Abstract
Identification of novel targets for the treatment of basal-like breast cancer is essential for improved outcomes in patients with this disease. This study investigates the association of MMP7 expression and MMP7 promoter methylation with subtype and outcome in breast cancer patient cohorts. Immunohistochemical analysis was performed on a breast cancer tissue microarray and validated in independent histological samples. MMP7 expression significantly correlated with patient age, tumor size, triple-negative (TN) status, and recurrence. Analysis of publically available datasets confirmed MMP7 gene expression as a prognostic marker of breast cancer metastasis, particularly metastasis to the brain and lungs. Methylation of the MMP7 promoter was assessed by methylation-specific PCR in a panel of breast cancer cell lines and patient tumor samples. Hypomethylation of the MMP7 promoter significantly correlated with TN status in DNA from patient tumor samples, and this association was confirmed using The Cancer Genome Atlas (TCGA) dataset. Evaluation of a panel of breast cancer cell lines and data from the Curtis and TCGA breast carcinoma datasets revealed that elevated MMP7 expression and MMP7 promoter hypomethylation are specific biomarkers of the basal-like molecular subtype which shares considerable, but not complete, overlap with the clinical TN subtype. Importantly, MMP7 expression was identified as an independent predictor of pathological complete response in a large breast cancer patient cohort. Combined, these data suggest that MMP7 expression and MMP7 promoter methylation may be useful as prognostic biomarkers. Furthermore, MMP7 expression and promoter methylation analysis may be effective mechanisms to distinguish basal-like breast cancers from other triple-negative subtypes. Finally, these data implicate MMP7 as a potential therapeutic target for the treatment of basal-like breast cancers.
Keywords: MMP7, Triple negative, Basal-like, Breast cancer, Metastasis, Promoter methylation
Introduction
Matrix metalloprotease 7 (MMP7, Matrilysin, PUMP-1) is a secreted proteolytic enzyme capable of degrading components of the extracellular matrix (ECM). Members of the matrix metalloprotease family (MMPs) have well-established roles in the growth and invasion of cancer cells and metastatic progression [1, 2]. In addition to ECM remodeling, MMPs regulate the activity or availability of many cell adhesion molecules, as well as cytokines, growth factors, and their respective receptors to aid tumor formation and progression [3–7]. Most MMPs are produced by stromal cells, predominantly fibroblasts, rather than epithelial cells [8]. However, MMP7 is among the select few MMPs produced by epithelial cells [8]. The expression of MMP7 within breast carcinoma cells makes it an intriguing potential biomarker and molecular target for novel anti-cancer therapies.
Elevated expression of MMP7 and a correlation between MMP7 expression and an unfavorable outcome is well established in a number of cancers including adenocarcinomas of the colon and pancreas [9–12]. In colorectal cancer elevated MMP7 expression is a prognostic marker of overall survival [13, 14], associated with resistance to chemotherapy [14, 15], and increased metastasis to the liver [16]. In addition, mutations in the MMP7 promoter are associated with the increased risk of colorectal cancer [17–19]. In the mammary gland, MMP7 is expressed in the normal ductal and lobular epithelia and elevated expression of MMP7 at the mRNA level has been observed in some breast carcinomas [8, 20]. Previous studies have suggested that expression of MMP7 in breast cancer may be positively regulated by active epithelial growth factor receptor 2 (HER2) implying that MMP7 may be an important factor in the growth and metastasis of HER2+ breast cancers [21, 22]. In contrast, our group recently analyzed MMP7 expression in a panel of breast cancer cell lines and found that a subgroup of triple-negative (i.e., those lacking expression of the estrogen receptor [ER], progesterone receptor [PR], and HER2) breast cancer cell lines that closely resemble the basal-like breast cancer subtype expressed significantly higher levels of MMP7 relative to representative luminal (ER+/PR+/HER2−), HER2+, or TN cell lines that are distinct from the basal-like intrinsic subtype [23]. Whether the pattern of MMP7 expression observed in breast cancer cell lines is consistent with MMP7 expression in human tumors is not yet known. Although there is significant overlap between the clinically defined TN and molecularly defined basal-like subtypes, with approximately 75 % of TN cancers exhibiting a basal-like gene expression profile and 75 % of basal-like cancers exhibiting ER/PR/HER2 negativity [24, 25], these subtypes are not synonymous. While categorization of patient samples into clinical breast cancer subtypes can be readily accomplished by immunohistochemical (IHC) staining for ER, PR, and HER2 with or without accompanying fluorescence in situ hybridization for HER2 amplification, categorizing breast cancers into the basal-like or other intrinsic molecular subtypes (Luminal A, Luminal B, HER2+, or normal-like) requires quantitation of a large number of genes such as those included in the PAM50 gene signature [26]. Classification of breast cancers into their intrinsic subtypes has important prognostic and predictive value as some subtypes (i.e., basal-like) have a poorer prognosis, are more prone to distant metastasis and are more sensitive to specific chemotherapy regimens [27–29]. Biomarkers that can help identify the basal-like subtype, without the requirement for larger-scale molecular profiling, or increase the precision of current classifiers would greatly improve our ability to predict patient response to current therapeutic strategies and speed the design of novel targeted agents.
In the present study, we examine MMP7 expression by immunohistochemistry in a cohort of 157 breast cancer patients with a median clinical follow-up of 5.9 years and validate the subtype specificity of MMP7 expression in an additional 80 patient samples. To identify a potential mechanism underlying the subtype-specific expression of MMP7, we also examine the relationship between methylation of CpG sites in the MMP7 promoter and breast cancer subtype in genomic DNA from 48 breast cancer patient samples and confirm these data using the large TCGA breast cancer cohort. Examination of our breast cancer patient data demonstrates a significant correlation between MMP7 positivity and the TN breast cancer clinical subtype and reveals MMP7 as a potential prognostic marker. To further examine the efficacy of MMP7 as a biomarker of breast cancer subtype, we utilized information from publicly available breast cancer patient databases and discovered that MMP7 is a biomarker of the basal-like molecular subtype of breast cancer. Finally, we identify MMP7 expression as an independent predictor of complete pathological response in a large breast cancer patient cohort. These data suggest that MMP7 may prove useful as an efficient means to identify basal-like breast cancers, as well as a potential therapeutic target for the treatment of this disease.
Materials and methods
Tissue samples
With approval from the University Hospitals Case Medical Center (UHCMC) Institutional Review Board (IRB), tissue microarrays (TMA) were constructed from archived tumor tissue of 157 newly diagnosed patients who underwent primary surgery for invasive breast carcinoma at UHCMC between 1998 and 2003. These patients were chosen because both tumor tissue and clinical follow-up data through the UHCMC Tumor Registry were available. The final selection of patient tumor samples for the TMA was made using prior immunohistochemistry for expression of ER, PR, and HER2 to maintain equal representation of the luminal (ER+/PR+/HER2−), HER2+ , and triple-negative (ER−/PR−/HER2−) breast cancer subtypes. Whenever possible, consecutive samples were obtained. The constructed TMAs consisted of 8 by 10 arrays of 1.5–2 mm cores from archived formalin-fixed, and paraffin-embedded surgical pathology blocks. Each de-identified tumor core was given an anonymous identification number linked to an IRB-approved database containing clinicopathological information and clinical follow-up data.
To validate the association between MMP7 staining and the TN subtype, 80 independent recently diagnosed, de-identified, formalin-fixed, paraffin-embedded tumor sections of known ER, PR, and HER2 status, but without available clinical outcomes data, were obtained from the University Hospitals Case Medical Center Department of Pathology archives.
For analysis of the methylation status of CpG sites in the MMP7 promoter, breast cancer genomic DNA was obtained from the Department of Pathology at Technical University in Munich, Germany. Genomic DNA was extracted from 48 formalin-fixed, paraffin embedded, de-identified primary breast cancer samples representing the luminal (14 samples), HER2+ (19 samples), and TN (15 samples) subtypes. Genomic DNA extraction was performed using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany) following microdissection to ensure a tumor cell content of >70 % in all samples. 41 of the 48 DNA samples analyzed by methylation-specific (MS) PCR yielded data suitable for quantitation.
Immunohistochemistry
MMP7 antibody from R&D Systems (#AF907) was used to determine expression of MMP7 by immunohistochemistry. An optimal working protocol and antibody specificity were first confirmed using paraformaldehyde-fixed, paraffin-embedded tumor sections from xenografted breast cancer cell lines that either endogenously express (HCC1187) or do not express (MCF7) MMP7 as determined previously by qRT-PCR and western blot analyses. Using these control sections, we observed positive staining only in HCC1187 tumor tissue and no staining in either MCF7 or normal IgG control serial sections (data not shown). Patient breast tumor sections were then re-hydrated and endogenous peroxidases quenched with peroxidase blocking reagent (DAKO). No antigen retrieval step was performed. Sections were then blocked at room temperature for 1 h with 1.5 % normal donkey serum diluted in PBS containing 5 % bovine serum albumin and incubated overnight at 4 °C with primary antibody diluted 1:20 in PBS. Secondary detection was completed using the Vectastain Elite ABC Kit for Goat IgG (Vector Laboratories) according to manufacturer’s recommendations using 3,3′-diaminobenzidine (DAKO). Sections were counterstained with Gill’s #3 Hematoxylin (Fisher), dehydrated and mounted. H&E staining of adjacent sections was performed by the Case Western Reserve University Tissue Procurement and Histology Core Facility. MMP7 staining was scored on a 0 to 3+ intensity scale by two independent pathologists (C.N.B. and F.A.K.). An individual case was considered to be positive for MMP7 expression if at least 10 % of the tumor cells showed positive staining (≥1+).
Cell culture
All cell lines were acquired from American Type Culture Collection. MCF7 and MDA-MB-468 cells were cultured in DMEM supplemented with 10 % FBS while T47D, MDA-MB-231, HCC1187, and HCC1143 cells were grown in RPMI supplemented with 10 % FBS. All culture media were supplemented with 1 % l-glutamine, 1 % penicillin, and 1 % streptomycin.
Methylation-specific PCR
For analysis of MMP7 promoter methylation, genomic DNA was harvested from breast cancer cell lines or breast cancer patient tumor samples using the Qiagen DNeasy kit per the manufacturer’s recommendations. Isolated genomic DNA was then bisulfate converted using the Epitect Bisulfite kit (Qiagen). Methylation-specific PCR was performed using the following primer sets: MMP7 unmethylated (sense) 5′-GATGGTAGTTATGTGATTTATTG-3′, MMP7 unmethylated (antisense) 5′-TCCCACCTCCT AAAACAACA-3′, MMP7 methylated (sense) 5′-ACGG TAGTTATGCGATTTATC-3′, MMP7 methylated (anti-sense) 5′-CCCGCCTCCTAAAACAACG-3′. These primer sets detect differential methylation of CpG sites in the 5′UTR and coding sequence of MMP7 exon 1 [30]. PCR conditions used an initial denaturation step at 95 °C for 10 m followed by 30 cycles of: 95 °C for 30 s, 51 °C for 30 s, and 72 °C for 1 m followed by a final 7 m extension phase at 72 °C. The resulting PCR products were resolved on 5 % agarose gels. The intensities of the bands generated using the unmethylated- and methylated-specific primer sets were calculated using Image J software [31].
