Abstract
The MRE11/RAD50/NBS1 (MRN) complex is a major sensor of DNA double strand breaks, whose role in controlling faithful DNA replication and preventing replication stress is also emerging. Inactivation of the MRN complex invariably leads to developmental and/or degenerative neuronal defects, the pathogenesis of which still remains poorly understood. In particular, NBS1 gene mutations are associated with microcephaly and strongly impaired cerebellar development, both in humans and in the mouse model. These phenotypes strikingly overlap those induced by inactivation of MYCN, an essential promoter of the expansion of neuronal stem and progenitor cells, suggesting that MYCN and the MRN complex might be connected on a unique pathway essential for the safe expansion of neuronal cells. Here, we show that MYCN transcriptionally controls the expression of each component of the MRN complex. By genetic and pharmacological inhibition of the MRN complex in a MYCN overexpression model and in the more physiological context of the Hedgehog-dependent expansion of primary cerebellar granule progenitor cells, we also show that the MRN complex is required for MYCN-dependent proliferation. Indeed, its inhibition resulted in DNA damage, activation of a DNA damage response, and cell death in a MYCN- and replication-dependent manner. Our data indicate the MRN complex is essential to restrain MYCN-induced replication stress during neural cell proliferation and support the hypothesis that replication-born DNA damage is responsible for the neuronal defects associated with MRN dysfunctions.
The MRE11/RAD50/NBS1 (MRN) complex is a major sensor of DNA double strand breaks (DSBs) that exerts essential roles in DNA repair processes and in the DNA damage response (DDR).1 Hypomorphic mutations of MRE11, NBS1 and RAD50 genes in humans are responsible for the Ataxia-Telangiectasia like disorder (A-TLD), Nijmegen Breakage Syndrome (NBS) and NBS like disorder (NBSLD), respectively.1 These syndromes share common cellular phenotypes related to their DNA repair defect (i.e., hypersensitivity to DSB inducers, altered cell cycle checkpoints and chromosomal instability), and may or may not include combined immunodeficiency, germ cell defects and cancer predisposition.2, 3, 4 Similar to other 'DDR-defective syndromes', MRN genetic defects invariably result in neurological abnormalities, such as progressive cerebellar ataxia in A-TLD patients, and modest-to-severe microcephaly in NBS patients and in the unique NBSLD case described so far.2, 3, 4 Of relevance, craniofacial and digital abnormalities and gastrointestinal atresias are also sporadic features of NBS.2 The essential role of the MRN complex in neural development has been modeled in mice. Indeed, the conditional knockout of the Nbs1 gene in the CNS leads to microcephaly, severe ataxia and dramatically impaired cerebellar development, owing to a proliferation arrest of granule cell progenitors (GCPs) and apoptosis of postmitotic neurons.5 Nonetheless, the molecular bases of the neurological phenotypes observed in the MRN-defective syndromes are still poorly understood.
So far, very few papers have addressed how MRN complex expression is controlled, most often reporting on its deregulation in cancer cells. Indeed, MRE11 and RAD50 are repressed by p63 and p73 in response to anticancer drugs,6 while FOXM1 and c-myc stimulate and hypoxia represses NBS1 expression.7, 8, 9 MRN components are generally thought to be constitutively expressed in mammalian tissues. However, it has been noted that NBS1 expression levels are higher in organs where DNA DSBs occur physiologically or at sites of high proliferative activity,10 suggesting the MRN complex might have an important role in cell proliferation. Intriguingly, constitutive MRN knockout mice are unviable, indicating all three components are involved in pathway/s whose integrity is essential for mammalian development.1 All these observations fit with the emerging role of the MRN complex in controlling faithful DNA replication by restarting stalled and collapsed replication forks.11, 12, 13, 14, 15, 16
Embryonic and postnatal cerebellar development and GCPs expansion is largely dependent on the Hedgehog (Hh) pathway and its downstream target Mycn.17 Together with c-MYC and L-MYC, MYCN belongs to a family of transcription factors and key regulators of mammalian development. Both c-Myc- and Mycn-deficient mice die at early embryonic stages18 and show severe hypoplasia in several organs including the nervous system.19, 20, 21 Their role in directing neural stem and precursor cells fate, cycling and metabolism is clearly established.22, 23 Underlying its pivotal role in CNS development and pre- and post-natal cerebellar maturation, Mycn conditional KO in the nervous system severely impairs brain development and results in microcephaly with extreme cerebellar hypoplasia,24 all phenotypes shared with the CNS-restricted Nbs1 knockout.5 Intriguingly, MYCN haploinsufficiency, due to heterozygous mutations or deletions, causes the Feingold syndrome, an inherited human disease characterized by microcephaly, digital anomalies, facial dysmorphisms, gastrointestinal atresias and variable learning disabilities.25, 26, 27 Thus, MYCN and NBS1 inactivation lead to overlapping phenotypes both in mice and humans.
As many other proto-oncogenes, MYC overexpression induces replication stress and a DDR by multiple mechanisms, which include direct binding on and boosting of DNA replication factories.28, 29, 30, 31 Interestingly, c-MYC promotes the expression of DDR proteins such as CHK1 and WRN helicase, both of which are necessary for MYC-dependent cancer cell proliferation/survival,32, 33 indicating that restraining replication stress is necessary and intrinsically promoted by MYC. Whether MYC-dependent replication stress might occur in more physiological conditions and independent of MYC oncogenic activity, and how is this eventually controlled is presently unknown.
The substantial overlap of the cerebellar phenotypes of MYCN and NBS1 genetic defects in humans and mice prompted us to explore the functional connections between MYCN and the MRN complex in neural cells. We report here that the MRN complex is induced by MYCN and is essential to restrain MYCN-dependent replication stress during neural cell proliferation.
Results
MYCN transcriptionally regulates MRN complex expression
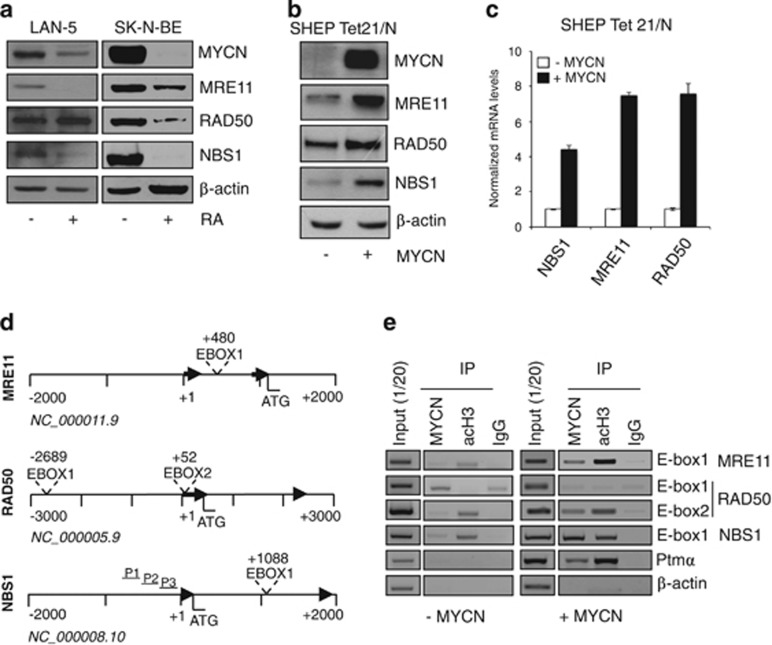
Proliferating and retinoic acid (RA)-differentiated human LAN-5 cells have been previously used as a neuron-like model to assess the function of DNA repair pathways.34 In these and similar settings, where MYCN repression is required for cell growth inhibition and morphological differentiation induced by RA35, 36 (Supplementary Figure S1A), we reproducibly observed the downregulation of MRE11 and NBS1 (Figure 1a), while a sharp RAD50 decrease occurred only in some models. Owing to the coregulation of MYCN and MRN complex during RA-induced differentiation of neuron-like cells, we used both overexpression and knockdown approaches to address whether MYCN directly controls MRN complex expression. Transient (in SK-N-SH) or inducible MYCN overexpression (in the SHEP Tet21/N cell model, since now on MYCN+ and MYCN− cells) clearly increased the expression of the three components of the MRN complex at the protein and mRNA levels (Figures 1b and c and Supplementary Figure S1B). Conversely, MYCN knockdown caused MRN complex reduction in LAN-1 cells (Supplementary Figure S1C), suggesting that MYCN might directly control MRE11, RAD50 and NBS1 transcription in neuron-like cells. Unique canonical CACGTG MYC binding sequences (E-boxes) are located in the intron 1 of both MRE11 and NBS1 human genes, while two E-boxes are present in human RAD50 gene (Figure 1d). A similar organization also occurred in the homologous genes, in mice (Supplementary Figure S2A). Importantly, MYCN bound transcriptionally active H3-acetylated chromatin encompassing the E-boxes in human MRE11 and NBS1 genes, in addition to its known target pro-thymosin-α (Figure 1e). It also bound RAD50 E-box2, but not RAD50 E-box1 and actin promoter (Figure 1e). Moreover, in a luciferase reporter assay, MYCN activated a NBS1 promoter fragment containing the wild-type intron-1 E-Box (Supplementary Figure S1D), but not its mutant version, further supporting a direct transcriptional regulation of the MRN complex.
