Abstract
Clustered regularly interspaced short palindromic repeat (CRISPR) has arisen as a frontrunner for efficient genome engineering. However, the potentially broad therapeutic implications are largely unexplored. Here, to investigate the therapeutic potential of CRISPR/Cas9 in a diverse set of genetic disorders, we establish a pipeline that uses readily obtainable cells from affected individuals. We show that an adapted version of CRISPR/Cas9 increases the amount of utrophin, a known disease modifier in Duchenne muscular dystrophy (DMD). Furthermore, we demonstrate preferential elimination of the dominant-negative FGFR3 c.1138G>A allele in fibroblasts of an individual affected by achondroplasia. Using a previously undescribed approach involving single guide RNA, we successfully removed large genome rearrangement in primary cells of an individual with an X chromosome duplication including MECP2. Moreover, removal of a duplication of DMD exons 18–30 in myotubes of an individual affected by DMD produced full-length dystrophin. Our findings establish the far-reaching therapeutic utility of CRISPR/Cas9, which can be tailored to target numerous inherited disorders.
Introduction
Many bacteria and archaea use clustered regularly interspaced short palindromic repeat (CRISPR)-Cas systems, which are adaptive immune systems, to fight off foreign DNA in the form of bacterial phages and/or plasmids.1 Specifically, the type II CRISPR/Cas system works through RNA-directed endonuclease cleavage of the invading genomic sequence. The invading sequence is captured and inserted directly into the genome of the host organism between CRISPR regions.2, 3, 4 After transcription and processing of these loci, RNA-guided endonucleases are made with the capability to target foreign nucleic acids on the basis of complementarity with the RNA.5
Since the realization of the potential power of a programmable nuclease in editing mammalian genomes, the CRISPR/Cas9 system has been developed as a technology for multiple biological contexts.6, 7 Regardless of the platform, this system requires a mammalian-optimized Cas9 and a chimeric single guide RNA (sgRNA), which is made up of CRISPR RNAs (crRNAs) and a trans-activating CRISPR RNA (tracrRNA).6, 7, 8, 9 The guide sequences are generally 17–20 bp long.10 Target sequences must be adjacent to a protospacer-adjacent motif (PAM) for Streptococcus pyogenes Cas9 (SpCas9) in the form of 5′-NGG-3′.11 Cas9 target recognition is dictated by the Watson-Crick base pairing between an RNA guide and its DNA target.3, 4 Once present in cells, Cas9 and the sgRNA form a complex, bind to the target sequence, and make a double-stranded break (DSB) in the target. The break is repaired via the cellular process of non-homologous end joining (NHEJ), an error-prone process that introduces insertions and deletions (indels) into the target sequence. Targeted mutations can also be introduced by co-transfection of single- or double-stranded DNA templates to promote homology-directed repair (HDR). To date, SpCas9 has been used broadly for achieving efficient genome editing in a variety of species and cell types, including human cell lines, bacteria, zebrafish, yeast, mouse, fruit fly, roundworm, rat, common crops, pig, and monkey (reviewed in Hsu et al.12).
Another application of the CRISPR/Cas9 tool is to regulate gene expression. This approach uses a catalytically inactive or “dead” Cas9 (dCas9), which when bound to DNA elements can repress transcription by sterically hindering the RNA polymerase machinery,13 most likely by stalling transcriptional elongation. Alternative strategies, such as converting Cas9 into a synthetic transcriptional activator by fusing it to multiple copies of VP16 activator,14, 15, 16 have been developed. Studies from several groups suggest that using a sgRNA to target Cas9 activators to a particular endogenous gene promoter leads to only modest transcriptional upregulation.15, 16, 17 Furthermore, the domains used in CRISPR/Cas9-based activation, such as the VP16 decamer,18 act as recruiters for multiple components of the pre-initiation complex19 and most importantly do not enzymatically affect the epigenetic form of the chromatin.20 Instead, targeting a promoter with multiple sgRNAs might be a better alternative for increasing activation due to synergistic effects.15, 16, 17 Taken together, the CRISPR/Cas9 genome-engineering system has provided opportunities that have already revolutionized science in all areas of biomedical research.
Although CRISPR/Cas9 has been widely used as a research tool, its potential in far-reaching therapeutic applications has largely been unexplored. Most genetic disorders are associated with a life-threatening or a life-limiting disease trajectory, for which current clinical management is mainly supportive in nature. Recently, CRISPR/Cas9 has been employed for loss-of-function mutations in homozygous autosomal-recessive disorders (e.g., cystic fibrosis and sickle-cell disease)21, 22 and X-linked recessive disorders (e.g., Duchenne muscular dystrophy [DMD (MIM: 310200)])23, 24, 25 to correct the causative mutation and/or restore the open reading frame. In order to further explore the versatile and potentially broad therapeutic applications of the CRISPR/Cas9 technology, we developed a pipeline in which genome-engineering strategies use widely accessible cells from individuals affected by various genetic conditions. We show that this system can be employed for developing treatment strategies that modulate expression of genes that are known to play a critical role in disease pathogenesis. Furthermore, we demonstrate that CRISPR/Cas9 can be utilized to therapeutically target autosomal-dominant, heterozygous, gain-of-function mutations and large chromosomal rearrangements.
Material and Methods
Mapping the Duplication Junction
A series of probes near the junction were designed, and qPCR followed by sequencing was used for mapping out the exact breakpoint of the duplication. Primers for the MECP2 duplication were 5′-CCCACAGAGTAGAGTGGAGCAG-3′ (forward) and 5′-TTAGACAGAGTCTCACTCCATCACC-3′ (reverse). Primers for the duplication of DMD (MIM: 300377) exons 18–30 were 5′-CAGCATCATGACCTGTTTCAATC-3′ (forward) and 5′-TTGTTAGAGGGCAGCAAGTTTGT-3′ (reverse).
Cell Culture
Primary fibroblasts from individuals with achondroplasia (MIM: 100800), methyl-CpG binding protein 2 (MECP2 [MIM: 300005]) duplication syndrome (MIM: 300260), and DMD involving a duplication of DMD exons 18–30 were obtained from skin tags and established at the Hospital for Sick Children. They were maintained in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS), L-glutamine, and 1× penicillin and streptomycin (all from Life Technologies). Immortalized DMD myoblasts were obtained from the Institut de Myologie26 and maintained in Skeletal Muscle Cell Growth Media (Promocell) at 37°C with 5% CO2 incubation. The research ethic boards of each institution approved all of the experiments.
sgRNA Design
All UTRN (MIM: 128240) guides were selected on the basis of their proximity (50–500 bp) to the UTRN A or B transcription start site (TSS) and subcloned into dCas9-VP160 (Addgene 48240).27 Each plasmid contained a single locus-specific sgRNA in conjunction with a catalytically inactive SpCas9. FGFR3 sgRNAs g1 and g2 were chosen on the basis of their proximity to the mutation intended for editing and were subcloned into pSpCas9(BB)-2A-GFP (Addgene 48138).28 Guides targeting the MECP2 duplication were designed on the basis of the lowest possible off-target hits within the genome,29 as well as their position within the duplication. The guides targeting the DMD duplication were designed on the basis of the most active sgRNAs as computationally predicted by the online Benchling Tool described by Doench et al.30 All sgRNAs with a predicted activity score greater than 0.75 were next analyzed by the CRISPR Design tool29 and ranked according to the least possible number of potential off-target sites. The best predicted sgRNAs (Table S1) were then subcloned into the lentiCRISPR v.2 vector (Addgene 52961).31
Electroporation
For UTRN upregulation experiments, 3 μg of DNA composed of combinations of sgRNAs cloned into a dCas9-VP160 plasmid was electroporated into DMD myoblasts. An equal amount of original dCas9-VP160 plasmid served as a control. Electroporation was conducted on 2 × 105 myoblasts per condition with the NEON Transfection System (Life Technologies), which double pulsed cells at 950 V and a pulse width of 30 ms. Culture medium was replaced the following day, and cells were harvested for protein analysis 4 days after electroporation.
