Abstract
The use of massively parallel sequencing of maternal cfDNA for non-invasive prenatal testing (NIPT) of aneuploidy is widely available. Recently, the scope of testing has increased to include selected subchromosomal abnormalities, but the number of samples reported has been small. We developed a calling pipeline based on a segmentation algorithm for the detection of these rearrangements in maternal plasma. The same read depth used in our standard pipeline for aneuploidy NIPT detected 15/18 (83%) samples with pathogenic rearrangements > 6 Mb but only 2/10 samples with rearrangements < 6 Mb, unless they were maternally inherited. There were two false-positive calls in 534 samples with no known subchromosomal abnormalities (specificity 99.6%). Using higher read depths, we detected 29/31 fetal subchromosomal abnormalities, including the three samples with maternally inherited microduplications. We conclude that test sensitivity is a function of the fetal fraction, read depth, and size of the fetal CNV and that at least one of the two false negatives is due to a low fetal fraction. The lack of an independent method for determining fetal fraction, especially for female fetuses, leads to uncertainty in test sensitivity, which currently has implications for this technique’s future as a clinical diagnostic test. Furthermore, to be effective, NIPT must be able to detect chromosomal rearrangements across the whole genome for a very low false-positive rate. Because standard NIPT can only detect the majority of larger (>6 Mb) chromosomal rearrangements and requires knowledge of fetal fraction, we consider that it is not yet ready for routine clinical implementation.
Introduction
Unbalanced chromosomal rearrangements, including those in microdeletion and microduplication syndromes, are associated with a range of adverse phenotypes and are individually rare. Although the overall incidence is unknown, it is thought that the combined incidence might approach that of Down syndrome (trisomy 21 [MIM 190685]).1, 2 The majority of cases occur randomly, but some, for example, those in DiGeorge syndrome (22q11.2 deletion [MIM: 188400]), Cri du chat syndrome (5p deletion [MIM: 123450]), and Charcot-Marie-Tooth type 1A disease (17p11.2 duplication [MIM: 118220]), are recurrent. Unlike Down syndrome, these other rearrangement disorders do not have a universal prenatal screening program, although they might be found more commonly in fetuses with an increased nuchal translucency or other fetal abnormalities.1, 3, 4 Currently, prenatal diagnosis of such rearrangements requires an invasive procedure, such as chorionic villus sampling or amniocentesis followed by karyotyping or microarray analysis.
Since 2011, the use of massively parallel sequencing (MPS) of cell-free DNA (cfDNA) in maternal plasma for non-invasive prenatal testing (NIPT) of fetal aneuploidies has become available in more than 60 countries.5 Most national and international organizations now recognize NIPT as a highly sensitive screening test that can reduce the need for invasive testing when it is used in high-risk pregnancies.6, 7, 8 Over the same time period, there has been a move to replace traditional karyotyping following invasive testing with microarray analysis9, which increases detection of pathogenic chromosomal rearrangements to include microdeletion and microduplication syndromes.1, 2, 4 There are concerns that widespread implementation of NIPT stands to decrease the detection of these other pathogenic rearrangements.10 However, in principle, sequencing of cfDNA can also be used for detecting other unbalanced chromosomal rearrangements prenatally, and a number of proof-of-concept studies using a variety of sequencing depths and bioinformatics approaches have detected a range of fetal subchromosomal abnormalities in maternal plasma.11, 12, 13, 14, 15 Indeed, several commercial providers have expanded their NIPT platform to include a panel of syndromes characterized by recurrent microdeletions and microduplications.
The statistical power of the methods published to date is a function of the read depth and the size of the fetal copy-number variants (CNVs). Using one billion reads, Srinivasan et al. detected fetal CNVs as small as 300 kb,14 whereas Chen et al. claimed that their pipeline can detect all fetal CNVs bigger than 10 Mb with just two to eight million reads.13 Until now, NIPT of subchromosomal abnormalities has been reported only in a small number of cases of affected pregnancies, although a larger series using spiked samples has been reported.16 The lack of data makes it difficult to accurately determine the test sensitivity and specificity and, more importantly, the positive and negative predictive values, which are crucial if this is to be implemented in clinical practice.
Algorithms for detecting subchromosomal abnormalities can be categorized into two main groups: the targeted approach, which looks for abnormalities in known locations,15, 16 and the whole-genome approach, which can be applied in situations where the location and size of the fetal CNVs are not known13, 14 and where read counts higher or lower than those of the reference set can indicate the presence of CNVs. Here, we present results for a series of maternal plasma samples from pregnancies with known subchromosomal abnormalities occurring across the genome and explore the potential for routine implementation.
Material and Methods
Maternal blood samples were collected from women undergoing invasive procedures for clinical indications in 40 maternity clinics around the UK as part of the RAPID (Rapid Accurate Prenatal Non-invasive Diagnosis) project. The study was approved by the local research ethics committee (01/0095). Prior to invasive testing, women were asked to consent to the withdrawal of 20–30 ml of maternal blood to be placed in EDTA or in cell-stabilizing blood-collection tubes when there was an anticipated transit delay of more than 24 hr. Samples were centrifuged (3,000 rpm [1,000 × g] for 15 min followed by 14,000 rpm for 10 min) for separating the plasma, which was then stored in 2 ml aliquots at −80°C. For the purpose of this study, 4 ml plasma was defrosted and cfDNA was extracted with the QIAsymphony DSP Virus/Pathogen Midi Kit and a custom protocol on the QIAsymphony (Qiagen) and then sequenced with the Illumina HiSeq 2500 system with single-end 50 bp reads. After sequencing, reads were aligned to the human reference genome (UCSC Genome Browser hg19) with Bowtie (see Web Resources). Only reads that uniquely mapped with no mismatches were retained.