Figure 1.
The MRN complex is transcriptionally controlled by MYCN. (a) Western blot (WB) analysis of MYCN and MRN protein components in the LAN5 and SK-N-BE neuron-like cell models. (b, c) Analysis of the expression of the indicated proteins (b) and transcripts (c) in the SHEP Tet 21/N inducible-MYCN overexpression model. Transcripts expression were normalized on GAPDH levels and given as fold induction compared with MYCN uninduced controls. (d) Organization of the promoters of the MRE11, RAD50 and NBS1 human genes. Typical E-Boxes (CACGTG) are present in the human MRE11, RAD50 and NBS1 promoters at the indicated positions. Black arrows indicate exons one and two, and nucleotide positions are given from the first nucleotide of exon 1, for each gene. P1, P2 and P3 indicate GC-reach sequences of the NBS1 promoter. (e) PCR amplification of immunoprecipitated chromatin prepared from MYCN induced or uninduced SHEP Tet 21/N (MYCN+/−) cells. Chromatin was incubated with anti-IgG (IgG), anti-acetyl histone 3 (acH3) or with anti-MYCN antibodies. Prothymosin alpha (Ptmα) and β-actin primers were used as positive and negative controls, respectively
The MRN complex is required for MYCN-dependent proliferation
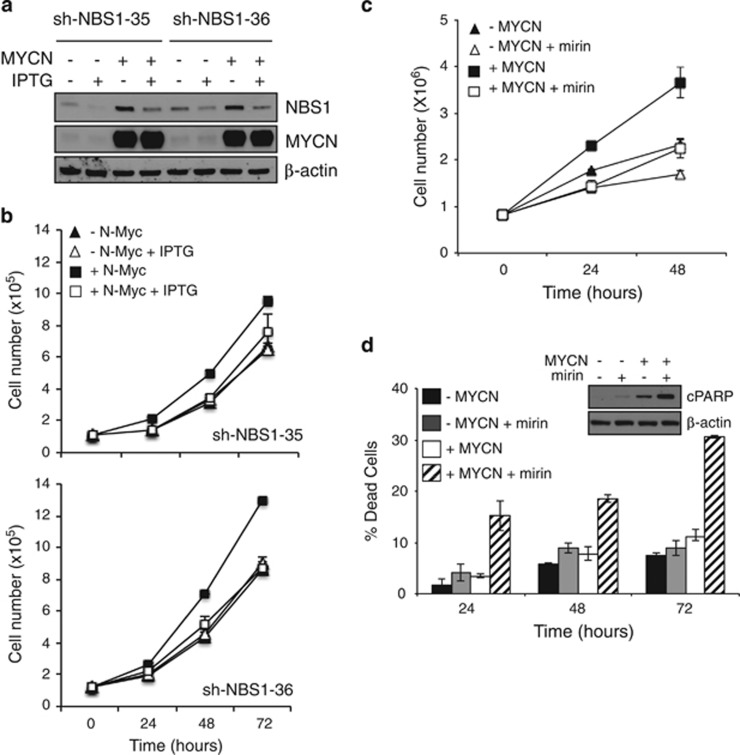
Subsequently, we addressed the role of the MRN complex in MYCN-dependent proliferation in neuron-like cultures. Partial NBS1 knockout with two different shRNAi impaired cell proliferation selectively in MYCN+ cells (Figures 2a and b, Supplementary Figure S3A). Moreover, inhibition of the MRN complex activity with the recently described MRE11 pharmacological inhibitor mirin37 impaired Tet21/N cell proliferation, with a much stronger effect on MYCN+ cells (Figure 2c). Interestingly, mirin caused no significant death, but altered cell cycle profile and impaired BrdU incorporation, in MYCN− cells (Figure 2d and Supplementary Figures S3B and C), suggesting it might impair cell proliferation by delaying cell cycle progression in this setting. Notably, however, mirin did not impair BrdU incorporation and mainly induced cell death in MYCN+ cells, as indicated by death cell count and PARP1 cleavage (Figure 2d), in addition to cell cycle modifications (Supplementary Figures S3B and C). Therefore, MRN complex function seems to be required for a safe progression through the cell cycle in neuron-like cell models and to prevent cell death in particular in a MYCN-dependent context.
Figure 2.
