Abstract
Recurrent deletions and duplications at chromosomal region 16p11.2 are a major genetic contributor to autism but also associate with a wider range of pediatric diagnoses, including intellectual disability, coordination disorder, and language disorder. In order to investigate the potential genetic basis for phenotype variability, we assessed the parent of origin of the 16p11.2 copy-number variant (CNV) and the presence of additional CNVs in 126 families for which detailed phenotype data were available. Among de novo cases, we found a strong maternal bias for the origin of deletions (59/66, 89.4% of cases, p = 2.38 × 10−11), the strongest such effect so far observed for a CNV associated with a microdeletion syndrome. In contrast to de novo events, we observed no transmission bias for inherited 16p11.2 CNVs, consistent with a female meiotic hotspot of unequal crossover driving this maternal bias. We analyzed this 16p11.2 CNV cohort for the presence of secondary CNVs and found a significant maternal transmission bias for secondary deletions (32 maternal versus 14 paternal, p = 1.14 × 10−2). Of the secondary deletions that disrupted a gene, 82% were either maternally inherited or de novo (p = 4.3 × 10−3). Nine probands carry secondary CNVs that disrupt genes associated with autism and/or intellectual disability risk variants. Our findings demonstrate a strong bias toward maternal origin of 16p11.2 de novo deletions as well as a maternal transmission bias for secondary deletions that contribute to the clinical outcome on a background sensitized by the 16p11.2 CNV.
Introduction
Duplication and deletion of an ∼550 kbp region on chromosome 16p11.2 accounts for ∼1% of autism cases, representing one of the most common contributors to autism spectrum disorder (ASD) in the human population.1, 2 Unlike many other syndromic disorders, such as Smith-Magenis or Prader-Willi syndromes, detailed studies of individuals with the 16p11.2 copy-number variant (CNV) have revealed marked phenotypic variability.3, 4, 5, 6, 7, 8, 9, 10, 11 Phenotypic studies indicate different phenotypes associated with the CNV, and opposite phenotypes are sometimes associated with 16p11.2 deletion and duplication. For example, the deletion has been associated with seizures,6 obesity,12 intellectual disability,4 and macrocephaly,3 whereas the duplication has been associated with schizophrenia,13 reduced BMI,14 and microcephaly.3 Although it is clear that the 16p11.2 CNV confers a strong risk for neurodevelopmental disease,15, 16, 17, 18 it is likely that other factors, including genetic background, are key in determining the severity of the phenotypic outcome.19, 20
Recently, a cohort of over 120 families, including at least one proband carrying a 16p11.2 CNV, was assembled as part of the Simons Variation in Individuals Project (Simons VIP).21 This collection is one of the largest cohorts for the 16p11.2 CNV and is distinctive in its comprehensive phenotypic assessment of participants. It offers a useful resource for studying genetic differences on a background sensitized by a known pathogenic CNV and for studying how these differences affect phenotype severity. In this analysis, carriers of the 16p11.2 CNV are either probands or other family members who are heterozygous for the deletion or duplication, irrespective of diagnostic ascertainment or inheritance status. The goal of this study was 2-fold: (1) to provide genetic detail regarding the extent and transmission characteristics of the CNV in these families and (2) to investigate the presence of CNVs in addition to the role of the 16p11.2 CNV in modifying the severity of the phenotype. For clarity and to distinguish from the ascertained 16p11.2 CNV, we will refer to the rare additional CNVs (present in <0.1% of control individuals) as secondary CNVs. In this study, we assess the parent of origin and mechanism of unequal crossing over for the 16p11.2 de novo CNVs and examine transmission bias for secondary CNVs within these families.
Subjects and Methods
Samples
DNA samples were derived from peripheral blood obtained from 482 individuals from 141 families affected by the 16p11.2 CNV as part of the Simons VIP. Exclusion criteria included the presence of any additional pathogenic CNVs or other neurogenetic or neurological diagnoses unrelated to 16p11.2.21 More than 80% of probands were of full European ancestry (Table S1). We utilized the Simons VIP September 30, 2014 release of phenotypic information for these individuals. All procedures for clinical assessment and blood extraction were approved by the institutional review boards of participating institutions, and informed consent was obtained for participation in this research.
Phenotypic Assessment
As part of participation in the Simons VIP,21 standardized assessments, including psychiatric, neurocognitive, behavioral, motor, and neurologic evaluation, were conducted at three Simons VIP clinical sites; additionally, detailed medical histories were collected through interviews and review of medical records for each participant. Psychiatric and neurodevelopmental conditions were diagnosed by experienced, licensed clinicians using all available information, including clinical observation, caregiver history, and records review, and in accordance with DSM-IV-TR (Diagnostic and Statistical Manual of Mental Disorders, 4th edition, text revision) criteria.22 Diagnostic foci included ASD, attention deficit hyperactivity disorder (ADHD), communication disorders, anxiety disorders, mood disorders, intellectual disability, tic disorders, elimination disorders, learning disorders, and behavioral disorders, totaling 27 diagnostic codes. Full-scale intelligence quotient (FSIQ) was determined by the developmentally appropriate cognitive measure (Mullen Scales of Early Learning23), the Differential Abilities Scale, Second Edition,24 or the Wechsler Abbreviated Scales of Intelligence.25 For our phenotype analysis, we defined the FSIQ decrement as the average of the FSIQ of the parents subtracted from the FSIQ of the proband (Table 1).
Table 1.
Clinical Characteristics of Screened Probands
|
Probands with Deletions (40 F and 50 M) |
Probands with Duplications (16 F and 20 M) |
|||||
|---|---|---|---|---|---|---|
| Median | Range | Number Reported | Median | Range | Number Reported | |
| Age (years) | 7.83 | 0.83–20.75 | 89/90 | 5.83 | (1.42–23.42) | 35/36 |
| FSIQ | 86 | 46–122 | 87/90 | 77 | (28–114) | 33/36 |
| FSIQ decrement | 25.5 | −9–68.5 | 49/90 | 22 | (6.5–89) | 23/36 |
| Social Responsiveness Scale score | 74.5 | 37–90 | 78/90 | 76 | (42–90) | 25/36 |
| Autism Diagnostic Interview Revised score | 11 | 2–30 | 60/90 | 11.5 | (2–26) | 18/36 |
| Head circumference (cm) | 54 | 45–59.7 | 86/90 | 51.05 | (44.4–58) | 34/36 |
| BMI | 19.2 | 13.35–37.13 | 86/90 | 16.17 | (13.36–28.37) | 34/36 |
| Number of diagnosesa | 3 | 1–5 | 80/90 | 2 | (1–5) | 32/36 |
Abbreviations are as follows: F, female; M, male.
This includes ASD, ADHD, communication disorders, anxiety disorders, mood disorders, intellectual disability, tic disorders, elimination disorders, learning disorders, and behavioral disorders, totaling 27 diagnostic codes. 21/89 deletion-carrying probands and 8/35 duplication-carrying probands for whom data was reported have been diagnosed with clinical autism.