Quantitative real-time PCR
Total RNA was extracted and used to generate cDNA as previously described [23]. Quantitative real-time PCR (qRT-PCR) was conducted using an Applied Biosystems (ABI) Step-One real-time PCR instrument by the comparative Ct method. MMP7 expression was assessed using Hs01042793_m1 (ABI). GAPDH (Hs99999905_m1) was used as an endogenous control. Data were averaged from at least three independent experiments, each conducted in triplicate. Statistical differences in gene expression were assessed by ANOVA.
Datasets
Data from the Bos [32], Curtis [33], and Hatzis [34] datasets were retrieved from Oncomine (www.oncomine.com). Additional data for the Curtis dataset (EGAS00000000083) were downloaded from the European Bioinformatics Institute’s website, while additional data for the Hatzis study (GSE25066) were retrieved from the gene expression omnibus. Data from the TCGA invasive breast carcinoma dataset were downloaded from the cancergenome.nih.gov website.
Statistics
Bivariate comparisons between MMP7 staining and other clinicopathological data within the TMA were conducted by Fisher’s exact probability test. For statistical analysis of survival and recurrence among patients in the TMA, Kaplan-Meier curves were generated and compared by the log-rank test. Univariate and stepwise multivariate analyses were carried out using the Cox proportional hazards model. For analysis of MMP7 promoter methylation in the publically available TCGA dataset of invasive breast carcinoma, the methylation values from Illumina Human Methylation Array probe MMP7 cg25511807 from 463 patients were retrieved along with TN and PAM50 subtype status [35]. The Illumina probe cg25511807 detects the methylation state of CpG sites in the MMP7 5′-UTR and partially overlaps the sequence used for the construction of the methylation-specific PCR sense-strand primers used in this study. Inverse methylation values, representing the relative amount of the source sequence in the unmethylated state, were calculated and used for statistical comparison of MMP7 promoter methylation between breast cancer subtypes. For co-expression analyses, the Pearson correlation coefficient between expression of each gene and MMP7 expression was determined for the Curtis, Hatzis, and TCGA cohorts. The list of correlated genes in each dataset was restricted to the top 1 % using the absolute value of the Pearson correlation coefficients, and those genes that significantly correlate with MMP7 in all three studies were identified. Venn diagrams were generated using the Venny tool (http://bioinfogp.cnb.csic.es/tools/venny/). To identify predictive factors of pathological complete response (pCR), an odds ratio (OR) was calculated for each risk parameter, including MMP7 expression (separated into high and low expression by median expression value), in the Hatzis dataset. To study the additive predictive value of MMP7 expression, a multiple logistic regression model was fitted from all risk factors excluding MMP7 expression, while a second model was fitted using all available factors including MMP7 expression. Both models were generated in accordance with the Akaike information criterion using backward stepwise variable selection. The two models were compared by the likelihood ratio test and by comparison of the areas under the curve (AUC) for their respective receiver operator characteristic curves. Student’s t test was used for direct statistical comparisons between two groups when appropriate; analysis of data across multiple groups was conducted by one-way ANOVA. Statistical significance was assumed if P < 0.05.
Results
MMP7 positivity correlates with an earlier age and increased tumor size at time of diagnosis and triple-negative status
The clinicopathological variables of the study cohort for TMA analysis, as a whole and grouped by ER, PR, and HER2 status, are summarized in Table 1. Importantly, in order to attain equal representation of the three primary clinical breast cancer subtypes in the TMA, eligible patients were divided into groups based on ER, PR, and HER2 status prior to their selection for use in the TMA. As a result, the percentage of ER and PR-positive samples is lower, while abundance of HER2+ and TN samples on the TMA is higher than would be expected if consecutive patient samples were used. Of the samples on the array, 48.8 % are ER-positive, 47.1 % are positive for PR, and 33.1 % show HER2 amplification.
Table 1.
Clinicopathologic characteristics of the TMA patient cohort
| Characteristic | All patients n = 157 (100 %) |
ER+/PR+/ HER2− patients n = 49 (31.2 %) |
ER+/PR+/ HER2+ patients n = 20 (12.7 %) |
ER−/PR−/ HER2+ patients n = 29 (18.5 %) |
ER−/PR−/ HER2− patients n = 47 (29.9 %) |
Other patients n = 12 (7.6 %) |
|---|---|---|---|---|---|---|
| Age, mean ± SD | 56.9 ± 14.7 | 61.8 ± 15.5 | 53.8 ± 10.3 | 55.0 ± 16.4 | 54.5 ± 14.6 | 55.7 ± 9.0 |
| Median follow-up time (years) | 5.9 (0.2–12.9) | 6.1 (0.7–11.1) | 6.2 (0.2–8.6) | 4.7 (0.6–9.3) | 5.2 (0.4–8.7) | 6.4 (3.7–12.9) |
| Patient race | ||||||
| Caucasian | 108 (68.8 %) | 44 (89.8 %) | 14 (70.0 %) | 14 (48.3 %) | 29 (61.7 %) | 7 (58.3 %) |
| African–American | 36 (22.9 %) | 4 (8.2 %) | 3 (15.0 %) | 12 (41.4 %) | 14 (29.8 %) | 3(25.0 %) |
| Unknown | 13 (8.3 %) | 1 (2.0 %) | 3 (15.0 %) | 3 (10.3 %) | 4 (8.5 %) | 2 (16.7 %) |
| Tumor size | ||||||
| T1 (<2 cm) | 55 (35.0 %) | 23 (46.9 %) | 4 (20.0 %) | 9 (31.0 %) | 14 (29.8 %) | 5 (41.7 %) |
| T2 (>2 cm, <5 cm) | 50 (31.8 %) | 14 (28.6 %) | 8 (40.0 %) | 10 (34.5 %) | 15 (31.9 %) | 3 (25.0 %) |
| T3 (>5 cm) | 15 (9.6 %) | 2 (4.1 %) | 1 (5.0 %) | 6 (20.7 %) | 5 (10.6 %) | 1 (8.3 %) |
| T4 (extension to the chest wall or skin) |
4 (2.5 %) | 2 (4.1 %) | 1 (5.0 %) | 0 (0.0 %) | 0 (0.0 %) | 1 (8.3 %) |
| Unknown | 33 (21.0 %) | 8 (16.3 %) | 6 (30.0 %) | 4 (13.8 %) | 13 (27.7 %) | 2 (16.7 %) |
| Nodal status | ||||||
| Negative | 60 (38.2 %) | 23 (46.9 %) | 5 (25.0 %) | 13 (44.8 %) | 14 (29.8 %) | 5 (41.7 %) |
| N1 (1–3 lymph nodes) | 56 (35.7 %) | 17 (34.7 %) | 9 (45.0 %) | 7 (24.1 %) | 19 (40.4 %) | 4 (33.3 %) |
| N2 (4–9 lymph nodes) | 5 (3.2 %) | 1 (2.0 %) | 0 (0.0 %) | 3 (10.3 %) | 1 (2.1 %) | 0 (0.0 %) |
| N3 (10 or more lymph nodes) | 2 (1.3 %) | 0 (0.0 %) | 0 (0.0 %) | 2 (6.9 %) | 0 (0.0 %) | 0 (0.0 %) |
| Unknown | 34 (21.7 %) | 8 (16.3 %) | 6 (30.0 %) | 4 (13.8 %) | 13 (27.7 %) | 3 (25.0 %) |
| M stage | ||||||
| M0 | 120 (76.4 %) | 41 (83.7 %) | 14 (70.0 %) | 23 (79.3 %) | 34 (72.3 %) | 8 (66.7 %) |
| M1 | 3 (1.9 %) | 0 (0.0 %) | 0 (0.0 %) | 1 (3.4 %) | 0 (0.0 %) | 2 (16.7 %) |
| Unknown | 34 (21.7 %) | 8 (16.3 %) | 6 (30.0 %) | 5 (17.2 %) | 13 (27.7 %) 2 | (16.7 %) |