The MRN complex is required for MYCN-dependent proliferation in neuron-like cells. (a) WB analysis of the expression of the indicated proteins and (b) cells counts performed in MYCN+/− cells in which NBS1 was downregulated by two different IPTG-inducible shRNAs (shNBS1-35 and sh-NBS-36). (c) Cell counts performed on the MYCN+/- cells treated with mirin or vehicle (DMSO). (d) Cell death was monitored by trypan blue exclusion assay in MYCN+/− cells with or without mirin treatment and by detection of the cleaved form of PARP1 via immunoblot (cPARP in the inset)
The MRN complex controls MYCN-dependent replication stress
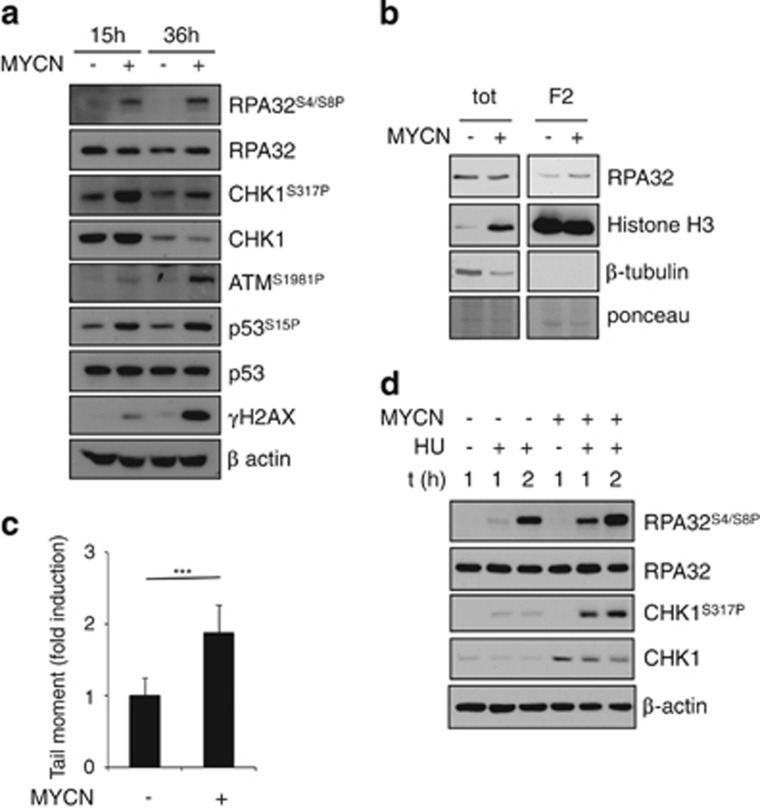
MYC protein overexpression has been consistently shown to cause DNA damage, DDR and genetic instability.38, 39, 40, 41 This seems to be largely, although not exclusively, due to the induction of replication stress. Indeed, it increases the density of early replicating origins and triggers elevated replication fork stalling or collapse.30 Several transcription-dependent and -independent mechanisms have been shown to contribute to this end.28, 29 We have recently shown that MYCN expression in neuron-like models also induce a time-dependent activation of ATM, H2AX and p53S15 phosphorylation, possibly as consequence of a replication stress-dependent DDR.42 Consistent with this hypothesis, MYCN induced accumulation of RPA and CHK1 phosphorylation (Figure 3a), known reporters of the replication stress-dependent ATR activation, and was also associated with slightly increased amounts of chromatin-bound RPA (Figure 3b). In time, also ATM, H2AX and p53S15 phosphorylation increased in MYCN+ cells, consistent with the previously reported activation of a DDR42 and with the appearance of DNA damage (Figure 3c). Moreover, impairing regeneration of nucleotide pools via hydroxyurea induced a replication stress response (indicated by RPA and CHK1 phosphorylation) more promptly and more efficiently in MYCN+ compared with MYCN− cells (Figure 3d), further supporting the idea that MYCN increases replication stress in neuron-like models.
Figure 3.
MYCN induces replication stress. (a) WB analysis of the expression of the indicated proteins and phosphoepitopes in MYCN+/− cells at various time points. (b) WB analysis of RPA32 protein expression in total extracts and in the chromatin-bound fractions, in MYCN+/− cells. Histone H3 and β tubulin were used as positive and negative controls of chromatin fraction, respectively. The ponceau staining was used as the loading control. (c) Neutral comet assay of MYCN+/− cells in basal condition. (d) WB analysis of the expression of the indicated protein in MYCN+/− cells untreated or treated with hydroxyurea (HU, 1 mM). (***P<0.005)
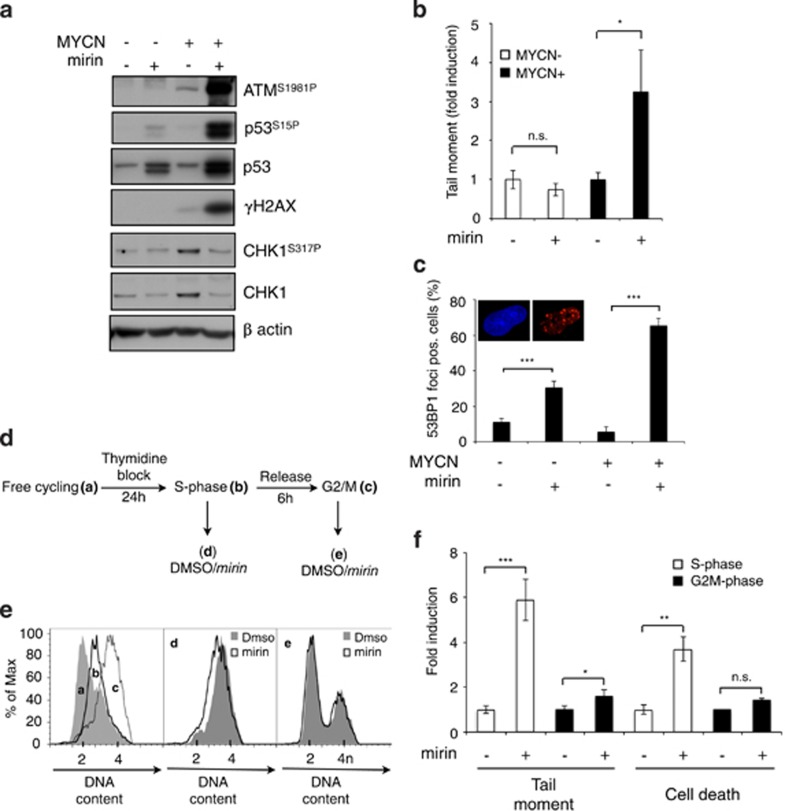
Because the MRN complex is involved in restarting stalled and collapsed replication forks,11, 12 we tested whether it might be required to protect cells from MYCN-dependent replication stress and DNA damage. Indeed, MRN inhibition via mirin was clearly associated with increased signs of DDR (including ATM, H2AX and p53S15 phosphorylation) even at time points in which they are barely detectable in untreated MYCN+ cells (Figure 4a). Consistently, mirin treatment caused the accumulation of DNA DSB in MYCN+, but not in MYCN− cells (Figure 4b). Supporting the occurrence of replication stress-associated DNA lesions, mirin also caused accumulation of multiple 53BP1 foci in a larger number of MYCN+ compared with MYCN− cells (Figure 4c). Not unexpectedly, we failed to detect phosphorylation of CHK1 upon mirin treatment (Figure 4a and Supplementary Figure S3E), which is coherent with the reported requirement of MRE11 exonuclease function for RPA foci formation and ATR-CHK1 pathway activation.13, 43, 44 Indeed, mirin also inhibited CHK1 phosphorylation induced by hydroxyurea in MYCN+ cells (Supplementary Figure S3D). Consistent with the absence of activation of the ATR-CHK1-dependent checkpoint despite replication stress, mirin did not lead to inhibition of DNA synthesis in MYCN+ cells (Supplementary Figure S3B). Overall, these data suggest that the MRN complex prevents MYCN-dependent DNA damage by controlling MYCN-dependent replication stress. To directly prove that induction of DNA damage by mirin is related to replication stress, we synchronized MYCN+ cells in the S phase of the cell cycle, by thymidine block; upon release, we exposed the cells to mirin either immediately (S-phase cells) or after 6 h, when most of the cells were in the G2/M phase (Figures 4d and e). Exposure of S-phase-enriched MYCN+ cells to mirin resulted in both high levels of DNA DSBs and cell death, while this did not occur if cells were treated in the G2/M phase (Figure 4f) confirming that mirin-induced and MYCN-dependent DNA damage and cell death require DNA replication.
Figure 4.