Nucleoporation
5 × 105 achondroplasia fibroblasts were nucleoporated with a total of 3 μg of DNA with program U-023 on the Amaxa system and the Primary Fibroblast Kit (Lonza). Cells were sorted by fluorescence-activated cell sorting 4 days after nucleoporation, and DNA was collected on the same day.
Lentivirus Production and Transduction
Production of lentiCRISPR and transduction into target cells were performed as described by Sanjana et al.31 with a slight modification. For production of the lentiCRISPR, 293T cells (ATCC) at 80% confluency in a 10 cm petri dish were transfected with 10 μg of transfer lentiCRISPR plasmid, 5 μg of the envelope (pCMV-VSV-G, Addgene) plasmid, and 7.5 μg of packaging (psPAX2, Addgene) plasmid via the calcium phosphate transfection method. 60 hr after transfection, supernatant was collected, centrifuged at 3,000 rpm for 10 min, and filtered through a 0.45 μm low-binding filter (Whatman).31 Fibroblasts affected by MECP2 duplication syndrome were plated on a 24-well plate until they were 60% confluent and then transduced with 1 ml lentiCRISPR plasmid containing MECP2 sgRNA A1 or A2. Each transduction condition was performed in triplicate, and proliferation medium was added to make the final volume 1.5 ml. Fibroblasts with the DMD duplication were co-transduced with Ad-MyoD (Vector Biolabs) at 100 MOI in DMEM with 1% FBS (for inducing their differentiation into myoblasts) and with a lentiCRISPR vector containing DMD sgRNA 1, similarly to above. 3 days after transduction, 2 μg/ml puromycin was added for selecting cells containing the lentiCRISPR-sgRNA constructs. DNA was collected 7 days after puromycin selection, and proteins were collected 7 days after differentiation.
Western Blot
The cells were lysed with RIPA buffer (50 mM Tris HCl [pH 7.4], 150 nM NaCl, 1 mM EDTA, 1% deoxycholate, 1% NP40, and 1% Triton X-100 supplemented with phosphatase and protease inhibitor cocktails [Roche]), and the protein concentration was measured by bicinchoninic acid (BCA) assay. 25 μg of protein lysates were resolved by western blot on 3%–8% Tris-acetate gels, transferred to nitrocellulose membranes, and probed for utrophin (MANCHO7 7E3, DSHB), dystrophin (MAB1692, Millipore), β-dystroglycan (MANDAG clone 7D11, DSHB), α-dystroglycan (kindly provided by Kevin Campbell), and β-tubulin (SantaCruz).
On-Target Editing Analysis
DNA transfected with Cas9 and FGFR3 sgRNA 1 or 2 was collected with the DNA Blood and Tissue Extraction Kit (QIAGEN). At the primary PCR step, DNA was amplified with primers FGFR3 5′-GCCCTCTAGACTCACTGGCGTTACT-3′ (forward) and FGFR3 5′-TGCCCCAAAGTACCCTAGGCTCTACAT-3′ (reverse) under the following conditions: initial denaturation at 95°C for 15 min, 15 cycles of denaturation at 95°C for 30 s, annealing at 58°C for 30 s, and extension at 72°C for 30 s. Finally, there was a further extension for 7 min at 72°C before the samples were cooled and stored at 4°C. The secondary PCR was performed to incorporate barcodes with the universal reverse primer 5′-CCACTACGCCTCCGCTTTCCTCTC TATGGGCAGTCGGTGATTCTGTTACCTGTCGCTTGA-3′ and either sgRNA 1 forward primer 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGAAGAGGATTCGATTCTTTGCAGCCGAGGAG-3′ or sgRNA 2 forward primer 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAGCTAAGGTAACGATTCTTTGCAGCCGAGGAG-3′. The PCR conditions were 95°C for 15 min, 15 cycles of denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Finally, there was a further extension for 7 min at 72°C before the samples were cooled and stored at 4°C. A purified fusion amplicon library was submitted to the sequencing facility at The Centre for Applied Genomics Facility (TCAG) at The Hospital for Sick Children (Toronto) for quality control, whereby the size of the amplicon library was checked on a Bioanalyzer 2100 instrument (Agilent) and quantified by Qubit (Invitrogen). Sequencing was performed at TCAG on an Ion Torrent Proton with a PI chip V3. Each potential on-target site was evaluated after corresponding sequencing reads were aligned to the human reference genome (UCSC Genome Browser hg19). The proportion of reads that matched the reference genome and the proportion of those with insertions, deletions, and substitutions near predicted cleavage sites were used for estimating the on-target editing activity of a corresponding sgRNA. A custom Python script was written for assessing read support for evidence of DNA editing given a BAM file, an anchor position P, and target length L (8 nucleotides). All reads overlapping position P were extracted from the BAM file. Reads that did not extend at least 8 bp beyond anchor point P were discarded. For each read, the 8 bases extending past position P were extracted, and the overall frequencies were calculated.
Off-Target Analysis
Off-target analysis was conducted as follows for all lentivirus-delivery-based gene-editing experiments. Primers targeting loci corresponding to each sgRNA’s top 20 off-target hits, as computed by the CRISPR Design tool,29 were designed and used to amplify DNA from each gene-editing experiment with custom GeneRead DNaseq Targeted Panels (QIAGEN). ∼200 bp amplicons were purified with magnetic beads, and library preparation was conducted with sample-specific barcodes and the Ion Torrent Library Preparation Kit (Life Technologies) at TCAG at The Hospital for Sick Children. Sequencing was performed with the Ion Torrent Proton. Each potential off-target site was evaluated after corresponding sequencing reads were aligned to the human reference genome (hg19). A custom Python script was utilized similarly as described above, and the proportion of reads that matched the reference genome and the proportion of those with insertions, deletions, and substitutions near predicted cleavage sites were used for estimating the off-target activity of a corresponding sgRNA. The top 12 off-target hits computed by COSMID,32 which were not predicted by the CRISPR Design tool,29 were assayed with the GeneArt Genomic Cleavage Detection Kit (Life Technologies) according to the manufacture’s protocol.