| Clinical stage | ||||||
| I | 29 (18.5 %) | 14 (28.6 %) | 1 (5.0 %) | 4 (13.8 %) | 6 (12.8 %) | 4 (33.3 %) |
| II | 96 (61.1 %) | 26 (53.1 %) | 18 (90.0 %) | 17 (58.6 %) | 30 (63.8 %) | 5 (41.7 %) |
| III | 26 (16.6 %) | 8 (16.3 %) | 1 (5.0 %) | 6 (20.7 %) | 10 (21.3 %) | 1 (8.3 %) |
| IV | 4 (2.5 %) | 1 (2.0 %) | 0 (0.0 %) | 1 (3.4 %) | 0 (0.0 %) | 2 (16.7 %) |
| Unknown | 2 (1.3 %) | 0 (0.0 %) | 0 (0.0 %) | 1 (3.4 %) | 1 (2.1 %) | 0 (0.0 %) |
| Tumor grade | ||||||
| 1 | 5 (3.2 %) | 5 (10.2 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) |
| 2 | 36 (22.9 %) | 27 (55.1 %) | 3 (15.0 %) | 4 (13.8 %) | 2 (4.3 %) | 0 (0.0 %) |
| 3 | 87 (55.4 %) | 10 (20.4 %) | 12 (60.0 %) | 20 (69.0 %) | 37 (78.7 %) | 2 (16.7 %) |
| 4 | 14 (8.9 %) | 3 (6.1 %) | 1 (5.0 %) | 3 (10.3 %) | 6 (12.8 %) | 1 (8.3 %) |
| Unknown | 15 (9.6 %) | 4 (8.2 %) | 4 (20.0 %) | 2 (6.9 %) | 2 (4.3 %) | 3 (25.0 %) |
| Hormone-receptor status | ||||||
| ER+ | 76 (48.4 %) | 49 (100 %) | 20 (100 %) | 0 (0.0 %) | 0 (0.0 %) | 7 (58.3 %) |
| PR+ | 74 (47.1 %) | 49 (100 %) | 20 (100 %) | 0 (0.0 %) | 0 (0.0 %) | 5 (41.7 %) |
| Unknown | 1 (0.6 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 1 (8.3 %) |
| HER2 status | ||||||
| HER2+ | 52 (33.1 %) | 0 (0.0 %) | 20 (100 %) | 29 (100 %) | 0 (0.0 %) | 3 (25.0 %) |
| Unknown | 7 (4.5 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 7 (58.3 %) |
| Treatment | ||||||
| Hormone therapy | 57 (36.6 %) | 37 (75.5 %) | 13 (65.0 %) | 0 (0.0 %) | 2 (4.3 %) | 4 (33.3 %) |
| Chemotherapy | 118 (75.2 %) | 32 (65.3 %) | 18 (90.0 %) | 21 (72.4 %) | 36 (76.6 %) | 11 (91.7 %) |
| Radiation therapy | 86 (54.8 %) | 26 (53.1 %) | 9 (45.0 %) | 12 (41.4 %) | 32 (68.1 %) | 7 (58.3 %) |
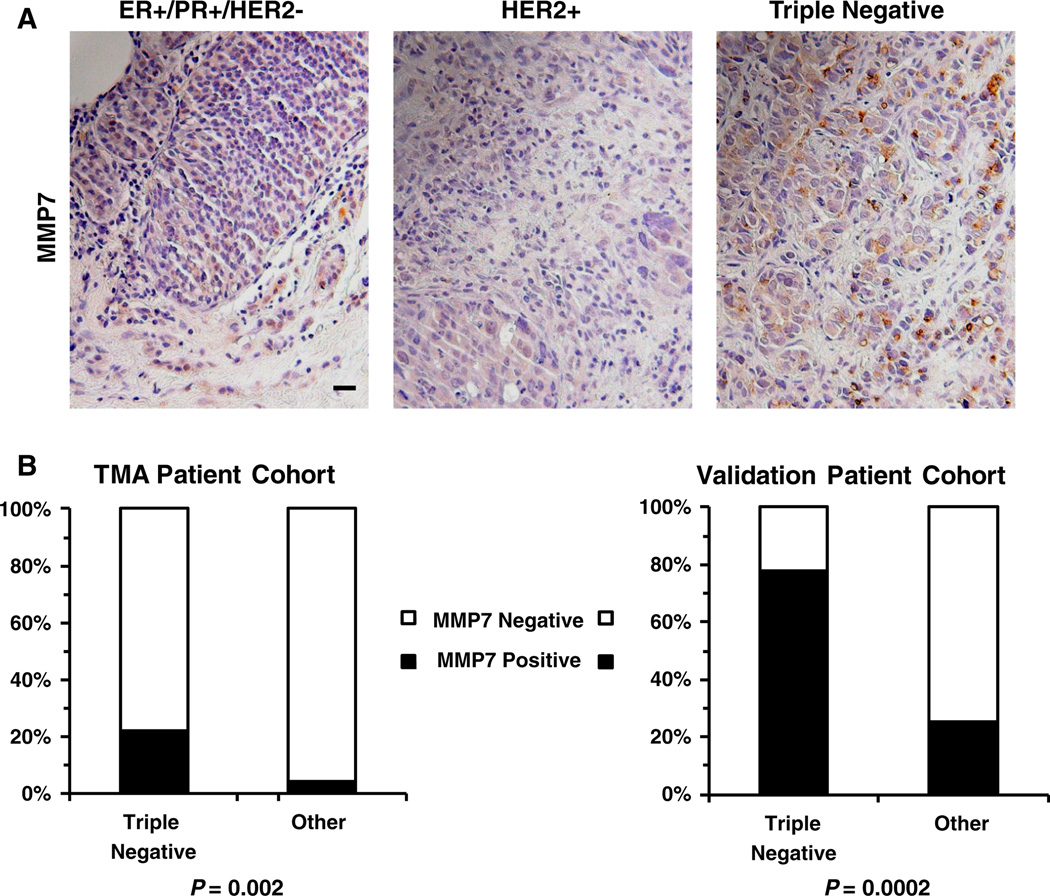
Of the 143 breast cancers in the TMA for which MMP7 scores and receptor status could be acquired (Table 2 and Fig. 1), only 4.12 % (4 of 97) of the luminal (Fig. 1a, left panel) and HER2+ (Fig. 1a, middle panel) samples showed MMP7 positivity (defined as a score of ≥1+ with at least 10 % of the tumor cells staining for MMP7). In contrast, 21.7 % (10 of 46) of the TN samples showed positive cytoplasmic and/or pericellular staining for MMP7 (Fig. 1 a, right panel; Supplemental Figure 1). The results of these analyses are summarized in Fig. 1b (left panel). To confirm the association between MMP7 positivity and TN status, we stained an additional 80 paraffin-embedded breast cancer sections for MMP7. The ER, PR, and HER2 status for these samples is known, but no further clinicopathological or outcome data are available for this cohort. Of the 18 TN samples in the validation cohort, 14 (77.8 %) showed positive (≥1+, >10 %) staining for MMP7, while only 25.4 % of the non-TN samples were positive for MMP7 staining (Fig. 1b, right panel). The discrepancy between the percentages of samples staining positive for MMP7 in the TMA cohort (21.7 %) versus the validation cohort (77.8 %) can likely be attributed to heterogeneity of MMP7 staining [36]. This heterogeneity may be driven by location-specific expression MMP7 as a greater concentration of MMP7 is expected along the invasive, leading edge of the tumor relative to the tumor interior [37–39]. The larger histological sections used in the validation cohort are more likely to contain areas of leading edge, and therefore, more likely to have areas of MMP7 positivity, than the smaller TMA tissue cores. An example of the heterogeneous MMP7 staining in a TN sample is depicted in Supplemental Figure 1. Despite the discrepancies arising from staining heterogeneity and sample coverage, staining for MMP7 is highly correlative with TN status (Fig. 1b) in both the TMA cohort (P = 0.002) and validation cohort (P = 0.0002).
Table 2.
Clinicopathologic characteristics of the TMA patient cohort stratified by MMP7 status
| MMP7 positive |
MMP7 negative |
p value | |
|---|---|---|---|
| Characteristic | n = 16 | n = 136 | |
| Age, mean ± SD | 48.4 ± 6.5 | 58.2 ± 15.1 | 0.0118 |
| Median follow-up time (years) |
5.7 (0.4–9.3) | 6 (0.2–12.9) | – |
| Patient race | |||
| Caucasian | 9 | 97 | 0.7330 |
| African–American | 4 | 29 | |
| Unknown | 3 | 10 | |
| Tumor size | |||
| T1 or T2 (<5 cm) | 8 | 93 | 0.0268 |
| T3 or T4 (>5 cm) | 5 | 13 | |
| Unknown | 3 | 30 | |
| Nodal status | |||
| Negative | 5 | 53 | 0.5589 |
| N1+ | 8 | 52 | |
| Unknown | 3 | 31 | |
| M Stage | |||
| M0 | 12 | 103 | 0.2977 |
| M1 | 1 | 2 | |
| Unknown | 3 | 31 | |
| Clinical stage | |||
| I or II | 9 | 111 | 0.0804 |
| III or IV | 6 | 24 | |
| Unknown | 1 | 1 | |
| Tumor grade | |||
| 1 or 2 | 2 | 38 | 0.2338 |
| 3 or 4 | 12 | 85 | |
| Unknown | 2 | 13 | |
| Hormone-receptor status | |||
| ER+ | 1 | 73 | 0.0003 |
| PR+ | 1 | 71 | 0.0003 |
| Unknown | 0 | 1 | |
| HER2 status | |||
| HER2+ | 4 | 46 | 0.7715 |
| Unknown | 2 | 5 | |
| Triple-negative status | |||
| Triple negative | 10 (62.5 %) | 36 (26.5 %) | 0.0016 |
| Unknown | 2 (12.5 %) | 5 (3.7 %) |
Fig. 1.
a. Representative photographs demonstrating the absence of MMP7 in ER+/PR+/HER2− (left panel) and HER2+ (middle panel) breast cancer samples and cytoplasmic and pericellular MMP7 staining in a representative TN sample (right panel). Scale bar = 20 µm. b. Bar graphs summarizing the percentage of TN samples versus those from the other subtypes (ER+/PR+/HER2− and HER2+ groups combined) that stained positively for MMP7 in the 152 patient TMA cohort (left panel) as well as in an 80 patient validation cohort (right panel). Samples were considered positive for MMP7 if at least 10 % of the tumor scored positive (1+). MMP7 staining was significantly more prevalent in the TN samples in both the TMA cohort (P = 0.002) and the validation cohort (P = 0.0002)
The association between MMP7 staining and clinicopathological prognostic factors was examined (Table 2). Patients whose tumors stained positive for MMP7 were significantly younger at the time of diagnosis (P <0.02, mean age of 48.4 years) than those that were not positive for MMP7 (mean age of 58.2 years) as would be expected for patients with TN tumors, which tend to present at an earlier age [40]. Tumors positive for MMP7 were also significantly larger at time of diagnosis (P <0.03). There was also a trend toward MMP7 positive tumors associating with higher tumor stage at time of diagnosis that did not reach statistical significance (P = 0.08). As mentioned previously, there was significant correlation between MMP7 positivity and TN status (P = 0.002) and a significant association between MMP7 staining and ER (P = 0.003) and PR (P = 0.003) negativity. No trend was observed between HER2 expression and MMP7 staining (P = 0.7715).
