Abstract
Ion channel proteins are required for both the establishment of resting membrane potentials and the generation of action potentials. Hundreds of mutations in genes encoding voltage-gated ion channels responsible for action potential generation have been found to cause severe neurological diseases. In contrast, the roles of voltage-independent “leak” channels, important for the establishment and maintenance of resting membrane potentials upon which action potentials are generated, are not well established in human disease. UNC80 is a large component of the NALCN sodium-leak channel complex that regulates the basal excitability of the nervous system. Loss-of-function mutations of NALCN cause infantile hypotonia with psychomotor retardation and characteristic facies (IHPRF). We report four individuals from three unrelated families who have homozygous missense or compound heterozygous truncating mutations in UNC80 and persistent hypotonia, encephalopathy, growth failure, and severe intellectual disability. Compared to control cells, HEK293T cells transfected with an expression plasmid containing the c.5098C>T (p.Pro1700Ser) UNC80 mutation found in one individual showed markedly decreased NALCN channel currents. Our findings demonstrate the fundamental significance of UNC80 and basal ionic conductance to human health.
Main Text
Among mammalian neurons from different brain regions, there is considerable variability in basal excitability and resting membrane potentials (from approximately −85 mV to −50 mV) upon which action potentials are generated. Many mutations in voltage-gated ion channels responsible for the generation of action potentials have been found to cause severe diseases, including migraine, episodic muscle paralysis, ataxia, epilepsy, autism, and sudden cardiac death (see Jurkat-Rott et al., Kullmann and Waxman, and Catterall et al.1, 2, 3 for reviews). In contrast, the molecular mechanisms and the role of basal excitability in human diseases are not well understood.
The resting membrane potential is primarily determined by ion channels that are open and “leaky” at rest. The basal Na+ leak is mainly established through the NALCN Na+-leak channel. In mouse hippocampal neurons, NALCN is responsible for ∼70% of the total basal Na+ leak.4 NALCN is a member of the 24-transmembrane domain (24-TM) ion channel superfamily, which includes the ten voltage-gated Ca2+ channels and ten Na+ channels.5 It forms a voltage-independent, Na+-permeable, non-selective, non-inactivating cation channel. NALCN’s unique properties allow it to generate background sodium-leak currents. The balance between Na+ influx through NALCN and K+ efflux through other channels is predicted to generate a large dynamic range of neuronal excitability. In mammalian brains, the NALCN complex also contains UNC79 and UNC80, homologs of C. elegans Unc-79 and Unc-80.6, 7, 8 In situ hybridization analysis, RNA sequencing (RNA-seq), and expression sequence-tag (EST) database searches suggest that NALCN (MIM: 611549), UNC79, and UNC80 (MIM: 612636) are widely expressed in the brain.4 Both UNC79 and UNC80 are large proteins (∼3,000 amino acids) but do not have recognizable domains. NALCN and UNC79 do not directly interact with each other; the three components are brought together by UNC80’s association with both UNC79 and NALCN.6, 7 UNC80 is required for the channel’s regulation by extracellular Ca2+ through a G-protein-dependent pathway and by neuropeptides, such as substance P, through a G-protein-independent pathway that also requires the Src family of kinases.5
In worms and flies, mutations in Nalcn, Unc-79, and Unc-80 lead to uncoordinated movements, abnormal circadian rhythms, and altered sensitivity to anesthetics.9, 10, 11, 12, 13 In mice, knocking out Nalcn or Unc79 leads to severe apnea and neonatal lethality.4, 5, 7 In humans, inherited homozygous loss-of-function NALCN mutations cause infantile hypotonia with psychomotor retardation and characteristic facies (IHPRF [MIM: 615419]).14, 15 De novo heterozygous NALCN mutations are also found in individuals with arthrogryposis, hypotonia, intellectual disability, and developmental delay.16, 17
Despite its widespread expression in the brain, the in vivo function of UNC80 is largely unknown, and no mutations in mammals have been reported. We report the identification and characterization of UNC80 mutations as the cause of severe neurological phenotypes in four female individuals from three unrelated families.
The four subjects have similar phenotypes characterized by severe hypotonia of neonatal onset and that persists until adolescence, motor delays, lack of independent ambulation, constipation, encephalopathy, seizures, absent speech, and severe intellectual disability. The clinical features and genetic results of all subjects are summarized in Table 1 and Figure 1. Research protocols were approved in each country via institutional research boards and regional ethics committees, in keeping with national guidelines and the principles laid out in the Declaration of Helsinki. For the Norwegian family (F3), molecular analyses were performed in accordance with the Norwegian National Biotechnology Act. All parents provided written informed consent for study participation for themselves and their children, as well as for publication of clinical information, molecular findings, and photographs.
Table 1.