Results
Therapeutic Use of CRISPR/Cas9 for Modulating Gene Expression
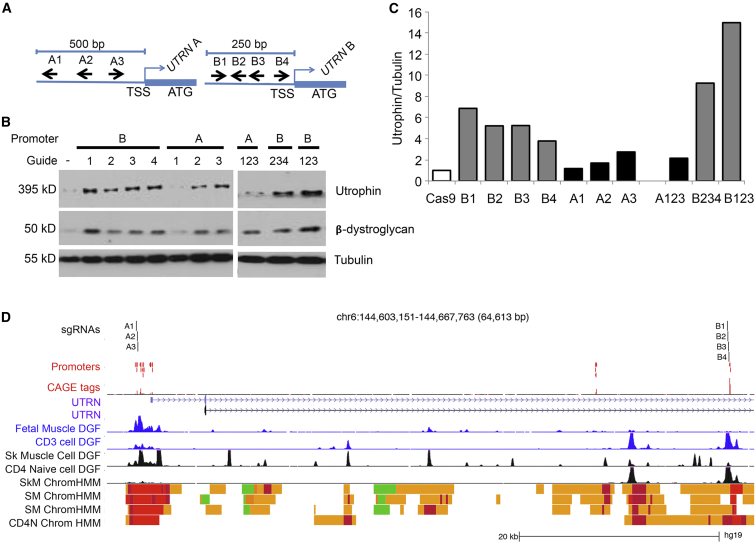
In order to explore the feasibility of utilizing a modified version of CRISPR/Cas9 for therapeutic approaches based on modulating gene expression, we aimed to upregulate utrophin in skeletal-muscle cells of an individual with DMD. DMD is an X-linked recessive neuromuscular disorder associated with muscle degeneration causing progressive weakness. Pathogenic mutations in DMD lead to an absence of the protein product, dystrophin, resulting in a disruption of the dystrophin-glycoprotein complex (DGC) at the sarcolemma. Increased amounts of utrophin have previously been shown to compensate in part for the loss of dystrophin.33 If successful, this approach would be beneficial to all individuals affected by DMD, independently of their primary genetic mutation. Here, we transfected myoblasts of a DMD-affected individual (who carries a deletion of DMD exons 45–52) with a catalytically inactive SpCas9 fused to ten tandem repeats of a transcriptional transactivator VP16 (dCas9-VP160)27 guided to either the UTRN A or UTRN B promoter (Figure 1A). Remarkably, we demonstrated that several sgRNAs targeting either promoter A34 or B35 upregulated utrophin amounts such that they were 1.7- to 2.7-fold or 3.8- to 6.9-fold, respectively, higher than basal amounts (Figures 1B and 1C). Furthermore, we demonstrated that the combination of guides targeting promoter B region, but not A, further increased the amount of utrophin.
Figure 1.
Utilizing CRISPR/Cas9 to Modulate Expression of UTRN, a Disease-Modifying Gene in DMD Myoblasts
(A) Schematic diagram (not to scale) of sgRNAs targeting regions upstream of UTRN A (A1–A3) and B (B1–B4) TSSs.
(B) CRISPR/Cas9-mediated transcriptional activation of UTRN in DMD myoblasts. Amounts of utrophin, β-dystroglycan, and tubulin were analyzed by western blot 4 days after transfection with dCas9-VP160 plasmid containing each sgRNA.
(C) The amount of utrophin was normalized to that of tubulin by densitometric analysis of four different experiments.
(D) Location of sgRNAs in relation to UTRN TSS, DNase I hypersensitivity footprints, and chromatin-state maps. sgRNAs are plotted above experimentally determined TSSs obtained from a FANTOM5 assay of over 300 primary tissues. The maximum signal at each promoter region is shown below the TSSs (CAGE tags). Digital DNase Footprinting (DGF) assays for fetal muscle and primary CD3 cells are shown in blue (ENCODE). DGF assays for skeletal-muscle cells, skeletal muscle, and naive CD4 cells are shown in black. Chromatin-state maps from the Roadmap Epigenomic Consortium are shown for skeletal-muscle cells (SkM), skeletal muscle (SM), and naive CD4 cells (CD4N). Red indicates TSSs, and yellow indicates enhancer states. The A guides all fall within muscle promoter regions. The B guides fall into an enhancer region immediately upstream of an annotated promoter region. In CD4 cells, this region is considered an active promoter. At promoter B, the DGF footprint in muscle cells is weak in comparison to that in CD4 cells. Data were plotted according to positions from the UCSC Genome Browser. FANTOM5, DGF, and chromatin-state data were obtained from UCSC “Track Hubs.”
We then sought to investigate the transcriptional mechanisms that could explain the different potential of our A and B guides to upregulate UTRN. We analyzed publicly available data describing experimentally captured TSSs (FANTOM536), high-resolution DNase I hypersensitivity mapping (Digital Genomic Footprinting [DGF]37), and chromatin states from the Roadmap Epigenomics Consortium38 and ENCODE.39 We observed that in fetal and adult skeletal muscle, there was a prominent DNase I footprint at the A promoter, but not at the B promoter (Figure 1D; Figure S1), which is preferentially used in T lymphocytes. These suggest that in muscle cells, the potential to open chromatin and activate the B promoter is greater than the potential at the already active A promoter.
Nevertheless, despite possible differences in the promoter usage in muscle cells, even a modest ∼1.7-fold increase in the amount of utrophin (in comparison to basal amounts), as observed with sgRNA A2, was accompanied by restored amounts of β-dystroglycan, providing evidence for the functional relevance of this strategy. Our data demonstrate that this adapted, sgRNA CRISPR/Cas9 approach is capable of targeting expression levels of disease-modifying genes that can be explored for various human disorders.
Allele-Specific Disruption of a Dominant-Negative, Gain-of-Function Mutation
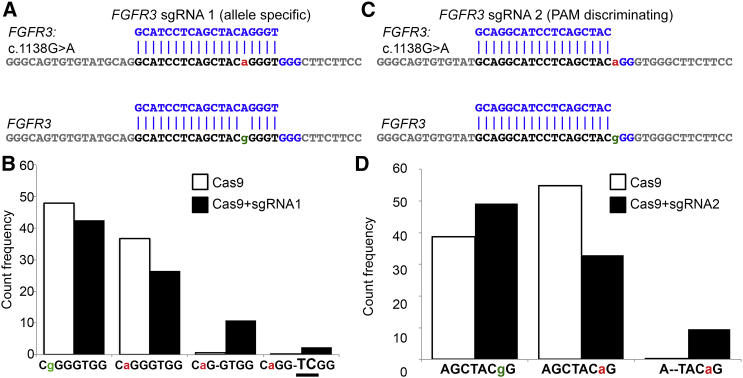
We next wanted to interrogate whether CRISPR/Cas9 can be employed for achieving NHEJ-mediated allele-specific disruption of a dominant-negative disease-causing allele. We therefore assessed fibroblasts of an individual with achondroplasia, which is the most common cause of dwarfism in humans and is often associated with hydrocephalus, sleep apnea, and spinal stenosis. 98% of affected individuals carry the same pathogenic dominant-negative, gain-of-function mutation (c.1138G>A [p.Gly380Arg]) in fibroblast growth factor receptor 3 (FGFR3 [MIM: 134934]). Given the sequence surrounding the region of interest, we were able to design an allele-specific FGFR3 sgRNA 1 complementary to the pathogenic allele (Figure 2A). We then electroporated the achondroplasia fibroblasts with Cas9 and FGFR3 sgRNA 1 and used deep sequencing to analyze the FGFR3 exon 9 region. We detected a variety of indel types (Table S3), but 10.85% of the total reads carried a 1 nt deletion on the pathogenic allele (Figure 2B), consistent with its inactivation. Furthermore, the FGFR3 c.1138G>A mutation presents an unusual situation where the pathogenic SNP generates a unique PAM sequence. The wild-type (WT) allele contains a 5′-GGG-3′ PAM site, and the mutant FGFR3 c.1138G>A allele has a 5′-AGG-3′ variant. The PAM is necessary for Cas9 recognition of the target sequence, and the canonical PAM for SpCas9 is 5′-NGG-3′,9 where any nucleotide can constitute the first 5′ position. Interestingly, recent analyses of activity of a vast library of sgRNAs detected a PAM-specific bias,30 suggesting that unique differences coupled with a precise sgRNA sequence might confer a targeting discrimination. Hence, we utilized the 1 bp discrepancy in the PAM-recognition sequence to design PAM-discriminating sgRNA 2 (Figure 2C) in an attempt to achieve allele-specific targeting in achondroplasia. Interestingly, we found that the pathogenic A allele was targeted more frequently than the G allele in that up to 9.8% of total reads carried a 2 nt deletion (Figure 2D; Table S3). These data demonstrate that our designed sgRNA preferentially targets the mutant allele. Altogether, we provide evidence that both allele-specific and PAM-discriminating sgRNAs can be used for preferential targeting of disease-causing autosomal-dominant mutations.
