognosis: cohort study and literature review. J Pathol. 2010;222:350–366. doi: 10.1002/path.2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5:43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shenker BJ, Listgarten MA, Taichman NS. Suppression of human lymphocyte responses by oral spirochetes: a monocyte-dependent phenomenon. J Immunol. 1984;132:2039–2045. [PubMed] [Google Scholar]
- 31.Jewett A, Hume WR, Le H, Huynh TN, Han YW, Cheng G, Shi W. Induction of apoptotic cell death in peripheral blood mononuclear and polymorphonuclear cells by an oral bacterium, Fusobacterium nucleatum. Infect Immun. 2000;68:1893–1898. doi: 10.1128/iai.68.4.1893-1898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kinder Haake S, Lindemann RA. Fusobacterium nucleatum T18 aggregates human mononuclear cells and inhibits their PHA-stimulated proliferation. J Periodontol. 1997;68:39–44. doi: 10.1902/jop.1997.68.1.39. [DOI] [PubMed] [Google Scholar]
- 33.Shenker BJ, Datar S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect Immun. 1995;63:4830–4836. doi: 10.1128/iai.63.12.4830-4836.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shenker BJ, Matt WC. Suppression of human lymphocyte responsiveness by forskolin: reversal by 12-O-tetradecanoyl phorbol 13-acetate, diacylglycerol and ionomycin. Immunopharmacology. 1987;13:73–86. doi: 10.1016/0162-3109(87)90028-2. [DOI] [PubMed] [Google Scholar]
- 35.Huynh T, Kapur RV, Kaplan CW, Cacalano N, Kinder Haake S, Shi W, Sieling P, Jewett A. The role of aggregation in Fusobacterium nucleatum- induced immune cell death. J Endod. 2011;37:1531–1535. doi: 10.1016/j.joen.2011.06.034. [DOI] [PubMed] [Google Scholar]
- 36.Mima K, Sukawa Y, Nishihara R, Qian ZR, Yamauchi M, Inamura K, Kim SA, Masuda A, Nowak JA, Nosho K, et al. Fusobacterium nucleatum and T Cells in Colorectal Carcinoma. JAMA Oncol. 2015;1:653–661. doi: 10.1001/jamaoncol.2015.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flanagan L, Schmid J, Ebert M, Soucek P, Kunicka T, Liska V, Bruha J, Neary P, Dezeeuw N, Tommasino M, et al. Fusobacterium nucleatum associates with stages of colorectal neoplasia development, colorectal cancer and disease outcome. Eur J Clin Microbiol Infect Dis. 2014;33:1381–1390. doi: 10.1007/s10096-014-2081-3. [DOI] [PubMed] [Google Scholar]
- 38.Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogino S, Nosho K, Irahara N, Meyerhardt JA, Baba Y, Shima K, Glickman JN, Ferrone CR, Mino-Kenudson M, Tanaka N, et al. Lymphocytic reaction to colorectal cancer is associated with longer survival, independent of lymph node count, microsatellite instability, and CpG island methylator phenotype. Clin Cancer Res. 2009;15:6412–6420. doi: 10.1158/1078-0432.CCR-09-1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 42.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 43.Speetjens FM, Kuppen PJ, Morreau H, van der Burg SH. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;135:711–712; author reply 712-713. doi: 10.1053/j.gastro.2008.04.040. [DOI] [PubMed] [Google Scholar]
- 44.Curiel TJ. Tregs and rethinking cancer immunotherapy. J Clin Invest. 2007;117:1167–1174. doi: 10.1172/JCI31202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Disis ML, Bernhard H, Jaffee EM. Use of tumour-responsive T cells as cancer treatment. Lancet. 2009;373:673–683. doi: 10.1016/S0140-6736(09)60404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alexander J, Watanabe T, Wu TT, Rashid A, Li S, Hamilton SR. Histopathological identification of colon cancer with microsatellite instability. Am J Pathol. 2001;158:527–535. doi: 10.1016/S0002-9440(10)63994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shia J, Ellis NA, Paty PB, Nash GM, Qin J, Offit K, Zhang XM, Markowitz AJ, Nafa K, Guillem JG, et al. Value of histopathology in predicting microsatellite instability in hereditary nonpolyposis colorectal cancer and sporadic colorectal cancer. Am J Surg Pathol. 2003;27:1407–1417. doi: 10.1097/00000478-200311000-00002. [DOI] [PubMed] [Google Scholar]