Clinical Characteristics of Individuals with UNC80 Mutations
| Subject |
Family 1 |
Family 2 |
Family 3 |
|
|---|---|---|---|---|
| IV.1 | V.5 | II.1 | II.3 | |
| Age at last review | 4 years | 4 years | 15 years | 9 years |
| Gender | female | female | female | female |
| Country of parental origin | Iraq | Morocco | Norway | Norway |
| Parental consanguinity | + | + | − | − |
| Gestational age | 41 weeks | 41 weeks | 38 weeks | 40 weeks |
| Birth weight (percentile) | 3,000 g (25th) | 3,158 g (50th) | 2,960 g (25th) | 3,070 g (25th) |
| OFC at birth (percentile) | 33 cm (25th) | unavailable | 32 cm (10th) | 35 cm (75th) |
| Hypotonia first noted | birth | birth | birth | 3 months |
| Persistent hypotonia | + | + | + | + |
| Feeding difficulties | + | + | − | + |
| Age at G-tube insertion (degree of dependency) | 2 years (dependent) | 3.5 years (dependent) | 15 years (supplemental use) | 10 months (completely dependent) |
| Current age | 4 years | 4 years | 15 years | 10 years |
| Current height (percentile) | 72 cm (<<3rd) | 91.5 cm (<3rd) | 144 cm (<<3rd) | 125 cm (3rd)a |
| Current weight (BMI) | 7.4 kg (14.3) | 12.9 kg (15.4) | 40.2 kg (17) | 23.5 kg (15.7)a |
| Current OFC (percentile) | 43 cm (<<3rd, or −4 SD) | 47 cm (2nd, or −1 SD) | 51.5 cm (3rd) | 50.5 cm (10th)a |
| Ophthalmology | punctate keratopathy, normal visual evoked potentials | alternating esotropia, normal visual evoked potentials | esotropia, normal vision | esotropia, normal vision, normal visual evoked potentials |
| Hypotonic facies | + | + | + | + |
| Small hands and feet | + | − | + | + |
| Severe global developmental delay and intellectual disability | + | + | + | + |
| Communication | no speech | no speech | no speech, babbles socially, expressive body language | no speech, babbles socially, expressive body language |
| Psychosocial | poor eye contact, minimal interest in surroundings | sociable, studies faces with interest | sociable, content, interested in surroundings | sociable, content, interested in surroundings |
| Gross motor | unable to sit, non-ambulatory | sits without support, non-ambulatory | sits with support, pulls to stand, walks with a walker | sits without support, stands and walks with support |
| Fine motor | transfers objects across midline | brings food to mouth, manipulates toys | brings food to mouth, manipulates toys | Manipulates objects |
| Muscle weakness | +, MRC grade 3/5 | difficult to assess due to severe hypotonia | +, MRC grade 4/5 | +, MRC grade 4/5 |
| Seizures, age of onset | +, 3 months (none since 3 years old) | +, 3 years and 10 months | +, 6 months | +, 3 years |
| Seizure types | generalized tonic-clonic | generalized tonic-clonic | myoclonic, atonic, generalized tonic-clonic, atypical absences | atonic, atypical absences |
| Current anti-convulsive therapy | clonazepam | valproate, rivotril | valproate, lamotrigine, levetiracetam, vagal nerve stimulator | valproate |
| Severe constipation | + | + | + | + |
| Tactile aversion | − | − | + | + |
| Hypersensitivity for stimuli | − | + | + | + |
| Sleep disturbance | +, obstructive sleep apnea | + | − | − |
| Brain MRI | thin corpus callosum | no structural anomalies | no structural anomalies | no structural anomalies |
| EEG | encephalopathic background with occasional spike and wave discharges | encephalopathic background pattern with frequent multifocal discharges | encephalopathic background activity with frequent spike and wave discharges | encephalopathic background activity with frequent spike and wave discharges |
| Other | non-specific myopathic changes on muscle biopsy | not reported | mild scoliosis, euthyroid | milk protein intolerance in infancy, euthyroid |
| Mutationb | chr2:210783340C>T | chr2:210824431G>C | chr2:210832310T>A | chr2:210685105delA |
| UNC80 (GenBank: NM_032504.1) | c.5098C>T | c.7607G>C | c.7757T>A | c.2033delA |
| Exon number | 32 | 50 | 51 | 13 |
| Predicted effect on protein | p.Pro1700Ser | p.Arg2536Thr, predicted to cause aberrant splicing | p. Leu2586∗ | p.Asn678Thrfs∗15 |
| Type of mutation | missense | missense | stop | frameshift insertion |
Abbreviations are as follows: +, present; −, not present; OFC, occipital frontal circumference; MRC, Medical Research Council scale for muscle strength.
Measurements for individual II.3 were taken at the last examination at the age of 9 years, rather than at the current age.
Based on UCSC Genome Browser GRCh37.