MMP7 staining correlates with decreased recurrence-free survival
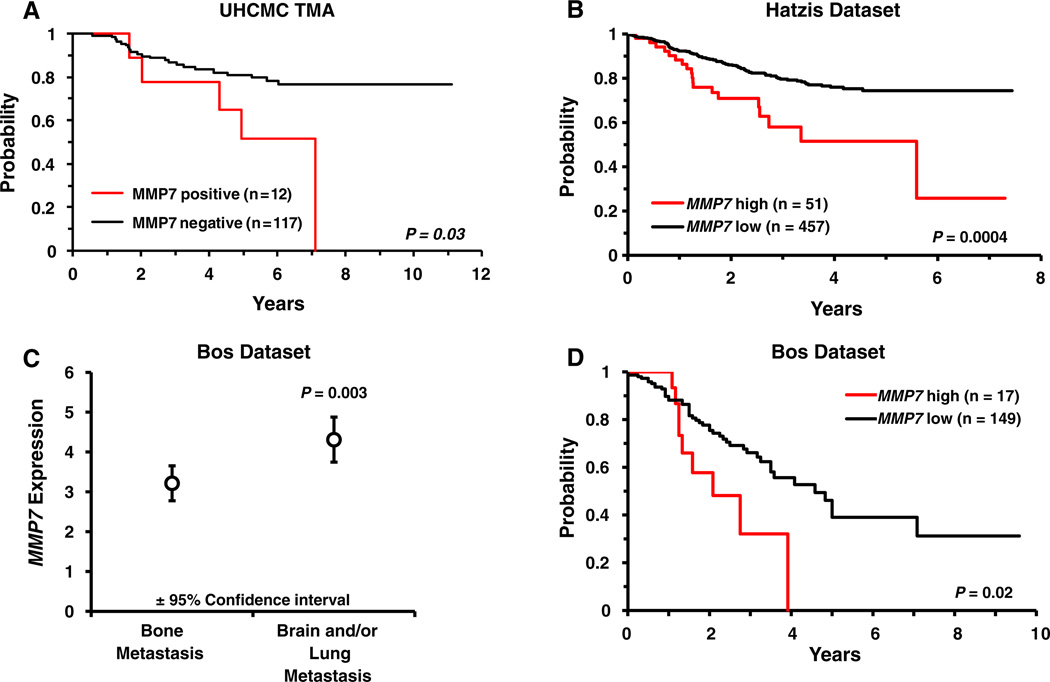
Clinical follow-up data, with a median follow-up time of 5.9 years, including time of recurrence and cause of death were available for up to 157 patients enrolled in the TMA study. There was no difference in over-all survival between MMP7 negative and MMP7 positive (≥1+, >10 %) groups (P = 0.72; data not shown). On the other hand, within the 129 patients for which recurrence information and MMP7 status were known, the MMP7 positive group (n = 12) had a significantly higher likelihood of recurrence (Fig. 2a, P = 0.03) compared to the MMP7 negative group (n = 117). Of the 12 MMP7 positive cases, five (41.7 %) experienced recurrence during the study follow-up period compared to only 18.8 % (22 of the 117) of the cases that showed no staining for MMP7. While MMP7 positivity was not found to be an independent predictor of recurrence by multivariate analysis (Supplemental Figure 2A), it is important to note that the association between MMP7 staining and recurrence cannot simply be attributed to MMP7 acting as a surrogate for TN status as TN status is not significantly associated with recurrence in this patient cohort (P = 0.65; Supplemental Figure 2B). To determine if MMP7 gene expression could serve as a prognostic marker, we utilized the publicly available Hatzis dataset [34] which contains metastatic recurrence information for 508 patients with invasive breast cancer. The Hatzis patient cohort was stratified along the 90th percentile of MMP7 expression to maintain the same ratio of MMP7 high to MMP7 low samples as in our TMA patient cohort wherein ~10 % (12 of 129) of the cohort scored positive for MMP7 expression. Elevated MMP7 expression significantly (P = 0.0004) correlated with shorter time to metastatic recurrence (Fig. 2b). TN breast cancers show a proclivity for metastasis at particular sites, notably brain and lung compared to other sites such as bone [41]. Analyzing data from the Bos patient cohort [32] we found that MMP7 expression was significantly elevated (P = 0.003) in the primary tumors of women whose cancers metastasized to the brain and/or lungs compared to those who developed bone metastases (Fig. 2c). We also assessed whether MMP7 expression could function as a prognostic marker of breast cancer metastasis to the brain and/or lungs in the Bos cohort. Stratifying the patient population by MMP7 expression along the 90th percentile revealed that elevated MMP7 expression significantly correlated (P = 0.02) with shorter time to the development of brain and/or lung metastases (Fig. 2d). MMP7 expression did not correlate with bone metastasis in these patients (P = 0.995, data not shown).
Fig. 2.
a Kaplan–Meier curves demonstrating the recurrence probability for the UHCMC TMA patient population stratified by MMP7 staining. Positive MMP7 staining is associated with significantly shorter time to recurrence (P = 0.03). b In the Hatzis patient cohort, the time to metastatic recurrence is significantly shorter (P = 0.0004) in patients with elevated MMP7 expression. c In the Bos patient cohort, mean expression of MMP7 is significantly higher (P = 0.003) in primary tumors of breast cancer patients that had metastases to the brain and/or lung versus patients that had bone metastases. d Kaplan– Meier curves demonstrating a significantly shorter time to metastasis to the brain and/or lung (P = 0.02) in association with elevated MMP7 expression in the Bos cohort
Hypomethylation of CpG sites in the MMP7 promoter correlates with TN status
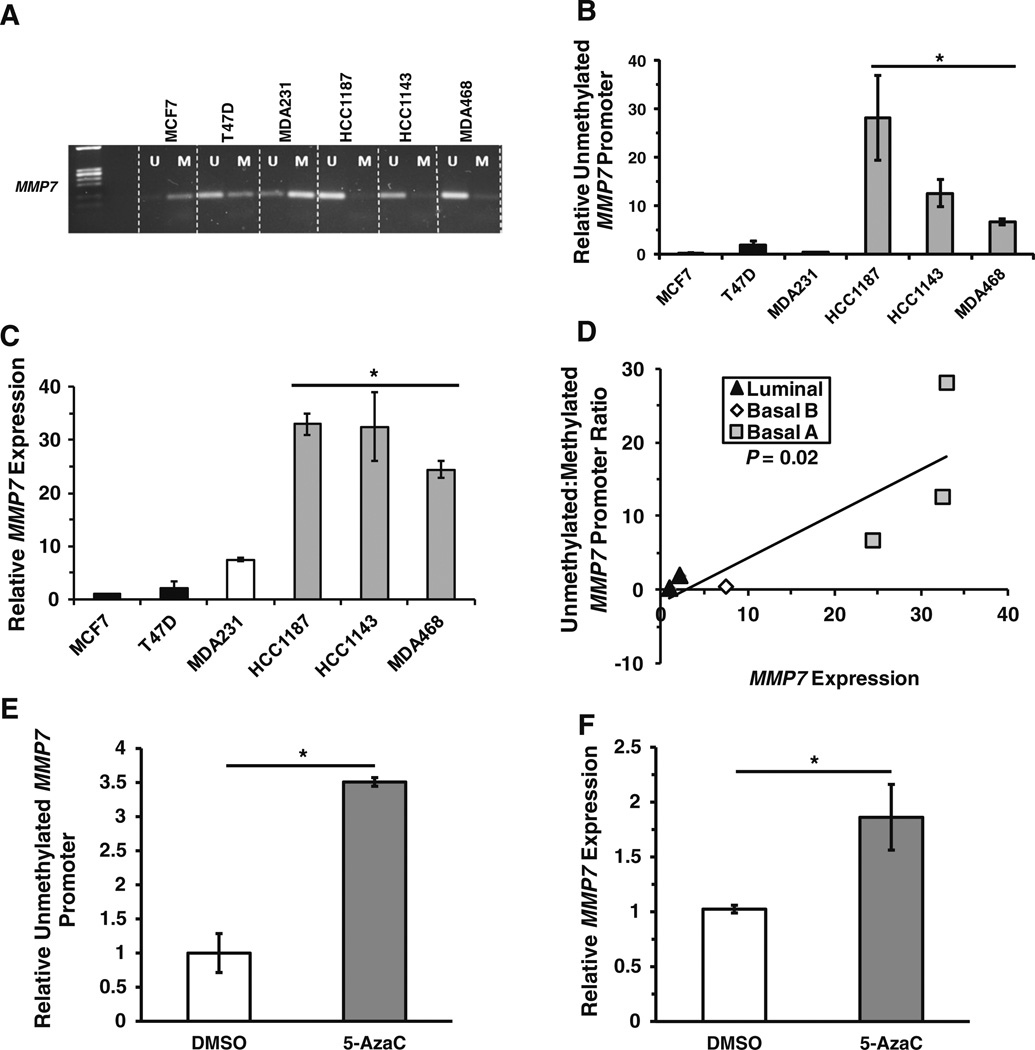
We previously reported that MMP7 mRNA and protein are elevated in a sub-group of TN breast cancer cell lines [23]. MMP7 expression has been demonstrated to be regulated by methylation status of CpG sites in the MMP7 promoter in pancreatic cancer cell lines [30]. To determine if epigenetic regulation is a key mechanism controlling MMP7 expression in breast cancer, we analyzed a number of breast cancer cell lines by methylation specific-PCR (MS-PCR) using a set of primers that detect the methylation status of CpG sites in the 5′ UTR and the coding region of exon1 of MMP7. Three of the four TN breast cancer cell lines analyzed (HCC1187, HCC1143, and MDA-MB-468) had significantly less methylation of the CpG sites than did the ER+ cell lines MCF7 and T47D or a fourth TN cell line, MDA-MB-231 (Fig. 3a, b). We utilized qRT-PCR to assess MMP7 expression in these cell lines and found a significant correlation (P = 0.02) between hypomethylation of the MMP7 promoter and elevated MMP7 expression (Fig. 3c, d). We then treated MCF7 cells, which have a highly methylated MMP7 promoter and concomitant low MMP7 expression, with the DNA methyltransferase inhibitor 5-Aza-2′-deoxycytidine (5-AzaC). Treatment with 5-AzaC significantly increased the amount of unmethylated MMP7 promoter (~3.5-fold; P < 0.05; Fig. 3e; Supplemental Figure 3A), as well as MMP7 expression (~2.0-fold; P < 0.05; Fig. 3f). Hence, epigenetic regulation appears to be a key determinant of MMP7 expression. Importantly, the three TN cell lines in which the MMP7 promoter is primarily in the hypomethylated state also have a gene expression profile most similar to the basal-like molecular breast cancer subtype [42, 43] that is shared by approximately 75 % of TN breast cancers [25, 44]. In contrast, the MDA-MB-231 TN cell line in which the MMP7 promoter is primarily in the hypermethylated state, has an expression signature that is similar to claudin-low tumors, a distinct TN subtype [32, 33, 35].
