Abstract
Background:
In metastatic colorectal cancer (mCRC), BRAFV600E mutation has been variously associated to specific clinico-pathological features.
Methods:
Two large retrospective series of mCRC patients from two Italian Institutions were used as training-set (TS) and validation-set (VS) for developing a nomogram predictive of BRAFV600E status. The model was internally and externally validated.
Results:
In the TS, data from 596 mCRC patients were gathered (RAS wild-type (wt) 281 (47.1%); BRAFV600E mutated 54 (9.1%)); RAS and BRAFV600E mutations were mutually exclusive. In the RAS-wt population, right-sided primary (odds ratio (OR): 7.80, 95% confidence interval (CI) 3.05–19.92), female gender (OR: 2.90, 95% CI 1.14–7.37) and mucinous histology (OR: 4.95, 95% CI 1.90–12.90) were independent predictors of BRAFV600E mutation, with high replication at internal validation (100%, 93% and 98%, respectively). A predictive nomogram was calculated: patients with the highest score (right-sided primary, female and mucinous) had a 81% chance to bear a BRAFV600E-mutant tumour; accuracy measures: AUC=0.812, SE:0.034, sensitivity:81.2% specificity:72.1%. In the VS (508 pts, RAS wt: 262 (51.6%), BRAFV600E mutated: 49 (9.6%)), right-sided primary, female gender and mucinous histology were confirmed as independent predictors of BRAFV600E mutation with high accuracy.
Conclusions:
Three simple and easy-to-collect characteristics define a useful nomogram for predicting BRAF status in mCRC with high specificity and sensitivity.
Keywords: BRAF, nomogram, metastatic colorectal cancer
In the last years, significant improvements in the treatment of metastatic colorectal cancer (mCRC) progressively increased the survival expectancy of the overall patients' population to over 2 years (Heinemann et al, 2014; Lenz et al, 2014; Loupakis et al, 2014). A major contribution to these achievements was given by the introduction of RAS testing and the opportunity of treating wild-type (wt) patients with anti-epidermal growth factor receptor (EGFR) monoclonal antibodies (Atreya et al, 2015). Although RAS status ascertainment is recommended by all major guidelines (Van Cutsem et al, 2014; Clinical Practice Guidelines in Oncology (NCCN Guidelines), 2015), the predictive role towards anti-EGFRs of V600E activating mutation of BRAF is still debated (Di Nicolantonio et al, 2008; Laurent-Puig et al, 2009; Loupakis et al, 2009; Souglakos et al, 2009; De Roock et al, 2010) To this extent, studies have not been conclusive maybe due to the low incidence of BRAFV600E mutation (<10% of mCRC; Davies et al, 2002) and to the intrinsic limitations of retrospective subgroup analyses. Nevertheless, all the published series recognised that BRAFV600E mutation is a strong negative prognostic determinant in mCRC and BRAF-mutated metastatic patients have an extremely poor life-expectancy of around 12 months (Richman et al, 2009; Souglakos et al, 2009; Saridaki et al, 2010; Tie et al, 2011; Tran et al, 2011; Yokota et al, 2011; Saridaki et al, 2013; Yaeger et al, 2014).
BRAF-mutated CRCs constitute a distinct subgroup with specific characteristics as underlined by their peculiar gene expression signature (Popovici et al, 2012). The presence of a BRAF mutation has also been associated to specific clinico-pathological features (Samowitz et al, 2006; Roth et al, 2010; Tie et al, 2011; Tran et al, 2011; Yokota et al, 2011; Clancy et al, 2013; Saridaki et al, 2013; Gonsalves et al, 2014; Yaeger et al, 2014). In some published series, BRAF mutation occurred more frequently in older patients and in females, and showed a higher rate of nodal and peritoneal metastases and a lower rate of lung involvement. BRAF-mutant CRCs were also more frequently right-sided, poorly differentiated, mucinous, microsatellite instable and T4-staged. In addition, patients bearing a BRAF-mutant tumour often had a poor performance status (PS) and multiple metastatic sites at diagnosis. However, the association between these features and BRAF mutation was only preliminarily described and up today, no clear and definitive comprehensive data are available, especially in terms of multivariate modelling.
Moving from such considerations, we tested the specific contribution of each clinico-pathological feature for predicting BRAF mutational status in RAS-wt mCRC in a large training-set (TS) population. On those basis, we built a nomogram to predict the likelihood of BRAF mutation occurrence and validated it in a confirmatory external data set (Iasonos et al, 2008).
Materials and methods
A specific database including the variables previously associated to the presence of BRAF mutation in CRC patients was built (Samowitz et al, 2006; Roth et al, 2010; Tie et al, 2011; Tran et al, 2011; Yokota et al, 2011; Clancy et al, 2013; Saridaki et al, 2013; Gonsalves et al, 2014; Yaeger et al, 2014). The following characteristics were selected: age, ECOG-PS, time to metastatic presentation (i.e. synchronous vs metachronous), primary tumour site (i.e. right-sided, from caecum up to transverse colon included vs left-sided, from splenic flexure to rectum), resection of the primary tumour, mucinous histology (as indicated in pathological report), number of metastatic sites, peritoneal, lung, distant lymph nodes as metastatic sites, tumour grading, RAS and BRAF mutational status.
The analysis was conducted as follows: (1) to determine (and confirm) the independent prognostic role for survival of BRAF mutation in our series of mCRC patients; (2) to identity the clinico-pathological predictive factors of the presence of BRAF mutation (predictive nomogram) in RAS-wt patients; (3) to measure the predictive accuracy of the generated nomogram; (4) to internally and externally validate the predictive nomogram. Thus, a step-by-step protocol was followed according to the methodological approach for building a nomogram according to Iasonos et al (2008), with respect to the Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK) criteria for the conduction of a retrospective study in the context of an unselected population (Simon et al, 2009).