Figure 2.
Targeted Elimination of a Dominant-Negative, Gain-of-Function Allele in the Receptor Gene FGFR3
(A) Position of FGFR3 sgRNA 1 in relation to that of the pathogenic FGFR3 c.1138G>A allele and the WT FGFR3 allele. Lowercase letters denote the distinguishing bases between the affected (a, red) and WT (g, green) alleles.
(B) Quantification of major indels associated with alleles A and G in cells transfected with Cas9 (white bars) or Cas9 and sgRNA 1 (black bars).
(C) Position of FGFR3 sgRNA 2 in relation to that of the pathogenic FGFR3 c.1138G>A allele and the WT FGFR3 allele.
(D) Quantification of major indels associated with allele A and G in cells transfected with Cas9 (white bars) or Cas9 and sgRNA 2 (black bars).
Targeted Removal of a Large Duplicated Chromosomal Rearrangement
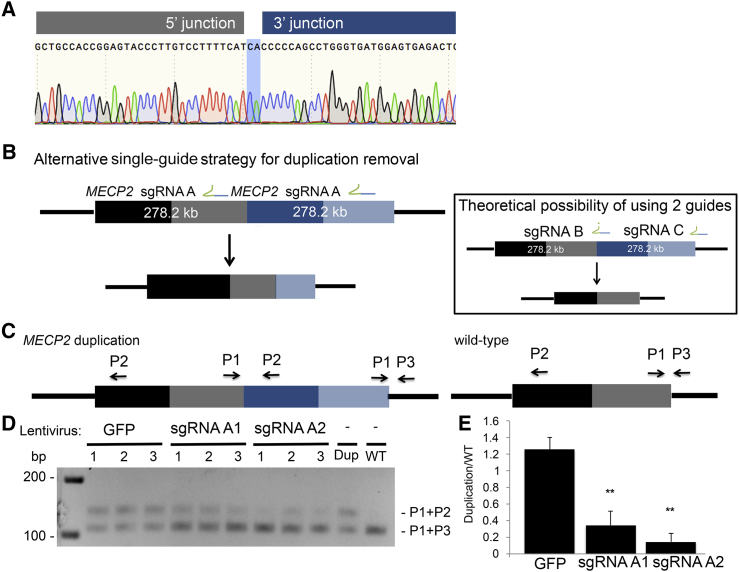
With the development of powerful genome-analysis platforms, there is growing evidence of the prevalence of copy-number variations (CNVs) associated with numerous genetic conditions.40 However, to date no therapeutic strategies have been developed to target these large genomic rearrangements, such as that in MECP2 duplication syndrome. This is a rare condition associated with intellectual disability and macrocephaly. It is caused by a variably sized X chromosome CNV that includes a duplication of MECP2. In order to explore whether the CRISPR/Cas9 technology could be utilized to target this CNV, we first determined the exact orientation and breakpoint junction sequence of the duplication in a male individual with this disorder to be a chrX: 153,420,649–153,142,419 (hg19) duplication with a CA insertion at the breakpoint junction (Figure 3A). In this study, we explored an alternative therapeutic strategy of using the CRISPR/Cas9 system in conjunction with only one sgRNA to remove a duplication (Figure 3B). The sgRNA will bind to two places within a duplication, leading to the formation of two DSBs and hence the removal of the intervening sequence, which equates to the total size of the duplication. Duplication removal could theoretically be achieved with two sgRNAs, one of which targets the duplication junction (Figure 3B, inset); however, it is dependent on the availability of PAM-recognition sequences within this region. Furthermore, the one-sgRNA approach provides an opportunity to evaluate the entire duplication sequence in order to design RNA guides with the least possible off-targets, thereby alleviating the sequence restrictions presented by the two-guide approach. Moreover, using one instead of two sgRNAs is therapeutically appealing given the limited loading capacity of potential in vivo delivery vehicles such as adeno-associated virus 9 (AAV9).
Figure 3.
Targeted Removal of a 278 kb X Chromosome Duplication Containing MECP2
(A) Electropherogram of the X chromosome duplication junction; highlighted in blue is the insertion of CA at the junction.
(B) Overview of the single-guide strategy for removing the MECP2 duplication. The first copy of MECP2 is denoted by black and gray bars, whereas the second copy is depicted by dark and light-blue bars. The inset describes an alternate, less favorable duplication-removal strategy using two sgRNAs. The relative positions of MECP2 sgRNAs A1 and A2 are shown on the duplicated region.
(C) Removal of the duplicated region was detected with a PCR strategy using three primers positioned to the duplicated locus. P1 and P3 are universal to the region of interest, whereas P2 only amplifies the duplication junction with P1.
(D) A Cas9 nuclease guided by sgRNA 1 or 2 or a GFP control was delivered to affected fibroblasts via lentiviral particles. Three-primer PCR demonstrated an accumulation of the bottom band (corresponding to the WT single-copy amplicon) and a decrease in the top band (corresponding to the duplicated copy).
(E) Densitometric analysis depicting a decrease in the ratio between the duplicated band and the WT band. ∗∗p < 0.01 (Student’s t test from three independent experiments).
In the affected fibroblasts, the duplicated copy of MECP2 could be precisely removed with a single sgRNA given its tandem head-to-tail orientation. In order to test this hypothesis, we designed two sgRNAs within the 278 kb duplication while excluding any known coding regions or regulatory elements (Figure 3A). We tested the activity of these guides in primary fibroblasts of a healthy control individual by using MECP2 sgRNAs A1 and A2 with SpCas9, and we were able to show that the deletion process of a 114 kb fragment occurred through precise end joining of the two ends upon the two staggered Cas9-mediated DSBs (Figures S2B–S2D). Next, we transduced primary dermal fibroblasts from a male with MECP2 duplication syndrome with lentiCRISPR containing either MECP2 sgRNA A1 or A2. We isolated DNA 10 days after infection and employed a three-primer PCR detection strategy where primers 1 and 3 (P1 + P3) were universal to the region of interest and amplified the WT, single-copy junction and primer 2 (P2) was specific to the junction of the duplicated region (Figure 3C). We were able to detect a loss of the amplicon specific to the duplicated region and an accumulation of the amplicon corresponding to a single WT copy, demonstrating that the entire duplication was successfully removed after treatment (p < 0.01; Figures 3D and 3E). Our results establish that a single-sgRNA approach provides a highly efficient therapeutic strategy for removing chromosomal duplications and that this strategy can be explored for a number of different disorders caused by CNVs.