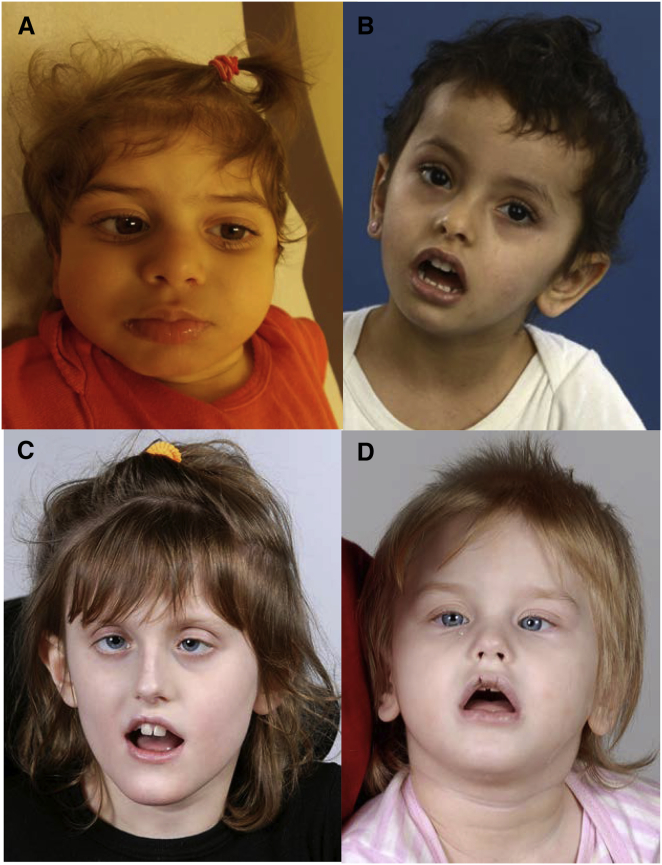
Figure 1.
Clinical Photographs of All Four Subjects
(A) Subject F1-IV.1 at the age of 4 years.
(B) Subject F2-V.5 at the age of 3 years.
(C) Subject F3-II.1 at the age of 9 years.
(D) Subject F3-II.3 at the age of 3 years.
For family 1, the whole-exome sequencing (WES) library preparation, exon capture, and sequencing were performed at the Genome Québec Innovation Center (Montréal) as previously described.18 Genomic DNA was captured with the SureSelect Human 50 Mb All Exon V5 kit (Agilent Technologies). Sequencing was performed on an Illumina HiSeq 2000 (Illumina) with paired-end 100-bp reads. A mean coverage of 134× was obtained, and 97% of the bases were covered at more than 10×. WES trio average coverage was 55×. Read alignment, variant calling, and annotation were done with a pipeline based on the Burrows-Wheeler Aligner (BWA), SAMtools, ANNOVAR, and custom annotation scripts, and reads were aligned to the reference human genome (UCSC Genome Browser hg19).
For family 2, DNA capture was performed with the SeqCap EZ Exome v.3.0 (Roche Nimblegen). Sequencing was performed on an Illumina HiSeq 2000 (Illumina) with paired-end 100-bp reads. A mean coverage of 76× was obtained, and 94% of the bases were covered at more than 10×. Read alignment to the reference human genome (hg19) and variant calling were done with a pipeline based on BWA-MEM v.0.7 and the Genome Analysis Toolkit (GATK v.3.1-1-g07a4bf8). Variant annotation and prioritizing were done with Cartagenia NGS Bench (Cartagenia).
Four individuals from family 3 were tested and included in the Center for Mendelian Genomics project at the Baylor College of Medicine (BCM) in Houston, Texas. The WES method used at the BCM-HGSC (Human Genome Sequencing Center) has previously been described.19 In brief, DNA was prepared for Illumina paired-end libraries, and capture was performed with the in-house-developed BCM-HGSC Core design and sequenced on the Illumina HiSeq 2500 platform (Illumina). Data produced was processed through the HGSC-developed Mercury pipeline, available in the cloud,20 to produce variant call format files (.vcf) with the Atlas2 variant calling method.21, 22, 23 Variants were annotated with the in-house-developed “Cassandra”24 annotation pipeline, based on ANNOVAR.25 Variants of interest were selected on the basis of inheritance pattern, rarity, and evaluation of possible genotype-phenotype correlation by knowledge of gene function, pathway, expression pattern, and results from other model organisms. Alamut v.2.4.6 (Interactive Biosoftware) was used in evaluation of pathological relevance. Additional support was sought by application of computational prediction tools (PhyloP, SIFT, PolyPhen-2, LRT, and MutationTaster). All UNC80 variants were confirmed by Sanger sequencing. Unaffected family members were also Sanger sequenced to confirm that the variant segregated with disease. Sanger sequencing was performed on genomic DNA from peripheral blood. Primers for Sanger sequencing were designed with Primer3 software and sequenced on an ABI 3730 sequencer (Applied Biosystems, Life Technologies), and sequence data was analyzed with SeqScape v.2.7 (Life Technologies) and 4Peaks.