Patients' population
Consecutive mCRC patients with available clinical and pathological data (including RAS, BRAF mutational status) referred to the Unit of Oncology, Azienda Ospedaliero-Universitaria Pisana (Pisa, Italy) from February 2000 to October 2014, were retrospectively gathered (TS). Using the same database, data of patients with overlapping entry criteria, referred to the Department of Oncology, Azienda Ospedaliero-Universitaria Santa Maria della Misericordia (Udine, Italy) in the same time frame were gathered for the validation set (VS).
End point
The aim was to generate a predictive nomogram according to clinical and pathological factors for the identification of RAS-wt patients more likely to carry the BRAF mutation.
Mutational analyses
DNA was extracted from a single formalin-fixed-paraffin-embedded block. Haematoxylin-eosin slides were revised by expert pathologists who macrodissected proper representative areas, to obtain an amount of neoplastic cells of at least 50%. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) with overnight proteinase K digestion and DNA concentration was determined by NanoDrop 2000c spectrophotometer (Nanodrop Technologies Inc., Wilmingon, DE, USA). KRAS (exons 2, 3 and 4), NRAS (exons 2, 3 and 4) and BRAFV600E mutational status was tested by means of Pyrosequencing on the PyroMarkQ96 ID instrument (Qiagen) with commercially available kits (Diatech Pharmacogenetics, Italy). Sensitivity (detectable percentage of mutant alleles) of the Pyrosequencing technique is around 5%.
Statistics
Descriptive statistics was used to summarise pertinent study information. Follow-up was analysed and reported according to Shuster (1991). The correlation between variables was analysed according to χ2. A multivariate Cox proportional hazard model was developed using stepwise regression (forward selection, enter/remove limits P=0.10 and P=0.15) to identify independent predictors of the presence of BRAF mutation; the odds ratio (OR) and the 95% confidence intervals (95% CI) were estimated for each variable. The assessment of interactions between significant investigation variables was taken into account when developing the multivariate model. Overall survival (OS) was calculated by the Kaplan–Meier product limit method from the date of diagnosis of metastatic disease until death due to cancer or death for any cause. The hazard ratio (HR) and 95% CI were estimated for each variable using the Cox multivariate model. The log-rank test was used to assess differences between subgroups. Significance was defined at the P<0.05 level. The SPSS (21.0), and MedCalc (14.12.0) licensed statistical programs were used for all analyses.
Internal validation
To address the over-fitting of multivariate model and to validate the results, a cross-validation technique that evaluates the replication stability of the Cox multivariate model in predicting the presence of BRAF mutation was investigated, using a resampling procedure considering those variables independent at the multivariate analysis (Iasonos et al, 2008). This technique generates a number of simulation data sets (at least 100, each ∼80% of the original size), by randomly selecting patients from the original sample, to establish the consistency of the model across less-powered patient samples (Iasonos et al, 2008).
Predictive score assessment
The log-ORs obtained from the Cox model were used to derive weighting factors of a predictive index, aimed at identifying differential probability of the presence of BRAF mutation. Coefficients estimates were ‘normalised' dividing by the smallest one and rounding the resulting ratios to the nearest integer value.
External validation
The predictive accuracy of the derived nomogram predictive of BRAF mutation was evaluated in the context of the VS. The sample size of the VS was calculated based on the predictive performance of the model estimated in the TS, to have a similar predictive performance between the two populations with a null hypothesis of 0.65, a power of 95% and an alpha-error of 5%. To assess the prognostic value of BRAF mutation, a multivariate model for OS was derived in the VS as well. A χ2 comparison between the predictive performances at the ROC analyses of the nomogram in the TS and in the VS was thereafter carried out.
Results
Data for overall 1104 advanced CRC patients were gathered (TS: 596, VS: 508 patients, respectively). Patients' characteristics are reported in Table 1. Overall, the two populations were similar, although the VS cohort included more patients with ECOG-PS⩾2, on site primary tumour, higher histological grade and number of metastatic sites. Median age was 65 (range 25–92) and 67 years (range 32–85) in the TS and VS, respectively. Median follow-up was 24 (range 0–163) and 20 months (range 1–165) in the TS and VS, respectively.
Table 1. Patients' characteristics in the TS and VS (overall population, N=1104).
| TS N (%) | VS N (%) | P-value | |
|---|---|---|---|
| Number of patients | 596 (100) | 508 (100) | – |
|
Gender | |||
| Male | 359 (60.2) | 318 (62.6) | 0.46 |
| Female | 237 (39.8) | 190 (37.4) | |
|
Age (years) | |||
| <65 | 287 (48.2) | 224 (44.1) | 0.20 |
| ⩾65 | 309 (51.8) | 284 (55.9) | |
|
ECOG-PS | |||
| 0–1 | 468 (78.5) | 433 (85.2) | <0.0001 |
| ⩾2 | 10 (1.7) | 75 (14.8) | |
| Missing | 118 (19.8) | 0 (0) | |
|
Driver mutation | |||
| Wt | 227 (38.1) | 213 (41.8) | 0.96 |
| BRAF mutant (V600E) | 54 (9.1) | 49 (9.6) | |
| RAS mutant | 315 (52.8) | 246 (48.6) | |
|
Number of metastatic sites | |||
| 1 | 378 (63.4) | 216 (42.5) | <0.0001 |
| >1 | 218 (36.6) | 292 (57.5) | |
|
Peritoneal metastases | |||
| Yes | 130 (21.8) | 107 (21.1) | 0.82 |
| No | 466 (78.2) | 401 (78.9) | |
|
Lung metastases | |||
| Yes | 147 (24.7) | 152 (29.9) | 0.06 |
| No | 449 (75.3) | 356 (70.1) | |
|
Synchronous metastases | |||
| Yes | 412 (69.1) | 367 (72.2) | 0.29 |
| No | 184 (30.9) | 141 (27.8) | |
|
Distant lymph-node metastases | |||
| Yes | 117 (19.6) | 110 (21.7) | 0.45 |
| No | 479 (80.4) | 398 (78.3) | |
|
Mucinous histology | |||
| Yes | 110 (18.5) | 76 (15.0) | <0.0001 |
| No | 395 (66.3) | 416 (81.9) | |
| Missing | 91 (15.3) | 16 (3.1) | |
|
Primary tumour site | |||
| Right | 202 (33.9) | 162 (31.9) | 0.52 |
| Left | 394 (66.1) | 346 (68.1) | |
|
Primary tumour resected | |||
| Yes | 466 (78.2) | 344 (67.8) | <0.0001 |
| No | 130 (21.8) | 164 (32.2) | |
|
Tumour grading | |||
| G1–2 | 248 (41.6) | 143 (28.1) | <0.0001 |
| G3–4 | 195 (32.7) | 221 (43.5) | |
| Missing | 153 (25.7) | 144 (28.3) | |
Abbreviations: N=number; PS=performance status; TS=training set; VS=validation set; wt=wild type.