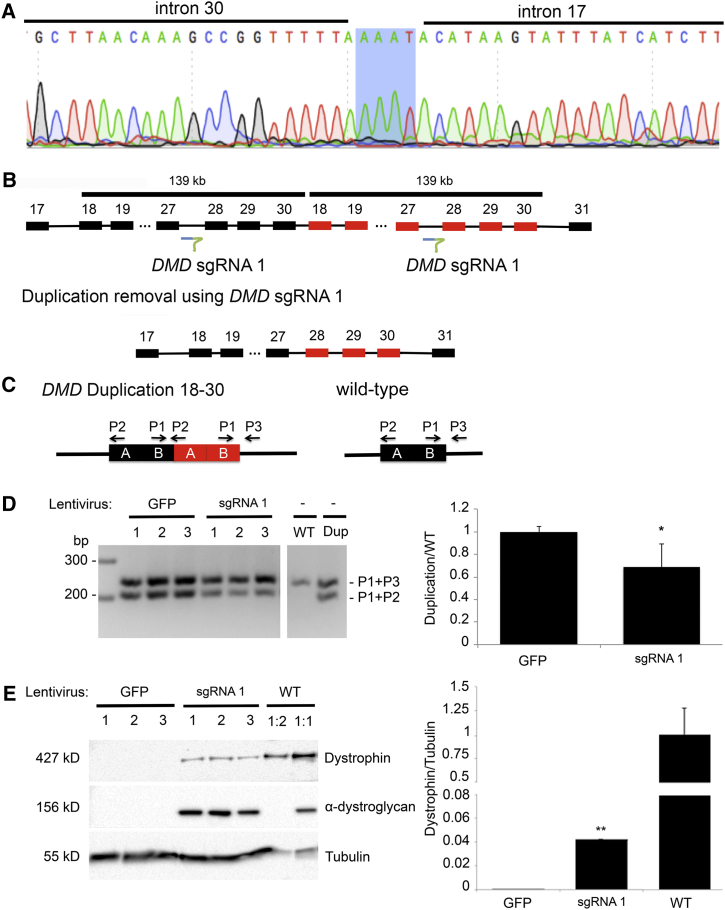
Given our successful removal of a large chromosomal duplication, we next investigated whether a similar, single-sgRNA approach could be applied to large exonic duplications in DMD. Treatment strategies that target duplications in DMD have not been extensively studied to date even though duplications of one or more exons compose approximately 10% of the DMD mutation spectrum.41 We first determined the junction of a duplication of exons 18–30 in a DMD-affected individual to be a chrX: 32,552,206–32,413,149 (hg19) tandem duplication with an AAAT insertion at the breakpoint junction (Figure 4A). We co-transduced the affected fibroblasts with adeno-MyoD to induce transdifferentiation of fibroblasts into myoblasts and lentiviral-vector-containing DMD sgRNA 1. To assess for evidence of duplication removal on a molecular level, we employed a three-primer PCR strategy as previously described and illustrated in Figure 4C. In the WT control, we detected a higher band, corresponding to the amplification product of P1 + P3, whereas the duplication control showed two bands corresponding to the P1 + P3 product and the duplication-junction-specific amplification product of P1 + P2 at a ratio of 1:1. After lentiCRISPR treatment with sgRNA 1, but not with LentiGFP, the ratio became skewed toward the top band (p < 0.05; Figure 4D), indicating a conversion of the duplicated allele toward the WT single copy. We next explored whether the molecular transition toward the WT allele leads to functional restoration of protein. Remarkably, we detected full-length dystrophin (4.42%) in transdifferentiated myotubes treated with the single DMD sgRNA 1, and this was accompanied by restoration of α-dystroglycan, a critical component of the DGC (Figure 4E). All together, our data demonstrate that CRISPR/Cas9-mediated removal of duplications results in production of full-length functional dystrophin in myotubes, which opens up entirely new treatment strategies for individuals affected by DMD duplications.
Figure 4.
Genome-Editing Strategies for Individuals with Duplication of DMD Exons 18–30
(A) Electropherogram of the junction of the duplication of DMD exons 18–30; highlighted in blue is the insertion of AAAT at the junction.
(B) Schematic of the position of DMD sgRNA 1 and the duplication-removal strategy.
(C) Schematic of the three-primer duplication-removal strategy.
(D) Targeted deletion of a 139 kb duplication in DMD. PCR was performed on DNA from three replicate experiments in which affected myoblasts were transduced with LentiGFP or lentiCRISPR Cas9 nuclease with DMD sgRNA 1. The top band was amplified with universal primers (P1 + P3) to both an allele with the duplication and a control. The bottom band is specific to alleles harboring the duplication (P1 + P2). A decrease in the bottom band, indicating removal of the duplicated region, was only observed when Cas9 and sgRNA 1 were present.
(E) Western blot with antibodies against dystrophin, α-dystroglycan, and tubulin as a loading control. The amount of dystrophin was normalized to that of tubulin by densitometric analysis. ∗p < 0.05, ∗∗p < 0.01 (Student’s t test from three independent experiments).
Discussion
Recent development of genome-editing technologies based on the RNA-guided CRISPR-associated endonuclease Cas9 has generated enormous excitement across many fields, including biological research, biotechnology, and clinical medicine. Despite being a nascent technology, Cas9 has been successfully used for generating an increasing number of cellular and animal models for a variety of basic research, and it has also been applied in biotechnology. Furthermore, the CRISPR/Cas9 system can be exploited for the development of genome-engineering therapies, which carries the potential to revolutionize medical management in the future. Our current study defines a pipeline in which genome-engineering strategies use easily accessible cells from affected individuals and provides evidence of the versatility of the CRISPR/Cas9 system, which can be employed for various genetic conditions.
One of the opportunities to therapeutically utilize the CRISPR/Cas9 technology is to affect disease phenotype and progression by modulating expression of genes that are known to play a critical role in disease pathogenesis. An example of a gene that ameliorates the progression of disease is UTRN, which has been known to partly compensate for the loss of dystrophin in DMD (reviewed in Fairclough et al.42). DMD is a life-limiting, progressive neuromuscular disorder, and currently the only therapies available are corticosteroids, which slightly improve the phenotype of individuals affected by DMD and are associated with a number of significant side effects (reviewed in Bushby et al.43). Utrophin’s ability to modify disease progression has been established in multiple experiments using the dystrophin-negative mdx mouse, and it is suggested that increasing levels of UTRN mRNA over 2-fold more than basal levels is sufficient to have functional benefit.42, 44 In addition, small molecules that target upregulation of utrophin are currently in early stages of clinical trials.33 Here, we provide an alternative, CRISPR/Cas9-mediated therapeutic approach for upregulating utrophin and using sgRNAs to target two UTRN promoters. Our approach upregulated the amount of utrophin such that it was 1.7- to 6.9-fold more than the basal amount and restored the amount of β-dystroglycan in muscle cells of individuals with DMD. Interestingly, we found that we could successfully target both UTRN A and B promoters with single sgRNAs and a combination of three sgRNAs; however, we found that upregulation by guides targeting promoter B was more robust.