CNV Detection
SNP microarray data were generated from the Illumina HumanOmniExpress v.1 (104 probands, 280 family members) and v.2 (26 probands, 72 family members) microarray platforms. Each microarray contains over 715,000 probes and has the power to detect CNVs >100 kbp with >95% sensitivity (Figure S1). CNVs were detected with the cnvPartition algorithm (see Web Resources). We chose this algorithm because its performance (as determined by the cnvPartition score) had previously been optimized by comparison against CNVs detected by deep whole-genome sequence data.26 For both array designs, we generated a cluster definition file from only the individuals who did not carry the 16p11.2 CNV by using the Illumina Genome Studio software (see Web Resources). Samples in the extremes for call rate and autosomal LogR SD were manually inspected. We assessed one family with an individual carrying a 16p11.2 triplication and removed this family (Simons VIP family 14752), as well as families in which the proband did not have the expected 16p11.2 CNV identified in the clinic (14905 and 14925), from subsequent analysis. Familial relationships were assessed with the program KING,27 and samples that did not match expected pedigree membership were removed (Table S1). The analysis showed that the probands were unrelated, except for two probands (14710.x7 and 14877.x7) who have a possible third-degree relationship. To ensure accurate comparisons between OmniExpress platforms, we required a minimum of seven probes within unique regions for both platforms and excluded the call if it contained >50% segmental duplication. Calls with the same state in the same individuals within 500 kbp of one another were merged if appropriate after manual inspection, and all calls identified as de novo were manually inspected. A subset of the calls >100 kbp were validated with an array comparative genomic hybridization (CGH) platform (Tables S2 and S3). After this curation, 102 probands and 264 family members were analyzed on the HumanOmniExpress v.1 platform, and 24 probands and 68 family members were analyzed on the HumanOmniExpress v.2 platform. Secondary CNVs intersecting genes associated with autism risk variants were defined with the SFARI gene list (June 2015, see Web Resources). We used the two-sided binomial test in this study and all genomic coordinates are listed in the UCSC Genome Browser (hg19), unless otherwise indicated.
CNV Inheritance and Validation
For each CNV call in a proband, we genotyped parents and siblings (if present), computed the median log ratio across these regions, and used this information to genotype across the family. We further validated a subset of large (>100 kbp) CNVs by using a custom array CGH platform (Table S3). We utilized a previously designed custom 12-plex NimbleGen array with a total of 135,000 probes targeted to genomic hotspots for CNV detection.28 The hotspot array consists of a high density of probes (approximately 2.6 kbp apart) targeting 107 genomic hotspot regions and a probe spacing of approximately 36 kbp in the genomic backbone. Array hybridization experiments and analysis were performed as described previously.28 All signal intensities from the array CGH experiments were loaded onto a UCSC Genome Browser mirror and manually visualized. 26/34 secondary CNVs >100 kbp called by the SNP microarray were validated by array CGH. The eight events that did not validate had insufficient coverage on the array CGH platform (≤5 probes spanning the region).
Analysis of Control Cohorts
To assess the population frequency of each secondary CNV, we used two sets of curated control samples. Set I focuses on larger CNVs from 19,584 previously published control individuals15 whose ethnicity is similar to that of our case individuals (79.2% with known ethnicity are of European descent). Set II is a curated set of 4,092 samples from the Welcome Trust Case Control Consortium (WTCCC, see Web Resources) and analyzed with a custom Illumina 1.2 million SNP microarray. Because of a higher density of probes, set II has increased sensitivity for smaller events as compared to set I. Set II CNVs were recalled with the cnvPartition algorithm in order to improve the comparison with the case individual calls. We called CNVs on 2,920 samples from the WTCCC 58C cohort and 2,698 samples from the WTCCC UKBS (UK Blood Service) cohort. The ethnicity of the UKBS cohort is 100% European ancestry. Although the ethnicity of the 58C cohort is not available, this is a 1958 British Birth Cohort and therefore likely to primarily contain individuals of European descent. Control individuals were not ascertained specifically for neurological disorders, but all control samples were obtained from adult individuals providing informed consent, so severe developmental phenotypes should be exceedingly rare.
Samples with a SNP call rate <0.98 and/or an autosomal LogR SD ≤0.37 were removed.15 We utilized an outlier detection method for skewed data29 to identify and remove additional samples with an excess of calls and/or an excess of larger calls (>100 kbp or >500 kbp). Finally, we applied this outlier method to exclude CNVs within these size ranges when their median LogR value subtracted from their mean LogR value was greater than 0.2 or less than −0.15, which are known characteristics of false-positive calls. 4,092 samples passed quality control (2,025 samples from the 58C cohort and 2,067 from the UKBS cohort). Similarly to our analysis of case CNVs, we required at least seven unique probes for each CNV call. Calls with the same CNV state and mapping within 500 kbp of one another were manually inspected and merged if appropriate. To assess the frequency of CNVs in case individuals, we computed the number of state-matched events that have a 50% reciprocal overlap with a control event in both set I and set II. Because of the probe density, set II offered greater sensitivity for assessing the frequency of smaller CNVs in case individuals. In addition, set II uses the same technology as the case platforms, and CNV calls were made with the same algorithm. We only considered secondary CNVs as rare if there were sufficient probes to call the variant in either set I or set II and the estimated control frequency was <0.1% (Table S3).
De Novo 16p11.2 CNV Parent-of-Origin Analysis
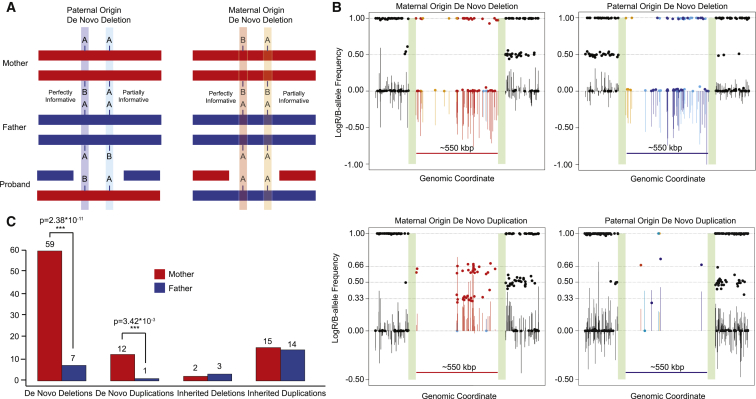
We used the signal intensity data (LogR) to confirm the presence of the 16p11.2 CNV and B-allele frequency (BAF) across the critical region to infer the parent of origin for 79 families in which a de novo 16p11.2 CNV had been identified (Figures 1A and 1B, Tables S4 and S5). This included 64 individuals from the Simons VIP (58 deletions, 6 duplications) as well as 15 individuals from the Simons Simplex Collection (SSC) who were previously assessed with SNP microarrays30 (8 deletions, 7 duplications, Table S6). In total, 34 quads (families for which data from both parents, a sibling, and a proband are available), 22 trios (families for which data for both parents and a proband [but no sibling] are available), and 10 probands with data available from only one parent were used to assess de novo deletion cases (Table S4). A total of 8 quads and 5 trios were used to assess de novo duplication cases (Table S5). We restricted this analysis to probes mapping within the 16p11.2 critical region (112 for the OmniExpress arrays). For deletions, only two genotypes are possible for each probe (A or B), with corresponding BAFs of 0 or 1, whereas for duplications, four genotypes (AAA, AAB, ABB, and BBB) with corresponding BAFs of 0, 1/3, 2/3, and 1, respectively, are possible (see Supplemental Appendix). For case individuals for whom we had SNP microarray data from both parents (trios), we computed the probability that the unaffected haplotype came from the mother versus from the father by using parental SNP genotypes. In the case individuals with a 16p11.2 deletion previously confirmed as de novo and for whom only one parent’s data was available, we estimated the probability of the genotypes for the unobserved parent by using the known allele frequencies for particular probes from the 1000 Genomes Project.31 To test the fidelity of this approach for families affected by deletions and with incomplete data, we estimated the false discovery rate by removing a parent from a subset of the families for which we had information from both parents (Table S7). Using this approach, we found that 78/88 parent-of-origin estimates matched our inferences (a false discovery rate of 11.4%).