%: rate; P-value: χ2 test.
In the TS, data from 596 advanced CRC patients were gathered (RAS wt: 281 (47.1%); BRAF mutant: 54 (9.1%)). BRAF mutation was more frequent in female (13.1% vs 6.4%, P=0.005), right-sided primary (16.3% vs 8.0%, P<0.0001), mucinous tumours (19.1% vs 6.8%, P<0.0001), poorly differentiated tumours (16.4% vs 4.7%, P<0.0001), patients with peritoneal metastases (14.6% vs 7.5%, P=0.01) and distant lymph-node metastases (15.4% vs 7.5%, P=0.008; Supplementary Table 1).
In the overall sample, age ⩾65-years-old, ECOG-PS⩾2, unresected primary tumour, multiple metastatic sites and BRAF mutation were independent prognostic factors for poorer OS with a trend towards significance for mucinous histology (Table 2). BRAF mutation had the higher prognostic power at the multivariate analysis (HR: 2.98, 95% CI 1.96–4.52, P<0.0001). Thus, the identification of clinical and pathological predictors of the presence of BRAF mutation in the context of RAS-wt patients was justified.
Table 2. Uni- and multivariate analyses for OS.
|
TS (N=596) |
VS (N=508) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
|||||
| HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | HR (95% CI) | P-value | |
|
Gender | ||||||||
| F vs M | 1.14 (0.92–1.42) | 0.222 | – | – | 1.17 (0.95–1.46) | 0.15 | – | – |
|
Age (cut-off: 65 years) | ||||||||
| ⩾65 vs <65 | 1.67 (1.35–2.07) | 0.0001 | 1.86 (1.38–2.49) | <0.0001 | 1.21 (0.98–1.49) | 0.08 | – | – |
|
ECOG-PS | ||||||||
| ⩾2 vs <2 | 2.12 (1.70–2.63) | 0.0001 | 1.54 (1.14–2.07) | 0.004 | 1.11 (0.83–1.47) | 0.49 | – | – |
|
Synchronous metastases | ||||||||
| yes vs no | 1.31 (1.03–1.66) | 0.026 | – | – | 1.05 (0.85–1.30) | 0.66 | – | – |
|
Primary site | ||||||||
| right vs left | 1.52 (1.22–1.88) | 0.0001 | – | – | 1.48 (1.18–1.86) | 0.001 | 1.33 (1.05–1.69) | 0.02 |
|
Primary resected | ||||||||
| no vs yes | 1.97 (1.53–2.53) | 0.0001 | 2.27 (1.52–3.39) | <0.0001 | 1.11 (0.95–1.38) | 0.23 | – | – |
|
Mucinous histology | ||||||||
| yes vs no | 1.38 (1.05–1.82) | 0.018 | 1.43 (0.97–2.08) | 0.065 | 1.26 (0.97–1.63) | 0.09 | – | – |
|
Number of metastatic sites | ||||||||
| >1 vs 1 | 2.08 (1.67–2.59) | 0.0001 | 1.83 (1.38–2.43) | <0.0001 | 1.94 (1.56–2.42) | <0.0001 | 1.76 (1.38–2.25) | <0.0001 |
|
Peritoneal metastases | ||||||||
| yes vs no | 1.69 (1.31–2.18) | 0.0001 | – | – | 1.37 (1.06–1.76) | 0.01 | – | – |
|
Lung metastases | ||||||||
| yes vs no | 1.05 (0.82–1.35) | 0.646 | – | – | 0.99 (0.74–1.21) | 0.53 | – | – |
|
Distant lymph-node metastases | ||||||||
| yes vs no | 1.54 (1.18–2.02) | 0.002 | – | – | 1.92 (1.50–2.47) | <0.0001 | 1.31 (0.99–1.74) | 0.06 |
|
Tumour grading | ||||||||
| 3–4 vs 1–2 | 2.34 (1.83–3.01) | 0.0001 | – | – | 1.23 (0.98–1.56) | 0.08 | – | – |
|
RAS | ||||||||
| mut vs wt | 1.20 (0.97–1.49) | 0.088 | – | – | 1.01 (0.82–1.25) | 0.91 | – | – |
|
BRAF | ||||||||
| mut vs wt | 2.56 (1.84–3.56) | 0.0001 | 2.98 (1.96–4.52) | <0.0001 | 2.72 (1.92–3.85) | <0.0001 | 2.33 (1.61–3.35) | <0.0001 |
Abbreviations: CI=confidence intervals; HR=Hazard Ratio; N=number; OS=Overall Survival; PS=performance status; TS=training set; VS=validation set; wt=wild type.