The MRN complex controls MYCN-dependent replication stress. (a) WB analysis of the expression of the indicated proteins and phosphoepitopes in MYCN+/− cells treated or untreated with mirin (15 h). (b) Neutral comet assay of MYCN+/− cells treated or untreated with mirin (15 h). Data are given as fold incuction with respect to mirin-untreated controls. (c) Quantitation of the cells showing more than three 53BP1 foci in MYCN+/− cells treated or untreated with mirin (24 h). Representative images of 53BP1 immunofluorescent staining positive nuclei are given in the insets. (d–f) Effect of mirin treatment in MYCN+ synchronized cells. Free cycling MYCN+ cells (a) were synchronized in the S-phase of the cell cycle by thymidine treatment (b), washed and treated with mirin either immediately, and thus in S-phase (b) or after 6 h from thymidine release, and thus in G2/M (c), as confirmed by cell cycle FACS analysis (e); d and e curves represent the cell cycle distribution of S-phase (b) and G2/M phase (c) cells treated for 5 h with DMSO or mirin. (f) DNA damage and cell death were monitored by neutral comet assay and trypan blue exclusion assay after 5 and 24 h of mirin treatment, respectively. (*P<0.05; **P< 0.01; ***P< 0.005)
The Mycn-MRN axis prevents the deleterious effects of replication stress during Hh- and Mycn-dependent GCP proliferation
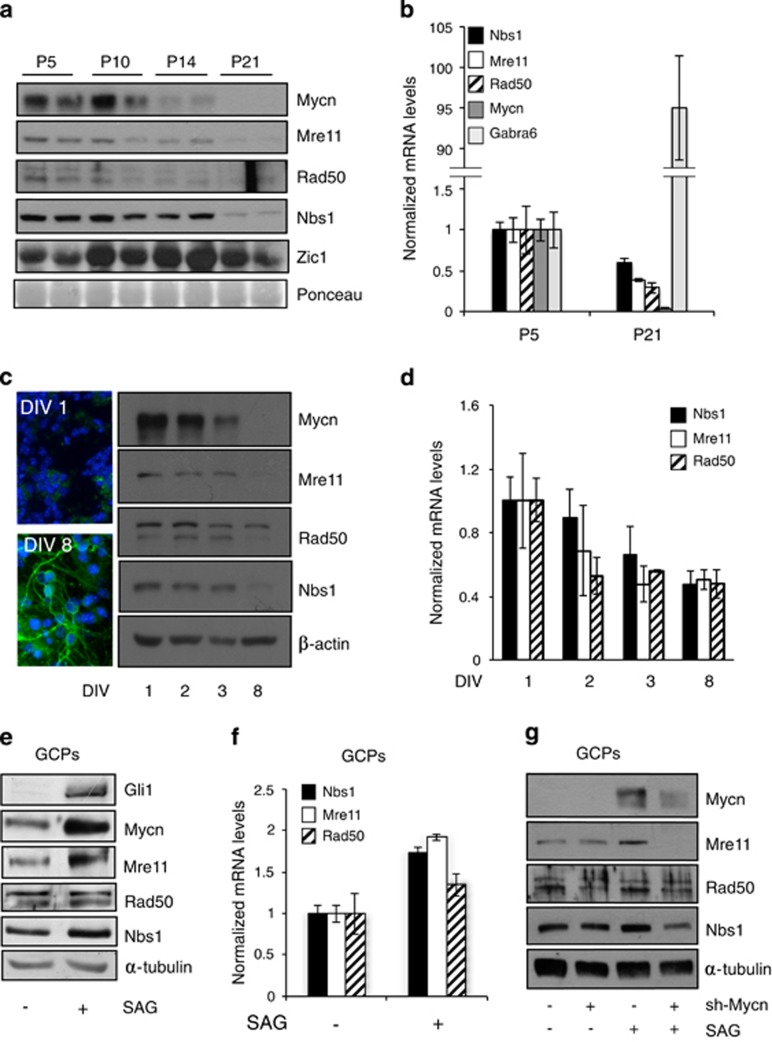
In mice, an Hh-dependent proliferative expansion of the external granule cell layer of the cerebellum characterizes the first 5–10 days post partum, supported by the expression of the Hh target Mycn.17 Shortly after, Hh signal declines, external granule cell layer cells migrate through the molecular and Purkinje layers towards the internal granule cell layer, and their differentiation progressively takes over, leading to completion of the cerebellar development by the end of postnatal week 3. Accordingly, Mycn expression declines and differentiation markers such as Zic1 and Gabra6 rise up (Figures 5a and b). Concerning the three partners of the MRN complex, their protein and mRNA expression remains high during the Hh-driven external granule cell layer expansion and progressively declines during the differentiation phase, to reach very low levels by post-natal day 21, when cerebellar maturation is complete (Figures 5a and b). Isolated GCPs cultured in the absence of Hh pathway stimulation spontaneously stop proliferation and undergo differentiation. This is underlined by a sharp decrease in Mycn expression and acquisition of morphological features and markers of differentiation, such as β3-tubulin expression (Figure 5c inset). Also, in isolated GCP primary cultures, growth arrest and differentiation were accompanied by a progressive decline in the MRN complex expression, paralleled by a reduction in the expression of Mre11, Rad50 and Nbs1 transcripts (Figures 5c and d). In these settings, Hh pathway activation via the Smoothened agonist (SAG) induces the Gli1 transcription factor which stimulates cell proliferation and prevents differentiation by transcriptionally regulating the expression of a large number of targets,17 among which is Mycn (Figure 5e). Hh pathway activation also increased Mre11 and Nbs1 expression and to a lesser extent also Rad50 expression (Figures 5e and f). Importantly, Mycn knockdown impaired SAG-dependent induction of the MRN complex (Figure 5g), indicating that Mycn controls MRN complex expression levels during Hh-dependent GCPs proliferation.
Figure 5.
Mycn promotes MRN complex expression in Hh-induced mouse cerebellar GCPs. (a and b) WB and real-time Q-PCR analysis of the expression of the indicated proteins/transcripts in mouse cerebella at the indicated post-natal (P) days; Zic1 and gamma-aminobutyric acid receptor subunit alpha-6 (Gabra6) were used as differentiation markers and the ponceau staining was used as the loading control. For Q-PCR, the levels of mRNAs were normalized to Hprt expression and reported as fold variation compared with P5 levels. (c) β-tubulin immunofluorescent staining and WB analysis of Mycn and MRN complex proteins in GPC cultures (DIV: days in vitro). (d) Q-PCR assessment of the expression of the indicated transcripts normalized as above and expressed as fold variation compared with the levels at DIV 1. (e–g) WB (e and g) and real-time Q-PCR analysis (f) of the expression of the indicated proteins/transcripts in GPCs treated with DMSO or SAG for 48 h without (e–g) or with (g) lentiviral-transduced anti-Mycn shRNAi
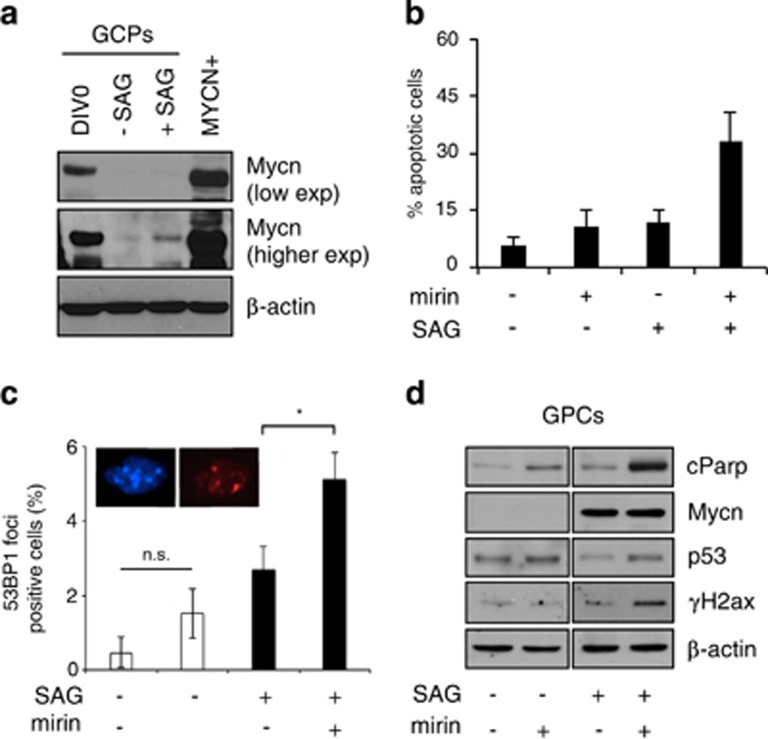
Interestingly, MYCN expression obtained in MYCN+ neuron-like cells was not dramatically different from that observed in freshly isolated GCPs (DIV0, Figure 6a), while both of them were far above the levels obtained in cultured SAG-treated GCPs (Figure 6a). Very importantly, mirin induced cell death selectively in SAG-treated, but not in non-proliferating, GCPs (Figure 6b), suggesting that MRN function is indispensable even in the more physiological context of Hh-induced and Mycn-dependent GCPs proliferation. Of relevance, mirin treatment yielded accumulation of multiple 53BP1 foci and typical signs of DDR (including p53 accumulation and H2AX phosphorylation) in Hh-induced and Mycn-dependent GCPs proliferation, but not in non-proliferating GCPs (Figures 6c and d) supporting the hypothesis that MRN complex induction by Mycn is required to prevent the accumulation of DNA damage due to Mycn-dependent replication stress during the physiological expansion of the cerebellar external granule cell layer.