In this study, 31 test samples with known unbalanced chromosomal rearrangements were selected from the RAPID project sample set. The clinical outcomes were determined by conventional karyotyping, fluorescence in situ hybridization (FISH), microarray, or molecular techniques such as multiplex ligation-dependent probe amplification (MLPA) for clinical indications. The samples in our study had fetal CNVs ranging in size from less than 3 Mb to 42 Mb on a number of different chromosomes. There were also three cases of unbalanced translocations. Table 1 details the test samples, including the method used for determining the karyotype and the gestational age at the time of sample collection. In addition to sequencing the test samples, we also sequenced 534 known euploid samples (hereafter referred to as the reference set) according to the same parameters. For these 534 samples, invasive testing had been performed for clinical indications, and results were available after karyotyping (506 samples) or rapid aneuploidy screening via FISH or quantitative fluorescence PCR (QF-PCR) (28 samples).
Table 1.
Tested Samples Ordered by CNV Size, Sequencing Detection, and Fetal-Fraction Estimation
| Sample No. | Outcome | CNV Size (Mb)a | How Outcome Was Determined | Gestation (Weeks + Days) | CNV Position (kb) |
Sequencing Detection |
Implied Fetal Fraction |
||
|---|---|---|---|---|---|---|---|---|---|
| 12-Plex | Deeper | From CNV | From Chr Y | ||||||
| 1470 | 47,XY,+idic(9)(pter→q21.1::q21.1→pter) | ∼42 | karyotype | 12 + 2 | dup9: 1–72,100 | yes | not done | 7.5% (15.1%)b | 9.7% |
| 8048 | 47,XX,+del(9)(q11) | ∼42 | karyotype | 27 + 1 | dup9: 1-71,200 | yes | not done | 31.8% | – |
| 7851 | 47,XY,+der(22)t(9;22)(p13;q11.2)mat | chr9: ∼42 | karyotype | 11 + 6 | dup9: 1–72,600 | yes | not done | 7.2% | 6.8% |
| chr22: ∼3 | dup22: 17,300–21,000 | no | 8.2% | ||||||
| 5216 | 46,XX,del(13)(q22) | ∼40 | karyotype | 11 + 5 | del13: 83,600–115,000 | no | yes | 4.6% | – |
| 12317 | 46,XX,del(4)(p15.2) | ∼26 | karyotype | 24 + 2 | del4: 1–25,200 | yes | not done | 11.7% | – |
| 2092 | 46,XX,der(4)ins(14;4)(q13:q25q21.3)mat | chr4: ∼25 | karyotype | 11 + 5 | del4: 85,900–109,500 | yes | not done | 13.8% | – |
| 10853 | 46,XY,del(2)(p23p25.1) | ∼25 | karyotype | 24 + 1 | del2: 11,000–28,400 | yes | not done | 16.8% | 18% |
| 11660 | 46,XX,der(7)t(4;7)(p15.3;q34)pat | chr4: ∼17 | karyotype | 13 + 2 | dup4: 1–16,800 | yes | not done | 8.2% | – |
| chr7: ∼20 | chr7 not detected | no | |||||||
| R-01071 | 46,XY,del(4)(p15.32) | ∼18 | karyotype | 21 + 0 | del4: 1–15,600 | no | yes | 4.6% | 5.3% |
| 13122 | 46,XX,dup(21)(q21.2q22.2) | ∼16 | karyotype | 20 + 0 | dup21: 25,800–41,400 | yes | not done | 9.2% | – |
| 21 | 46,XX,del(5)(p15.1)dn | ∼15 | karyotype | 14 + 0 | del5: 1–15,500 | yes | not done | 19.2% | – |
| 12279 | 46,XY,del(18)(p11.1).arr 18p11.32p11.21(149,080–14,007,190)×1 | 13.8 | karyotype and microarray | 18 + 2 | del18: 1–18,700 | yes | not done | 7.8% | 7.7% |
| dup18: 18,700–78,000 | 9.0% | ||||||||
| R-01025 | 46,XX,del(9)(p23)dn.ish del(9)(34H2−,RP11-165F24−) | ∼13 | karyotype and FISH | 17 + 3 | del9: 1–16,800 | yes | not done | 11.9% | – |
| 12344 | arr 15q26.1q26.3(91,836,928–102,481,320)×1 | 11 | microarray | 28 + 4 | dup15: 34,400–90,100 | yes | not done | 14.8% | 15.8% |
| del15: 90,100–102,300 | |||||||||
| 1144 | 46,XX,der(18)t(12:18)(p13.1;q23)pat | chr12: ∼12 | karyotype | 17 + 1 | dup12: 1–9,400 | yes | not done | 8.8% | – |
| chr18: ∼5 | del18: 72,300–78,000 | 8.0% | |||||||
| 10855 | 46,XX,del(1)(q41q42) | ∼12 | karyotype | 13 + 2 | not detected | no | no | – | – |
| R-00940 | 46,XX,der(7)(pter→q36.1::?q36.1→q36.1::p21→pter)dn.ish der(7)?dup(7)(q36.1→q36.1)(RP11-4340++)del(7)(q36.3→qter)(RP11-6903−,c3K23−)dup(7)(p21→pter)(c109A6+) | ∼10 | karyotype and FISH | 12 + 5 | dup7: 1–16,500 | yes | not done | 7.7% | – |
| del7: 152,700–159,100 | 11.5% | ||||||||
| 9639 | 46,XY,del(6)(p25) | ∼7 | karyotype | 21 + 2 | del6: 1–7,400 | yes | not done | 15.8% | 20% |
| R-01716 | 46,XX,add(6)(p25).ish del(6)(p25.3p25.3)(CTB-62I11−,RP11-118B18−).arr[hg18] 6p25.3p25.1(77,025–5,820,602)×1 | 5.7 | karyotype, FISH, and microarray | 21 + 0 | del6: 1–6,400 | yes | not done | 17.3% | – |
| R-00875 | 46,XX,del(5)(p15.33)dn.ish del(5)(p15.33)(c84C11−,RP11-348B13−,RP11-129I19−,RP11-473F9+,RP11-259D10+) | ∼4.5 | karyotype and FISH | 17 + 3 | del5: 1–10,500 | yes | not done | 9.7% | – |
| 10256 | 46,XY,ish del(8)(q24.3q24.3)(RP11-65A5−).arr 8q24.3(140,186,782–144,969,635)×1 dn | 4.8 | karyotype and microarray | 13 + 2 | del8: 140,300–145,100 | no | yes | 9.8% | 9.2% |
| 11600 | 46,XX dup within 17p12 | ∼1.5 | karyotype and QF-PCR | 11 + 2 | dup17: 14,600–16,000 | no | yes | 10.2% | – |
| 8383 | 46,XY,dup(17)(p11.2p11.2).arr 17p11.2(16,637,872-20,294,010)×3 | 3.7 | karyotype and microarray | 22 + 0 | dup17: 16,900–20,800 | no | yes | 7.0% | 5.8% |
| 11590 | 46,XX dup within 17p12 inherited from the mother | ∼1.5 | karyotype and QF-PCR | 11 + 0 | dup17: 14,200–15,900 | yes | not done | – | – |