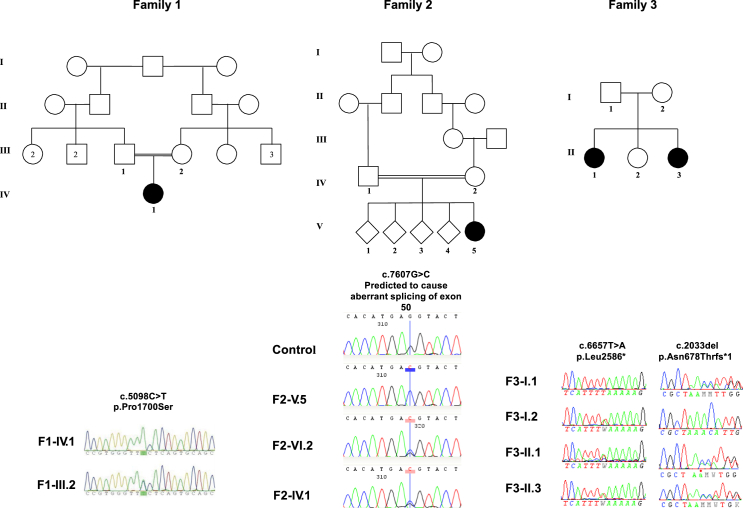
Subject F1-IV.1 was found to be homozygous for a c.5098C>T missense variant (p.Pro1700Ser) in exon 32 of UNC80 (GenBank: NM_032504.1). The variant is predicted to be deleterious and is present at a position completely conserved among all animals (Figure S3). The subject’s father was unable to be tested, but her mother was confirmed to be a carrier of the variant (Figure 2). Subject F2-V.5 had a homozygous missense variant in UNC80, c.7607G>C (p.Arg2536Thr). G7607 is the last nucleotide of exon 50. The c.7607G>C variant disrupts the “AG” exonic sequence commonly found in exon-intron junctions upstream of the conserved splice-donor sequence “GT.” The variant is predicted by five independent methods to affect splicing efficiency (Figure S4). Splicing studies of UNC80 were not performed because UNC80 is not expressed in peripheral leukocytes and no other tissue was available from this individual. The variant (p.Arg2536Thr) is also predicted to neutralize a charged residue highly conserved in all animals (Figure S5), suggesting that the variant disrupts protein function even in transcripts that are correctly spliced. Splicing analysis in neurons and functional characterization of the corresponding protein are required to determine the molecular consequence of this variant. Both healthy parents of subject F2-V.5 were heterozygous carriers, and all four healthy siblings were either heterozygous or bi-allelic wild-type (WT) at this nucleotide position (Figure 2). Subject F3-II.1 and subject F3-II.3 were both found to be compound heterozygous for the following UNC80 variants: c.2033delA (p.Asn678Thrfs∗15) and c.7757T>A (p.Leu2586∗) (GenBank: NM_032504.1). Both parents were found to be heterozygous carriers of one of the variants (Figure 2). None of the variants were present in publically available databases (dbSNP, 1000 Genomes, and the ExAC Browser), and the c.5098C>T variant was found in the heterozygous state in one individual in the database of over 3,000 exomes at the Center for Mendelian Genomics.
Figure 2.
Pedigrees and Chromatograms from All Three Families
Affected individuals are shaded in black.
The three mutations in subjects F2-V.5, F3-II.1, and F3-II.3 are predicted to lead to disruption (in F2-V.5, due to aberrant splicing) or truncation (in F3-II.1 and F3-II.3) of UNC80 and are most likely loss-of-function mutations. To determine whether the c.5098C>T missense variant (p.Pro1700Ser) in subject F1-IV.1 affects expression and function of UNC80, we introduced the variant into mouse Unc80 cDNA, which encodes a protein with 97% identity to the human UNC80 and has been functionally characterized. The mouse Unc80 cDNA (GenBank: FJ210934) was cloned into the EcoRI and NotI sites of vector pcDNA3.1. The p.Pro1769Ser variant (equivalent to human p.Pro1700Ser) was introduced with the Gibson assembly method. The sequences of the primers used for mutagenesis are as follows: 5′-GTTCGACCCTCCGTGGGTCtCTCAGTGCAGCGGGAGTG-3′ (forward), 5′-CACTCCCGCTGCACTGAGAGACCCACGGAGGGTCGAAC-3′ (reverse). The mutation was confirmed with Sanger sequencing.