The different potentials of our A and B guides to activate UTRN expression could be explained by the epigenomic landscape surrounding the promoter regions (Figure 1D; Figure S1). Although the relationship between genomic features and gene expression is complex, shorter genes generally correlate with higher transcript levels.45 For this reason, we speculate that if both the A and B promoters had similar promoter-proximal polymerase II kinetics, the B promoter, which is located more than 50 kb downstream of the A promoter, could result in more-efficient transcription of UTRN. Given that a small increase in utrophin is sufficient to restore β-dystroglycan levels and this phenomenon is true for other genetic diseases,46, 47 it will be important to further establish the mechanisms behind the differential ability to upregulate the transcription and translation of genes of interest.
Future experiments focusing on both promoters could reveal new insights into determining the preferred target promoter for developing CRISPR/Cas9-based therapeutic strategies for DMD. Importantly, previous data have shown that the efficiency of CRISPR/Cas9-mediated gene upregulation is very gene specific.27 Thus, this approach could serve as an alternative to current pharmacological drug-development strategies, which modulate pathways associated with disease pathogenesis. Furthermore, in vivo targeting using AAVs with specific tissue tropism (e.g., AAV9 for striated muscles) will add to the specificity of CRISPR-mediated upregulation because delivery can be tailored to particular cell types of interest.
CRISPR-mediated targeted removal of a dominant allele is distinct from the editing of homozygous and X-linked recessive mutations because the designed guide RNA would require allele specificity. This can be particularly challenging given that the difference between two alleles is often only one nucleotide, as in the case of the FGFR3 c.1138G>A mutation. Here, we have shown that one can achieve allelic specificity by designing allele-specific and PAM-discriminating sgRNAs. Allele-discriminating guides based on a single SNP in the guide target itself have been previously reported,48, 49 but ours is an example of an allele-specific PAM-discriminating guide. The pathogenic FGFR3 c.1138G>A mutation is unique in that the 5′ N of the 5′-NGG-3′ PAM is the discriminating nucleotide between the two alleles. The striking preferential targeting of the mutant allele indicates that the first position in the PAM can dramatically affect sgRNA activity, an observation previously reported by two different groups on the basis of analysis of diverse sgRNA libraries.30, 50 Moreover, Gagnon et al.50 have suggested a preferential targeting of the 5′-AGG-3′ PAM over the 5′-GGG-3′ PAM, which is in agreement with the data presented here. It should be emphasized, however, that recent studies evaluating activities of almost 2,000 sgRNAs in zebrafish in vivo demonstrated preferential utilization of either the G or C nucleotide in the variable position of the NGG PAM,51 further highlighting current limitations in bioinformatic predictions of the most active sgRNAs. Therefore, a possible use of an individual PAM-discriminating sgRNA should be considered on a case-by-case basis in situations where allele-specific guide RNAs cannot be designed or display similar activities targeting both alleles. Nevertheless, we provide evidence that PAM-discriminating sgRNAs can correct dominant-negative disease-associated sequence variants, and this might serve as an alternative approach for allele-specific targeting of disease-causing mutations.
An increasing number of genetic disorders are caused by chromosomal rearrangements and CNVs. However, treatments targeting the underlying causes of these disorders are currently not available. Although deletion of genomic, single-copy DNA has been shown with the use of zinc fingers52 and two guides in the CRISPR/Cas9 system,25, 53 removal of duplications has not yet been demonstrated. Furthermore, it has been unclear whether this type of genetic correction would restore a fully functional gene. Here, we developed a strategy that employs the CRISPR/Cas9 system to remove duplicated regions within the genome. Our strategy uses only one sgRNA, which creates two DSBs because of the nature of a tandem (head-to-tail) duplication (Figure 3B). Because we are targeting a sequence within a duplication, the sgRNA target will be detected twice, leading to the formation of two DSBs and hence the removal of the intervening sequence, which equates to the total size of the duplication. There are several advantages to this strategy. First, the design of RNA guides is not limited to specific sequences near the breakpoints. This allows for the selection from a large pool of guide RNAs targeting any portion of the duplicated sequence, therefore greatly reducing possible off-target effects. Second, given the limited loading capacity of potential in vivo delivery vehicles such as AAV9, strategies using the least amount of CRISPR components will be critical for the development of further therapeutic applications. We first used this strategy to successfully remove a large X chromosome rearrangement containing MECP2, indicating that this approach can target several chromosomal-duplication syndromes. Importantly, off-target analysis showed no significant hits in the top 60 sites predicted by the CRISPR Design tool29 (Table S2) and top 12 hits predicted by COSMID32 (these hits were validated by next-generation sequencing and the GeneArt Genomic Cleavage Detection Kit [Figure S3]), suggesting that the accuracy and safety of our system lend themselves to a viable strategy for future therapeutic developments. It is important to note that there were discrepancies between the off-target sites identified by the CRISPR Design tool and those identified by COSMID, further emphasizing that there is a need for new non-biased off-target analyses, such as GUIDE-seq54 and/or high-throughput, genome-wide, translocation sequencing (HTGTS) methods.55, 56
To determine whether our previously undescribed duplication-removal strategy has broader applicability, we applied this approach to individuals affected by DMD. To date, treatments that specifically target duplications in DMD have not been extensively studied even though duplications of one or more exons compose approximately 10% of the DMD mutation spectrum.41 Recent therapeutic strategies undertaken by other groups include gene-replacement therapies, which deliver truncated but functional microdystrophin genes.57, 58 Other strategies utilize exon skipping, where antisense oligonucleotides complementary to regions of premature DMD mRNA are used to induce skipping of one59, 60 or more61 exons and hence restore the open reading frame to produce a shorter dystrophin protein. Similarly, previous studies from other laboratories have demonstrated that the CRISPR/Cas9 system can be utilized to restore the reading frame of large deletions in DMD.25 However, one potential shortcoming of these approaches is that the shorter dystrophin product ameliorates the disease phenotype only to the extent of making it similar to that of individuals affected by Becker muscular dystrophy, where a truncated yet functional dystrophin protein is detected.62 Thus, our data are of particular importance given that removal of a duplication restores the full-length dystrophin, which represents new therapeutic opportunities for DMD-affected individuals with duplications.
An important consideration in establishing a treatment for DMD is determining how much dystrophin is necessary for ameliorating the disease phenotype. It is estimated that in humans, about 20% of truncated dystrophin is sufficient to cause a less severe phenotype and maintain ambulation.63, 64 Furthermore, studies in mdx mice suggest that approximately 5% of full-length dystrophin can improve disease pathology and that >20% is needed for fully protecting muscle fibers from exercise-induced damage.65, 66, 67 One potential challenge for this treatment strategy is the delivery vehicle for Cas9 and sgRNAs. In this study, we used lentiviral vectors because they easily infect primary cell cultures. However, future in vivo studies will include more clinically feasible vehicles such as AAVs. Nonetheless, although it is difficult to extrapolate our in vitro data to potential in vivo situations, our data demonstrating 4.42% of full-length dystrophin accompanied by restoration of components of the DGC are promising as we currently continue to explore the in vivo therapeutic feasibility of this approach.