- 48.Jenkins MA, Hayashi S, O’Shea AM, Burgart LJ, Smyrk TC, Shimizu D, Waring PM, Ruszkiewicz AR, Pollett AF, Redston M, et al. Pathology features in Bethesda guidelines predict colorectal cancer microsatellite instability: a population-based study. Gastroenterology. 2007;133:48–56. doi: 10.1053/j.gastro.2007.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tougeron D, Fauquembergue E, Rouquette A, Le Pessot F, Sesboüé R, Laurent M, Berthet P, Mauillon J, Di Fiore F, Sabourin JC, et al. Tumor-infiltrating lymphocytes in colorectal cancers with microsatellite instability are correlated with the number and spectrum of frameshift mutations. Mod Pathol. 2009;22:1186–1195. doi: 10.1038/modpathol.2009.80. [DOI] [PubMed] [Google Scholar]
- 50.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23:609–618. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 51.Sinicrope FA, Rego RL, Ansell SM, Knutson KL, Foster NR, Sargent DJ. Intraepithelial effector (CD3+)/regulatory (FoxP3+) T-cell ratio predicts a clinical outcome of human colon carcinoma. Gastroenterology. 2009;137:1270–1279. doi: 10.1053/j.gastro.2009.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, Schreiber H. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 2007;67:425; author reply 426. doi: 10.1158/0008-5472.CAN-06-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65:3437–3446. doi: 10.1158/0008-5472.CAN-04-4262. [DOI] [PubMed] [Google Scholar]
- 55.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 56.Dong J, Zhao YP, Zhou L, Zhang TP, Chen G. Bcl-2 upregulation induced by miR-21 via a direct interaction is associated with apoptosis and chemoresistance in MIA PaCa-2 pancreatic cancer cells. Arch Med Res. 2011;42:8–14. doi: 10.1016/j.arcmed.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 57.Frampton AE, Giovannetti E, Jamieson NB, Krell J, Gall TM, Stebbing J, Jiao LR, Castellano L. A microRNA meta-signature for pancreatic ductal adenocarcinoma. Expert Rev Mol Diagn. 2014;14:267–271. doi: 10.1586/14737159.2014.893192. [DOI] [PubMed] [Google Scholar]
- 58.Ma MZ, Kong X, Weng MZ, Cheng K, Gong W, Quan ZW, Peng CH. Candidate microRNA biomarkers of pancreatic ductal adenocarcinoma: meta-analysis, experimental validation and clinical significance. J Exp Clin Cancer Res. 2013;32:71. doi: 10.1186/1756-9966-32-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mace TA, Collins AL, Wojcik SE, Croce CM, Lesinski GB, Bloomston M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. J Surg Res. 2013;184:855–860. doi: 10.1016/j.jss.2013.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nagao Y, Hisaoka M, Matsuyama A, Kanemitsu S, Hamada T, Fukuyama T, Nakano R, Uchiyama A, Kawamoto M, Yamaguchi K, et al. Association of microRNA-21 expression with its targets, PDCD4 and TIMP3, in pancreatic ductal adenocarcinoma. Mod Pathol. 2012;25:112–121. doi: 10.1038/modpathol.2011.142. [DOI] [PubMed] [Google Scholar]
- 61.Wang P, Zhuang L, Zhang J, Fan J, Luo J, Chen H, Wang K, Liu L, Chen Z, Meng Z. The serum miR-21 level serves as a predictor for the chemosensitivity of advanced pancreatic cancer, and miR-21 expression confers chemoresistance by targeting FasL. Mol Oncol. 2013;7:334–345. doi: 10.1016/j.molonc.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Farrell JJ, Toste P, Wu N, Li L, Wong J, Malkhassian D, Tran LM, Wu X, Li X, Dawson D, et al. Endoscopically acquired pancreatic cyst fluid microRNA 21 and 221 are associated with invasive cancer. Am J Gastroenterol. 2013;108:1352–1359. doi: 10.1038/ajg.2013.167. [DOI] [PubMed] [Google Scholar]