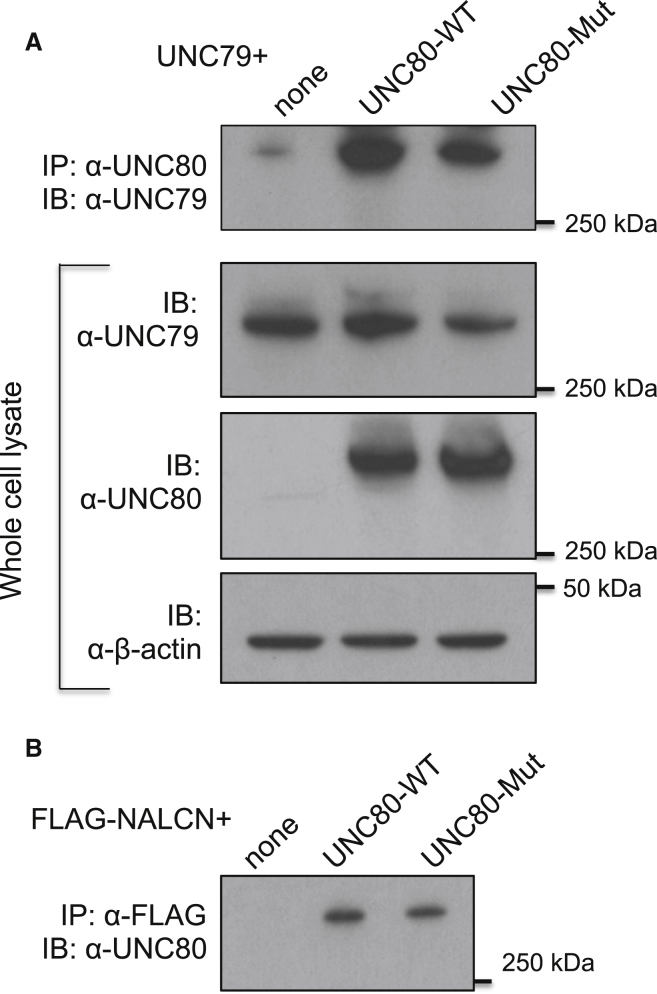
We transfected the WT and variant Unc80 into human HEK293T cells (from ATCC, maintained in DMEM [GIBCO] supplemented with 10% FBS [Atlanta Biologicals] and 1× Glutamax [GIBCO]; transfection done with Lipofectamine 2000 as the transfection reagent). The Unc80 variant yielded an amount of protein comparable to that yielded by the WT Unc80 (Figure 3A, lower two panels), suggesting that this missense variant does not disrupt production of the protein. The variant protein also retained its ability to associate with UNC79 (Figure 3A) and NALCN (Figure 3B), which was tested by co-immunoprecipitation.
Figure 3.
Variant UNC80 Associates with UNC79 and NALCN
(A) Cells were transfected with UNC79 alone or together with WT or variant Unc80 as indicated in each lane. Immunoprecipitates (IP) with an anti-UNC80 antibody were blotted (IB) with anti-UNC79 to allow us to probe the association between UNC79 and UNC80 (upper panel). Total cell lysates (lower three panels) were blotted with anti-UNC79 or anti-UNC80 antibodies for comparison of protein amounts. Immunoblotting with anti-β-actin (lower panel) was used as a control for sample loading.
(B) Cells were transfected with FLAG-tagged NALCN alone or together with the WT or variant Unc80, as indicated. Immunoprecipitates with anti-FLAG were blotted with anti-UNC80 to allow us to probe the association between UNC80 and NALCN. For immunoprecipitation, cells were solubilized in lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 1% NP-40, 1× PIC [Roche]) and immunoprecipitated with anti-UNC80 (2 μg/ml)7 or anti-FLAG (5 μg/ml, no. F3165 [Sigma]). The immune complexes were analyzed by western blot with anti-UNC80 (2.2 μg/ml) or anti-UNC79 (3 μg/ml). Cell lysates (inputs) were also analyzed by western blot with anti-UNC80 or anti-β-actin (1:1,000, no. 4970S [Cell Signaling]) as indicated.