Figure 1.
Maternal Origin of 16p11.2 De Novo CNVs
(A) We assigned SNPs on the unaffected critical region haplotype to either a paternal or maternal haplotype by using B-allele frequency (BAF) data. Informative or partially informative markers for parent of origin are shown.
(B) LogR (lines) and BAF (dots) plots for all de novo deletion and duplication categories. Colors correspond to the inferred parent of origin of the 16p11.2 CNV from each type of SNP marker highlighted in (A). Green bars indicate the location of segmental duplications associated with BP4 and BP5—collapsed here for ease of display.
(C) Approximately 90% of de novo 16p11.2 deletions and duplications originate on the maternal haplotype, a significant maternal bias (p = 2.38 × 10−11 deletions, 3.42 × 10−3 duplications, two-sided binomial test). Such a bias was not observed for inherited 16p11.2 CNVs.
Mechanism of Unequal Crossover and Recombination Analyses
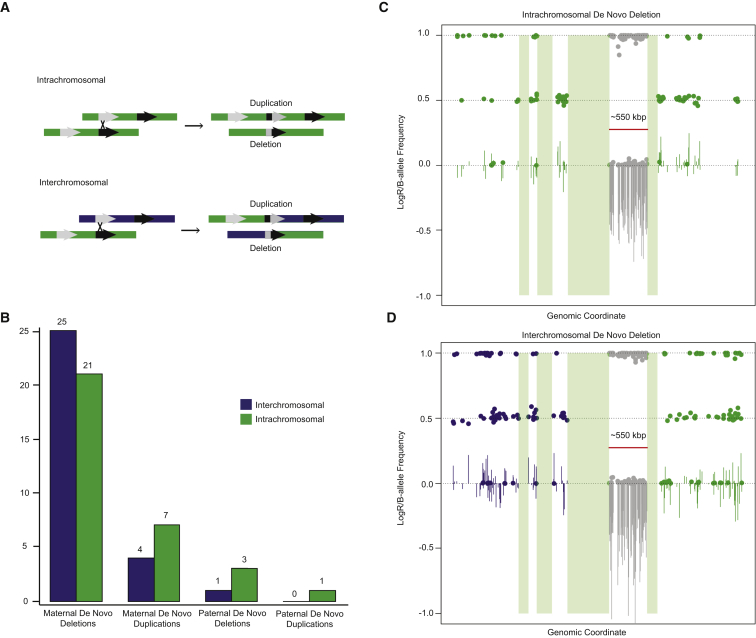
To determine the mechanism of unequal crossover of de novo 16p11.2 CNV events, we phased the haplotypes in the unique regions flanking the 16p11.2 critical region in the proband by using the sibling (if present) or dbSNP (if absent)32 (see Supplemental Appendix for details and calculation). An exchange of flanking SNP markers suggests an interchromosomal mechanism (nonallelic homologous recombination [NAHR] during meiosis I), whereas maintenance of the haplotype phase (i.e., no exchange) suggests an intrachromosomal or interchromatidal mechanism (most likely NAHR during meiosis II). Because this method cannot differentiate between intrachromosomal and interchromatidal mechanisms, we refer to both as an intrachromosomal mechanism. Male and female recombination rates for the critical region were obtained from Kong et al.33 The genetic distance between the leftmost and rightmost markers in our analysis is 6.20 cM for the female versus 0.45 cM for the male, which corresponds to a probability of crossover of 6.2% and 0.45%, respectively. We also used the recombination rate data to estimate the average difference between male and female recombination rates within the 16p11.2 critical region, and in 550 kbp regions genome wide, for comparison. We sampled 10,000 regions of 550 kbp (the size of the 16p11.2 critical region), excluding regions containing segmental duplications or gaps and the sex chromosomes, and determined that the region ranks in the 87th percentile for mean difference between male and female recombination rates genome wide (Figure S2).
Results
Characterization of 16p11.2 CNVs in the Simons VIP Cohort
We confirmed the presence or absence of the 16p11.2 deletion or duplication by using a SNP microarray (Illumina OmniExpress) in a total of 459 individuals from 126 families in which a proband with either a duplication (n = 36) or deletion (n = 90) had been identified (Table 2). For 81% of the probands (102/126), DNA was available from at least one parent, and 60% (76/126) had DNA available from both parents (Table S1). We confirmed the presence of the canonical breakpoint 4 to breakpoint 5 (BP4–BP5) deletion or duplication for most of the probands (125/126), corrected familial transmission status for one Simons VIP family (14784, Figure S3), and confirmed the presence of a de novo deletion in a set of monozygotic twins (in family 14824, Figure S4). In one severely affected proband (14720.x7), we identified a larger 2 Mbp deletion extending from BP2 to beyond BP5 (Figure S5).6 In addition to the phenotype information for the 70 case individuals with available DNA who were screened, phenotype information (but no DNA) is available for a larger set of 150 probands and their family members. Among Simons VIP case individuals for whom both parents were also screened for the 16p11.2 CNV (109/150) (either via clinical microarray, fluorescence in situ hybridization, and/or the present analysis), 90% of deletion cases (65/72) were de novo or mosaic in the germline. In contrast, only 24% (9/37) of duplication cases were confirmed as de novo.
Table 2.
Number of 16p11.2 CNV Carriers and Non-carrier Family Members Analyzed
| Male Probands | Female Probands | Mother | Father | Sibling | Other Family Membera | Total | Trios | Quads | |
|---|---|---|---|---|---|---|---|---|---|
| 16p11.2 Deletion Carriers | |||||||||
| De novob | 37 | 24 | 0 | 0 | 1 | 0 | 62 | 22 | 27 |
| Inherited | 3 | 6 | 0 | 0 | 4 | 0 | 13 | 1 | 2 |
| Unknown | 10 | 10 | 3 | 4 | 0 | 0 | 27 | NA | NA |
| Total | 50 | 40 | 3 | 4 | 5 | 0 | 102 | 23 | 29 |
| 16p11.2 Duplication Carriers | |||||||||
| De novo | 3 | 5 | 0 | 2 | 0 | 0 | 10 | 3 | 3 |
| Inherited | 16 | 7 | 2 | 4 | 6 | 13 | 48 | 9 | 9 |
| Unknown | 1 | 4 | 9 | 8 | 0 | 4 | 26 | NA | NA |
| Total | 20 | 16 | 11 | 14 | 6 | 17 | 84 | 12 | 12 |
| Non-carriers | |||||||||
| Total | NA | NA | 94 | 69 | 73 | 27 | 263 | NA | NA |
Abbreviation is as follows: NA, not applicable.
Other family members include grandparents, half-siblings, aunts, uncles, and cousins.
Includes one proband with a confirmed deletion resulting from germline mosaicism.
Maternal Parent of Origin of the 16p11.2 CNV
We observe a striking maternal bias for the parent of origin of 16p11.2 de novo deletions (Figure 1). 89.4% (59/66) occur on the maternal haplotype, representing a significant departure from expectation (p = 2.38 × 10−11) (Figure 1C). A similar result was observed for duplications (12 maternal versus 1 paternal, p = 3.42 × 10−3). For individuals with inherited 16p11.2 CNVs and for whom we have information from both parents, we observed no significant parental transmission biases for either duplication (15/29 maternal, p = 1) or deletion (2/5 maternal, p = 1) cases (Figure 1C, Table S8). We also used the microarray data to assess the relative proportion of interchromosomal (between homologs) and intrachromosomal (within homolog) NAHR events by phasing haplotypes of the unique regions flanking the critical region (see Subjects and Methods and Supplemental Appendix). We observed no preference for a particular mechanism of crossover for either maternal events (29 inter- versus 28 intrachromosomal, p = 1) or paternal events (1 inter- versus 4 intrachromosomal, p = 0.375) (Figure 2). If we restrict the analysis to families for which we have high-confidence phasing information as a result of the presence of unaffected siblings, there is a trend toward maternal interchromosomal unequal crossover events for deletions (19 inter- versus 8 intrachromosomal, p = 0.052) (Tables S9 and S10).