- 63.Nosho K, Igarashi H, Ito M, Mitsuhashi K, Kurihara H, Kanno S, Yoshii S, Mikami M, Takahashi H, Kusumi T, et al. Clinicopathological and molecular characteristics of serrated lesions in Japanese elderly patients. Digestion. 2015;91:57–63. doi: 10.1159/000368820. [DOI] [PubMed] [Google Scholar]
- 64.Igarashi H, Kurihara H, Mitsuhashi K, Ito M, Okuda H, Kanno S, Naito T, Yoshii S, Takahashi H, Kusumi T, et al. Association of MicroRNA-31-5p with Clinical Efficacy of Anti-EGFR Therapy in Patients with Metastatic Colorectal Cancer. Ann Surg Oncol. 2015;22:2640–2648. doi: 10.1245/s10434-014-4264-7. [DOI] [PubMed] [Google Scholar]
- 65.Aoki H, Nosho K, Igarashi H, Ito M, Mitsuhashi K, Naito T, Yamamoto E, Tanuma T, Nomura M, Maguchi H, et al. MicroRNA-31 expression in colorectal serrated pathway progression. World J Gastroenterol. 2014;20:12346–12349. doi: 10.3748/wjg.v20.i34.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito M, Mitsuhashi K, Igarashi H, Nosho K, Naito T, Yoshii S, Takahashi H, Fujita M, Sukawa Y, Yamamoto E, et al. MicroRNA-31 expression in relation to BRAF mutation, CpG island methylation and colorectal continuum in serrated lesions. Int J Cancer. 2014;135:2507–2515. doi: 10.1002/ijc.28920. [DOI] [PubMed] [Google Scholar]
- 67.Nosho K, Igarashi H, Nojima M, Ito M, Maruyama R, Yoshii S, Naito T, Sukawa Y, Mikami M, Sumioka W, et al. Association of microRNA-31 with BRAF mutation, colorectal cancer survival and serrated pathway. Carcinogenesis. 2014;35:776–783. doi: 10.1093/carcin/bgt374. [DOI] [PubMed] [Google Scholar]
- 68.Schwarzenbach H, Nishida N, Calin GA, Pantel K. Clinical relevance of circulating cell-free microRNAs in cancer. Nat Rev Clin Oncol. 2014;11:145–156. doi: 10.1038/nrclinonc.2014.5. [DOI] [PubMed] [Google Scholar]
- 69.Li Z, Rana TM. Therapeutic targeting of microRNAs: current status and future challenges. Nat Rev Drug Discov. 2014;13:622–638. doi: 10.1038/nrd4359. [DOI] [PubMed] [Google Scholar]
- 70.Mitsuhashi K, Yamamoto I, Kurihara H, Kanno S, Ito M, Igarashi H, Ishigami K, Sukawa Y, Tachibana M, Takahashi H, et al. Analysis of the molecular features of rectal carcinoid tumors to identify new biomarkers that predict biological malignancy. Oncotarget. 2015;6:22114–22125. doi: 10.18632/oncotarget.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Virtue A, Wang H, Yang XF. MicroRNAs and toll-like receptor/interleukin-1 receptor signaling. J Hematol Oncol. 2012;5:66. doi: 10.1186/1756-8722-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Honda T, Takahashi N, Miyauchi S, Yamazaki K. Porphyromonas gingivalis lipopolysaccharide induces miR-146a without altering the production of inflammatory cytokines. Biochem Biophys Res Commun. 2012;420:918–925. doi: 10.1016/j.bbrc.2012.03.102. [DOI] [PubMed] [Google Scholar]
- 74.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 75.Iliopoulos D, Jaeger SA, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506. doi: 10.1016/j.molcel.2010.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pencheva N, Tavazoie SF. Control of metastatic progression by microRNA regulatory networks. Nat Cell Biol. 2013;15:546–554. doi: 10.1038/ncb2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hansen TF, Kjær-Frifeldt S, Christensen RD, Morgenthaler S, Blondal T, Lindebjerg J, Sørensen FB, Jakobsen A. Redefining high-risk patients with stage II colon cancer by risk index and microRNA-21: results from a population-based cohort. Br J Cancer. 2014;111:1285–1292. doi: 10.1038/bjc.2014.