Fig. 3.
a A representative agarose gel displaying the bands produced by methylation-specific PCR (MS-PCR) using primers specific for unmethylated (U) or methylated (M) MMP7 promoter in a panel of breast cancer cell lines. b and c Bar graphs summarizing the mean ratio ± SD of unmethylated:methylated MMP7 promoter band intensities from three independent MS-PCR experiments (b) and the relative mean MMP7 expression ± SD from three independent quantitative real-time PCR (qRT-PCR) experiments (c). d A scatterplot illustrating the significant correlation (P = 0.02) between methylation status of the MMP7 promoter as determined by MS-PCR and MMP7 expression measured by qRT-PCR. e and f Representative bar graphs summarizing the relative amount of unmethylated MMP7 promoter (e) or MMP7 expression (f) in MCF7 cells treated with DMSO or 2.5 µM 5-Aza-2′-deoxycytidine for 72 h. *P < 0.05
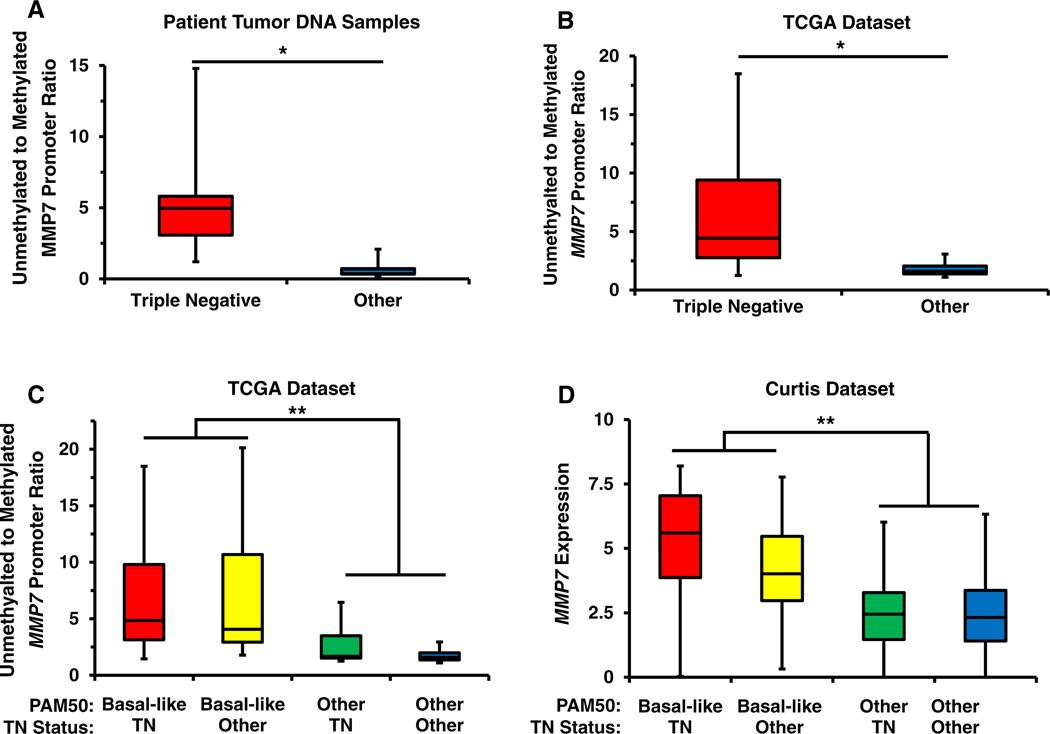
To expand our investigation of the relationship between MMP7 methylation status and breast cancer subtype to clinical breast cancer samples, we analyzed the methylation state of MMP7 in genomic DNA from 48 breast cancer samples using the same methylation-specific PCR primer sets. The characteristics of the patient population and the relative unmethylated:methylated (U:M) MMP7 promoter ratios are summarized in Table 3. Genomic DNA from 14 luminal, 19 HER2+, and 15 TN tumors was analyzed by MS-PCR (Table 3 and Supplemental Figure 3B). The CpG sites in the MMP7 promoter were predominantly in the methylated state in luminal breast cancers with an average U:M MMP7 promoter ratio of 0.34 and a maximum ratio of 0.82. Similarly, the HER2+ samples were primarily in the methylated state with an average U:M ratio of 0.71, a minimum value of 0.11 and a maximum U:M ratio of 2.08. In contrast, these CpG sites were predominantly in the unmethylated state in the TN samples with an average U:M ratio of 5.84, a maximum U:M value of 14.80, and a minimum value of 1.20. Overall, the analyzed CpG sites were significantly (P < 0.0001) more hypomethylated in the TN samples than in the other two subtypes (Fig. 4a). Combined, these results demonstrate a significant correlation between the methylation of CpG sites in the MMP7 promoter and the TN subtype and suggest that methylation of this genomic region may be a key regulatory mechanism controlling its transcriptional activity.
Table 3.
The MMP7 promoter is hypomethylated in TN breast tumor samples
| Subtype | Sample | Tumor (%) | ER (%) | PR (%) | HER2 | Unmethylated: methylated MMP7 promoter ratio |
|
|---|---|---|---|---|---|---|---|
| ER+/PR+/HER2− | 1 | 80 | 60 | 10 | Negative | N/A | |
| 2 | 75 | Positive | Positive | Negative | 0.23 | ||
| 3 | 85 | 80 | 75 | Negative | 0.28 | ||
| 4 | 90 | 90 | 60 | Negative | 0.12 | ||
| 5 | 80 | 90 | 70 | Negative | 0.26 | ||
| 6 | 80 | 70 | 70 | Negative | 0.23 | Average: 0.34 | |
| 7 | 95 | 80 | 80 | Negative | 0.32 | Min: 0.12 | |
| 8 | 65 | 45 | 55 | Negative | 0.82 | Max: 0.82 | |
| 9 | 70 | 80 | 70 | Negative | N/A | ||
| 10 | 60 | 80 | 60 | Negative | N/A | ||
| 11 | 70 | 100 | 50 | Negative | 0.70 | ||
| 12 | 90 | Positive | Positive | Negative | 0.30 | ||
| 13 | 70 | 80 | 50 | Negative | 0.13 | ||
| 14 | 80 | positive | positive | Negative | 0.40 | ||
| Triple negative | 15 | 100 | 0 | 0 | Negative (2+ , not ampl.) | 3.08 | |
| 16 | 85 | 0 | 0 | Negative | 2.50 | ||
| 17 | 70 | 0 | 0 | Negative | 9.22 | ||
| 18 | 85 | 0 | 0 | Negative | 3.56 | ||
| 19 | 85 | 0 | 0 | Negative | N/A | ||
| 20 | 80 | 0 | 0 | Negative | 3.04 | ||
| 21 | 80 | 0 | 0 | Negative | N/A | Average: 5.84 | |
| 22 | 60 | 0 | 0 | Negative | 3.86 | Min: 1.20 | |
| 23 | 80 | 0 | 0 | Negative | 9.53 | Max: 14.80 | |
| 24 | 80 | 0 | 0 | Negative | 9.22 | ||
| 25 | 80 | 0 | 0 | Negative | 5.38 | ||
| 26 | 80 | 0 | 0 | Negative | 14.80 | ||
| 27 | 65 | 0 | 0 | Negative (2+ , not ampl.) | 5.53 | ||
| 28 | 70 | 0 | 0 | Negative (2+ , not ampl.) | 4.97 | ||
| 29 | 55 | 0 | 0 | Negative (2+ , not ampl.) | 1.20 | ||
| HER2+ | 30 | 85 | 0 | 0 | Positive | N/A | |
| 31 | 65 | 0 | 0 | Positive | 1.20 | ||
| 32 | 60 | 0 | 0 | Positive | 0.62 | ||
| 33 | 75 | 0 | 0 | Positive | 0.79 | ||
| 34 | 80 | 0 | 0 | Positive | 0.32 | ||
| 35 | 80 | 0 | 0 | Positive | 0.44 | ||
| 36 | 75 | 0 | 0 | Positive | 0.64 | ||
| 37 | 80 | 0 | 0 | Positive | 0.11 | ||
| 38 | 85 | 0 | 0 | Positive | N/A | Average: 0.71 | |
| 39 | 65 | 0 | 0 | Positive | 1.48 | Min: 0.11 | |
| 40 | 65 | 0 | 0 | Positive | 2.08 | Max: 2.08 | |
| 41 | 80 | 0 | 0 | Positive (2+/ampl. Ratio 7:1) | 0.53 | ||
| 42 | 70 | 90 | 30 | Positive | 0.30 | ||
| 43 | 90 | Positive | Positive | Positive | 0.78 | ||
| 44 | 90 | 30 | 95 | Positive (2+/ampl. Ratio 2:1) | 0.52 | ||
| 45 | 75 | 60 | 10 | Positive | 1.15 | ||
| 46 | 90 | 100 | 85 | Positive | 0.20 | ||
| 47 | 75 | 95 | 95 | Positive | 0.47 | ||
| 48 | 85 | 80 | 0 | Positive (2+/ampl. Ratio 4:1) | 0.37 |
Fig. 4.
a Box-and-whisker plot summarizing the ratio of unmethylated:methylated MMP7 promoter band intensities as determined by MS-PCR using the DNA from 41 patients stratified by TN status. The MMP7 promoter is significantly less methylated in tumor DNA from patients with TN breast cancer. b Box-and-whisker plot summarizing the ratio of unmethylated:methylated MMP7 promoter as detected by Illumina methylation array probe cg25511807 in data from 463 patients in the TCGA invasive breast carcinoma dataset stratified by TN status. The MMP7 promoter is significantly less methylated in the tumor DNA of patients that are classified as TN. c Box-and-whisker plot summarizing the ratio of unmethylated:methylated MMP7 promoter in the TCGA patient cohort stratified by both basal-like gene expression signature, as defined by PAM50 analysis, and TN status. The MMP7 promoter is significantly hypomethylated in the basal-like subtype regardless of TN status but is only hypomethylated in those TN samples with a basal-like expression signature. d Box-and-whisker plot illustrating MMP7 expression in 1,969 patients from the Curtis dataset. MMP7 expression is significantly elevated in basal-like samples regardless of TN status. *P < 0.0001; **P < 0.001
MMP7 promoter methylation status and expression correlates with the basal-like breast cancer subtype
To confirm the differential methylation of the MMP7 promoter across breast cancer subtypes we analyzed the methylation status of the MMP7 promoter in data available from 463 breast cancer samples in the TCGA invasive breast carcinoma database. The Illumina methylation array probe utilized in this study (cg25511807) detects methylation of a region in the 5′UTR of MMP7 and partially overlaps the region analyzed by our MS-PCR assay. In agreement with our MS-PCR data, the CpG sites recognized by cg25511807 were significantly less methylated in TN samples in the TCGA data set versus the non-TN samples (P < 0.0001; Fig. 4b). Since we had observed a correlation between MMP7 promoter methylation status and the basal-like molecular subtype during our analysis of breast cancer cell lines, we explored this relationship in the TCGA patient dataset, wherein tumors have been subtyped according to PAM50 intrinsic signatures and, thus, can distinguish basal-like from other TN tumors. Interestingly, when the TCGA cohort was stratified by both basal-like status, as well as TN status, we discovered that an elevated U:M MMP7 promoter ratio significantly correlated (P = 0.001) with the basal-like molecular subtype and not with TN status (Fig. 4c). This result suggests that our earlier observations linking MMP7 promoter methylation to TN status were due to the 84 % overlap between the TN subtype (defined by lack of ER, PR, and HER2 expression) and the basal-like subtype (defined by PAM50 classification) and in the TCGA cohort.