Figure 2.
Mechanisms of Unequal Crossing Over
(A) Schematic shows intrachromosomal and interchromosomal NAHR events and the resulting products. Colors (green and purple) indicate different homologs.
(B) Counts of interchromosomal and intrachromosomal NAHR events by parent of origin and by deletion versus duplication status. None of the differences are significant based on a two-sided binomial test.
(C and D) LogR (lines) and BAF (dots) plots are shown for an intrachromosomal (C) and interchromosomal (D) de novo deletion across the 16p11.2 region. Green bars indicate the location of segmental duplications associated with BP1–BP5.
Secondary CNVs and Maternal Transmission Bias
We considered the presence of secondary rare CNVs (<0.1% frequency in control individuals) as a potential modifier of phenotype severity within the context of each family. The SNP microarray used to detect CNVs in this study has >95% sensitivity for detecting events >100 kbp throughout the genome, although we note that events as small as 2 kbp can be detected (Figure S1). Despite the Simons VIP exclusion criteria for additional pathogenic CNVs, 70% of assessed probands (88/126) carried at least one secondary CNV, and 35% (44/126) of probands had two or more secondary CNVs. The percentage of deletion and duplication probands carrying a secondary CNV is similar (69% and 69.5%, respectively), and no significant differences in secondary CNV presence were observed between males and females (65% and 75%, respectively) (Table 3 and S11). Overall, only five of the secondary CNVs were determined to be de novo (4 deletions and 1 duplication), although in 40% of the families (50/126), inheritance status could not be determined due to the absence of DNA from both parents. Over a third (50/126) of all probands carried a secondary CNV >100 kbp in size (Tables 3 and S11). 81 secondary CNVs disrupted an annotated exon of a gene. 11 of these corresponded to genes associated with autism risk variants (Table S12), consistent with their potential contribution to disease etiology in the nine individuals in whom they were found.
Table 3.
Secondary CNVs
|
Secondary Deletionsa |
No. of Probands |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Maternal | Secondary CNVs | >1 Secondary CNVs | >1 Secondary Deletion | >1 Secondary Duplication | Secondary Deletion > 100 kbp | Secondary Duplication > 100 kbp | >1 Secondary CNV > 100 kbp | |
| Probands with a 16p11.2 Deletion | |||||||||
| Total (90) | 31 | 19 (61.3%) | 62 | 28 | 10 | 11 | 19 | 19 | 6 |
| Males (50) | 21 | 14 (66.7%) | 32 | 13 | 4 | 6 | 11 | 10 | 5 |
| Females (40) | 10 | 5 (50%) | 30 | 15 | 6 | 5 | 8 | 9 | 1 |
| Probands with a 16p11.2 Duplication | |||||||||
| Total (36) | 19 | 13 (68.4%) | 25 | 16 | 6 | 6 | 9 | 8 | 5 |
| Males (20) | 13 | 7 (53.8%) | 13 | 8 | 5 | 3 | 4 | 4 | 4 |
| Females (16) | 6 | 6 (100%) | 12 | 8 | 1 | 3 | 5 | 4 | 1 |
Number of events in probands with inheritance information available from both the mother and father.
Among secondary CNVs for which inheritance could be unambiguously determined (i.e., both parents were screened), maternally inherited events predominated (52 maternal versus 35 paternal, p = 0.086). The maternal bias is strongest for the most likely pathogenic events. If we consider only secondary deletions, 70% are transmitted maternally (32 maternal versus 14 paternal, p = 1.14 × 10−2). This is significant both in terms of the number of events as well as the number of probands inheriting an event from a particular parent (29 maternal versus 10 paternal, p = 3.38 × 10−3). This effect remains significant if we restrict our analysis to secondary deletions intersecting an exon (13 maternal versus 4 paternal secondary CNVs, p = 4.9 × 10−2). These trends also hold for secondary deletions >100 kbp in length, although this finding does not reach significance as a result of sample size limitations. This maternal bias for deletions is observed for both 16p11.2 deletion and duplication individuals, irrespective of the gender of the proband (Table S11).
Phenotypic Features
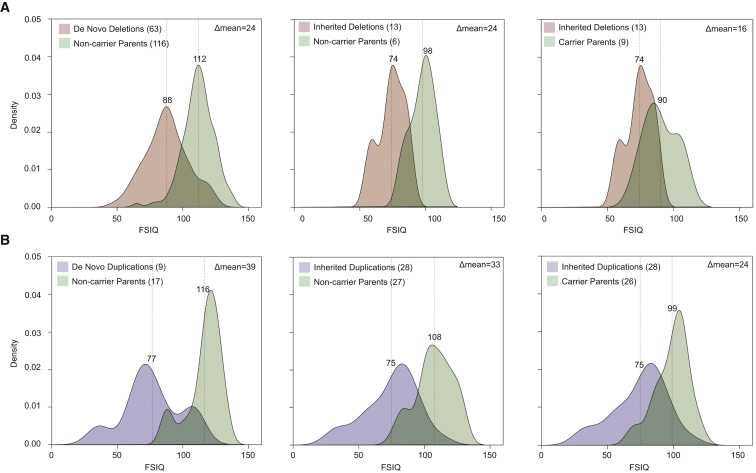
Carriers and non-carriers of the 16p11.2 CNV within the same family vary dramatically in their phenotypic presentation (e.g., FSIQ difference,5, 6 Figure 3, Tables 1 and S13). We observe statistically significant differences between the FSIQ distributions of parents carrying the 16p11.2 deletion and of probands with the deletion (p = 6 × 10−3, Student’s t test). Similarly, the means of the FSIQ distributions of the parents and probands carrying the duplication are significantly different (p = 3.89 × 10−6, Student’s t test). Such differences between parents and children carrying the 16p11.2 CNV suggest that other genetic and non-genetic factors are contributing to the phenotype. We investigated the relationship between additional-CNV burden and severity of phenotype by using FSIQ, Social Responsiveness Scale (SRS) scores, Autism Diagnostic Interview Revised (ADI-R) scores, head circumference, and BMI as phenotypic metrics. We found a modest negative correlation between FSIQ and the number of secondary CNVs (R2 = 0.04, p = 0.03, Figure S6). This signal is driven primarily by secondary deletions and is consistent with previous findings on the overall burden of CNV deletions and reduced intelligence quotient (IQ).19 Although no other significant correlations are observed with other quantitative measurements, an examination of the clinical details for individuals carrying these secondary CNVs showed evidence of clinodactyly, scoliosis, hypopigmentation, and craniofacial abnormalities consistent with a more severe phenotypic outcome.
Figure 3.
Familial IQ Decrement in 16p11.2 Deletion and Duplication Families
(A) Density plots of FSIQ for deletion families (A) and duplication families (B) from the entire Simons VIP cohort. The significant decrement between parents carrying a 16p11.2 deletion and inherited-deletion probands (p = 6 × 10−3, Student’s t test) and between parents carrying a 16p11.2 duplication and inherited-duplication probands (p = 3.89 × 10−6, t test), shown in the third panels of (A) and (B), suggest that factors other than the 16p11.2 CNV contribute to FSIQ decrement.