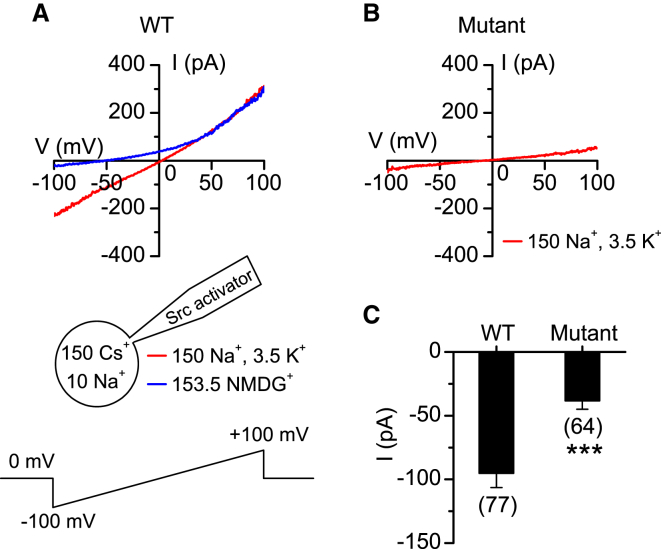
We next used electrophysiological assays to test whether the p.Pro1769Ser (mouse equivalent of p.Pro1700Ser) variant affects the function of UNC80. A unique property of UNC80 is its ability to scaffold Src kinase and to enhance NALCN currents through a Src-dependent pathway.6, 8 We recorded NALCN currents from HEK293T cells that we co-transfected with NALCN cDNA (human isoform,7 in a pTracer vector expressing GFP under a separate promoter) and WT or variant Unc80 by using patch clamp pipettes containing a peptide Src activator (Figure 4A, lower panel). Compared to those in cells transfected with WT Unc80, the sizes of the currents were significantly reduced in cells expressing the variant (Figures 4B and 4C).
Figure 4.
Altered Function in Variant UNC80
(A and B) Representative NALCN currents recorded in HEK293T cells co-transfected with NALCN and WT Unc80 (A) or the mutant version of Unc80 (B). Whole-cell currents were recorded with a voltage ramp from −100 mV to +100 mV in 1 s (Vh = 0 mV, illustrated in the lower panel of A). In cells with currents larger than 50 pA, NMDG was used to substitute Na+ and K+ to confirm that the cells had no non-specific leak, as demonstrated by the abolishment of inward currents with ion substitution.
(C) Summary of the current amplitudes recorded at −100 mV. Numbers of cells recorded are in parentheses. Data were presented as mean ± SEM. ∗p < 0.05. ∗∗∗p < 0.001. For patch clamp recordings, cells in a 35-mm dish (∼90% confluency) were transfected with 3 μg plasmids DNA with Lipofectamine 2000 for ∼40 hr and plated onto polylysine-coated coverslips. Recordings were done 40–48 hr after transfection. Cells with GFP fluorescence intensity within the top 20%–30% on each coverslip were selected for patch clamp recordings. Signal was acquired with an amplifier (Axopatch 200B or Multiclamp 700B) and a Digidata 1440A data acquisition system controlled by PClamp software (Molecular Device). The pipette solution contained 150 mM Cs, 120 mM Mes, 10 mM NaCl, 10 mM EGTA, 4 mM CaCl2, 0.3 mM Na2GTP, 2 mM Mg-ATP, 10 mM HEPES and 2 μM Src family kinase activator (pH 7.4; from Santa Cruz). Bath solutions contained 150 mM NaCl, 3.5 mM KCl, 1 mM MgCl2, 1.2 mM CaCl2, 20 mM glucose, and 10 mM HEPES (pH 7.4). In the NMDG bath, Na+ and K+ were replaced by NMDG+. Liquid junction potentials were corrected online.
In summary, our studies reveal a genetic cause for global developmental delay and neurological dysfunction and demonstrate the importance of UNC80 in humans. From a motor perspective, all four individuals were significantly delayed in sitting; one individual was still unable to achieve this at the age of 4 years. None of the individuals were able to ambulate independently. These symptoms are comparable with those observed in individuals with loss-of-function NALCN mutations.14, 15 Unlike individuals with NALCN mutations, our four UNC80 individuals show no true facial dysmorphism, although their faces are characterized by features related to severe hypotonia. All individuals in our study were described as encephalopathic, both clinically and on electroencephalogram (EEG) monitoring. Growth failure, severe constipation, and the feeding difficulties requiring G-tube insertion might be due, in part, to dysfunctional muscle coordination via neuronal compromise; however, standardized evaluation of swallowing function for all individuals would be required to confirm this. Subject F1-IV.1 also had severe sleep disturbance, including complete reversal of her sleep-wake cycle, which was resistant to treatment with melatonin. Subject F2-V.5 also had sleep disturbance, including difficulty falling asleep. In flies and mice, NALCN is essential for the depolarization of clock neurons in the morning and the maintenance of normal circadian rhythms.26 It is possible that abnormal UNC80 prevents activation of the NALCN complex and inhibits the ability of pacemaker neurons to maintain basal sleep-wake rhythm.
Compared to phenotypes associated with mutations found in known neuronal genes, the neurological phenotypes we describe in these four UNC80 individuals are at the severe end of the spectrum. This underscores the pivotal importance of leak ion channel genes and the associated basal excitability in human health and disease.