Figure 6.
The MRN complex controls replication stress during HH-induced mouse cerebellar GCPs proliferation. (a) WB analysis of Mycn protein levels in freshly isolated GPC (DIV0), SAG treated (48 h) GPCs and MYCN+ cells. (b) Tunel assays in GPCs cultures pre-treated with SAG and then incubated with mirin for 18 h. (c) Quantitation of the cells showing more than three 53BP1 foci in GPCs cultures pre-treated with SAG and then incubated with mirin (6 h). (d) WB analysis of the expression of the indicated proteins and phosphoepitopes in cells treated as in panel c. (*P<0.05)
Discussion
Prompted by the striking similarities in the CNS phenotypes associated with MYCN and NBS1 genetic defects, we investigated on the functional links between MYCN and the MRN complex. The expression of the MRN components is generally believed to be ubiquitous and largely constitutive. However, here, we provide evidence that Mycn and the MRN complex are coregulated during mouse cerebellar development and in the Hh-dependent proliferation of primary cerebellar GCPs. Moreover, MYCN transcriptionally controls the MRN complex expression in human neuron-like models, and its depletion impairs Hh-induced regulation of the MRN complex in primary GCPs. Interestingly, MYCN coordinates the expression of a complex pattern of S-phase promoting factors, including MCM proteins and other ORC components, cyclin/cdk complexes, their regulatory kinases and phosphatases (i.e., WEE1 and CDC25) and CDK inhibitors.45 In addition, it also promotes the expression of DNA repair factors involved in S-phase transition, including RAD51, CHK1 and BLM.45, 46, 47 Therefore, our data on the MRN complex regulation might be part of a much broader picture suggesting that a main orchestrator of neural progenitor cell expansion, such as MYCN, may require and directly impose a high rate of DNA repair activity.
The MRN complex function has been mainly characterized in the context of DNA DSBs repair where it has a pivotal role in sensing DNA lesions, activating ATM/ATR-dependent DDR, tethering broken ends and allowing their rejoining.48 MRE11 nucleolitic activities appear to be crucial even for the choice of which DNA DSB repair pathway should come into play.43, 49, 50 Most recent evidence, however, indicates an essential role of the MRN complex also in faithful DNA replication. Indeed, it is required to stabilize and restart stalled and collapsed forks.11, 12 MRE11 nucleolitic activity is essential for survival in response to replication fork toxins in fission yeast.51 Its exonuclease activity is involved in resecting newly replicated DNA at collapsed forks,15, 52 suggesting that also at these sites, the MRN complex is necessary for licensing homologous recombination, allowing replication restart. Consistently, NBS1 KO is associated with the accumulation of aberrant replication intermediates.16 Thus, it emerges that the MRN complex directly participates in processing the 'stressed' forks, more than just being involved in repairing DNA DSBs generated at these locations.
All these mechanistic insights may be used to interpret how MRN dysfunctions may lead to the dramatic CNS phenotypes reported in mutation carriers and in the CNS-restricted NBS1 knockout mouse. It has been argued that the waves of rapid and sustained cellular proliferation of neural progenitor cells (i.e., cerebellar GCPs) would be associated with relevant levels of replication stress and DNA damage strictly requiring the protective role of the MRN complex. This hypothesis, however, still remains largely speculative because direct evidences of replication stress and/or DNA damage occurring in physiologically proliferating neuronal precursors are scant and their mechanisms of generation unknown. Filling these gaps, we now show that the replication mode imposed by a prominent regulator of neuronal progenitor cell expansion, like MYCN, is a source of endogenous stress requiring the MRN complex activity to prevent DNA DSBs accumulation and eventually cell death. Indeed, MYCN-dependent replication is associated with appearance of known markers of replication stress such as increased levels of chromatin-bound RPA, CHK1 and RPA phosphorylation, and with DNA damage and DDR activation. Genetic and pharmacological inactivation of the MRN complex more strongly impairs MYCN-dependent, rather than MYCN-independent, proliferation. Moreover, MRE11 pharmacological inhibition results in DNA damage, DDR and cell death not only in a MYCN overexpression model, but also in the more physiological context of Mycn-dependent and SAG-induced murine GCPs expansion. Importantly, DNA damage unveiled by mirin and cell death failed to occur if treatment started after completion of S-phase, confirming they both depend on DNA replication.
Our data clearly indicate that the MRN complex is induced by MYCN and is necessary to restrain the deleterious effects of the replication stress. By contextualizing our observations directly in continuous and primary neuronal cells, our data are in line with and extend the observations of Brunh et al.16 reporting that the accumulation of aberrant replication intermediates is responsible for DDR activation, p53-dependent cell cycle slow down and cell death in NBS1-defective primary mouse embryo fibroblasts. Interestingly, MRE11 pharmacological inhibition by mirin prevented activation of the ATR-CHK1-dependent checkpoint, allowed cell cycle progression in the presence of unrepaired DNA damage and caused cell death, in MYCN overexpressing neuron-like cells. The apparent inconsistency in the final outcome of MRN complex inactivation in the two models might be related to the different methodological approaches that were used: that is, depletion by NBS1 KO versus MRE11 pharmacological inhibition. Indeed, it has been shown that mirin and mirin-derived selective inhibitors of MRE11 exonuclease activity prevent end resection, RPA and RAD51 coating, and impair repair via homologous recombination, leading to DNA DSBs accumulation.49 In contrast, the selective inhibition of MRE11 endonuclease activity (which works upstream of the exonuclease activity) allows for DNA DSB repair via alternative pathways, limiting the accumulation of DNA damage. Although the effects of these MRN inhibitors have not yet been studied at stressed/collapsed replication forks, we speculate that mirin and other MRE11 exonuclease inhibitors may allow engagement of the MRN complex on collapsed forks, while inhibiting its end-resection ability. This would impair RPA-coating, ATR activation and fork restart via homologous recombination. As a consequence, high levels of DNA DSB would accumulate, eventually inducing cell death, all of which we observed upon mirin treatment in our MYCN-dependent replication stress model. Selective inhibition of MRE11 endonuclease activity might be predicted to impair the engagements of the MRN complex on the collapsed forks, leaving room for alternative pathways to intervene. Similar to the absence of MRN components, this, however, could also lead to the accumulation of replication intermediates, checkpoint activation and cell cycle slowdown, as reported for NBS1 deficiency by Bruhn et al.16
While our data clearly demonstrate that MYCN upregulates the expression of the MRN complex to restrain the replication stress it induces during neural cell proliferation, they also open to the possibility that replication stress and the need for high MRN complex activity is intrinsically associated with neural progenitor cell expansion, perhaps also in CNS territories not dependent on MYCN for their proliferation. Therfore, they support the hypothesis that replication-born DNA damage is responsible for the major neuronal defects associated with MRN dysfunctions.