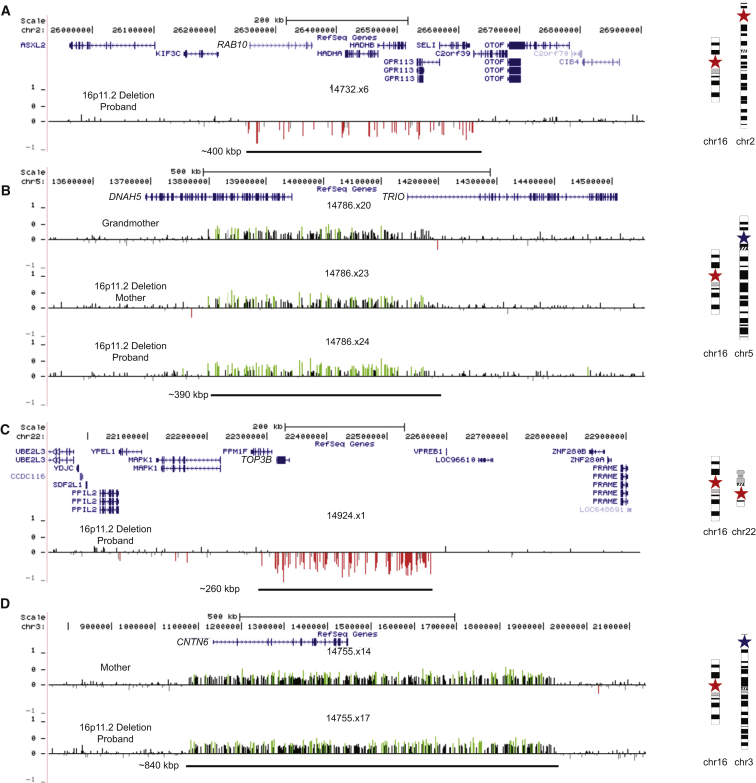
Among the secondary CNVs were several deletions and duplications corresponding to genes strongly implicated in synaptic function and/or risk of autism. Nine individuals, for example, had rare deletions or duplications in genes implicated in autism as defined by a curated list of genes associated with autism risk (see Web Resources), including CACNA2D3 (MIM: 606399), TRIO (MIM: 601893), and KATNAL2 (MIM: 614697) (Table S12). In a proband with a 16p11.2 deletion, we validated an additional private ∼400 kbp deletion that affects six genes, including RAB10 (MIM: 612672)—a gene important in vesicular transport and membrane trafficking in neurons.34 This proband is among the most severely affected females in our cohort. She exhibits autism (SRS score = 90), intellectual disability (FSIQ = 54), pediatric seizures, anxiety, obsessive compulsive disorder (OCD), and phobia, along with structural defects of the brain, including enlarged ventricles and abnormal cerebellar vermis and corpus callosum (Figure 4A). Because DNA is not available for either parent, the inheritance status for both deletions is unknown. The severity of this proband’s phenotype is similar to that of the male proband with severe intellectual disability (NVIQ = 29) who carried an atypical deletion of 16p11.2 encompassing more than 50 genes (Figure S5). We also discovered a secondary duplication disrupting DNAH5 (MIM: 603335) and TRIO that was transmitted from grandmother, to daughter, to son. Transmission of this CNV was associated with a characteristic facies. Although the mother and son both carry the 16p11.2 deletion, the severity of the phenotype, based on FSIQ, increased from generation to generation (Figure 4B), and the son manifests other features such as gynecomastia, clinodactyly, and scoliosis.
Figure 4.
Examples of Secondary Large CNVs
Microarray signal intensity data shown for various deletions and duplications.
(A) Data for 400 kbp gene-rich deletion of RAB10 in a 16p11.2-deletion female proband (14732.x6) with FSIQ = 54, SRS = 90, autism, intellectual disability, pediatric seizures, anxiety, OCD, and phobia. The CNV was private and not observed in 19,584 control individuals; parental DNA was not available for analysis.
(B) Data for 390 kbp duplication disrupting DNAH5 and TRIO in a grandmother (14786.x20), mother (14786.x23), and male proband (14786.x24). The mother and proband also carry the 16p11.2 deletion. From grandmother, to daughter, to grandson, the FSIQ decreases from 99 to 89 to 63, respectively. This CNV was private and not observed in 4,092 control individuals.
(C) Data for 260 kbp deletion of TOP3B in a 16p11.2 deletion female (14924.x1) with non-verbal IQ (NVIQ) = 109; SRS = 80; autism; language, learning, and articulation disorder; and ADHD. The CNV was observed in 24 of 19,584 control individuals; parental DNA was not available for analysis.
(D) Data for maternally inherited 840 kbp duplication of CNTN6 in a 16p11.2 deletion female (14755.x17) with FSIQ = 75, SRS = 90, intellectual disability, and enuresis. The CNV was observed in only 1 of 4,092 control individuals. Stars on chromosome ideograms designate the presence and approximate position of the deletion (red) or duplication (blue).
In a high-functioning female proband with autism and a 16p11.2 deletion, an ∼250 kbp additional deletion of TOP3B (MIM: 603582) was validated (Figure 4C). TOP3B has been strongly implicated in neurodevelopmental disorders and is thought to be important in the co-recruitment of FMRP to mRNPs.35 Although this event is found in 24 of 19,584 control individuals (0.123%), this same deletion in the homozygous state was found to segregate with schizophrenia or intellectual disability in three Northern Finnish families.36 We discovered an 840 kbp duplication harboring the autism risk locus, contactin-6 (CNTN6 [MIM: 607220]), that was transmitted from a mother (Broader Autism Phenotype Questionnaire score = 124) to her daughter (Figure 4D). In this particular case, the autistic daughter inherited the 16p11.2 deletion from her father. Hence, this is a case in which a 16p11.2 deletion is transmitted from the father and a secondary event from the mother. We also observed a smaller ∼50 kbp de novo deletion disrupting BIRC6 (MIM: 605638) in this proband. BIRC6 inhibits apoptosis by facilitating the degradation of apoptotic proteins by ubiquitination,37 and previous studies have identified three de novo variants in this gene in individuals with an ASD diagnosis.38, 39 In this family, it is highly unlikely that the decrement in IQ can be solely attributed to the 16p11.2 deletion event given that the FSIQ of the father carrying the 16p11.2 deletion and the FSIQ of his proband daughter, who also carries the 16p11.2 deletion, differ by more than 28 points. Finally, we note that two 16p11.2 duplication carriers have rare independent deletions in CTNNA3 (MIM: 607667) (Figure S7)—a locus previously associated with autism40, 41 and for which rare deletions have been reported in individuals with ASD.42
Discussion
Our results show that most recurrent rearrangements between BP4 and BP5 in chromosome 16p11.2 originate maternally. Specifically, nearly 90% of de novo deletions and duplications arise on maternal haplotypes, and an approximately equal proportion of inter- and intrachromosomal rearrangements is consistent with unequal crossover events during meiosis I and II, respectively. This observation stands in stark contrast to the 75%–80% of de novo CNVs identified in other studies that originate paternally.43, 44 Excluding genomic disorders associated with imprinted loci, a maternal parent-of-origin bias has been reported for two genomic disorders to date: the NF1 region on 17q11.2 and the 22q11.2 microdeletion associated with velocardiofacial and DiGeorge syndromes.45, 46 Neither of these regions, however, demonstrates such a high level of maternal bias as that which we have observed for the 16p11.2 CNV. For 16p11.2, we observe no correlation with advanced maternal age (p = 0.43, Student’s t test) (Tables S4 and S5), and there is no compelling evidence of imprinted genes within the critical region.47, 48 Importantly, no bias is observed in maternal or paternal transmission for inherited events, arguing against selection at the level of the germline or early embryogenesis.