To explore the relationship between MMP7 gene expression and the basal-like molecular and TN clinical subtypes, we utilized multiple large, publicly available datasets [33, 35]. When we examined the expression of MMP7 in 1,969 samples of the Curtis cohort for which expression data and subtype information were available (Fig. 4d), we found that basal-like molecular classification but not TN status correlated with higher MMP7 expression (P = 0.001). These results were confirmed by analyzing MMP7 expression in TCGA samples grouped by basal-like and TN status (P = 0.05, Supplemental Figure 4A). Importantly, we also observed significant correlation between MMP7 promoter methylation status and MMP7 expression in the TCGA cohort (P < 0.00001, Supplemental Figure 4B).
While there is significant overlap between the basal-like molecular and TN clinical subtypes, as mentioned previously, this overlap is incomplete. The majority of TN cancers express a basal-like gene signature, however, a minority of TN cancers belong to one of the other intrinsic subtypes (luminal A, luminal B, HER2, or normal-like) [45, 46] or belong to the claudin-low subtype which is molecularly distinguishable from other basal-like cancers [47]. To determine if expression of MMP7 is correlated with the expression of other genes associated with a particular breast cancer molecular subtype, we utilized the Curtis, Hatzis, and TCGA datasets to identify genes with the highest level of coordinate expression with MMP7. The top 1 % of genes, as determined by the Pearson correlation coefficient for expression of each gene with MMP7, from the three datasets were compared for overlapping features (Supplemental Figure 5). Twenty six genes were highly co-expressed with MMP7 in all three datasets. Comparison of the set of genes coordinately expressed with MMP7 to gene sets in the Molecular Signature Database (http://www.broadinstitute.org/gsea/msigdb/index.jsp) revealed that the MMP7-correlated genes are enriched for genes that are up-regulated in the basal subtype of breast cancer and genes that are down-regulated in luminal breast cancers (Supplemental Figure 6). Among the coordinately expressed genes are the basal cytokeratins KRT6B and KRT5 and a number of other genes (SFRP1, FOXC1, CXCL1, PRNP, and CDH3) previously found to be part of a core basal-like breast cancer signature [48]. Notably, the list of highly correlated genes included the gene for the transcription factor FOXC1 and the Wnt-related genes, SFRP1 and FZD7. FOXC1 and the wnt pathway are the key regulators of basal-like breast cancer phenotypes and both induce MMP7 expression [23, 49]. Absent from the list of genes coordinately regulated with MMP7 are genes associated with the luminal, HER2+, normal-like or claudin-low subtypes. These data indicate that MMP7 expression is highly correlated with the core set of basal genes and breast tumors expressing elevated MMP7 are likely to have a basal-like molecular signature.
Four genes (EGFR, VIM, KRT5, and KRT14) have previously been described as potential biomarkers of the basal-like breast cancer subtype [50–54]. We determined if the expression frequency of MMP7 in basal-like cancers was greater than these established markers. In the Curtis cohort, 16.7 % (331 of 1,986) of the samples are classified as basal-like. We used this 16.7 % cut-off to analyze the overlap between high expression (upper 16.7 %) of each marker and the basal-like subtype. Perfect concordance between high expression of a given gene and the basal-like subtype results in an expression frequency of 1. MMP7 had the highest frequency of all the biomarkers we analyzed. With a value of 0.58, MMP7 had the greatest overlap with the basal-subtype with EGFR and KRT5 having frequencies of 0.56 and 0.55, respectively (Supplemental Figure 7, upper panel). The overlap between the basal-subtype and KRT14 and VIM was even lower at 0.27 and 0.35, respectively. These results were confirmed in the TCGA cohort (17.5 % of the samples are basal-like in this cohort) where the frequency of MMP7 expression in the basal-like samples was 0.57. EGFR frequency in the TCGA cohort was higher than MMP7 at 0.74, but KRT5 (0.51), KRT14 (0.52), and VIM (0.31) were again expressed at a lower frequencies than MMP7 in the basal-like tumors (Supplemental figure 7, lower panel). These results suggest that both MMP7 promoter methylation status and MMP7 expression correlate with the basal-like molecular subtype independently of TN status and may outperform other biomarkers of the basal-like intrinsic subtype.
Elevated MMP7 expression is a predictor of pathological complete response
Basal-like breast cancers are more sensitive to cytotoxic chemotherapy than other breast cancer subtypes [29]. To assess MMP7 expression as a predictor of pCR to chemotherapy we utilized the Hatzis et al. dataset which includes pCR information for 454 patients treated with standard taxane-based chemotherapy regimens [34]. As expected, univariate analyses revealed that lower clinical T-stage, higher grade, ER, and PR negativity, HER2 positivity, higher node status, younger age, triple-negative status, and the basal-subtype all are associated with higher pCR in this cohort (Supplemental Figure 8). In addition, elevated MMP7 status is significantly associated with pCR in this patient population (P = 4.3 × 10−6).
To further investigate whether MMP7 may be a predictive factor for pCR, we generated two multivariate models predictive of pCR. The first model excludes MMP7 as a possible predictive factor and is comprised of grade, age, and basal-subtype (by PAM50 classification) as independent predictors of pCR. For the second model, MMP7 was included as a possible predictive factor and was found to be an additional independent predictive factor of pCR along with the three parameters identified by the initial model (Table 4). These two models were compared by the likelihood ratio test and the addition of MMP7 expression significantly improved the model (P = 0.009956). In addition, the two models were compared using their receiver operator characteristics and the addition of MMP7 expression increased the area under the curve value from 0.745 to 0.762 (Supplemental Figure 9). Thus, inclusion of MMP7 expression improves prediction of chemotherapeutic response in this cohort. Furthermore, both PAM50 and MMP7 expression are independent predictors of pCR and inclusion of both factors significantly improves prediction of pCR. Thus, MMP7's ability to predict response may extend beyond its ability to act as a surrogate marker of the basal-like subtype.
Table 4.
Predictive models of complete pathological response excluding or including MMP7 expression as a predictive factor
| Characteristic | Estimate | Std. Error | z value | P value | OR | 95 % CI |
|---|---|---|---|---|---|---|
| Model 1: Logistic regression model predicting pCR excluding MMP7 expression | ||||||
| Grade (3–4 vs. 1–2) | 1.2296 | 0.3496 | 3.517 | 0.000437 | 3.419876 | 1.763–7.01 |
| Age (>=50) | −0.375 | 0.2553 | −1.469 | 0.141762 | 0.687259 | 0.414–1.13 |
| PAM50 (Basal vs. others) | 1.0752 | 0.2821 | 3.811 | 0.000138 | 2.930536 | 1.7–5.152 |
| Model 2: Logistic regression model predicting pCR including MMP7 expression | ||||||
| Grade (3–4 vs. 1–2) | 1.2801 | 0.354 | 3.616 | 0.000299 | 3.597169 | 1.839–7.437 |
| Age (>=50) | −0.3774 | 0.2576 | −1.465 | 0.142875 | 0.685623 | 0.412–1.133 |
| PAM50 (Basal vs. others) | 0.7629 | 0.3077 | 2.48 | 0.013143 | 2.144579 | 1.179–3.952 |
| MMP7 Expression (high vs. low) | 0.741 | 0.2912 | 2.545 | 0.010941 | 2.097993 | 1.193–3.751 |
Likelihood ratio test (Model 1 vs. Model 2): P = 0.009956
Hatzis et al. Dataset
Discussion
Herein, we report that MMP7 staining correlates with the TN clinical subtype (P = 0.03) and disease recurrence (P = 0.03) in a 157 sample TMA. In addition, MMP7 positivity correlates with the lack of ER and PR (P = 0.0003, respectively), an earlier age at diagnosis (P = 0.0118), and increased tumor size (P = 0.0268). The association between MMP7 expression and TN status was validated in a second patient cohort and the prognostic value of MMP7 was confirmed in multiple publicly-available datasets. To investigate if epigenetic factors contribute to the differential expression of MMP7 in the various clinical breast cancer subtypes, we used methylation-specific PCR to determine the methylation status of CpG sites in the MMP7 promoter across a panel of breast cancer cell lines and a cohort of 48 tumor samples. These data were confirmed by in silico analysis of the publicly available TCGA dataset. Furthermore, these analyses led to the discovery that MMP7 promoter hypomethylation and expression correlate with the more specific basal-like molecular subtype rather than the broader TN clinical subtype. Lastly, we identified MMP7 as an independent predictor of favorable response to chemotherapy in a large breast cancer patient cohort [34]. Combined, our data indicate that MMP7 may be a valuable biomarker for distinguishing the basal-like molecular breast cancer subtype and predicting response to standard chemotherapy. Given its role as an extracellular enzyme, MMP7 may also serve as a target for novel treatments of this disease.
Breast cancers can be classified histologically by their expression of ER, PR, and HER2 or by gene expression signatures into one of five intrinsic subtypes (basal-like, luminal A, luminal B, HER2+, normal-like). TN tumors are defined by their lack of ER, PR, or HER2 expression and this classification is a rapid, albeit generalized, method for distinguishing highly aggressive tumors. The majority, but not all, of these tumors overlap with the more precisely defined basal-like molecular subtype. Likewise, basal-like tumors tend to be TN. However, there is not complete synonymy between the TN and basal-like subtypes [24, 25] and the identification of biomarkers that improve classification of these tumor types should bolster patient prognostication and potentially accelerate the development of novel therapeutics.
Currently, classification of breast cancers as basal-like by methods such as PAM50 (Prosigna®) requires the analysis of a number of molecular markers by qRT-PCR or gene expression microarray. Several biomarkers detectable by IHC, including the intermediate filaments cytokeratin 5, cytokertin 14, and vimentin, have been proposed as potential identifiers of the basal-like subtype especially when combined with TN status [50–54]. While these structural proteins may serve to distinguish some basal-like tumors from other tumor types, they do not hold potential as therapeutic targets. In addition to cytokeratins and vimentin, epithelial growth factor receptor (EGFR) is a marker of the basal-like breast cancers that is expressed in approximately 50 % [51, 52] of this subtype. Targeting EGFR is a promising avenue for treatment of some basal-like breast cancers, however, not all basal-like/TN breast cancers are positive for EGFR [51, 52]. In addition, the likelihood of developing resistance to EGFR-directed therapies is high as has been observed in lung cancer [55, 56]. Thus, other therapeutic targets for basal-like cancers must be identified. Our results suggest that MMP7 may serve not only as an important biomarker for identification of basal-like breast cancers and predictor of response to conventional chemotherapy but also as a promising target for novel therapies in the treatment of BLBC. IHC analysis revealed that 77.8 % of the histological sections from TN breast cancers stained positively for MMP7. Additional studies are needed to determine if staining for MMP7 or analysis of MMP7 promoter methylation status among the TN samples is specific for those with a basal-like molecular subtype. However, our analyses of publically available datasets suggest that MMP7 and promoter methylation status are specific markers of the basal-like molecular subtype regardless of receptor status.