Materials and Methods
GPCs and neuron-like cell cultures
GPCs were isolated and grown as described.53 Human neuron-like cells were as follows: LAN-5 were acquired from Deutsche Sammlung Von Mikroorganismen und Zellkulturen (Braunschweig, Germany; www.dsmz.de); SK-N-BE were obtained from European Collection of Cell Cultures, (Porton Down, UK; https://www.phe-culturecollections.org.uk/collections/ecacc); SK-N-SH and LAN-1 were a kind gift of Dr. Carol J. Thiele, CMBS, NCI, Bethesda, MD, USA. These cells were grown in standard conditions54 and validated by genetic search of MYCN amplification, p53 loss (for LAN-1) or other genetic aberrations. SHEP Tet21/N cells, received from Dr. Schwabb, DKFZ, Heidelberg, Germany, were cultured as reported55 and validated for MYCN inducibility in the presence or absence of doxycycline (2 μg/ml). Differentiation of LAN-5 and SK-N-BE cells was obtained by RA treatment (5 μM, Sigma Aldrich, St. Louis, MO, USA) for 4 and 15 days, respectively.35 Hh pathway activation was obtained treating GPC culture with SAG (0.2 μM, Alexis, Enzo Life Sciences, Inc, Farmingdale, NY, USA) for the indicated times. The MRE11 inhibitor mirin (Sigma Aldrich) was used at 40 μM concentration. S-phase synchronization was obtained by treating cells with thymidine (2.5 mM, Sigma Aldrich) for 24 h. Hydroxyurea (Sigma Aldrich) was used at 1 mM concentration for the indicated times.
Transfection, lentivirus infection and luciferase assay
MYCN-expressing plasmid35 was transfected in SK-N-SH line with Lipofectamine Plus Reagent (Invitrogen Corporation, San Diego, CA, USA). RNA interference experiments were all performed using validated small interfering RNA sequences (Sigma Aldrich) transfected by Dharmafect 2 Reagent (Thermo Fisher Scientific, Waltham, MA, USA) or by means shRNA lentiviral transduction particles (Santa Cruz Biothecnology, Dallas, TX, USA and Sigma Aldrich). IPTG-inducible NBS1 knockdown was obtained by infecting SHEP TET21/N cells with Mission Custom shRNA lentiviral transduction particles (pLKO-puro-IPTG-3xLacO-NBS1, Sigma Aldrich). Details on siRNA and shRNA constructs are given as Supplementary materials. The NBS1 promoter luciferase reporter construct (NBSluc-WT and NBSluc-mut) was a kind gift of Yu-Chi Chiang.7 Luciferase assay was conducted on triplicate samples via a Dual luciferase reporter assay (Promega Corporation, Madison, WI, USA) and measured by a Glomax multidetection luminometer (Promega Corporation).
Cell proliferation, cell death analysis and cell cycle analysis
MTS assay was performed using the MTS kit (Promega Corporation) according to the manufacturer's instructions. For BrdU incorporation assays, BrdU pulse was applied to MYCN+/− cells for 2 h, and BrdU-staining was assayed by the 5-Bromo-2′-deoxy-uridine Labeling and Detection Kit I (Roche Diagnostics, Indianapolis, IN, USA). Cell proliferation and cell death were evaluated by plating an equal number of cells and counting alive and death cells by trypan blue exclusion test at the indicated time points and different conditions. For TUNEL staining, fixed cells were labeled using the in situ Cell Death Detection Kit (Roche Diagnostics). At least 200 cells/sample were counted and each experiment was performed at least three times. For cell cycle analysis, cells were harvested, washed in succession with 10% FBS/PBS, PBS, 50% FBS/0.1%NaN3/PBS, fixed in 70% ethanol ON at −20 °C, washed twice in 0.1%NaN3/PBS and resuspended in propidium iodide staining solution (50 μg/ml propidium solution/PBS, RNase 40 μg/ml) for 30 min at room temperature. We acquired at least 20 000 events. Fluorescence was measured using a FACSCanto II (BD Biosciences, San Josè, CA, USA) and the percentages of cells in different phases of the cell cycle were determined using the FlowJo V9.3.2 computer software (TreeStar, Ashland, OR, USA).
Comet assay
Neutral comet assay was conducted as described56 with minor modifications. Briefly, cell suspensions were mixed with low melting point agarose (Bio-Rad, Hercules, CA, USA) at a final concentration of 0.5%, cast on a microscope slide precoated with 1% agarose (Bio Optica, Milano, Italy). After solidification, slides were immersed in cold neutral lysing solution (NaCl 2.5 M, EDTA 0.1 M, Tris-HCl 0.01 M, N-Lauroyl-sarcosine 0.34 M, pH 9.5) for 2 h at 4 °C. After lysis, the slides were placed on a horizontal gel electrophoresis unit (Bio-Rad) filled with fresh electrophoretic buffer (sodium acetate 0.3 M and Tris-HCl 0.1M, pH 8). Electrophoresis was carried out for 30' at fixed Volts and Ampere (40 mA). Slides were stained with ethidium bromide and tail moment was measured by the Tritek Comet Score TM freeware 1.6.1.13 Software (Tritek Corp., Sumerduck, VA, USA). At least 200 cell/sample were counted and each experiment was performed at least three times.
Immunofluorescence
Cells were fixed in 4% formaldehyde/PBS for 10 min at RT, permeabilized and blocked in 3% goat serum in PBS and incubated with the primary antibodies for 1 h at room temperature. Primary antibodies anti-β-tubulin (#MAB1637, Millipore, Billerica, MA, USA) and 53BP1 (NB100-304, Novus Biologicals, Littleton, CO, USA) were detected with AlexaFluor (Invitrogen Corporation) secondary antibodies.
RNA and protein preparation, chromatin fractionation Q-PCR and western blot
mRNA was extracted using TRIzol reagent (Invitrogen Corporation) and quantitative reverse transcription-PCR was performed as described.54 Primer sequences are given as Supplementary materials. Total protein extraction and western blot protocol have been previously described.54 Chromatin fractionation was performed as described.57 Primary antibodies were as follows: MYCN (B8.4.B), SC53993, p53 (DO-1) SC-126, CHK1 (G4), SC8408, Tubulin, (TU-02), SC-8035, and β-Actin (I-19) SC-1616 Santa Cruz Biotechnology; phospho-CHK1 (S317), DR1025, Calbiochem (San Diego, CA, USA); RAD50 (13B3/2C6), ab89, MRE11 (12D7) ab214, ZIC1 ab72694, Histone H3, ab1791, phospho-ATM (Ser 1981), ab81292, Rad50, ab499, and Mre11, ab6511, Abcam (Cambridge, UK); NBS1 (1C3) GTX70222, Gene Tex (Irvine, CA, USA); p53 (1C12), #2524, phospho-p53 (Ser 15), #9284, and phospho- histone H2AX (ser 139), #2577, Cell Signaling Technology (Danvers, MA, USA); PARP p85 fragment, #G7341, Promega Corporation; RPA32, A300-244 A, phospho-RPA32 S4/S8, A300-245 A, Bethyl Laboratories (Montgomery, TX, USA); Nbs1 (Y112), NB110-57272, Novus Biologicals; Immunoreactive bands were visualized by enhanced chemoluminescence (Advansta Inc., Menlo Park, CA, USA).