The most likely explanation for this maternal bias is different male and female recombination rates at 16p11.2. Examination of data from published recombination maps33, 49 reveals a clear hotspot of female recombination within the critical region (Figure S8). Females have a significantly higher mean recombination rate within this region than males do (0.82 versus 0.083, p = 0.01, Student’s t test), and this particular region ranks in the 87th percentile for mean difference between male and female recombination genome wide (Figure S2). The maximum recombination rate for females for the 16p11.2 critical region is 13.24, whereas for males it is 1.27, a more than 10-fold difference. A much milder excess of female recombination is also noted for the 22q11.2 microdeletion (1.2- to 2.8-fold), commensurate with a more subtle maternal bias for this genomic disorder (56% maternal).45 The 16p11.2, 22q11.2, and 17q11.2 CNVs all lie close to the centromere of their respective chromosomes, consistent with higher female recombination rates in pericentromeric regions.33 Thus, it is likely that gender-specific recombination hotspots are a much more general predictor of female and male biases for NAHR.
We observe not only a maternal parent-of-origin bias for de novo 16p11.2 deletions, but also that mothers transmit a significantly greater number of secondary deletions to probands than do fathers. Such a transmission disequilibrium has been observed for small CNVs and single-nucleotide variants (SNVs) in individuals with ASD,50, 51 and this effect might result from a higher female tolerance toward additional mutations. We extend this putative female protective effect to secondary CNVs in 16p11.2 families. It is striking that of the nine probands with a secondary CNV disrupting a gene from a curated list associated with autism risk (see Web Resources), six are female, including two with multiple events, suggesting that females might be more tolerant of severe mutations. We do not observe this bias for secondary duplications, most likely because duplications are generally less deleterious than deletions.
Our results suggest that genetic background plays a role in the observed phenotypic heterogeneity and that dosage imbalances at other loci contribute, especially in the case of 16p11.2 duplication carriers. It is interesting that, compared to that of their parents who also carry the 16p11.2 CNV, the FSIQ decrement for probands with an inherited 16p11.2 duplication is greater than the difference observed for transmission of the deletion (Figure 3). Such a difference, along with the statistically significant differences between the mean FSIQ of parents carrying the 16p11.2 CNV and the mean FSIQ of probands carrying the same CNV, suggests that additional factors are contributing to the severity of the phenotype. Our finding of a modest negative correlation between FSIQ and secondary CNVs, as well as the increased phenotypic severity of such individuals, argues in favor of additional rare gene disruptive mutations. These findings are consistent with studies focused on different genomic disorders that have shown that individuals with more than one large CNV tend to have a lower IQ than individuals with only a single CNV.19 Similarly, a recent study of an Estonian population cohort reported that a greater proportion of individuals carrying large CNVs (>250 kbp) failed to graduate high school than did individuals without such events. When CNVs exceeded 1 Mbp in size, there was a significant risk for intellectual disability.52
There are some clear limitations of this study. The number of complete families with a de novo mutation and parental phenotypic information is insufficient, especially for duplications. Investigation of a larger sample of 16p11.2 CNVs in conjunction with more detailed phenotypic data is necessary in order to confirm the observed trends. The Simons VIP is not a population cohort, but rather was clinically ascertained and subject to inclusion and exclusion criteria. Importantly, the Simons VIP was screened for large, most likely pathogenic CNVs, thus depleting the number of individuals with large secondary CNVs. A population-based cohort of sufficient size would prove most valuable if large-scale genetic screening were followed by detailed phenotypic assessment of individuals with particular genotypes.53 Because we focused on CNVs (typically >50 kbp), we did not assess other potentially deleterious mutations (e.g., SNVs or small CNVs).
The importance of secondary mutational hits at other loci affecting phenotype severity has been established in several disorders, and a model has been developed to explain the phenotype variability associated with pathogenic CNVs.19, 54, 55 Importantly, 11 of the secondary CNVs have already been implicated as risk factors for autism and developmental delay (e.g., 240 kbp deletion of the TOP3B locus on chromosome 22q11.22).36 Our results extend observations of secondary mutational hits to the 16p11.2 CNV and suggest that full-genome sequencing of individuals carrying the 16p11.2 CNV will ultimately be required to more precisely predict the severity of disease within the context of families. This is an important consideration because once the 16p11.2 CNV is discovered, such individuals are routinely excluded from further exome and genome sequencing analyses.17, 56, 57 The presence of additional risk factors discovered by either sequencing or diagnostic microarray will be important for projecting the disease trajectory and the diverse outcomes associated with this pathogenic CNV.
Conflicts of Interest
E.E.E. is on the scientific advisory board of DNAnexus and is a consultant for the Kunming University of Science and Technology as part of the 1000 China Talent Program.
Acknowledgments
We thank F. Hormozdiari, K. Steinman, and T. Brown for useful discussion and edits to the manuscript. We thank all of the families at the participating Simons VIP and SSC sites, as well as the Simons VIP and SSC consortia. We appreciate access to phenotypic data on SFARI Base. Approved researchers can obtain the Simons VIP population dataset described in this study by contacting the Simons Foundation Autism Research Initiative. A full list of the investigators who contributed to the generation of the WTCCC data is available from http://www.wtccc.org.uk/. M.H.D. was supported by US National Institute of Mental Health grant no. 1F30MH105055-01 and by the Simons Foundation, and X.N. was supported by a US National Science Foundation Graduate Research Fellowship (grant no. DGE-1256082). This work was supported by the Simons Foundation Autism Research Initiative grant no. 294112 (E.E.E.) and NIH grant no. R01MH101221 (E.E.E.). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Published: December 31, 2015
Footnotes
Supplemental Data include a Supplemental Appendix, eight figures, and fifteen tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.017.
Accession Numbers
Underlying SNP microarray data are available from the National Database for Autism Research under http://dx.doi.org/10.15154/1226522, as well as through SFARI Base to approved researchers. Underlying calls for the recalled WTCCC set (set II) are available in dbVar under accession number nstd122. CNV calls for the 19,584 control individuals (set I) are available in dbVar under accession number nstd100.