Recent estimates suggest that about 400 million people worldwide are affected by orphan diseases, and most of them are caused by primary genetic abnormalities.68 Although orphan-drug development has made some progress over the last few years, most genetic disorders lack efficient treatments and are often associated with a life-threatening or life-limiting disease trajectory. The CRISPR/Cas9 system provides a rare opportunity to employ a technology that can not only target the underlying primary disease-causing genetic abnormalities but also alter genetic modifiers that play a critical role in the pathogenesis of a certain disease. Here, we have developed a pipeline in which genome-engineering strategies use easily accessible cells from affected individuals and provide evidence of the versatility of the CRISPR/Cas9 system for various genetic conditions. As a mutation-independent approach, we demonstrated the feasibility of modulating disease-modifying genes, such as UTRN in DMD. Furthermore, we found that targeted elimination of disease-causing alleles can be employed as a therapeutic strategy for disorders caused by dominant-negative mutations. Finally, we demonstrated that individually tailored single RNA guides are able to remove large duplicated genomic rearrangements in two different genetic disorders. As outlined here, proof-of-concept studies utilizing affected individuals’ cells are critical in laying the foundation for further research into the application of these therapeutic strategies for safe and efficient postnatal, in vivo treatments for numerous inherited disorders.
Acknowledgments
We would like to thank Drs. Jennifer Doudna, Steve Lin, Aravinda Chakravarti, Hal Dietz, and Janet Rossant for critical insights into our studies, as well as Minggao Liang and Wilson Sung for their bioinformatics input. We also thank Drs. Feng Zhang and Rudolf Jaenisch and their laboratories for the CRISPR/Cas9 backbone constructs used in this study. We thank the R.D.C. lab members and The Centre for Applied Genomics staff for excellent technical assistance, as well as the platform for immortalization of human cells of the Institute of Myology in Paris. This work is supported by SickKids Restracomp and Eric Hani Fellowship (to D.W.), AFM-Telethon (to D.U.K), Cure CMD (to D.U.K. and R.D.C.), a tier II Natural Sciences and Engineering Research Council Canada Research Chair (436194-2013 to M.D.W), the Duchenne Children’s Trust (to R.D.C.), the Women's Auxiliary of the The Hospital for Sick Children (Chair in Clinical and Metabolic Genetics to R.D.C.), and the SickKids Foundation (to R.D.C.).
Published: December 10, 2015
Footnotes
Supplemental Data include three figures and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.012.
Web Resources
The URLs for data presented herein are as follows:
Benchling, https://www.benchling.com
CRISPR Design, http://crispr.mit.edu
OMIM, https://www.omim.org
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Barrangou R., Fremaux C., Deveau H., Richards M., Boyaval P., Moineau S., Romero D.A., Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 2.Bolotin A., Quinquis B., Sorokin A., Ehrlich S.D. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 3.Mojica F.J., Díez-Villaseñor C., García-Martínez J., Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 4.Pourcel C., Salvignol G., Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 5.Wiedenheft B., Sternberg S.H., Doudna J.A. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 6.Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A., Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deltcheva E., Chylinski K., Sharma C.M., Gonzales K., Chao Y., Pirzada Z.A., Eckert M.R., Vogel J., Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mojica F.J., Díez-Villaseñor C., García-Martínez J., Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 12.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert L.A., Larson M.H., Morsut L., Liu Z., Brar G.A., Torres S.E., Stern-Ginossar N., Brandman O., Whitehead E.H., Doudna J.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mali P., Aach J., Stranges P.B., Esvelt K.M., Moosburner M., Kosuri S., Yang L., Church G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez-Pinera P., Ousterout D.G., Brunger J.M., Farin A.M., Glass K.A., Guilak F., Crawford G.E., Hartemink A.J., Gersbach C.A. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat. Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maeder M.L., Linder S.J., Cascio V.M., Fu Y., Ho Q.H., Joung J.K. CRISPR RNA-guided activation of endogenous human genes. Nat. Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beerli R.R., Dreier B., Barbas C.F., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memedula S., Belmont A.S. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol. 2003;13:241–246. doi: 10.1016/s0960-9822(03)00048-4. [DOI] [PubMed] [Google Scholar]
- 20.Hilton I.B., D’Ippolito A.M., Vockley C.M., Thakore P.I., Crawford G.E., Reddy T.E., Gersbach C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang X., Wang Y., Yan W., Smith C., Ye Z., Wang J., Gao Y., Mendelsohn L., Cheng L. Production of Gene-Corrected Adult Beta Globin Protein in Human Erythrocytes Differentiated from Patient iPSCs After Genome Editing of the Sickle Point Mutation. Stem Cells. 2015;33:1470–1479. doi: 10.1002/stem.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schwank G., Koo B.K., Sasselli V., Dekkers J.F., Heo I., Demircan T., Sasaki N., Boymans S., Cuppen E., van der Ent C.K. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 23.Li H.L., Fujimoto N., Sasakawa N., Shirai S., Ohkame T., Sakuma T., Tanaka M., Amano N., Watanabe A., Sakurai H. Precise correction of the dystrophin gene in duchenne muscular dystrophy patient induced pluripotent stem cells by TALEN and CRISPR-Cas9. Stem Cell Reports. 2015;4:143–154. doi: 10.1016/j.stemcr.2014.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long C., McAnally J.R., Shelton J.M., Mireault A.A., Bassel-Duby R., Olson E.N. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ousterout D.G., Kabadi A.M., Thakore P.I., Majoros W.H., Reddy T.E., Gersbach C.A. Multiplex CRISPR/Cas9-based genome editing for correction of dystrophin mutations that cause Duchenne muscular dystrophy. Nat. Commun. 2015;6:6244. doi: 10.1038/ncomms7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mamchaoui K., Trollet C., Bigot A., Negroni E., Chaouch S., Wolff A., Kandalla P.K., Marie S., Di Santo J., St Guily J.L. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle. 2011;1:34. doi: 10.1186/2044-5040-1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng A.W., Wang H., Yang H., Shi L., Katz Y., Theunissen T.W., Rangarajan S., Shivalila C.S., Dadon D.B., Jaenisch R. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doench J.G., Hartenian E., Graham D.B., Tothova Z., Hegde M., Smith I., Sullender M., Ebert B.L., Xavier R.J., Root D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: A Web-based Tool for Identifying and Validating CRISPR/Cas Off-target Sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tinsley J.M., Fairclough R.J., Storer R., Wilkes F.J., Potter A.C., Squire S.E., Powell D.S., Cozzoli A., Capogrosso R.F., Lambert A. Daily treatment with SMTC1100, a novel small molecule utrophin upregulator, dramatically reduces the dystrophic symptoms in the mdx mouse. PLoS ONE. 2011;6:e19189. doi: 10.1371/journal.pone.0019189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dennis C.L., Tinsley J.M., Deconinck A.E., Davies K.E. Molecular and functional analysis of the utrophin promoter. Nucleic Acids Res. 1996;24:1646–1652. doi: 10.1093/nar/24.9.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burton E.A., Tinsley J.M., Holzfeind P.J., Rodrigues N.R., Davies K.E. A second promoter provides an alternative target for therapeutic up-regulation of utrophin in Duchenne muscular dystrophy. Proc. Natl. Acad. Sci. USA. 1999;96:14025–14030. doi: 10.1073/pnas.96.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J., Roadmap Epigenomics Consortium Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Consortium E.P., ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tuffery-Giraud S., Béroud C., Leturcq F., Yaou R.B., Hamroun D., Michel-Calemard L., Moizard M.P., Bernard R., Cossée M., Boisseau P. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database: a model of nationwide knowledgebase. Hum. Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 42.Fairclough R.J., Wood M.J., Davies K.E. Therapy for Duchenne muscular dystrophy: renewed optimism from genetic approaches. Nat. Rev. Genet. 2013;14:373–378. doi: 10.1038/nrg3460. [DOI] [PubMed] [Google Scholar]