| 612 | 46,XX dup within 17p12 inherited from the mother | ∼1.5 | karyotype and QF-PCR | 12 + 6 | dup17: 14,200–15,900 | yes | not done | – | – |
| R-00983 | 46,XY.ish del(22)(q11.2q11.2)(RP11-1057H19−) | <3 | karyotype and FISH | 21 + 0 | not detected | no | no | – | 2.5% |
| R-01012 | 46,XX.ish del(22)(q11.2q11.2)(RP11-1057H19−) | <3 | karyotype and FISH | 20 + 4 | del22: 18,600–22,000 | no | yes | 8.2% | – |
| 12295 | 46,XX,del(22)(q11.2q11.2) | <3 | karyotype and BACs-on- Beads | 12 + 6 | del22: 19,100–23,300 | no | yes | 9.6% | – |
| 6876 | 46,XY.mlpa 22q11.2(P290)×3 inherited from the mother | <3 | karyotype and MLPA | 14 + 1 | dup22: 19,100–22,000 | yes | not done | – | 18% |
| 13067 | 46,XY,del(22)(q11.2q11.2) | <3 | karyotype | 28 + 1 | del22: 19,200–22,000 | no | yes | 23.8% | 23.5% |
| 2493 | 46,XY.ish del(22)(q11.2q11.2)(TUPLE1−)dn | <3 | karyotype and FISH | 22 + 2 | del22: 19,200–22,000 | no | yes | 13.4% | 17.3% |
Abbreviations are as follows: BAC, bacterial artificial chromosome; Chr, chromosome; CNV, copy-number variant; FISH, fluorescence in situ hybridization; MLPA, multiplex ligation-dependent probe amplification; and QF-PCR, quantitative fluorescence PCR.
The ∼ symbols mean that we can only provide an upper limit for the size estimate because of the limits of karyotyping.
This sample has an extra isodicentric chromosome 9; therefore, there are four total fetal copies of the region 9pter→9q21.1, and the fetal-fraction measurement is based on this. In parentheses is given the fetal-fraction measurement if only three copies of this region are present in the fetus.
The test samples were first sequenced at 12-plex (24 samples to a flow cell), which was the same read depth as that of our standard in-house pipeline for aneuploidy NIPT. For samples where no anomaly was detected, re-sequencing at a higher depth across multiple additional 1.5-plex runs (three samples per flow cell) was performed until the CNV was detected. The highest read depth was ∼120 million reads per sample.
Bias Correction
It is well known that the PCR process introduces a bias related to the GC content of the read count. To remove this bias, we used a two-step process, available in the RAPIDR software package.17 First, we binned read counts into 20 kb lots and removed bins with zero counts, unusually high counts, or counts in known CNV regions. After binning, we adjusted the counts in each bin by a factor of , where is the average read count in all bins, and is the average read count in each 0.5% of GC content.18
After this first correction step, we applied principal-component analysis (PCA) to remove some of the residual bias. PCA does not require prior knowledge of the cause of the bias; rather, it assumes that the systematic noise is from the largest components of variance in the principal components. Krumm et al. used a similar technique to find CNVs in whole-exome sequencing.19 To perform PCA, we normalized binned read counts across samples by computing the ratio of the binned count over the total read count for each sample. Given N samples and k bins, we constructed an N × k matrix X, in which each entry is the mean-centered binned-count ratio for bin k in sample N. PCA decomposes the covariance matrix , where W is a matrix of eigenvectors of . By using the matrix W determined from the reference set, we rotated the test-sample data, T, onto the same basis, T′ = TW, and reconstructed the test set by subtracting the top L components. For this analysis, we subtracted contributions from the first ten principal components.
Segmentation Algorithm
The goal of a segmentation algorithm is to segment an ordered data set, Y1:n = {Y1, Y2, … Yn}, with m − 1 change points, t1:m − 1 = {t1, … tm − 1}, such that the cost function, , is minimized. From a computational standpoint, the challenge is to effectively consider all change-point locations (for our problem, this translates to all possible locations for the CNV calls). We modeled our count data as a beta-binomial distribution and used the following cost function:
Here, pi is the proportion of counts in bin i (which we determine by taking the mean of the counts in bin i [Ri] and dividing it by the total counts for samples in the reference set), A is the ratio between the counts in the test sample and the counts in the reference set (A = sum(Yi:j)/sum(Ri:j)), is the dispersion parameter estimated for each sample with the binned counts in all chromosomes, F(A) in the cost function associated with the fetal fraction, and is another penalty term based on the Bayesian information criterion and is defined as , where k is the number of bins.
We based our search on the pruned exact linear time (PELT) algorithm,20 which is fast (close to linear time in the number of observations) and finds the near-optimal change points. The computational speed-up of PELT comes from using a dynamic programming approach and pruning the solution space in each step. We performed the analysis by using R software21 with the RAPIDR-Plus package.