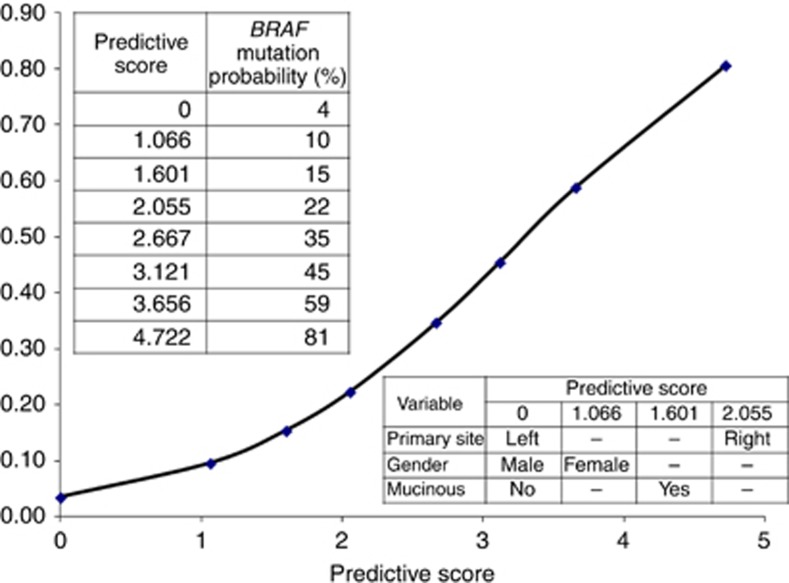
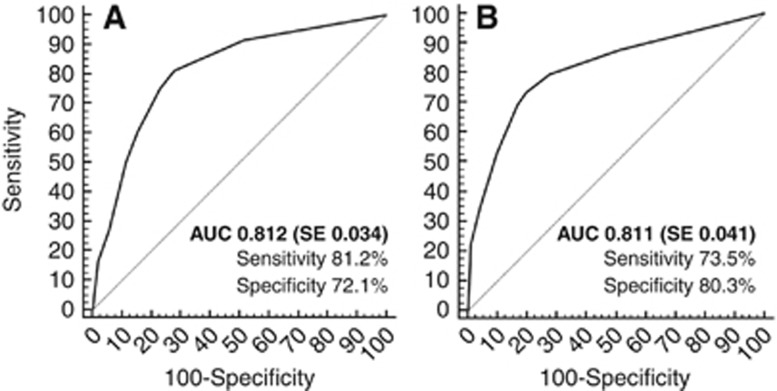
Patients' characteristics of the RAS0-wt population for the TS are reported in Supplementary Table 2. Female gender (OR: 2.90, 95% CI 1.14–7.37, P=0.025), right-sided primary site (OR: 7.80, 95% CI 3.05–19.92, P<0.0001) and mucinous histology (OR: 4.95, 95% CI 1.90–12.90, P<0.0001), resulted to be significant independent predictors of the presence of BRAF mutation in the TS (Table 3). These factors replicated at the internal cross-validation with a high rate, as follows: gender (93%), primary site (100%) and histology (98%). Figure 1 shows the probability of harbouring BRAF mutation according to the scoring index assigned to each patient combining the three independent variables. At the ROC analysis, the predictive accuracy of such nomogram was high (AUC: 0.812, standard error: 0.034), with a sensitivity of 81.2% and a specificity of 72.1% (Figure 2, panel A).
Table 3. Uni- and multivariate analyses for the presence of BRAF mutation in the population of RAS-wt patients.
|
TS (N=281) |
VS (N=262) |
|||||||
|---|---|---|---|---|---|---|---|---|
|
Univariate analysis |
Multivariate analysis |
Univariate analysis |
Multivariate analysis |
|||||
| OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | |
|
Gender | ||||||||
| F vs M | 2.78 (1.51–5.11) | 0.001 | 2.90 (1.14–7.37) | 0.025 | 2.31 (1.23–4.33) | 0.009 | 1.92 (0.92–3.97) | 0.08 |
|
Age (cut-off: 65 years) | ||||||||
| ⩾65 vs <65 | 1.03 (1.00–1.06) | 0.030 | – | 0.11 | 1.42 (0.76–2.68) | 0.27 | – | – |
|
ECOG-PS | ||||||||
| ⩾2 vs <2 | 2.09 (1.02–4.28) | 0.044 | – | 0.87 | 1.30 (0.54–3.12) | 0.55 | – | – |
|
Synchronous metastases | ||||||||
| yes vs no | 1.17 (0.63–2.19) | 0.606 | – | – | 1.34 (0.72–2.50) | 0.35 | – | - |
|
Primary site | ||||||||
| right vs left | 7.12 (3.74–13.56) | 0.0001 | 7.80 (3.05–19.92) | <0.0001 | 11.14 (5.50–22.56) | <0.0001 | 8.68 (4.18–18.02) | <0.0001 |
|
Primary resected | ||||||||
| no vs yes | 1.43 (0.68–3.02) | 0.339 | – | – | 1.29 (0.78–2.86) | 0.229 | – | - |
|
Mucinous histology | ||||||||
| yes vs no | 4.69 (2.33–9.46) | 0.0001 | 4.95 (1.90–12.90) | <0.0001 | 4.61 (2.34–9.07) | <0.0001 | 3.23 (1.49–7.02) | 0.003 |
|
Number of metastatic site | ||||||||
| >1 vs 1 | 1.21 (0.68–2.14) | 0.506 | – | – | 1.92 (1.03–3.60) | 0.04 | – | 0.34 |
|
Peritoneal metastases | ||||||||
| yes vs no | 2.19 (1.15–4.19) | 0.017 | – | 0.17 | 2.53 (1.27–5.02) | 0.01 | – | 0.15 |
|
Lung metastases | ||||||||
| yes vs no | 1.22 (0.59–2.51) | 0.586 | – | – | 1.12 (0.65–1.91) | 0.69 | – | – |
|
Distant lymp-node metastases | ||||||||
| yes vs no | 2.02 (1.05–3.88) | 0.009 | – | 0.64 | 3.30 (1.72–6.33) | <0.0001 | – | 0.19 |
|
Tumour grading | ||||||||
| 3–4 vs 1–2 | 4.54 (2.18–9.48) | 0.0001 | – | 0.46 | 1.63 (0.85–3.13) | 0.14 | – | – |
Abbreviations: CI=confidence intervals; N=number; OR=odds ratio; PS=performance status; TS=training set; VS=validation set; wt=wild type.