Acknowledgments
We would like to thank the participants and families for their contribution to this study. This work was selected for study by the Care4Rare Canada (Enhanced Care for Rare Genetic Diseases in Canada) Consortium Gene Discovery Steering Committee: Kym Boycott (lead; University of Ottawa), Alex MacKenzie (co-lead; University of Ottawa), J.M. (McGill University), Michael Brudno (University of Toronto), Dennis Bulman (University of Ottawa), and David Dyment (University of Ottawa). We also thank Hanne S. Sorte and Mari Ann Kulseth (Department of Medical Genetics, Oslo University Hospital) for excellent technical assistance. J.R.L. has stock ownership in 23andMe, is a paid consultant for Regeneron Pharmaceuticals, has stock options in Lasergen, is a member of the Scientific Advisory Board of Baylor Miraca Genetics Laboratories, and is a co-inventor on multiple US and European patents related to molecular diagnostics for inherited neuropathies, eye diseases, and bacterial genomic fingerprinting. The Department of Molecular and Human Genetics at Baylor College of Medicine (J.R.L.) derives revenue from the chromosomal microarray analysis and clinical exome sequencing offered in the Baylor Miraca Genetics Laboratory (http://www.bmgl.com). This work was supported, in part, by funding from the NIH to D.R. (NS055293 and NS074257). Funding was also received from Genome Canada, the Canadian Institutes of Health Research, the Ontario Genomics Institute, Ontario Research Fund, Genome Quebec, Children’s Hospital of Eastern Ontario Foundation, and The Hospital for Sick Children. The Baylor-Hopkins Center for Mendelian Genomics is supported by the National Human Genome Research Institute and the National Heart Lung and Blood Institute (U54HG006542).
Published: December 17, 2015
Footnotes
Supplemental Data include additional case histories, five figures, and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.004.
Contributor Information
Dejian Ren, Email: dren@sas.upenn.edu.
Grace Yoon, Email: grace.yoon@utoronto.ca.
Web Resources
The URLs for data presented herein are as follows:
1000 Genomes, http://browser.1000genomes.org
4Peaks, http://nucleobytes.com/4peaks
ExAC Browser, http://exac.broadinstitute.org/
GenBank, http://www.ncbi.nlm.nih.gov/genbank/
GeneMatcher, https://genematcher.org/
MutationTaster, http://www.mutationtaster.org/
NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS/
OMIM, http://www.omim.org/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
UCSC Genome Browser, http://genome.ucsc.edu
Supplemental Data
References
- 1.Jurkat-Rott K., Lerche H., Weber Y., Lehmann-Horn F. Hereditary channelopathies in neurology. Adv. Exp. Med. Biol. 2010;686:305–334. doi: 10.1007/978-90-481-9485-8_18. [DOI] [PubMed] [Google Scholar]
- 2.Kullmann D.M., Waxman S.G. Neurological channelopathies: new insights into disease mechanisms and ion channel function. J. Physiol. 2010;588:1823–1827. doi: 10.1113/jphysiol.2010.190652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catterall W.A., Dib-Hajj S., Meisler M.H., Pietrobon D. Inherited neuronal ion channelopathies: new windows on complex neurological diseases. J. Neurosci. 2008;28:11768–11777. doi: 10.1523/JNEUROSCI.3901-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu B., Su Y., Das S., Liu J., Xia J., Ren D. The neuronal channel NALCN contributes resting sodium permeability and is required for normal respiratory rhythm. Cell. 2007;129:371–383. doi: 10.1016/j.cell.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 5.Ren D. Sodium leak channels in neuronal excitability and rhythmic behaviors. Neuron. 2011;72:899–911. doi: 10.1016/j.neuron.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu B., Su Y., Das S., Wang H., Wang Y., Liu J., Ren D. Peptide neurotransmitters activate a cation channel complex of NALCN and UNC-80. Nature. 2009;457:741–744. doi: 10.1038/nature07579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu B., Zhang Q., Wang H., Wang Y., Nakayama M., Ren D. Extracellular calcium controls background current and neuronal excitability via an UNC79-UNC80-NALCN cation channel complex. Neuron. 2010;68:488–499. doi: 10.1016/j.neuron.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang H., Ren D. UNC80 functions as a scaffold for Src kinases in NALCN channel function. Channels (Austin) 2009;3:161–163. doi: 10.4161/chan.3.3.8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Humphrey J.A., Hamming K.S., Thacker C.M., Scott R.L., Sedensky M.M., Snutch T.P., Morgan P.G., Nash H.A. A putative cation channel and its novel regulator: cross-species conservation of effects on general anesthesia. Curr. Biol. 2007;17:624–629. doi: 10.1016/j.cub.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 10.Jospin M., Watanabe S., Joshi D., Young S., Hamming K., Thacker C., Snutch T.P., Jorgensen E.M., Schuske K. UNC-80 and the NCA ion channels contribute to endocytosis defects in synaptojanin mutants. Curr. Biol. 2007;17:1595–1600. doi: 10.1016/j.cub.2007.08.036. [DOI] [PubMed] [Google Scholar]