Web Resources
The URLs for data presented herein are as follows:
cnvPartition Algorithm, http://www.illumina.com/content/dam/illumina-marketing/documents/products/technotes/technote_cnv_plug_ins.pdf
Illumina Genome Studio Software, http://www.illumina.com/techniques/microarrays/array-data-analysis-experimental-design/genomestudio.html
National Database for Autism Research, https://ndar.nih.gov/
OMIM, http://www.omim.org/
SFARI Base, https://sfari.org/resources/sfari-base
Simons VIP, https://simonsvipconnect.org/
UCSC Genome Browser, http://genome.ucsc.edu
WTCCC2, http://www.wtccc.org.uk/ccc2/
Supplemental Data
References
- 1.Weiss L.A., Shen Y., Korn J.M., Arking D.E., Miller D.T., Fossdal R., Saemundsen E., Stefansson H., Ferreira M.A.R., Green T., Autism Consortium Association between microdeletion and microduplication at 16p11.2 and autism. N. Engl. J. Med. 2008;358:667–675. doi: 10.1056/NEJMoa075974. [DOI] [PubMed] [Google Scholar]
- 2.Kumar R.A., KaraMohamed S., Sudi J., Conrad D.F., Brune C., Badner J.A., Gilliam T.C., Nowak N.J., Cook E.H., Jr., Dobyns W.B., Christian S.L. Recurrent 16p11.2 microdeletions in autism. Hum. Mol. Genet. 2008;17:628–638. doi: 10.1093/hmg/ddm376. [DOI] [PubMed] [Google Scholar]
- 3.Shinawi M., Liu P., Kang S.-H.L., Shen J., Belmont J.W., Scott D.A., Probst F.J., Craigen W.J., Graham B.H., Pursley A. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J. Med. Genet. 2010;47:332–341. doi: 10.1136/jmg.2009.073015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenfeld J.A., Coppinger J., Bejjani B.A., Girirajan S., Eichler E.E., Shaffer L.G., Ballif B.C. Speech delays and behavioral problems are the predominant features in individuals with developmental delays and 16p11.2 microdeletions and microduplications. J. Neurodev. Disord. 2010;2:26–38. doi: 10.1007/s11689-009-9037-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moreno-De-Luca A., Evans D.W., Boomer K.B., Hanson E., Bernier R., Goin-Kochel R.P., Myers S.M., Challman T.D., Moreno-De-Luca D., Slane M.M. The role of parental cognitive, behavioral, and motor profiles in clinical variability in individuals with chromosome 16p11.2 deletions. JAMA Psychiatry. 2015;72:119–126. doi: 10.1001/jamapsychiatry.2014.2147. [DOI] [PubMed] [Google Scholar]
- 6.Zufferey F., Sherr E.H., Beckmann N.D., Hanson E., Maillard A.M., Hippolyte L., Macé A., Ferrari C., Kutalik Z., Andrieux J., Simons VIP Consortium. 16p11.2 European Consortium A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J. Med. Genet. 2012;49:660–668. doi: 10.1136/jmedgenet-2012-101203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maillard A.M., Ruef A., Pizzagalli F., Migliavacca E., Hippolyte L., Adaszewski S., Dukart J., Ferrari C., Conus P., Männik K., 16p11.2 European Consortium The 16p11.2 locus modulates brain structures common to autism, schizophrenia and obesity. Mol. Psychiatry. 2015;20:140–147. doi: 10.1038/mp.2014.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson E., Bernier R., Porche K., Jackson F.I., Goin-Kochel R.P., Snyder L.G., Snow A.V., Wallace A.S., Campe K.L., Zhang Y., Simons Variation in Individuals Project Consortium The cognitive and behavioral phenotype of the 16p11.2 deletion in a clinically ascertained population. Biol. Psychiatry. 2015;77:785–793. doi: 10.1016/j.biopsych.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qureshi A.Y., Mueller S., Snyder A.Z., Mukherjee P., Berman J.I., Roberts T.P.L., Nagarajan S.S., Spiro J.E., Chung W.K., Sherr E.H., Buckner R.L., Simons VIP Consortium Opposing brain differences in 16p11.2 deletion and duplication carriers. J. Neurosci. 2014;34:11199–11211. doi: 10.1523/JNEUROSCI.1366-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steinberg S., de Jong S., Mattheisen M., Costas J., Demontis D., Jamain S., Pietiläinen O.P.H., Lin K., Papiol S., Huttenlocher J., GROUP. Wellcome Trust Case Control Consortium 2 Common variant at 16p11.2 conferring risk of psychosis. Mol. Psychiatry. 2014;19:108–114. doi: 10.1038/mp.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reinthaler E.M., Lal D., Lebon S., Hildebrand M.S., Dahl H.-H.M., Regan B.M., Feucht M., Steinböck H., Neophytou B., Ronen G.M., 16p11.2 European Consortium. EPICURE Consortium. EuroEPINOMICS Consortium 16p11.2 600 kb Duplications confer risk for typical and atypical Rolandic epilepsy. Hum. Mol. Genet. 2014;23:6069–6080. doi: 10.1093/hmg/ddu306. [DOI] [PubMed] [Google Scholar]
- 12.Walters R.G., Jacquemont S., Valsesia A., de Smith A.J., Martinet D., Andersson J., Falchi M., Chen F., Andrieux J., Lobbens S. A new highly penetrant form of obesity due to deletions on chromosome 16p11.2. Nature. 2010;463:671–675. doi: 10.1038/nature08727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McCarthy S.E., Makarov V., Kirov G., Addington A.M., McClellan J., Yoon S., Perkins D.O., Dickel D.E., Kusenda M., Krastoshevsky O., Wellcome Trust Case Control Consortium Microduplications of 16p11.2 are associated with schizophrenia. Nat. Genet. 2009;41:1223–1227. doi: 10.1038/ng.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacquemont S., Reymond A., Zufferey F., Harewood L., Walters R.G., Kutalik Z., Martinet D., Shen Y., Valsesia A., Beckmann N.D. Mirror extreme BMI phenotypes associated with gene dosage at the chromosome 16p11.2 locus. Nature. 2011;478:97–102. doi: 10.1038/nature10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cooper G.M., Coe B.P., Girirajan S., Rosenfeld J.A., Vu T.H., Baker C., Williams C., Stalker H., Hamid R., Hannig V. A copy number variation morbidity map of developmental delay. Nat. Genet. 2011;43:838–846. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaminsky E.B., Kaul V., Paschall J., Church D.M., Bunke B., Kunig D., Moreno-De-Luca D., Moreno-De-Luca A., Mulle J.G., Warren S.T. An evidence-based approach to establish the functional and clinical significance of copy number variants in intellectual and developmental disabilities. Genet. Med. 2011;13:777–784. doi: 10.1097/GIM.0b013e31822c79f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanders S.J., Ercan-Sencicek A.G., Hus V., Luo R., Murtha M.T., Moreno-De-Luca D., Chu S.H., Moreau M.P., Gupta A.R., Thomson S.A. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70:863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coe B.P., Witherspoon K., Rosenfeld J.A., van Bon B.W.M., Vulto-van Silfhout A.T., Bosco P., Friend K.L., Baker C., Buono S., Vissers L.E.L.M. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat. Genet. 2014;46:1063–1071. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Girirajan S., Rosenfeld J.A., Coe B.P., Parikh S., Friedman N., Goldstein A., Filipink R.A., McConnell J.S., Angle B., Meschino W.S. Phenotypic heterogeneity of genomic disorders and rare copy-number variants. N. Engl. J. Med. 2012;367:1321–1331. doi: 10.1056/NEJMoa1200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu N., Ming X., Xiao J., Wu Z., Chen X., Shinawi M., Shen Y., Yu G., Liu J., Xie H. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N. Engl. J. Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simons Vip Consortium Simons Variation in Individuals Project (Simons VIP): a genetics-first approach to studying autism spectrum and related neurodevelopmental disorders. Neuron. 2012;73:1063–1067. doi: 10.1016/j.neuron.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 22.American Psychiatric Association. American Psychiatric Association . American Psychiatric Association; Washington, DC: 2000. Diagnostic and Statistical Manual of Mental Disorders: DSM-IV-TR. [Google Scholar]
- 23.Mullen E. Pearson Assessments; Circle Pines, MN: 1995. Mullen Scales of Early Learning, AGS Edition. [Google Scholar]