Segment Filtering
For samples with more than one segment per chromosome, we used a likelihood-ratio test to test whether each segment was likely to be a real fetal CNV. We summed the read count across the bins that lay within the location of the potential CNV and used a beta-binomial distribution to model the counts. The beta-binomial distribution is also known as a dispersed binomial distribution and can be parameterized with p (from the binomial distribution) and (the dispersion parameter). For each potential CNV segment, we used the reference set to find p and and then calculated the log-likelihood-ratio test statistics by using
where Y is the sum of counts within the CNV segment for the test sample, and A is a scale parameter, which takes a value of greater than 1 for duplications and less than 1 for deletions. Unlike the simple Z score test used by NIPT for aneuploidy, the likelihood-ratio test allows us to have different read depths between the reference set and the test set.
To call fetal CNVs, we used the following stringent set of criteria:
-
1.
S > 30. Because −2log (likelihood ratio) statistics asymptotically follow a chi-square distribution, this implies a p value cutoff of 2 × 10−8.
-
2.
1.015 < A < 1.3 or 0.7 < A < 0.985. Values of A close to 0.5 or 1.5 indicate a maternal deletion or duplication, respectively. A can be used for estimating the fetal fraction by FF = abs(1 − A) × 2. If A is very close to 1, it implies a low fetal fraction. This filtering criterion implies a fetal-fraction cutoff of 3%.
-
3.
Segment length > 1 Mb.
Estimating the Fetal Fraction
For male fetuses, we used reads mapped to chromosome Y to estimate the fetal fraction (Equation 5 from Rava et al.22). For samples with CNV calls identified by our algorithm, we used the estimate of A, as described in the previous section, to give an alternate estimate of the fetal fraction (Table 1).
Power Calculation
The sensitivity of our method is a function of the read depth, the size of the CNV, and the fetal fraction. We estimated the sensitivity as a function of fetal fraction by using estimates of the dispersion parameter from fitting the counts to the beta-binomial distribution. Unlike previous simulation studies,13, 15 which assumed no technical bias and an ideal binomial distribution, our simulation is more realistic because in the beta-binomial distribution, the dispersion parameter accounts for some of the residual bias in the data.
Results
Sequencing at Four to Ten Million Reads
Sequencing at 12-plex yielded between four and ten million reads per sample, and at this read depth, chromosomal rearrangements were detected in 20 out of 31 samples (Table 1). In samples with CNVs larger than 6 Mb, at least one abnormality was detected in 15 out of 18 samples (sensitivity = 83% [95% confidence interval (CI) = 61%–94%]). For samples with CNVs smaller than 6 Mb, five were detected, and three of them had duplications for which the mother was a carrier. It is clear from the plots of the normalized count ratios (Figure S1) that the CNVs for these three samples are maternal, given that the normalized count ratios are close to 1.5; however, using our current bioinformatics pipeline and standard depth of sequencing, we could not determine whether the fetus also carried the CNV. Of the ten other samples with CNVs smaller than 6 Mb, two were correctly identified (sensitivity = 20% [95% CI = 6%–51%]). Table 1 shows a summary of the results, including the locations of the CNV calls, for all samples.
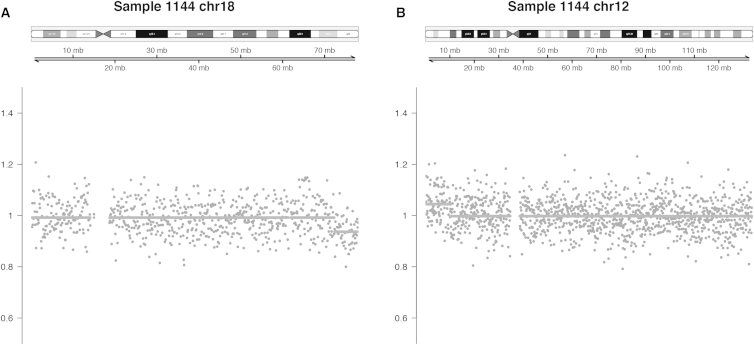
As an illustrative example, Figure 1 shows the normalized count ratios for sample 1144, which had an unbalanced translocation between chromosomes 12 and 18. Each point represents the read count in a 100 kb bin divided by the expected read count as determined from the reference set. The solid line is the output from the segmentation algorithm; the elevated segment in chromosome 12 represents a gain of the terminal short arm with a breakpoint at 12p13.1, and the depressed segment in chromosome 18 represents a loss of the terminal long arm with a breakpoint at 18q23. Plots of count ratios for the other samples can be found in the Supplemental Data.
Figure 1.
Plots of Count Ratios Illustrate a Microduplication and a Microdeletion in Sample 1144
Dots are the counts divided by the expected counts of the reference set in 100 kb bins; the solid line is the output from the segmentation algorithm.
(A) Microdeletion event in 18q23.
(B) Microduplication event in 12p13.1.
All segments called by our algorithm from the test samples were true positives, except for a discordant result for sample 12279. For this sample, karyotype and microarray results showed a deletion of the short arm of chromosome 18, which was also called by our pipeline, but our pipeline also found a duplication of the long arm of chromosome 18.
The analysis pipeline was also tested on a set of 534 samples that had no known chromosomal abnormalities and that had been sequenced with at least four million reads. After filtering, there were three false calls in two samples (specificity = 99.6% [95% CI = 98.6%–99.9%]). A false call can be located in any genomic position and is not confined to a specific region of interest. There were no false calls in off-target genomic locations in any of the samples with known microdeletions and/or microduplications.