- 11.Pierce-Shimomura J.T., Chen B.L., Mun J.J., Ho R., Sarkis R., McIntire S.L. Genetic analysis of crawling and swimming locomotory patterns in C. elegans. Proc. Natl. Acad. Sci. USA. 2008;105:20982–20987. doi: 10.1073/pnas.0810359105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeh E., Ng S., Zhang M., Bouhours M., Wang Y., Wang M., Hung W., Aoyagi K., Melnik-Martinez K., Li M. A putative cation channel, NCA-1, and a novel protein, UNC-80, transmit neuronal activity in C. elegans. PLoS Biol. 2008;6:e55. doi: 10.1371/journal.pbio.0060055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nash H.A., Scott R.L., Lear B.C., Allada R. An unusual cation channel mediates photic control of locomotion in Drosophila. Curr. Biol. 2002;12:2152–2158. doi: 10.1016/s0960-9822(02)01358-1. [DOI] [PubMed] [Google Scholar]
- 14.Köroğlu Ç., Seven M., Tolun A. Recessive truncating NALCN mutation in infantile neuroaxonal dystrophy with facial dysmorphism. J. Med. Genet. 2013;50:515–520. doi: 10.1136/jmedgenet-2013-101634. [DOI] [PubMed] [Google Scholar]
- 15.Al-Sayed M.D., Al-Zaidan H., Albakheet A., Hakami H., Kenana R., Al-Yafee Y., Al-Dosary M., Qari A., Al-Sheddi T., Al-Muheiza M. Mutations in NALCN cause an autosomal-recessive syndrome with severe hypotonia, speech impairment, and cognitive delay. Am. J. Hum. Genet. 2013;93:721–726. doi: 10.1016/j.ajhg.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chong J.X., McMillin M.J., Shively K.M., Beck A.E., Marvin C.T., Armenteros J.R., Buckingham K.J., Nkinsi N.T., Boyle E.A., Berry M.N., University of Washington Center for Mendelian Genomics De novo mutations in NALCN cause a syndrome characterized by congenital contractures of the limbs and face, hypotonia, and developmental delay. Am. J. Hum. Genet. 2015;96:462–473. doi: 10.1016/j.ajhg.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aoyagi K., Rossignol E., Hamdan F.F., Mulcahy B., Xie L., Nagamatsu S., Rouleau G.A., Zhen M., Michaud J.L. A Gain-of-Function Mutation in NALCN in a Child with Intellectual Disability, Ataxia, and Arthrogryposis. Hum. Mutat. 2015;36:753–757. doi: 10.1002/humu.22797. [DOI] [PubMed] [Google Scholar]
- 18.Tetreault M., Fahiminiya S., Antonicka H., Mitchell G.A., Geraghty M.T., Lines M., Boycott K.M., Shoubridge E.A., Mitchell J.J., Michaud J.L., Majewski J., Care4Rare Canada Consortium Whole-exome sequencing identifies novel ECHS1 mutations in Leigh syndrome. Hum. Genet. 2015;134:981–991. doi: 10.1007/s00439-015-1577-y. [DOI] [PubMed] [Google Scholar]
- 19.Yang Y., Muzny D.M., Reid J.G., Bainbridge M.N., Willis A., Ward P.A., Braxton A., Beuten J., Xia F., Niu Z. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N. Engl. J. Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid J.G., Carroll A., Veeraraghavan N., Dahdouli M., Sundquist A., English A., Bainbridge M., White S., Salerno W., Buhay C. Launching genomics into the cloud: deployment of Mercury, a next generation sequence analysis pipeline. BMC Bioinformatics. 2014;15:30. doi: 10.1186/1471-2105-15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Challis D., Yu J., Evani U.S., Jackson A.R., Paithankar S., Coarfa C., Milosavljevic A., Gibbs R.A., Yu F. An integrative variant analysis suite for whole exome next-generation sequencing data. BMC Bioinformatics. 2012;13:8. doi: 10.1186/1471-2105-13-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., 1000 Genomes Project Analysis Group The variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bainbridge M.N., Wiszniewski W., Murdock D.R., Friedman J., Gonzaga-Jauregui C., Newsham I., Reid J.G., Fink J.K., Morgan M.B., Gingras M.C. Whole-genome sequencing for optimized patient management. Sci. Transl. Med. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flourakis M., Kula-Eversole E., Hutchison A.L., Han T.H., Aranda K., Moose D.L., White K.P., Dinner A.R., Lear B.C., Ren D. A Conserved Bicycle Model for Circadian Clock Control of Membrane Excitability. Cell. 2015;162:836–848. doi: 10.1016/j.cell.2015.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.