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay was performed as described58 with minor modifications. Briefly, cross-linked chromatin was lysed in buffer L2 (50 mM Tris-HCl pH 8.0, 5 mM EDTA pH 8.0, 0,5% SDS, 150 mM NaCl plus protease inhibitors) and chromatin was sheared to 500–800 bp by sonication. Samples were centrifuged and supernatants were diluted fivefold with dilution buffer (50 mM Tris-HCl pH 8.0, 5 mM EDTA, 200 mM NaCl, 0.5% NP40 plus protease inhibitors). Chromatin extracts were immunoprecipitated overnight with anti-MYCN (Santa Cruz, sc-791), anti-acH3 (Millipore, 06-599) and normal IgG as a control. The antibody-bound chromatin was collected with pre-blocked Dynabeads protein A (Invitrogen Corporation) for 3 h at 4 °C. The supernatant of the no antibody sample was saved as Input chromatin. Chromatin immunoprecipitation complexes were washed three times for 5 min in RIPA buffer (1% NP-40, 0.1% SDS, 1 mM EDTA, 50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% deoxycholate plus protease inhibitors), once with LiCl buffer (Tris-HCl pH 8.0 10 mM, EDTA 1 mM, Deoxycholate 1%, NP40 1%, LiCl 250 mM) and twice with TE buffer (Tris-HCl pH 7.5 10 mM, EDTA 1 mM). Extraction from the beads was performed in 150 μl of elution buffer (1% SDS in TE) for 15 min at 65 °C. Cross-linking was reversed by heating DNA complexes at 65 °C for at least 5 h after addition of 200 mM NaCl, then digested with 100 ug/ml proteinase K for 2 h at 51 °C. DNA was extracted using Chromatin IP DNA purification kit (Active Motif, Carlsbad, CA, USA). For each PCR reaction, 1/20 of the eluted DNA was used. PCR primer sequences are given as Supplementary materials.
Statistical analysis
Statistical analysis was performed by a standard two-tailed Student's t test. P<0.05 was considered significant.
Acknowledgments
We are grateful to Dr. Yu-Chi Chiang for providing NBS1 promoter luciferase reporter constructs, to Drs. Pietro Pichierri and Annapaola Franchitto for helpful methodological suggestions, and to Dr. Marco Crescenzi and Gianluca Canettieri for careful reading of the manuscript. This work was partially supported by grants from Associazione Italiana per la Ricerca sul Cancro to GG (IG12116) and AG, AIRC 5XMILLE, MIUR FIRB and PRIN projects, Ministry of Health. MP is a recipient of a FIRC fellowship David Raffaelli.
Glossary
- MRN
MRE11/RAD50/NBS1
- DSBs
double strand breaks
- DDR
DNA damage response
- A-TLD
Ataxia-Telangiectasia like disorder
- NBS
Nijmegen Breakage Syndrome
- NBSLD
NBS like disorder
- GCPs
granule cell progenitors
- Hh
Hedgehog
- CNS
central nervous system
- RA
retinoic acid
- SAG
Smoothened agonist
- DIV
days in vitro
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by RA Knight
Supplementary Material
References
- 1Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nat Rev Mol Cell Biol 2011; 12: 90–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2Chrzanowska KH, Gregorek H, Dembowska-Baginska B, Kalina MA, Digweed M. Nijmegen breakage syndrome (NBS). Orphanet J Rare Dis 2012; 7: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Taylor AM, Groom A, Byrd PJ. Ataxia-telangiectasia-like disorder (ATLD)-its clinical presentation and molecular basis. DNA Repair (Amst) 2004; 3: 1219–1225. [DOI] [PubMed] [Google Scholar]
- 4Waltes R, Kalb R, Gatei M, Kijas AW, Stumm M, Sobeck A et al. Human RAD50 deficiency in a Nijmegen breakage syndrome-like disorder. Am J Hum Genet 2009; 84: 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Frappart PO, Tong WM, Demuth I, Radovanovic I, Herceg Z, Aguzzi A et al. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat Med 2005; 11: 538–544. [DOI] [PubMed] [Google Scholar]
- 6Lin YL, Sengupta S, Gurdziel K, Bell GW, Jacks T, Flores ER. p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet 2009; 5: e1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7Chiang YC, Teng SC, Su YN, Hsieh FJ, Wu KJ. c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J Biol Chem 2003; 278: 19286–19291. [DOI] [PubMed] [Google Scholar]
- 8Khongkow P, Karunarathna U, Khongkow M, Gong C, Gomes AR, Yague E et al. FOXM1 targets NBS1 to regulate DNA damage-induced senescence and epirubicin resistance. Oncogene 2014; 33: 4144–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. EMBO J 2006; 25: 4784–4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Wilda M, Demuth I, Concannon P, Sperling K, Hameister H. Expression pattern of the Nijmegen breakage syndrome gene, Nbs1, during murine development. Hum Mol Genet 2000; 9: 1739–1744. [DOI] [PubMed] [Google Scholar]
- 11Trenz K, Smith E, Smith S, Costanzo V. ATM and ATR promote Mre11 dependent restart of collapsed replication forks and prevent accumulation of DNA breaks. EMBO J 2006; 25: 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N et al. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 2009; 28: 2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Duursma AM, Driscoll R, Elias JE, Cimprich KA. A role for the MRN complex in ATR activation via TOPBP1 recruitment. Mol Cell 2013; 50: 116–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14Shiotani B, Nguyen HD, Hakansson P, Marechal A, Tse A, Tahara H et al. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell Rep 2013; 3: 1651–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011; 145: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Bruhn C, Zhou ZW, Ai H, Wang ZQ. The essential function of the MRN complex in the resolution of endogenous replication intermediates. Cell Rep 2014; 6: 182–195. [DOI] [PubMed] [Google Scholar]
- 17Oliver TG, Grasfeder LL, Carroll AL, Kaiser C, Gillingham CL, Lin SM et al. Transcriptional profiling of the Sonic hedgehog response: a critical role for N-myc in proliferation of neuronal precursors. Proc Natl Acad Sci USA 2003; 100: 7331–7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Hurlin PJ. Control of vertebrate development by MYC. Cold Spring Harb Perspect Med 2013; 3: a014332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev 1992; 6: 2235–2247. [DOI] [PubMed] [Google Scholar]
- 20Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP et al. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev 1992; 6: 2248–2257. [DOI] [PubMed] [Google Scholar]
- 21Sawai S, Shimono A, Wakamatsu Y, Palmes C, Hanaoka K, Kondoh H. Defects of embryonic organogenesis resulting from targeted disruption of the N-myc gene in the mouse. Development 1993; 117: 1445–1455. [DOI] [PubMed] [Google Scholar]
- 22Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum 2010; 9: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Wey A, Knoepfler PS. c-myc and N-myc promote active stem cell metabolism and cycling as architects of the developing brain. Oncotarget 2010; 1: 120–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev 2002; 16: 2699–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Cognet M, Nougayrede A, Malan V, Callier P, Cretolle C, Faivre L et al. Dissection of the MYCN locus in Feingold syndrome and isolated oesophageal atresia. Eur J Hum Genet 2011; 19: 602–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Marcelis CL, Hol FA, Graham GE, Rieu PN, Kellermayer R, Meijer RP et al. Genotype-phenotype correlations in MYCN-related Feingold syndrome. Hum Mutat 2008; 29: 1125–1132. [DOI] [PubMed] [Google Scholar]