Deeper Sequencing
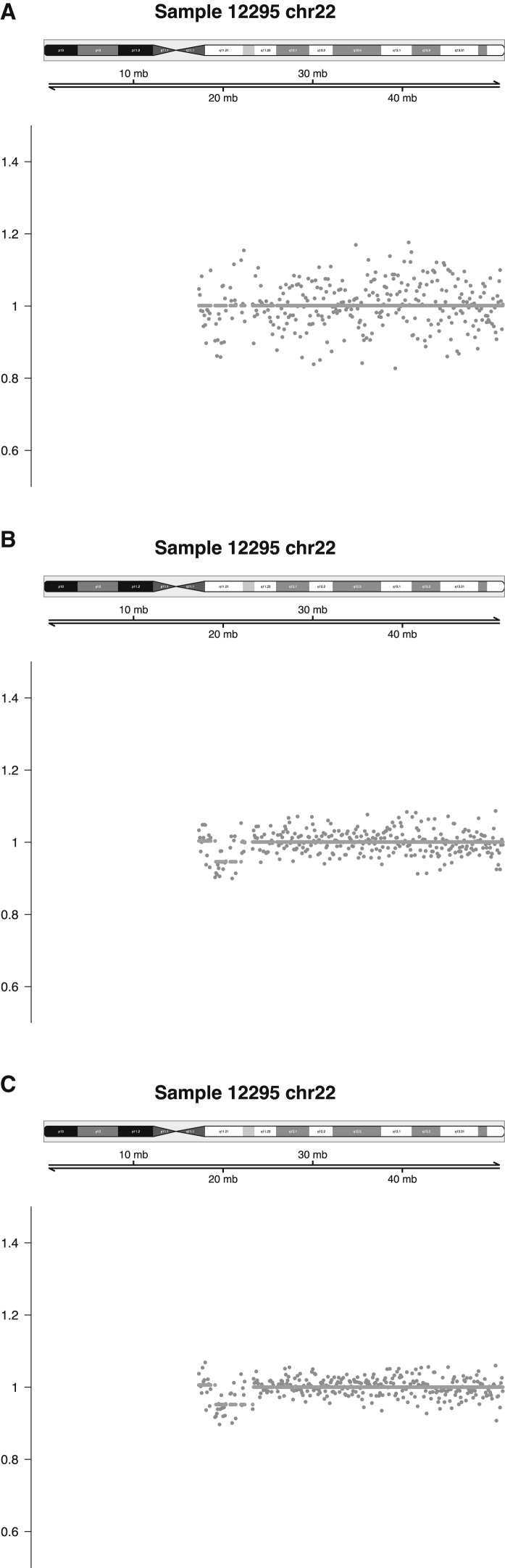
Deeper sequencing correctly identified the fetal CNV in 9 of 11 samples where the imbalance had not been detected by the standard shallow-sequencing pipeline. As expected, variance in the count ratios decreased as read depth increased, as demonstrated by one fetus with a 22q11.2 deletion, which was ultimately detected when the sample was sequenced to a depth of 32 million reads (Figure 2).
Figure 2.
Plots of Count Ratios for Sample 12295 Illustrate How Variance Decreases as Read Depth Increases
The three different read depths are 7 million (A), 32 million (B), and 71 million (C).
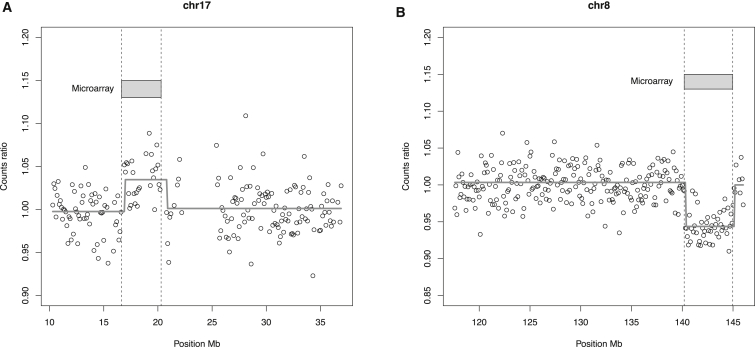
In addition to identifying the CNVs, the pipeline indicated locations that were highly accurate and matched well with positions given by microarray analysis (Table 1; Figure 3). For example, for sample 8383, our pipeline found a fetal CNV in chr17: 16,900,000–20,800,000, which matched the position obtained by genomic microarray analysis (chr17: 16,637,872–20,294,010) to within 500 kb. Similarly, for sample 10256, microarray analysis indicated a deletion in chr8: 140,186,782–144,969,635, and the location given by our pipeline was chr8: 140,300,000–145,100,000 (Table 1). Plots of the count ratios for samples 8383 and 10256 are shown in Figure 3.
Figure 3.
Comparison of the CNV Positions Derived from Microarray and Our Segmentation Algorithm
(A) Microduplication event of 3.7 Mb in 17p11.2.
(B) Microdeletion event of 4.8 Mb in 8q24.3.
CNVs in two samples were not detected by sequencing at a higher read depth. The first sample (R-00983) was from a male fetus with a 3 Mb deletion in 22q11.2. Using the reads mapped to chromosome Y, the pipeline estimated the fetal fraction to be 2.5% (Table 1). The second sample (10855) was from a female fetus with a deletion of around 12 Mb on chromosome 1, and estimation of the fetal fraction was not possible.
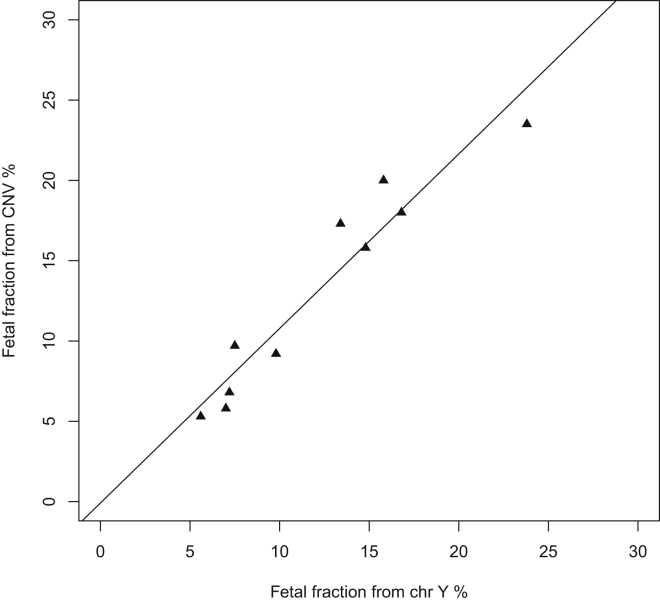
In male fetuses with positive CNV calls, the fetal-fraction estimates from CNV calls and from chromosome Y were highly correlated; linear regression yielded an r2 of 0.92 (Figure 4).