Figure 1.
Clinico-pathological nomogram for predicting BRAF mutational status. A predictive score is assigned to each variable and the sum of scores is converted to the probability of BRAF mutation occurrence.
Figure 2.
ROC curves in TS (panel A) and in VS (panel B) populations. Abbreviations: AUC=area under the curve; SE=standard error.
Patients' characteristics of the VS are reported in Table 1. In the VS, data from 508 mCRC patients were gathered (RAS wt: 262 (51.6%); BRAF mutant: 49 (9.6%)). Patients' characteristics of the RAS-wt population for the VS are reported in Supplementary Table 2. Right-sided primary site (OR: 8.68, 95% CI 4.18–18.02, P<0.0001) and mucinous histology (OR: 3.23, 95% CI 1.49–7.02, P=0.003) were confirmed as independent predictors of BRAF mutation in the VS, with a trend towards significance for female gender (OR: 1.92, 95% CI 0.92–3.97, P=0.081; Table 3). The predictive nomogram derived in the TS was then applied to the VS; at the ROC analysis, the predictive accuracy was high (AUC: 0.811, standard error: 0.041), with a sensitivity of 73.5% and a specificity of 80.3% (Figure 2, panel B). No significant difference between the predictive performance of the model in both patients' cohorts was found (P=1.0).
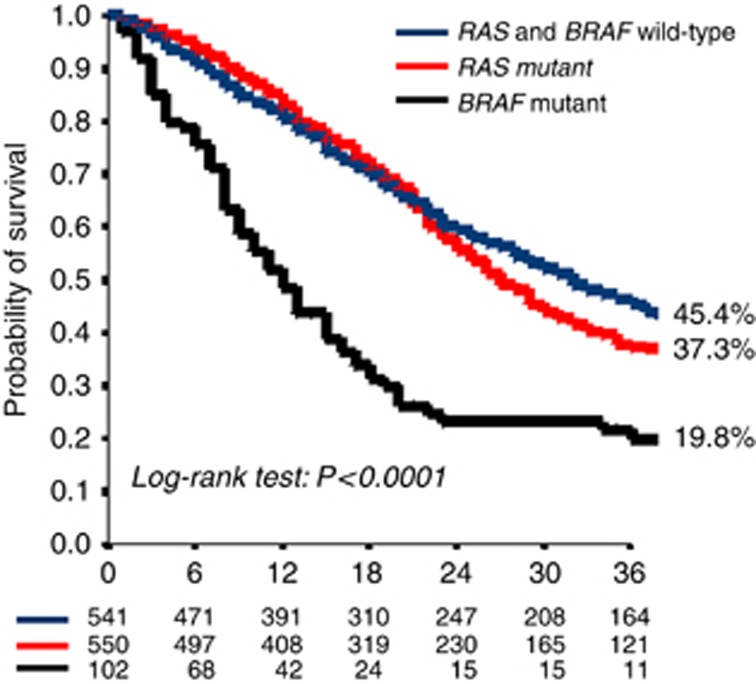
The Kaplan–Meier survival curves of patients in the TS and VS according to RAS and BRAF are shown in Figure 3. As expected, BRAF-mutant mCRC patients had a worse prognosis compared with RAS mutant, and RAS and BRAF-wt patients with an OS rate at 3 years of 19.8%, 37.3% and 45.5%, respectively (P<0.0001).
Figure 3.
Kaplan–Meier curves of the probability of OS in RAS and BRAF wt (blue), RAS-mutant (red) and BRAF-mutant (black) patients. A full colour version of this figure is available at the British Journal of Cancer journal online.
Discussion
BRAFV600E mutation occurs in 8–10% of mCRC and is associated with an extremely poor prognosis (Richman et al, 2009; Souglakos et al, 2009; Saridaki et al, 2010; Tie et al, 2011; Tran et al, 2011; Yokota et al, 2011; Saridaki et al, 2013; Yaeger et al, 2014). Despite this, the need of a routine assessment of BRAF mutational status for clinical practice is a matter of debate, because of the limited therapeutic implications outside of clinical trials (Van Cutsem et al, 2014; Clinical Practice Guidelines in Oncology (NCCN Guidelines), 2015). Conversely, the analysis of RAS mutational status is essential for defining resistance to anti-EGFR monoclonal antibodies (Atreya et al, 2015). Among RAS-wt patients the incidence of BRAF mutation is relatively higher (around 20%), because of the mutual exclusivity between RAS and BRAF mutations (Peeters et al, 2013). Recently, many retrospective series preliminary described some clinical features specifically associated with BRAF mutation (Samowitz et al, 2006; Roth et al, 2010; Tie et al, 2011; Tran et al, 2011; Yokota et al, 2011; Clancy et al, 2013; Saridaki et al, 2013; Gonsalves et al, 2014; Yaeger et al, 2014). Nevertheless, these studies were exploratory and included different stages (i.e. from I to IV), different settings, (i.e. first vs later lines of treatment), had an incomplete molecular assessment (in most of the cases only KRAS exon 2 was tested) and lacked of VSs. Hence, oncologists need to clarify and to properly measure the association between BRAFV600E mutational status and specific patients' and disease's characteristics in the context of the RAS-wt subgroup.