- 24.Eliot C. Second Edition. Harcourt Assessment; San Antonio, TX: 2007. Differential Abilities Scale. [Google Scholar]
- 25.Wechsler D. The Psychological Coorporation; San Antonio, TX: 1999. Wechsler Abbreviated Scale of Intelligence. [Google Scholar]
- 26.Sudmant P.H., Mallick S., Nelson B.J., Hormozdiari F., Krumm N., Huddleston J., Coe B.P., Baker C., Nordenfelt S., Bamshad M. Global diversity, population stratification, and selection of human copy-number variation. Science. 2015;349:aab3761. doi: 10.1126/science.aab3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Manichaikul A., Mychaleckyj J.C., Rich S.S., Daly K., Sale M., Chen W.-M. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girirajan S., Brkanac Z., Coe B.P., Baker C., Vives L., Vu T.H., Shafer N., Bernier R., Ferrero G.B., Silengo M. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 2011;7:e1002334. doi: 10.1371/journal.pgen.1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brys G., Hubert M., Struyf A. A robust measure of skewness. J. Comput. Graph. Stat. 2004;13:996–1017. [Google Scholar]
- 30.Sanders S.J., He X., Willsey A.J., Ercan-Sencicek A.G., Samocha K.E., Cicek A.E., Murtha M.T., Bal V.H., Bishop S.L., Dong S., Autism Sequencing Consortium Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Browning S.R., Browning B.L. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am. J. Hum. Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong A., Thorleifsson G., Gudbjartsson D.F., Masson G., Sigurdsson A., Jonasdottir A., Walters G.B., Jonasdottir A., Gylfason A., Kristinsson K.T. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 2010;467:1099–1103. doi: 10.1038/nature09525. [DOI] [PubMed] [Google Scholar]
- 34.Wang T., Liu Y., Xu X.-H., Deng C.-Y., Wu K.-Y., Zhu J., Fu X.-Q., He M., Luo Z.-G. Lgl1 activation of rab10 promotes axonal membrane trafficking underlying neuronal polarization. Dev. Cell. 2011;21:431–444. doi: 10.1016/j.devcel.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Xu D., Shen W., Guo R., Xue Y., Peng W., Sima J., Yang J., Sharov A., Srikantan S., Yang J. Top3β is an RNA topoisomerase that works with fragile X syndrome protein to promote synapse formation. Nat. Neurosci. 2013;16:1238–1247. doi: 10.1038/nn.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoll G., Pietiläinen O.P.H., Linder B., Suvisaari J., Brosi C., Hennah W., Leppä V., Torniainen M., Ripatti S., Ala-Mello S. Deletion of TOP3β, a component of FMRP-containing mRNPs, contributes to neurodevelopmental disorders. Nat. Neurosci. 2013;16:1228–1237. doi: 10.1038/nn.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hao Y., Sekine K., Kawabata A., Nakamura H., Ishioka T., Ohata H., Katayama R., Hashimoto C., Zhang X., Noda T. Apollon ubiquitinates SMAC and caspase-9, and has an essential cytoprotection function. Nat. Cell Biol. 2004;6:849–860. doi: 10.1038/ncb1159. [DOI] [PubMed] [Google Scholar]
- 38.Iossifov I., O’Roak B.J., Sanders S.J., Ronemus M., Krumm N., Levy D., Stessman H.A., Witherspoon K.T., Vives L., Patterson K.E. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Rubeis S., He X., Goldberg A.P., Poultney C.S., Samocha K., Cicek A.E., Kou Y., Liu L., Fromer M., Walker S., DDD Study. Homozygosity Mapping Collaborative for Autism. UK10K Consortium Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang K., Zhang H., Ma D., Bucan M., Glessner J.T., Abrahams B.S., Salyakina D., Imielinski M., Bradfield J.P., Sleiman P.M.A. Common genetic variants on 5p14.1 associate with autism spectrum disorders. Nature. 2009;459:528–533. doi: 10.1038/nature07999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyashita A., Arai H., Asada T., Imagawa M., Matsubara E., Shoji M., Higuchi S., Urakami K., Kakita A., Takahashi H., Japanese Genetic Study Consortium for Alzeheimer’s Disease Genetic association of CTNNA3 with late-onset Alzheimer’s disease in females. Hum. Mol. Genet. 2007;16:2854–2869. doi: 10.1093/hmg/ddm244. [DOI] [PubMed] [Google Scholar]
- 42.O’Roak B.J., Vives L., Girirajan S., Karakoc E., Krumm N., Coe B.P., Levy R., Ko A., Lee C., Smith J.D. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hehir-Kwa J.Y., Rodríguez-Santiago B., Vissers L.E., de Leeuw N., Pfundt R., Buitelaar J.K., Pérez-Jurado L.A., Veltman J.A. De novo copy number variants associated with intellectual disability have a paternal origin and age bias. J. Med. Genet. 2011;48:776–778. doi: 10.1136/jmedgenet-2011-100147. [DOI] [PubMed] [Google Scholar]
- 44.Kong A., Steinthorsdottir V., Masson G., Thorleifsson G., Sulem P., Besenbacher S., Jonasdottir A., Sigurdsson A., Kristinsson K.T., Jonasdottir A., DIAGRAM Consortium Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Delio M., Guo T., McDonald-McGinn D.M., Zackai E., Herman S., Kaminetzky M., Higgins A.M., Coleman K., Chow C., Jalbrzikowski M. Enhanced maternal origin of the 22q11.2 deletion in velocardiofacial and DiGeorge syndromes. Am. J. Hum. Genet. 2013;92:439–447. doi: 10.1016/j.ajhg.2013.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.López-Correa C., Dorschner M., Brems H., Lázaro C., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L. Recombination hotspot in NF1 microdeletion patients. Hum. Mol. Genet. 2001;10:1387–1392. doi: 10.1093/hmg/10.13.1387. [DOI] [PubMed] [Google Scholar]
- 47.Baran Y., Subramaniam M., Biton A., Tukiainen T., Tsang E.K., Rivas M.A., Pirinen M., Gutierrez-Arcelus M., Smith K.S., Kukurba K.R., GTEx Consortium The landscape of genomic imprinting across diverse adult human tissues. Genome Res. 2015;25:927–936. doi: 10.1101/gr.192278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luedi P.P., Dietrich F.S., Weidman J.R., Bosko J.M., Jirtle R.L., Hartemink A.J. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17:1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J., Fan H.C., Behr B., Quake S.R. Genome-wide single-cell analysis of recombination activity and de novo mutation rates in human sperm. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krumm N., Turner T.N., Baker C., Vives L., Mohajeri K., Witherspoon K., Raja A., Coe B.P., Stessman H.A., He Z.-X. Excess of rare, inherited truncating mutations in autism. Nat. Genet. 2015;47:582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jacquemont S., Coe B.P., Hersch M., Duyzend M.H., Krumm N., Bergmann S., Beckmann J.S., Rosenfeld J.A., Eichler E.E. A higher mutational burden in females supports a “female protective model” in neurodevelopmental disorders. Am. J. Hum. Genet. 2014;94:415–425. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Männik K., Mägi R., Macé A., Cole B., Guyatt A.L., Shihab H.A., Maillard A.M., Alavere H., Kolk A., Reigo A. Copy number variations and cognitive phenotypes in unselected populations. JAMA. 2015;313:2044–2054. doi: 10.1001/jama.2015.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stessman H.A., Bernier R., Eichler E.E. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156:872–877. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Girirajan S., Eichler E.E. Phenotypic variability and genetic susceptibility to genomic disorders. Hum. Mol. Genet. 2010;19(R2):R176–R187. doi: 10.1093/hmg/ddq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Girirajan S., Rosenfeld J.A., Cooper G.M., Antonacci F., Siswara P., Itsara A., Vives L., Walsh T., McCarthy S.E., Baker C. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat. Genet. 2010;42:203–209. doi: 10.1038/ng.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Iossifov I., Ronemus M., Levy D., Wang Z., Hakker I., Rosenbaum J., Yamrom B., Lee Y.-H., Narzisi G., Leotta A. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O’Roak B.J., Vives L., Fu W., Egertson J.D., Stanaway I.B., Phelps I.G., Carvill G., Kumar A., Lee C., Ankenman K. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.