Figure 4.
The Fetal-Fraction Estimates from CNVs Called by Our Pipeline Are Closely Correlated with the Fetal-Fraction Estimates from Chromosome Y
Power Calculation
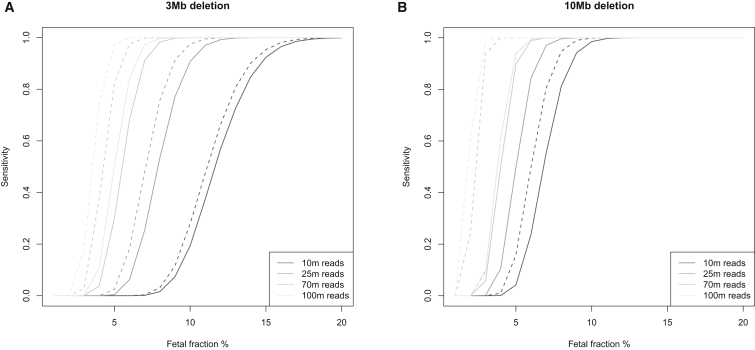
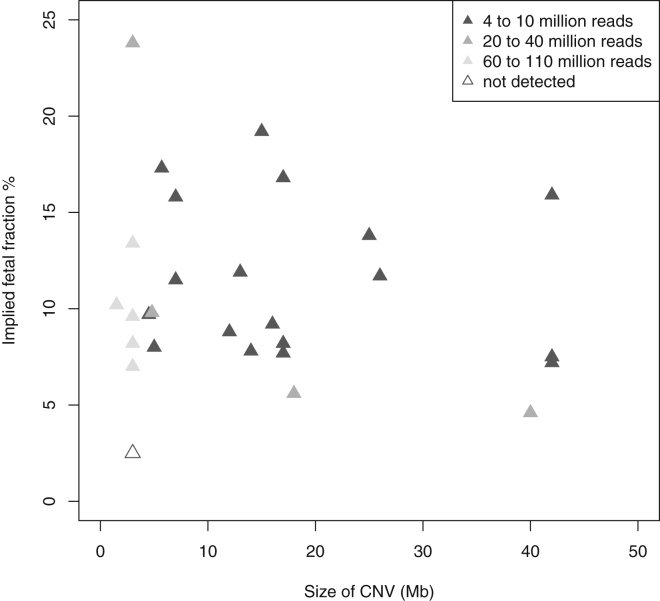
Results of a simulation study for the detection power for a 3 Mb CNV and a 10 Mb CNV at different fetal fractions and read depths are shown in Figure 5. Because the power estimation assumes a beta-binomial distribution for the counts and uses an estimate of the dispersion parameter that we derived from our data, our power estimation is specific to our laboratory, and others might find that they have different residual bias in the read count. After accounting for the residual bias, we found that the sensitivity of the test did not improve much with a higher read depth for a fetal fraction below 5%. On the other hand, CNVs as small as 1 Mb could be detected with 38 million reads if the fetal fraction was over 20%. Figure 6 illustrates empirically the impact of fetal fraction and CNV size in our test sensitivity by showing the number of reads required for detection in our dataset.
Figure 5.
Test Sensitivity for Deletions of Different Sizes
Deletions of 3 Mb (A) and 10 Mb (B). The dotted line assumes that the counts follow a binomial distribution, which is the theoretical optimal sensitivity that can be achieved with the read-counting method; the solid line assumes that the counts follow a beta-binomial distribution with = 10−8 for the 3 Mb deletion and = 3 × 10−8 for the 10 Mb deletion. is the over-dispersion parameter, and it captures the additional variance in the counts. The we used in the simulation was derived from our dataset.
Figure 6.
Number of Reads Required for Detecting Each CNV According to Size and Fetal Fraction
Fetal fraction were estimated from chromosome Y for male fetuses or from the CNV detected for female fetuses. Missed detection for female fetuses is not shown in this plot because we were unable to estimate the fetal fraction.
Discussion
In this study, we demonstrated that NIPT for aneuploidy can be used to detect the majority of pathogenic chromosomal rearrangements detectable by standard karyotyping. Applying our refined bioinformatics pipeline can achieve this without extra sequencing costs, given that we detected 15 out of 18 samples with fetal subchromosomal abnormalities larger than 6 Mb by using 12-plex sequencing (sensitivity = 83% [95% CI = 61–94%] and specificity = 99.6% [95% CI = 98.6%–99.9%]). Raising the read depth to 120 million reads increased the test sensitivity to 94% (95% CI = 74%–99%) for CNVs larger than 6 Mb and to 93% (95% CI = 77%–98%) for CNVs larger than 1.5 Mb. However, because our reference set was only sequenced at 12-plex, it is not possible to estimate the test specificity at the higher read depth. The cost of increasing the depth of sequencing would be high, and the false-positive rate is likely to increase. For fetuses with sonographic anomalies, an invasive test might still offer the most accurate and timely diagnosis because it is not possible to reliably exclude the possibility of a false-negative result even with deeper sequencing. One of the major perceived benefits of NIPT for aneuploidy is the significantly reduced requirement for invasive testing23 and the consequent improved safety, which women value most highly.24 Any change that increases the false-positive rate will negate to some degree this major benefit of NIPT screening. Given the rarity of these other rearrangements, the costs of introducing this testing (including economic costs, the potential loss of pregnancies resulting from increasing the invasive-testing rate, and the anxiety caused to parents) must be weighed against the possible benefits.