The data reported herein female sex, age ⩾65 years, worse ECOG-PS, right-sided primary tumour, mucinous histology, presence of nodal and peritoneal metastases and higher tumour grading, were associated to BRAFV600E mutation in the TS and these findings are consistent with most previous studies (Samowitz et al, 2006; Roth et al, 2010; Tie et al, 2011; Tran et al, 2011; Yokota et al, 2011; Clancy et al, 2013; Saridaki et al, 2013; Gonsalves et al, 2014; Yaeger et al, 2014). At the multivariate analysis, only female gender, right-sided primary and mucinous histology retained their significance as predictors of BRAFV600E mutation (OR: 2.90, 4.95 and 7.80 respectively). At internal cross-validation these three features were replicated with high rates (93%, 100% and 98%, respectively). These robust data allowed to build a nomogram for predicting the presence of BRAF mutation by combining the three independent variables. The probability to carry a BRAF-mutated tumour ranged from 4% to 81% with a predictive accuracy >80% and with high sensitivity and specificity (81.2% and 72.1%, respectively). In particular, a RAS-wt mCRC, not mucinous and originated from a left-sided primary occurring in male patients have an extremely poor likelihood to be BRAF mutant (4%). Conversely, female patients with mucinous histology and a right-sided primary RAS-wt tumour have a high probability to carry a BRAF-mutated cancer (81%). Finally, these data were replicated and validated in an external independent population. The predictive performance of the derived model in the context of the VS was impressively superimposable.
In a previous experience, Tie et al (Tie et al, 2011) reported a 50% incidence of BRAF mutation in KRAS-wt females aged ⩾70 years at diagnosis affected by a right-sided colon cancer. Nevertheless, the inclusion of stage I–IV patients, RAS testing limited to KRAS exon 2 mutations and the exclusion of mucinous histology from the model, attenuated the value of the determined correlations.
A weakness point of our study is the lack of data on microsatellite instability (MSI). Although the association between MSI-high status and BRAF mutation is well-established (Tran et al, 2011), the positive prognostic effect of MSI-high makes its occurrence in metastatic patients very uncommon (2–6% Richman et al, 2009; Goldstein et al, 2014). Given the above considerations and taking into account that MSI is not routinely tested in the metastatic setting (Van Cutsem et al, 2014), we could speculate that the inclusion of such variable in our nomogram would not have significantly affected the performance of the model. Also, we did not consider rare BRAF mutations other than V600E. In addition, we did not assess the status of other pathological characteristics potentially associated with BRAF mutational status (Schirripa et al, 2015; i.e. T and N pathological stage, vascular invasion, tumour budding, lymphocytic infiltrate and number of lymph nodes resected), because this information were not always available and were beyond the purpose of present study.
In the era of molecular characterisation, the present nomogram should not be considered a tool to replace the mutational analysis of CRC, but it could allow physicians to better estimate patients' prognosis where BRAF testing is not available or reimbursed because of regulatory restrictions.
Moreover, several studies are undergoing worldwide for exploring the efficacy of BRAF-targeting agents. We believe that by applying the proposed nomogram the molecular screening and therefore the overall accrual of those studies could be better implemented in terms of both costs and enrolment performance. Furthermore, the poor prognosis of BRAF-mutant patients makes their earliest identification essential to enable enrolment in clinical trials.
In addition, our nomogram may also potentially guide for prospective stratification of future randomized trials thus avoiding costly and time-consuming upfront testing procedures.
In addition, the identification of subgroups where BRAF mutation is very likely to occur would theoretically help to decrease the attrition bias of retrospective studies when tumour blocks are no longer available or difficult to retrieve. In the context of CRC, a clear example is given by the Analysis and Research in CAncers of the Digestive system (ARCAD) database: a large international effort for pooling data from major randomized trials, that nowadays includes >20 000 patients from >20 first-line mCRC trials (de Gramont et al, 2010; Lieu et al, 2014) and our nomogram may represent a valuable tool for guiding and interpreting the results of subgroup analyses.
From a broader perspective, what does a nomogram add to a multivariate model? We believe that while a simple multivariate model allows physicians to identify which are the independent predictors for the occurrence of a specific event, a nomogram, as the one herein proposed, may translate the statistical output of the identified predictors into a single numerical estimate of the probability of an event (Iasonos et al, 2008), which is, in this case, the chance to have a BRAF mutation. Given the nature of the required information (gender, primary site and histology) and the ease of the graphical interface (Figure 1) this nomogram is extremely valuable and ready-to-use.
Acknowledgments
SP, LC, IS and EB are supported by a grant of the Italian Association for Cancer Research (AIRC My First AIRC Grant No. 14282). This work was partially supported by ARCO No-profit Foundation and by a donation from Raddi family in loving memory of Bianca Lucarelli. FL and EB were responsible for study concept and design. MM, LS, MS, DR, LC, SP, LC, DI, MC and LF were responsible for data collection. FL, RM, CC, IS, GA, AF, GT and EB were responsible for analysis and interpretation of data. FL, RM and EB were responsible for drafting the manuscript. IS and EB were responsible for statistical analysis. All the authors approved the final version of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc).