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sempere LF, Preis M, Yezefski T, Ouyang H, Suriawinata AA, Silahtaroglu A, Conejo-Garcia JR, Kauppinen S, Wells W, Korc M. Fluorescence-based codetection with protein markers reveals distinct cellular compartments for altered MicroRNA expression in solid tumors. Clin Cancer Res. 2010;16:4246–4255. doi: 10.1158/1078-0432.CCR-10-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, Johnson DS, Chen Y, O’Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 81.Merline R, Moreth K, Beckmann J, Nastase MV, Zeng-Brouwers J, Tralhão JG, Lemarchand P, Pfeilschifter J, Schaefer RM, Iozzo RV, et al. Signaling by the matrix proteoglycan decorin controls inflammation and cancer through PDCD4 and MicroRNA-21. Sci Signal. 2011;4:ra75. doi: 10.1126/scisignal.2001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lu L, Byrnes K, Han C, Wang Y, Wu T. miR-21 targets 15-PGDH and promotes cholangiocarcinoma growth. Mol Cancer Res. 2014;12:890–900. doi: 10.1158/1541-7786.MCR-13-0419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14:e218–e228. doi: 10.1016/S1470-2045(12)70582-X. [DOI] [PubMed] [Google Scholar]
- 84.Wang D, Dubois RN. Eicosanoids and cancer. Nat Rev Cancer. 2010;10:181–193. doi: 10.1038/nrc2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Callaghan G, Ryan A, Neary P, O’Mahony C, Shanahan F, Houston A. Targeting the EP1 receptor reduces Fas ligand expression and increases the antitumor immune response in an in vivo model of colon cancer. Int J Cancer. 2013;133:825–834. doi: 10.1002/ijc.28076. [DOI] [PubMed] [Google Scholar]
- 86.Mao Y, Poschke I, Wennerberg E, Pico de Coaña Y, Egyhazi Brage S, Schultz I, Hansson J, Masucci G, Lundqvist A, Kiessling R. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mechanisms. Cancer Res. 2013;73:3877–3887. doi: 10.1158/0008-5472.CAN-12-4115. [DOI] [PubMed] [Google Scholar]
- 87.Naito T, Nosho K, Ito M, Igarashi H, Mitsuhashi K, Yoshii S, Aoki H, Nomura M, Sukawa Y, Yamamoto E, et al. IGF2 differentially methylated region hypomethylation in relation to pathological and molecular features of serrated lesions. World J Gastroenterol. 2014;20:10050–10061. doi: 10.3748/wjg.v20.i29.10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baba Y, Nosho K, Shima K, Huttenhower C, Tanaka N, Hazra A, Giovannucci EL, Fuchs CS, Ogino S. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139:1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Liao X, Lochhead P, Nishihara R, Morikawa T, Kuchiba A, Yamauchi M, Imamura Y, Qian ZR, Baba Y, Shima K, et al. Aspirin use, tumor PIK3CA mutation, and colorectal-cancer survival. N Engl J Med. 2012;367:1596–1606. doi: 10.1056/NEJMoa1207756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nosho K, Kure S, Irahara N, Shima K, Baba Y, Spiegelman D, Meyerhardt JA, Giovannucci EL, Fuchs CS, Ogino S. A prospective cohort study shows unique epigenetic, genetic, and prognostic features of synchronous colorectal cancers. Gastroenterology. 2009;137:1609–1620.e1-e3. doi: 10.1053/j.gastro.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, Giovannucci EL, Fuchs CS. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–96. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kaplan CW, Ma X, Paranjpe A, Jewett A, Lux R, Kinder-Haake S, Shi W. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect Immun. 2010;78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al. Treatment of HCV infection by targeting microRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 96.Cheng CJ, Bahal R, Babar IA, Pincus Z, Barrera F, Liu C, Svoronos A, Braddock DT, Glazer PM, Engelman DM, et al. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]