Because of the considerable uncertainty in the prevalence of clinically relevant microdeletion and microduplication syndromes, it is difficult to estimate the positive predictive value of our method. As a rough estimate, if we consider the incidence of all microdeletion and microduplication syndromes with CNVs larger than 6 Mb, excluding aneuploidies, to be 0.6% amongst fetuses with an indication,1 then our test, with a sensitivity of 83% and a specificity of 99.6%, would have a positive predictive value of 0.55. Expansion of NIPT to include detection of these other rearrangements stands to decrease the specificity and increase the invasive-testing rate. Our data, therefore, support the American Committee of Obstetricians and Gynecologists recommendation that routine cfDNA aneuploidy screening should not be expanded to include microdeletion syndromes.6
Our pipeline failed to detect three samples with CNVs > 6 Mb at 12-plex sequencing and, in one sample (12279), gave a true-positive call, as well as what initially appeared to be a false-positive call: a duplication of the whole long arm of chromosome 18, whereas invasive testing of amniotic fluid showed that the sample had only a non-mosaic deletion of the whole short arm of chromosome 18. Interestingly, this sample had also undergone NIPT for aneuploidy with a commercial company that reported it as highly likely to have trisomy 18. Given that different populations of cells are tested by NIPT (mainly cytotrophoblasts) and amniocentesis (amniocytes and fetal cells), the most likely explanation for this discrepant finding is confined placental mosaicism, and our finding potentially represents an isochromosome for 18q, i.e., the initial conception either carried the deletion of 18p and the deleted chromosome was duplicated via a U-type exchange mechanism in the placenta or carried the 18q isochromosome (both the deletion of 18p and the duplication of 18q), of which one arm was then deleted and lost in the fetus. Unfortunately, no placental material was available for confirming this hypothesis. The maternal karyotype was normal (46,XX), eliminating the possibility that a mosaic maternal rearrangement complicated our prenatal cfDNA result.
For samples with maternal duplications, our pipeline with the standard depth of sequencing detected the microduplication in all three samples but could not accurately determine whether the fetus had inherited the microduplication. In principle, the same counting statistics, when given the fetal fraction, should be able to determine fetal inheritance. If the fetus has inherited the microduplication, the expected count ratio is 1.5; if the fetus has not inherited the microduplication, the expected count ratio is reduced to 1.5 − fetal fraction/2. Achieveing sufficient accuracy would require knowledge of the fetal fraction and most likely a higher read depth.
Our standard sequencing depth for NIPT for aneuploidy gave 12 false negatives, which included three false negatives with CNVs larger than 6 Mb. We detected two of these samples with deeper sequencing and estimated their fetal fraction to be less than 5%, indicating that a low fetal fraction is likely to be the reason for failing to detect the rearrangement. Figure 6 further highlights this issue. Increasing the depth of sequencing increased the detection rate of smaller CNVs. However, our power analysis showed that it would be difficult for our test to achieve 100% sensitivity, even for large CNVs, if the fetal fraction is low. With 100 million reads, we can achieve 99% sensitivity for 10 Mb CNVs with a 5% fetal fraction, but if ∼5% of samples have fetal fractions less than 5%,22 then the test’s overall sensitivity will be reduced to ∼94%. Clearly, estimating the fetal fraction is crucial in the detection of CNVs, given that a low fetal fraction might lead to a negative result (Figure 6). For male fetuses, it might be possible to use the fetal fraction estimated from chromosome Y to constrain the minimum size of the detectable CNV. For female fetuses, alternative methods for estimating the fetal fraction will be needed. One such technique is to use the proportion of short reads to estimate the fetal fraction.25 The implication is that although standard NIPT with MPS might detect most of the large CNVs, a negative result cannot rule out the possibility that a subchromosomal CNV is present in the fetus because of a low fetal fraction.
Our method improves upon some of the previously published techniques for detecting subchromosomal copy-number changes in cfDNA. Some published methods bin the read counts into 1 Mb bins and only call a CNV if consecutive bins have Z scores above a certain value.14, 15 The requirement for consecutive bins to pass statistical tests is less powerful than summing the counts in all bins covered by the CNV and then performing a statistical test. Our method of binning into small, 100 kb bins and then performing segmentation to find potential CNV segments offers higher statistical power. A decision tree is another technique that has been referenced in the literature.26 Compared to our likelihood-ratio test, a decision tree is not statistically robust and lacks interpretability.
The approach we have taken in this study does not require prior knowledge of the fetal CNV’s location. The alternative approach is to test for a panel of known recurrent microdeletion and microduplication syndromes, as is now being done by a number of commercial companies.16, 26 By restricting the test to known regions, it might be possible to decrease the test-statistic cutoff for calling a CNV, thus improving sensitivity without sacrificing specificity, and this might offer easier interpretation of positive results given that there would be no variants of unknown significance. However, this assumes that we know the exact location of the CNV, and this might not be the case. Most unbalanced rearrangements are random, and a targeted approach would fail to detect many rearrangements.27 Of the 225 pathogenic cytogenetic rearrangements detected by traditional karyotyping in our laboratory over the past 15 years, only 62 (∼25%) would be detected by a targeted approach (E.K., unpublished data). Furthermore, the size of many of these rearrangements can be very variable. As of yet, no data are available to indicate what the limit of detection by sequencing approaches might be, so the negative predictive value cannot be determined, and confirming a negative result in a high-risk pregnancy would still require invasive testing.