- 43.Bushby K., Muntoni F., Urtizberea A., Hughes R., Griggs R. Report on the 124th ENMC International Workshop. Treatment of Duchenne muscular dystrophy; defining the gold standards of management in the use of corticosteroids. 2-4 April 2004, Naarden, The Netherlands. Neuromuscul. Disord. 2004;14:526–534. doi: 10.1016/j.nmd.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 44.Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J.M., Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 45.Chiaromonte F., Miller W., Bouhassira E.E. Gene length and proximity to neighbors affect genome-wide expression levels. Genome Res. 2003;13:2602–2608. doi: 10.1101/gr.1169203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gräslund T., Li X., Magnenat L., Popkov M., Barbas C.F., 3rd Exploring strategies for the design of artificial transcription factors: targeting sites proximal to known regulatory regions for the induction of gamma-globin expression and the treatment of sickle cell disease. J. Biol. Chem. 2005;280:3707–3714. doi: 10.1074/jbc.M406809200. [DOI] [PubMed] [Google Scholar]
- 47.Van Ry P.M., Minogue P., Hodges B.L., Burkin D.J. Laminin-111 improves muscle repair in a mouse model of merosin-deficient congenital muscular dystrophy. Hum. Mol. Genet. 2014;23:383–396. doi: 10.1093/hmg/ddt428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith C., Abalde-Atristain L., He C., Brodsky B.R., Braunstein E.M., Chaudhari P., Jang Y.Y., Cheng L., Ye Z. Efficient and allele-specific genome editing of disease loci in human iPSCs. Mol. Ther. 2015;23:570–577. doi: 10.1038/mt.2014.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yoshimi K., Kaneko T., Voigt B., Mashimo T. Allele-specific genome editing and correction of disease-associated phenotypes in rats using the CRISPR-Cas platform. Nat. Commun. 2014;5:4240. doi: 10.1038/ncomms5240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gagnon J.A., Valen E., Thyme S.B., Huang P., Akhmetova L., Pauli A., Montague T.G., Zimmerman S., Richter C., Schier A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moreno-Mateos M.A., Vejnar C.E., Beaudoin J.D., Fernandez J.P., Mis E.K., Khokha M.K., Giraldez A.J. CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat. Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee H.J., Kim E., Kim J.S. Targeted chromosomal deletions in human cells using zinc finger nucleases. Genome Res. 2010;20:81–89. doi: 10.1101/gr.099747.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Canver M.C., Bauer D.E., Dass A., Yien Y.Y., Chung J., Masuda T., Maeda T., Paw B.H., Orkin S.H. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tsai S.Q., Zheng Z., Nguyen N.T., Liebers M., Topkar V.V., Thapar V., Wyvekens N., Khayter C., Iafrate A.J., Le L.P. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 2015;33:187–197. doi: 10.1038/nbt.3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chiarle R., Zhang Y., Frock R.L., Lewis S.M., Molinie B., Ho Y.J., Myers D.R., Choi V.W., Compagno M., Malkin D.J. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147:107–119. doi: 10.1016/j.cell.2011.07.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frock R.L., Hu J., Meyers R.M., Ho Y.J., Kii E., Alt F.W. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat. Biotechnol. 2015;33:179–186. doi: 10.1038/nbt.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimura E., Li S., Gregorevic P., Fall B.M., Chamberlain J.S. Dystrophin delivery to muscles of mdx mice using lentiviral vectors leads to myogenic progenitor targeting and stable gene expression. Mol. Ther. 2010;18:206–213. doi: 10.1038/mt.2009.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y., Duan D. Novel mini-dystrophin gene dual adeno-associated virus vectors restore neuronal nitric oxide synthase expression at the sarcolemma. Hum. Gene Ther. 2012;23:98–103. doi: 10.1089/hum.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cirak S., Arechavala-Gomeza V., Guglieri M., Feng L., Torelli S., Anthony K., Abbs S., Garralda M.E., Bourke J., Wells D.J. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goemans N.M., Tulinius M., van den Akker J.T., Burm B.E., Ekhart P.F., Heuvelmans N., Holling T., Janson A.A., Platenburg G.J., Sipkens J.A. Systemic administration of PRO051 in Duchenne’s muscular dystrophy. N. Engl. J. Med. 2011;364:1513–1522. doi: 10.1056/NEJMoa1011367. [DOI] [PubMed] [Google Scholar]
- 61.Aoki Y., Yokota T., Nagata T., Nakamura A., Tanihata J., Saito T., Duguez S.M., Nagaraju K., Hoffman E.P., Partridge T., Takeda S. Bodywide skipping of exons 45-55 in dystrophic mdx52 mice by systemic antisense delivery. Proc. Natl. Acad. Sci. USA. 2012;109:13763–13768. doi: 10.1073/pnas.1204638109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jarmin S., Kymalainen H., Popplewell L., Dickson G. New developments in the use of gene therapy to treat Duchenne muscular dystrophy. Expert Opin. Biol. Ther. 2014;14:209–230. doi: 10.1517/14712598.2014.866087. [DOI] [PubMed] [Google Scholar]
- 63.Nicholson L.V., Bushby K.M., Johnson M.A., Gardner-Medwin D., Ginjaar I.B. Dystrophin expression in Duchenne patients with “in-frame” gene deletions. Neuropediatrics. 1993;24:93–97. doi: 10.1055/s-2008-1071521. [DOI] [PubMed] [Google Scholar]
- 64.Nicholson L.V., Johnson M.A., Bushby K.M., Gardner-Medwin D., Curtis A., Ginjaar I.B., den Dunnen J.T., Welch J.L., Butler T.J., Bakker E. Integrated study of 100 patients with Xp21 linked muscular dystrophy using clinical, genetic, immunochemical, and histopathological data. Part 1. Trends across the clinical groups. J. Med. Genet. 1993;30:728–736. doi: 10.1136/jmg.30.9.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Putten M., Hulsker M., Nadarajah V.D., van Heiningen S.H., van Huizen E., van Iterson M., Admiraal P., Messemaker T., den Dunnen J.T., ’t Hoen P.A., Aartsma-Rus A. The effects of low levels of dystrophin on mouse muscle function and pathology. PLoS ONE. 2012;7:e31937. doi: 10.1371/journal.pone.0031937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Putten M., Hulsker M., Young C., Nadarajah V.D., Heemskerk H., van der Weerd L., ’t Hoen P.A., van Ommen G.J., Aartsma-Rus A.M. Low dystrophin levels increase survival and improve muscle pathology and function in dystrophin/utrophin double-knockout mice. FASEB J. 2013;27:2484–2495. doi: 10.1096/fj.12-224170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Putten M., van der Pijl E.M., Hulsker M., Verhaart I.E., Nadarajah V.D., van der Weerd L., Aartsma-Rus A. Low dystrophin levels in heart can delay heart failure in mdx mice. J. Mol. Cell. Cardiol. 2014;69:17–23. doi: 10.1016/j.yjmcc.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 68.de Vrueh R., Baekelandt E.R., de Hann J.M. World Health Organization; 2013. Update on 2004 background paper: BP 6.19 rare diseases.http://www.who.int/medicines/areas/priority_medicines/BP6_19Rare.pdf [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.