To date, efforts to treat cancer, including invasive breast carcinoma, with compounds that inhibit protease activity of the MMP-family have not been successful [57]. However, non-inhibitor based methods to target MMPs may prove more useful in cancer treatment. In particular, treatment strategies that take advantage of the ability of the proteases to recognize and cleave selective amino acid sequences can be utilized to tailor anti-cancer compounds that are activated by specific MMPs. One such example is an MMP-activated photomolecular beacon that provides a molecular on-switch for photodynamic therapy that is controlled by the presence of MMP7 [58]. Other therapies that utilize MMP activation have been developed including MMP-activated cytotoxins that are capable of killing cancer cells only after an activating cleavage by MMPs [59, 60]. MMP7 may prove to be especially suited for this type of therapy given that it is specifically produced by cancer cells and not the surrounding stroma, unlike many other MMP-family members. Most importantly, MMP7-activated therapies may answer the need for targeted therapies that are effective against basal-like breast cancers.
Combined, our data demonstrate that elevated MMP7 expression and promoter hypomethylation may improve TN breast cancer classification and prognosis, and advocates for future studies to assess the efficacy of MMP7-activated anti-cancer compounds in the treatment of basal-like breast cancers.
Supplementary Material
Acknowledgments
This work was supported by grants from the Department of Defense W81XWH-09-1-0696 (to STS) and RO1 CA090398 and RO1 CA154384 (to RAK). The Tissue Procurement and Histology Core Facility of the Case Comprehensive Cancer Center (P30 CA043703) was instrumental in the development of the TMA. We thank Darcie Seachrist of the Department of Pharmacology at Case Western Reserve University for her assistance in acquiring images of the pathological samples. We would also like to thank Michael Goggins, M.D. of the Departments of Pathology, Oncology and Medicine at The Johns Hopkins Medical Institute for the primer sequences used to detect the MMP7 promoter methylation status.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10549-014-2989-4) contains supplementary material, which is available to authorized users.
Conflict of interest The authors declare that they have no conflicts of interest.
Contributor Information
Steven T. Sizemore, Department of Pharmacology, Case Western Reserve University School of Medicine, 10900 Euclid Ave., Cleveland, OH 44106-4965, USA
Gina M. Sizemore, Department of Pharmacology, Case Western Reserve University School of Medicine, 10900 Euclid Ave., Cleveland, OH 44106-4965, USA
Christine N. Booth, Department of Anatomic Pathology, Cleveland Clinic Foundation, Cleveland, OH 44195, USA
Cheryl L. Thompson, Department of Family Medicine and Community Health, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA
Paula Silverman, Department of Medicine, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA.
Gurkan Bebek, Center for Proteomics and Bioinformatics, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA; Genomic Medicine Institute, Cleveland Clinic Foundation, Cleveland, OH 44195, USA.
Fadi W. Abdul-Karim, Department of Anatomic Pathology, Cleveland Clinic Foundation, Cleveland, OH 44195, USA
Stefanie Avril, Department of Pathology, Technical University Munich, 81675 Munich, Germany.
Ruth A. Keri, Email: keri@case.edu, Department of Pharmacology, Case Western Reserve University School of Medicine, 10900 Euclid Ave., Cleveland, OH 44106-4965, USA; Division of General Medical Sciences-Oncology, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA; Department of Genetics, Case Western Reserve University School of Medicine, Cleveland, OH 44106, USA.
References
- 1.Hua H, Li M, Luo T, Yin Y, Jiang Y. Matrix metalloproteinases in tumorigenesis: an evolving paradigm. Cell Mol Life Sci. 2011;68(23):3853–3868. doi: 10.1007/s00018-011-0763-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kleiner DE, Stetler-Stevenson WG. Matrix metalloproteinases and metastasis. Cancer Chemother Pharmacol. 1999;43(Suppl):S42–S51. doi: 10.1007/s002800051097. [DOI] [PubMed] [Google Scholar]
- 3.Gialeli C, Theocharis AD, Kletsas D, Tzanakakis GN, Karamanos NK. Expression of matrix macromolecules and functional properties of EGF-responsive colon cancer cells are inhibited by panitumumab. Invest New Drugs. 2013;31(3):516–524. doi: 10.1007/s10637-012-9875-x. [DOI] [PubMed] [Google Scholar]
- 4.Noe V, Fingleton B, Jacobs K, Crawford HC, Vermeulen S, Steelant W, Bruyneel E, Matrisian LM, Mareel M. Release of an invasion promoter E-cadherin fragment by matrilysin and stromelysin-1. J Cell Sci. 2001;114(Pt 1):111–118. doi: 10.1242/jcs.114.1.111. [DOI] [PubMed] [Google Scholar]
- 5.Rucci N, Sanita P, Angelucci A. A Roles of metalloproteases in metastatic niche. Curr Mol Med. 2011;11(8):609–622. doi: 10.2174/156652411797536705. [DOI] [PubMed] [Google Scholar]
- 6.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14(2):163–176. [PMC free article] [PubMed] [Google Scholar]
- 7.Yu WH, Woessner JF, Jr, McNeish JD, Stamenkovic I. CD44 anchors the assembly of matrilysin/MMP-7 with heparin-binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev. 2002;16(3):307–323. doi: 10.1101/gad.925702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heppner KJ, Matrisian LM, Jensen RA, Rodgers WH. Expression of most matrix metalloproteinase family members in breast cancer represents a tumor-induced host response. Am J Path. 1996;149(1):273–282. [PMC free article] [PubMed] [Google Scholar]
- 9.Fang YJ, Pan ZZ, Li LR, Lu ZH, Zhang LY, Wan DS. MMP7 expression regulated by endocrine therapy in ERbeta-positive colon cancer cells. J Exp Clin Cancer Res. 2009;28:132. doi: 10.1186/1756-9966-28-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukuda A, Wang SC, Morris JPt, Folias AE, Liou A, Kim GE, Akira S, Boucher KM, Firpo MA, Mulvihill SJ, Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19(4):441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentner B, Wein A, Croner RS, Zeittraeger I, Wirtz RM, Roedel F, Dimmler A, Dorlaque L, Hohenberger W, Hahn EG, Brueckl WM. Differences in the gene expression profile of matrix metalloproteinases (MMPs) and their inhibitors (TIMPs) in primary colorectal tumors and their synchronous liver metastases. Anticancer Res. 2009;29(1):67–74. [PubMed] [Google Scholar]
- 12.Tonini G, Pantano F, Vincenzi B, Gabbrielli A, Coppola R, Santini D. Molecular prognostic factors in patients with pancreatic cancer. Expert Opin Ther Targets. 2007;11(12):1553–1569. doi: 10.1517/14728222.11.12.1553. [DOI] [PubMed] [Google Scholar]
- 13.Fang YJ, Lu ZH, Wang F, Wu XJ, Li LR, Zhang LY, Pan ZZ, Wan DS. Prognostic impact of ERβ and MMP7 expression on overall survival in colon cancer. Tumour Biol. 2010;31(6):651–658. doi: 10.1007/s13277-010-0082-0. [DOI] [PubMed] [Google Scholar]
- 14.Huang Y, Yu H, Lei H, Xie C, Zhong Y. Matrix metalloproteinase 7 is a usefulmarker for 5-fluorouracil-based adjuvant chemotherapy in stage II and stage III colorectal cancer patients. Med Oncol. 2014;31:824. doi: 10.1007/s12032-013-0824-0. [DOI] [PubMed] [Google Scholar]
- 15.Gallego R, Codony-Servat J, García-Albéniz X, Carcereny E, Longarón R, Oliveras A, Tosca M, Augé JM, Gascón P, Maurel J. Serum IGF-I, IGFBP-3, and matrix metalloproteinase-7 levels and acquired chemo-resistance in advanced colorectal cancer. Endocr Relat Cancer. 2009;16(1):1412–1420. doi: 10.1677/ERC-08-0250. [DOI] [PubMed] [Google Scholar]
- 16.Fang YJ, Lu ZH, Wang GQ, Pan ZZ, Zhou ZW, Yun JP, Zhang MF, Wan DS. Elevated expressions of MMP7, TROP2, and survivin are associated with survival, disease recurrence, and liver metastasis of colon cancer. Int J Colorectal Dis. 2009;24(8):875–884. doi: 10.1007/s00384-009-0725-z. [DOI] [PubMed] [Google Scholar]
- 17.Wu J, Guan X, Zhang K, Li YT, Bai P, Wu J. A/G polymorphism of matrix metalloproteinase 7 gene promoter region and cancer risk: a meta-analysis. Biomed Rep. 2013;1(5):792–796. doi: 10.3892/br.2013.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ke P, Wu ZD, Wen HS, Ying MX, Long HC, Qing LG. Current evidence on associations between the MMP-7 (−181A > G) polymorphism and digestive system cancer risk. Asian Pac J Cancer Prev. 2013;14(4):2269–2272. doi: 10.7314/apjcp.2013.14.4.2269. [DOI] [PubMed] [Google Scholar]
- 19.Liu D, Guo H, Li Y, Xu X, Yang K, Bai Yc. Association between polymorphisms in the promoter regions of matrix metalloproteinases (MMPs) and risk of cancer metastasis: a meta-analysis. PLoS ONE. 2012;7(2):e31251. doi: 10.1371/journal.pone.0031251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saarialho-Kere UK, Crouch EC, Parks WC. Matrix metalloproteinase matrilysin is constitutively expressed in adult human exocrine epithelium. J Invest Dermatol. 1995;105(2):190–196. doi: 10.1111/1523-1747.ep12317104. [DOI] [PubMed] [Google Scholar]
- 21.Yuan G, Qian L, Shi M, Lu F, Li D, Hu M, Yu M, Shen B, Guo N. HER2-dependent MMP-7 expression is mediated by activated STAT3. Cell Signal. 2008;20(7):1284–1291. doi: 10.1016/j.cellsig.2008.02.017. [DOI] [PubMed] [Google Scholar]
- 22.Yuan G, Qian L, Song L, Shi M, Li D, Yu M, Hu M, Shen B, Guo N. Heregulin-beta promotes matrix metalloproteinase-7 expression via HER2-mediated AP-1 activation in MCF-7 cells. Mol Cell Biochem. 2008;318(1–2):73–79. doi: 10.1007/s11010-008-9858-6. [DOI] [PubMed] [Google Scholar]