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Atreya CE, Corcoran RB, Kopetz S (2015) Expanded RAS: Refining the Patient Population. J Clin Oncol 33(7): 682–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy C, Burke JP, Kalady MF, Coffey JC (2013) BRAF mutation is associated with distinct clinicopathological characteristics in colorectal cancer: a systematic review and meta-analysis. Colorectal Dis 15(12): e711–e718. [DOI] [PubMed] [Google Scholar]
- Clinical Practice Guidelines in Oncology (NCCN Guidelines) (2015) Colon Cancer. Version 2. NCCN.org.
- Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W, Davis N, Dicks E, Ewing R, Floyd Y, Gray K, Hall S, Hawes R, Hughes J, Kosmidou V, Menzies A, Mould C, Parker A, Stevens C, Watt S, Hooper S, Wilson R, Jayatilake H, Gusterson BA, Cooper C, Shipley J, Hargrave D, Pritchard-Jones K, Maitland N, Chenevix-Trench G, Riggins GJ, Bigner DD, Palmieri G, Cossu A, Flanagan A, Nicholson A, Ho JW, Leung SY, Yuen ST, Weber BL, Seigler HF, Darrow TL, Paterson H, Marais R, Marshall CJ, Wooster R, Stratton MR, Futreal PA (2002) Mutations of the BRAF gene in human cancer. Nature 417(6892): 949–954. [DOI] [PubMed] [Google Scholar]
- de Gramont A, Haller DG, Sargent DJ, Tabernero J, Matheson A, Schilsky RL (2010) Toward efficient trials in colorectal cancer: the ARCAD Clinical Trials Program. J Clin Oncol 28(4): 527–530. [DOI] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, Penault-Llorca F, Rougier P, Vincenzi B, Santini D, Tonini G, Cappuzzo F, Frattini M, Molinari F, Saletti P, De Dosso S, Martini M, Bardelli A, Siena S, Sartore-Bianchi A, Tabernero J, Macarulla T, Di Fiore F, Gangloff AO, Ciardiello F, Pfeiffer P, Qvortrup C, Hansen TP, Van Cutsem E, Piessevaux H, Lambrechts D, Delorenzi M, Tejpar S (2010) Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol 11(8): 753–762. [DOI] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, Bardelli A (2008) Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 26(35): 5705–5712. [DOI] [PubMed] [Google Scholar]
- Goldstein J, Tran B, Ensor J, Gibbs P, Wong HL, Wong SF, Vilar E, Tie J, Broaddus R, Kopetz S, Desai J, Overman MJ (2014) Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H). Ann Oncol 25(5): 1032–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalves WI, Mahoney MR, Sargent DJ, Nelson GD, Alberts SR, Sinicrope FA, Goldberg RM, Limburg PJ, Thibodeau SN, Grothey A, Hubbard JM, Chan E, Nair S, Berenberg JL, McWilliams RR Alliance for Clinical Trials in O (2014) Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/Alliance N0147. J Natl Cancer Inst 106(7): 1–8. [Google Scholar]
- Heinemann V, von Weikersthal LF, Decker T, Kiani A, Vehling-Kaiser U, Al-Batran SE, Heintges T, Lerchenmuller C, Kahl C, Seipelt G, Kullmann F, Stauch M, Scheithauer W, Hielscher J, Scholz M, Muller S, Link H, Niederle N, Rost A, Hoffkes HG, Moehler M, Lindig RU, Modest DP, Rossius L, Kirchner T, Jung A, Stintzing S (2014) FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab as first-line treatment for patients with metastatic colorectal cancer (FIRE-3): a randomised, open-label, phase 3 trial. Lancet Oncol 15(10): 1065–1075. [DOI] [PubMed] [Google Scholar]
- Iasonos A, Schrag D, Raj GV, Panageas KS (2008) How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 26(8): 1364–1370. [DOI] [PubMed] [Google Scholar]
- Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, Rougier P, Lievre A, Landi B, Boige V, Ducreux M, Ychou M, Bibeau F, Bouche O, Reid J, Stone S, Penault-Llorca F (2009) Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol 27(35): 5924–5930. [DOI] [PubMed] [Google Scholar]
- Lenz H, Niedzwiecki D, Innocenti F, Blanke C, Mahony M, O'Neil B, Shaw J, Polite B, Hochster H, Atkins J, Goldberg R, Mayer R, Schilsky R, Bertagnolli M, Venook A (2014) CALGB/SWOG 80405: phase III trial of irinotecan/5-FU/ Leucovorin (FOLFIRI) or Oxaliplatin/5-FU/Leucovorin (MFOLFOX6) with bevacizumab (BV) or cetuximab (CET) for patients (PTS) with expanded ras analyses untreated metastatic adenocarcinoma of the colon or rectum (MCRC). Ann Oncol 25(Suppl. 5): v1–v41. [Google Scholar]
- Lieu CH, Renfro LA, de Gramont A, Meyers JP, Maughan TS, Seymour MT, Saltz L, Goldberg RM, Sargent DJ, Eckhardt SG, Eng C Aide et Recherche en Cancerologie Digestive F (2014) Association of age with survival in patients with metastatic colorectal cancer: analysis from the ARCAD Clinical Trials Program. J Clin Oncol 32(27): 2975–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loupakis F, Cremolini C, Masi G, Lonardi S, Zagonel V, Salvatore L, Cortesi E, Tomasello G, Ronzoni M, Spadi R, Zaniboni A, Tonini G, Buonadonna A, Amoroso D, Chiara S, Carlomagno C, Boni C, Allegrini G, Boni L, Falcone A (2014) Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N Engl J Med 371(17): 1609–1618. [DOI] [PubMed] [Google Scholar]