- 27van Bokhoven H, Celli J, van Reeuwijk J, Rinne T, Glaudemans B, van Beusekom E et al. MYCN haploinsufficiency is associated with reduced brain size and intestinal atresias in Feingold syndrome. Nat Genet 2005; 37: 465–467. [DOI] [PubMed] [Google Scholar]
- 28Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M et al. Non-transcriptional control of DNA replication by c-Myc. Nature 2007; 448: 445–451. [DOI] [PubMed] [Google Scholar]
- 29Rohban S, Campaner S. Myc induced replicative stress response: How to cope with it and exploit it. Biochim Biophys Acta 2014; 1849: 517–524. [DOI] [PubMed] [Google Scholar]
- 30Srinivasan SV, Dominguez-Sola D, Wang LC, Hyrien O, Gautier J. Cdc45 is a critical effector of myc-dependent DNA replication stress. Cell Rep 2013; 3: 1629–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Pusapati RV, Rounbehler RJ, Hong S, Powers JT, Yan M, Kiguchi K et al. ATM promotes apoptosis and suppresses tumorigenesis in response to Myc. Proc Natl Acad Sci USA 2006; 103: 1446–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32Murga M, Campaner S, Lopez-Contreras AJ, Toledo LI, Soria R, Montana MF et al. Exploiting oncogene-induced replicative stress for the selective killing of Myc-driven tumors. Nat Struct Mol Biol 2011; 18: 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33Moser R, Toyoshima M, Robinson K, Gurley KE, Howie HL, Davison J et al. MYC-driven tumorigenesis is inhibited by WRN syndrome gene deficiency. Mol Cancer Res 2012; 10: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Biton S, Dar I, Mittelman L, Pereg Y, Barzilai A, Shiloh Y. Nuclear ataxia-telangiectasia mutated (ATM) mediates the cellular response to DNA double strand breaks in human neuron-like cells. J Biol Chem 2006; 281: 17482–17491. [DOI] [PubMed] [Google Scholar]
- 35Giannini G, Cerignoli F, Mellone M, Massimi I, Ambrosi C, Rinaldi C et al. High mobility group A1 is a molecular target for MYCN in human neuroblastoma. Cancer Res 2005; 65: 8308–8316. [DOI] [PubMed] [Google Scholar]
- 36Thiele CJ, Reynolds CP, Israel MA. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature 1985; 313: 404–406. [DOI] [PubMed] [Google Scholar]
- 37Dupre A, Boyer-Chatenet L, Sattler RM, Modi AP, Lee JH, Nicolette ML et al. A forward chemical genetic screen reveals an inhibitor of the Mre11-Rad50-Nbs1 complex. Nat Chem Biol 2008; 4: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38Felsher DW, Bishop JM. Transient excess of MYC activity can elicit genomic instability and tumorigenesis. Proc Natl Acad Sci USA 1999; 96: 3940–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Kuzyk A, Mai S. c-MYC-induced genomic instability. Cold Spring Harb Perspect Med 2014; 4: a014373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Neiman PE, Kimmel R, Icreverzi A, Elsaesser K, Bowers SJ, Burnside J et al. Genomic instability during Myc-induced lymphomagenesis in the bursa of Fabricius. Oncogene 2006; 25: 6325–6335. [DOI] [PubMed] [Google Scholar]
- 41Ray S, Atkuri KR, Deb-Basu D, Adler AS, Chang HY, Herzenberg LA et al. MYC can induce DNA breaks in vivo and in vitro independent of reactive oxygen species. Cancer Res 2006; 66: 6598–6605. [DOI] [PubMed] [Google Scholar]
- 42Petroni M, Veschi V, Prodosmo A, Rinaldo C, Massimi I, Carbonari M et al. MYCN sensitizes human neuroblastoma to apoptosis by HIPK2 activation through a DNA damage response. Mol Cancer Res 2011; 9: 67–77. [DOI] [PubMed] [Google Scholar]
- 43Buis J, Wu Y, Deng Y, Leddon J, Westfield G, Eckersdorff M et al. Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 2008; 135: 85–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44Thompson R, Montano R, Eastman A. The Mre11 nuclease is critical for the sensitivity of cells to Chk1 inhibition. PloS One 2012; 7: e44021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45Valentijn LJ, Koster J, Haneveld F, Aissa RA, van Sluis P, Broekmans ME et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci USA 2012; 109: 19190–19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Chayka O, D'Acunto CW, Middleton O, Arab M, Sala A. Identification and pharmacological inactivation of the MYCN gene network as a therapeutic strategy for neuroblastic tumor cells. J Biol Chem 2015; 290: 2198–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proc Natl Acad Sci USA 2011; 108: 3336–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48Lafrance-Vanasse J, Williams GJ, Tainer JA. Envisioning the dynamics and flexibility of Mre11-Rad50-Nbs1 complex to decipher its roles in DNA replication and repair. Prog Biophys Mol Biol 2015; 117: 182–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49Shibata A, Moiani D, Arvai AS, Perry J, Harding SM, Genois MM et al. DNA double-strand break repair pathway choice is directed by distinct MRE11 nuclease activities. Mol Cell 2014; 53: 7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50Rahal EA, Henricksen LA, Li Y, Williams RS, Tainer JA, Dixon K. ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining. Cell Cycle 2010; 9: 2866–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51Williams RS, Moncalian G, Williams JS, Yamada Y, Limbo O, Shin DS et al. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 2008; 135: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52Ying S, Hamdy FC, Helleday T. Mre11-dependent degradation of stalled DNA replication forks is prevented by BRCA2 and PARP1. Cancer Res 2012; 72: 2814–2821. [DOI] [PubMed] [Google Scholar]
- 53Argenti B, Gallo R, Di Marcotullio L, Ferretti E, Napolitano M, Canterini S et al. Hedgehog antagonist REN(KCTD11) regulates proliferation and apoptosis of developing granule cell progenitors. J Neurosci 2005; 25: 8338–8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54Veschi V, Petroni M, Cardinali B, Dominici C, Screpanti I, Frati L et al. Galectin-3 impairment of MYCN-dependent apoptosis-sensitive phenotype is antagonized by nutlin-3 in neuroblastoma cells. PloS One 2012; 7: e49139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene 1996; 13: 803–812. [PubMed] [Google Scholar]
- 56Wojewodzka M, Buraczewska I, Kruszewski M. A modified neutral comet assay: elimination of lysis at high temperature and validation of the assay with anti-single-stranded DNA antibody. Mut Res 2002; 518: 9–20. [DOI] [PubMed] [Google Scholar]
- 57Wu J, Zhang X, Zhang L, Wu CY, Rezaeian AH, Chan CH et al. Skp2 E3 ligase integrates ATM activation and homologous recombination repair by ubiquitinating NBS1. Mol Cell 2012; 46: 351–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58Forloni M, Albini S, Limongi MZ, Cifaldi L, Boldrini R, Nicotra MR et al. NF-kappaB, and not MYCN, regulates MHC class I and endoplasmic reticulum aminopeptidases in human neuroblastoma cells. Cancer Res 70: 916–924. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.