- 23.Sizemore ST, Keri RA. The forkhead box transcription factor FOXC1 promotes breast cancer invasion by inducing matrix metalloprotease 7 (MMP7) expression. J Biol Chem. 2012;287(29):24631–24640. doi: 10.1074/jbc.M112.375865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bertucci F, Finetti P, Cervera N, Esterni B, Hermitte F, Viens P, Birnbaum D. How basal are triple-negative breast cancers? Int J Cancer. 2008;123(1):236–240. doi: 10.1002/ijc.23518. [DOI] [PubMed] [Google Scholar]
- 25.Valentin MD, da Silva SD, Privat M, Alaoui-Jamali M, Bignon YJ. Molecular insights on basal-like breast cancer. Breast Cancer Res Treat. 2012;134(1):21–30. doi: 10.1007/s10549-011-1934-z. [DOI] [PubMed] [Google Scholar]
- 26.Parker JS, Mullins M, Cheang MC, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z, Quackenbush JF, Stijleman IJ, Palazzo J, Marron JS, Nobel AB, Mardis E, Nielsen TO, Ellis MJ, Perou CM, Bernard PS. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, Thorsen T, Quist H, Matese JC, Brown PO, Botstein D, Lønning PE, Børresen-Dale AL. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98(19):10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weigelt B, Hu Z, He X, Livasy C, Carey LA, Ewend MG, Glas AM, Perou CM, Van’t-Veer LJ. Molecular portraits and 70-gene prognosis signature are preserved throughout the meta-static process of breast cancer. Cancer Res. 2005;65(20):9155–9158. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 29.Carey LA, Dees EC, Sawyer L, Gatti L, Moore DT, Collichio F, Ollila DW, Sartor CI, Graham ML, Perou CM. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. doi: 10.1158/1078-0432.CCR-06-1109. [DOI] [PubMed] [Google Scholar]
- 30.Sato N, Maehara N, Su GH, Goggins M. Effects of 5-aza-2′-deoxycytidine on matrix metalloproteinase expression and pancreatic cancer cell invasiveness. J Natl Cancer Inst. 2003;95(4):327–330. doi: 10.1093/jnci/95.4.327. [DOI] [PubMed] [Google Scholar]
- 31.Schneider CA, Rasband WS, Eliceiri KW. Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bos PD, Zhang XH, Nadal C, Shu W, Gomis RR, Nguyen DX, Minn AJ, van de Vijver MJ, Gerald WL, Foekens JA, Massague J. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459(7249):1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, Martin M, Cotrina J, Gomez H, Hubbard R, Chacon JI, Ferrer-Lozano J, Dyer R, Buxton M, Gong Y, Wu Y, Ibrahim N, Andreopoulou E, Ueno NT, Hunt K, Yang W, Nazario A, DeMichele A, O’Shaughnessy J, Hortobagyi GN, Symmans WF. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305(18):1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490(7418):61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Camp RL, Neumeister V, Rimm DL. A decade of tissue microarrays: progress in the discovery and validation of cancer biomarkers. J Clin Oncol. 2008;26(34):5630–5637. doi: 10.1200/JCO.2008.17.3567. [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa M, Furuya M, Kasuya Y, Nishiyama M, Sugiura T, Nikaido T, Momota Y, Ichinose M, Kimura S. CD151 dynamics in carcinoma-stroma interaction: integrin expression, adhesion strength and proteolytic activity. Lab Invest. 2007;87(9):882–892. doi: 10.1038/labinvest.3700657. [DOI] [PubMed] [Google Scholar]
- 38.Mitsui H, Suarez-Farinas M, Gulati N, Shah KR, Cannizzaro MV, Coats I, Felsen D, Krueger JG, Carucci JA. Gene Expression Profiling of the Leading Edge of Cutaneous Squamous Cell Carcinoma: IL-24-Driven MMP-7. J Invest Dermatol. 2013 doi: 10.1038/jid.2013.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Puthenedam M, Wu F, Shetye A, Michaels A, Rhee KJ, Kwon JH. Matrilysin-1 (MMP7) cleaves galectin-3 and inhibits wound healing in intestinal epithelial cells. Inflamm Bowel Dis. 2011;17(1):260–267. doi: 10.1002/ibd.21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Billar JA, Dueck AC, Stucky CC, Gray RJ, Wasif N, Northfelt DW, McCullough AE, Pockaj BA. Triple-negative breast cancers: unique clinical presentations and outcomes. Ann Surg Oncol. 2010;17(Suppl 3):384–390. doi: 10.1245/s10434-010-1260-4. [DOI] [PubMed] [Google Scholar]
- 41.Kennecke H, Yerushalmi R, Woods R, Cheang MC, Voduc D, Speers CH, Nielsen TO, Gelmon K. Metastatic behavior of breast cancer subtypes. J Clin Oncol. 2010;28(20):3271–3277. doi: 10.1200/JCO.2009.25.9820. [DOI] [PubMed] [Google Scholar]
- 42.Charafe-Jauffret E, Ginestier C, Monville F, Finetti P, Adelaide J, Cervera N, Fekairi S, Xerri L, Jacquemier J, Birnbaum D, Bertucci F. Gene expression profiling of breast cell lines identifies potential new basal markers. Oncogene. 2006;25(15):2273–2284. doi: 10.1038/sj.onc.1209254. [DOI] [PubMed] [Google Scholar]
- 43.Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, Speed T, Spellman PT, DeVries S, Lapuk A, Wang NJ, Kuo WL, Stilwell JL, Pinkel D, Albertson DG, Waldman FM, McCormick F, Dickson RB, Johnson MD, Lippman M, Ethier S, Gazdar A, Gray JW. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10(6):515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Palmieri D, Bronder JL, Herring JM, Yoneda T, Weil RJ, Stark AM, Kurek R, Vega-Valle E, Feigenbaum L, Halverson D, Vortmeyer AO, Steinberg SM, Aldape K, Steeg PS. Her-2 overexpression increases the metastatic outgrowth of breast cancer cells in the brain. Cancer Res. 2007;67(9):4190–4198. doi: 10.1158/0008-5472.CAN-06-3316. [DOI] [PubMed] [Google Scholar]
- 45.Perou CM. Molecular stratification of triple-negative breast cancers. Oncologist. 2011;16(Suppl 1):61–70. doi: 10.1634/theoncologist.2011-S1-61. [DOI] [PubMed] [Google Scholar]
- 46.Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist. 2013;18(2):123–133. doi: 10.1634/theoncologist.2012-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, Parker J, Ewend MG, Sawyer LR, Wu J, Liu Y, Nanda R, Tretiakova M, Ruiz-Orrico A, Dreher D, Palazzo JP, Perreard L, Nelson E, Mone M, Hansen H, Mullins M, Quackenbush JF, Ellis MJ, Olopade OI, Bernard PS, Perou CM. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genom. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crawford HC, Fingleton BM, Rudolph-Owen LA, Goss KJ, Rubinfeld B, Polakis P, Matrisian LM. The metalloproteinase matrilysin is a target of beta-catenin transactivation in intestinal tumors. Oncogene. 1999;18(18):2883–2891. doi: 10.1038/sj.onc.1202627. [DOI] [PubMed] [Google Scholar]
- 50.Brenton JD, Carey LA, Ahmed AA, Caldas C. Molecular classification and molecular forecasting of breast cancer: ready for clinical application? J Clin Oncol. 2005;23(29):7350–7360. doi: 10.1200/JCO.2005.03.3845. [DOI] [PubMed] [Google Scholar]
- 51.Cakir A, Gonul II, Uluoglu O. A comprehensive morphological study for basal-like breast carcinomas with comparison to nonbasal-like carcinomas. Diagn Pathol. 2012;7:145. doi: 10.1186/1746-1596-7-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nielsen TO, Hsu FD, Jensen K, Cheang M, Karaca G, Hu Z, Hernandez-Boussard T, Livasy C, Cowan D, Dressler L, Akslen LA, Ragaz J, Gown AM, Gilks CB, van de Rijn M, Perou CM. Immunohistochemical and clinical characterization of the basal-like subtype of invasive breast carcinoma. Clin Cancer Res. 2004;10(16):5367–5374. doi: 10.1158/1078-0432.CCR-04-0220. [DOI] [PubMed] [Google Scholar]
- 53.Rodriguez-Pinilla SM, Sarrio D, Honrado E, Moreno-Bueno G, Hardisson D, Calero F, Benitez J, Palacios J. Vimentin and laminin expression is associated with basal-like phenotype in both sporadic and BRCA1-associated breast carcinomas. J Clin Pathol. 2007;60(9):1006–1012. doi: 10.1136/jcp.2006.042143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sousa B, Paredes J, Milanezi F, Lopes N, Martins D, Dufloth R, Vieira D, Albergaria A, Veronese L, Carneiro V, Carvalho S, Costa JL, Zeferino L, Schmitt F. P-cadherin, vimentin and CK14 for identification of basal-like phenotype in breast carcinomas: an immunohistochemical study. Histol Histopathol. 2010;25(8):963–974. doi: 10.14670/HH-25.963. [DOI] [PubMed] [Google Scholar]
- 55.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, Chiang A, Yang G, Ouerfelli O, Kris MG, Ladanyi M, Miller VA, Pao W. Novel D761Y and common secondary T790 M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12(21):6494–6501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 56.Suda K, Onozato R, Yatabe Y, Mitsudomi T. EGFR T790 M mutation: a double role in lung cancer cell survival? J Thorac Oncol. 2009;4(1):1–4. doi: 10.1097/JTO.0b013e3181913c9f. [DOI] [PubMed] [Google Scholar]
- 57.Zucker S, Cao J, Chen WT. Critical appraisal of the use of matrix metalloproteinase inhibitors in cancer treatment. Oncogene. 2000;19(56):6642–6650. doi: 10.1038/sj.onc.1204097. [DOI] [PubMed] [Google Scholar]
- 58.Zheng G, Chen J, Stefflova K, Jarvi M, Li H, Wilson BC. Photodynamic molecular beacon as an activatable photosensitizer based on protease-controlled singlet oxygen quenching and activation. PNAS. 2007;104(21):8989–8994. doi: 10.1073/pnas.0611142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu S, Netzel-Arnett S, Birkedal-Hansen H, Leppla SH. Tumor cell-selective cytotoxicity of matrix metalloproteinase-activated anthrax toxin. Cancer Res. 2000;60(21):6061–6067. [PubMed] [Google Scholar]
- 60.Liu S, Wang H, Currie BM, Molinolo A, Leung HJ, Moayeri M, Basile JR, Alfano RW, Gutkind JS, Frankel AE, Bugge TH, Leppla SH. Matrix metalloproteinase-activated anthrax lethal toxin demonstrates high potency in targeting tumor vasculature. J Biol Chem. 2008;283(1):529–540. doi: 10.1074/jbc.M707419200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.