- Loupakis F, Ruzzo A, Cremolini C, Vincenzi B, Salvatore L, Santini D, Masi G, Stasi I, Canestrari E, Rulli E, Floriani I, Bencardino K, Galluccio N, Catalano V, Tonini G, Magnani M, Fontanini G, Basolo F, Falcone A, Graziano F (2009) KRAS codon 61, 146 and BRAF mutations predict resistance to cetuximab plus irinotecan in KRAS codon 12 and 13 wild-type metastatic colorectal cancer. Br J Cancer 101(4): 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Oliner KS, Parker A, Siena S, Van Cutsem E, Huang J, Humblet Y, Van Laethem JL, Andre T, Wiezorek J, Reese D, Patterson SD (2013) Massively parallel tumor multigene sequencing to evaluate response to panitumumab in a randomized phase III study of metastatic colorectal cancer. Clin Cancer Res 19(7): 1902–1912. [DOI] [PubMed] [Google Scholar]
- Popovici V, Budinska E, Tejpar S, Weinrich S, Estrella H, Hodgson G, Van Cutsem E, Xie T, Bosman FT, Roth AD, Delorenzi M (2012) Identification of a poor-prognosis BRAF-mutant-like population of patients with colon cancer. J Clin Oncol 30(12): 1288–1295. [DOI] [PubMed] [Google Scholar]
- Richman SD, Seymour MT, Chambers P, Elliott F, Daly CL, Meade AM, Taylor G, Barrett JH, Quirke P (2009) KRAS and BRAF mutations in advanced colorectal cancer are associated with poor prognosis but do not preclude benefit from oxaliplatin or irinotecan: results from the MRC FOCUS trial. J Clin Oncol 27(35): 5931–5937. [DOI] [PubMed] [Google Scholar]
- Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, Dietrich D, Biesmans B, Bodoky G, Barone C, Aranda E, Nordlinger B, Cisar L, Labianca R, Cunningham D, Van Cutsem E, Bosman F (2010) Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60-00 trial. J Clin Oncol 28(3): 466–474. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Albertsen H, Sweeney C, Herrick J, Caan BJ, Anderson KE, Wolff RK, Slattery ML (2006) Association of smoking, CpG island methylator phenotype, and V600E BRAF mutations in colon cancer. J Natl Cancer Inst 98(23): 1731–1738. [DOI] [PubMed] [Google Scholar]
- Saridaki Z, Papadatos-Pastos D, Tzardi M, Mavroudis D, Bairaktari E, Arvanity H, Stathopoulos E, Georgoulias V, Souglakos J (2010) BRAF mutations, microsatellite instability status and cyclin D1 expression predict metastatic colorectal patients' outcome. Br J Cancer 102(12): 1762–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saridaki Z, Tzardi M, Sfakianaki M, Papadaki C, Voutsina A, Kalykaki A, Messaritakis I, Mpananis K, Mavroudis D, Stathopoulos E, Georgoulias V, Souglakos J (2013) BRAFV600E mutation analysis in patients with metastatic colorectal cancer (mCRC) in daily clinical practice: correlations with clinical characteristics, and its impact on patients' outcome. PLoS One 8(12): e84604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirripa M, Cremolini C, Loupakis F, Morvillo M, Bergamo F, Zoratto F, Salvatore L, Antoniotti C, Marmorino F, Sensi E, Lupi C, Fontanini G, De Gregorio V, Giannini R, Basolo F, Masi G, Falcone A (2015) Role of NRAS mutations as prognostic and predictive markers in metastatic colorectal cancer. Int J Cancer 136(1): 83–90. [DOI] [PubMed] [Google Scholar]
- Shuster JJ (1991) Median follow-up in clinical trials. J Clin Oncol 9(1): 191–192. [DOI] [PubMed] [Google Scholar]
- Simon RM, Paik S, Hayes DF (2009) Use of archived specimens in evaluation of prognostic and predictive biomarkers. J Natl Cancer Inst 101(21): 1446–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souglakos J, Philips J, Wang R, Marwah S, Silver M, Tzardi M, Silver J, Ogino S, Hooshmand S, Kwak E, Freed E, Meyerhardt JA, Saridaki Z, Georgoulias V, Finkelstein D, Fuchs CS, Kulke MH, Shivdasani RA (2009) Prognostic and predictive value of common mutations for treatment response and survival in patients with metastatic colorectal cancer. Br J Cancer 101(3): 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie J, Gibbs P, Lipton L, Christie M, Jorissen RN, Burgess AW, Croxford M, Jones I, Langland R, Kosmider S, McKay D, Bollag G, Nolop K, Sieber OM, Desai J (2011) Optimizing targeted therapeutic development: analysis of a colorectal cancer patient population with the BRAF(V600E) mutation. Int J Cancer 128(9): 2075–2084. [DOI] [PubMed] [Google Scholar]
- Tran B, Kopetz S, Tie J, Gibbs P, Jiang ZQ, Lieu CH, Agarwal A, Maru DM, Sieber O, Desai J (2011) Impact of BRAF mutation and microsatellite instability on the pattern of metastatic spread and prognosis in metastatic colorectal cancer. Cancer 117(20): 4623–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Cervantes A, Nordlinger B, Arnold D (2014) Metastatic colorectal cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 25(Suppl. 3): iii1–iii9. [DOI] [PubMed] [Google Scholar]
- Yaeger R, Cercek A, Chou JF, Sylvester BE, Kemeny NE, Hechtman JF, Ladanyi M, Rosen N, Weiser MR, Capanu M, Solit DB, D'Angelica MI, Vakiani E, Saltz LB (2014) BRAF mutation predicts for poor outcomes after metastasectomy in patients with metastatic colorectal cancer. Cancer 120(15): 2316–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota T, Ura T, Shibata N, Takahari D, Shitara K, Nomura M, Kondo C, Mizota A, Utsunomiya S, Muro K, Yatabe Y (2011) BRAF mutation is a powerful prognostic factor in advanced and recurrent colorectal cancer. Br J Cancer 104(5): 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.