The main limitation of our technique is the lack of sensitivity for CNVs of less than 6 Mb. A previous study showed that microarray analysis detected clinically significant CNVs, ∼75% of which were smaller than 10 Mb, in up to 6.5% of fetuses with an ultrasound abnormality and normal karyotype.2 No comprehensive data currently give the true incidence of pathogenic CNVs < 6 Mb, and thus it is difficult to determine exactly what proportion of CNVs would be missed, but it is clear that a significant number would not be detected. Comparing the efficacy of the different diagnostic techniques and determining the limits of detection would require a very large prospective study comparing NIPT and microarray analysis. We have shown that deeper sequencing can improve sensitivity, albeit with a higher cost and the potential for detecting more variants of unknown significance. Another approach for increasing the detection rate for small CNVs is to use capture technology. This would require designing a capture panel around known common and/or recurrent CNV locations and using bioinformatics to identify breakpoints in the CNVs. This, too, is costly and would also limit detection to known CNVs rather than genome-wide CNVs, thereby limiting detection of pathogenic CNVs overall.
Conclusion
We have developed a calling algorithm that detects the majority of cytogenetically visible chromosomal rearrangements (>6 Mb) via standard sequencing used for trisomy NIPT. It has a modest false-positive rate of 0.4%, but reporting a negative result requires an accurate assessment of the fetal fraction. Microdeletions and microduplications of <6 Mb cannot reliably be detected unless they are maternally inherited or the depth of sequencing is increased significantly, when the false-positive rate is likely to increase. Even then, the negative predictive value cannot confidently exclude a very small rearrangement. At present, confidently excluding the presence of a pathogenic CNV requires analysis of fetal genetic material obtained from chorionic villus sampling or amniocentesis. Given that a significant benefit of using NIPT to screen for aneuploidy is the increased safety secondary to the reduced need for invasive testing, extending NIPT to include screening for subchromosomal rearrangements stands to reverse some of this benefit whilst not offering comprehensive detection of pathogenic rearrangements. The costs and benefits of extending NIPT to this indication should be seriously considered prior to routine implementation.
Acknowledgments
This manuscript presents independent research funded by the National Institute for Health Research (NIHR) under the Programme Grants for Applied Research (RP-PG-0707-10107; the Rapid Accurate Prenatal Non-invasive Diagnosis [RAPID] project), the Newlife Foundation for Disabled Children, and the NIHR Comprehensive Research Network. The NIHR Biomedical Research Centre at Great Ormond Street NHS Foundation Trust funded K.K.L. and partly funded L.S.C. V.P. is partly funded by a UK Medical Research Council grant MR/K018523/1. V.P. and K.L. are part-time employees and shareholders of Inivata (a company that analyzes circulating tumor DNA). The views expressed herein are those of the authors and not necessarily those of the National Health Service, the NIHR, or the UK Department of Health.
Published: December 17, 2015
Footnotes
Supplemental Data include two figures and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.016.
Web Resources
The URLs for data presented herein are as follows:
OMIM, http://omim.org/
RAPID project, www.rapid.nhs.uk
RAPIDR-Plus, http://ucl.github.io/RAPIDR/
UCSC Genome Browser, https://genome.ucsc.edu/
Supplemental Data
References
- 1.Wapner R.J., Martin C.L., Levy B., Ballif B.C., Eng C.M., Zachary J.M., Savage M., Platt L.D., Saltzman D., Grobman W.A. Chromosomal microarray versus karyotyping for prenatal diagnosis. N. Engl. J. Med. 2012;367:2175–2184. doi: 10.1056/NEJMoa1203382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shaffer L.G., Rosenfeld J.A., Dabell M.P., Coppinger J., Bandholz A.M., Ellison J.W., Ravnan J.B., Torchia B.S., Ballif B.C., Fisher A.J. Detection rates of clinically significant genomic alterations by microarray analysis for specific anomalies detected by ultrasound. Prenat. Diagn. 2012;32:986–995. doi: 10.1002/pd.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chitty L.S., Kagan K.O., Molina F.S., Waters J.J., Nicolaides K.H. Fetal nuchal translucency scan and early prenatal diagnosis of chromosomal abnormalities by rapid aneuploidy screening: observational study. BMJ. 2006;332:452–455. doi: 10.1136/bmj.38730.655197.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Callaway J.L.A., Shaffer L.G., Chitty L.S., Rosenfeld J.A., Crolla J.A. The clinical utility of microarray technologies applied to prenatal cytogenetics in the presence of a normal conventional karyotype: a review of the literature. Prenat. Diagn. 2013;33:1119–1123. doi: 10.1002/pd.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minear M.A., Lewis C., Pradhan S., Chandrasekharan S. Global perspectives on clinical adoption of NIPT. Prenat. Diagn. 2015;35:959–967. doi: 10.1002/pd.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.American College of Obstetricians and Gynecologists Committee Opinion No. 640: Cell-Free DNA Screening For Fetal Aneuploidy. Obstet. Gynecol. 2015;126:e31–e37. doi: 10.1097/AOG.0000000000001051. [DOI] [PubMed] [Google Scholar]
- 7.Gregg A.R., Gross S.J., Best R.G., Monaghan K.G., Bajaj K., Skotko B.G., Thompson B.H., Watson M.S. ACMG statement on noninvasive prenatal screening for fetal aneuploidy. Genet. Med. 2013;15:395–398. doi: 10.1038/gim.2013.29. [DOI] [PubMed] [Google Scholar]
- 8.Benn P., Borell A., Chiu R., Cuckle H., Dugoff L., Faas B., Gross S., Johnson J., Maymon R., Norton M. Position statement from the Aneuploidy Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat. Diagn. 2013;33:622–629. doi: 10.1002/pd.4139. [DOI] [PubMed] [Google Scholar]
- 9.American College of Obstetricians and Gynecologists Committee on Genetics Committee Opinion No. 581: the use of chromosomal microarray analysis in prenatal diagnosis. Obstet. Gynecol. 2013;122:1374–1377. doi: 10.1097/01.AOG.0000438962.16108.d1. [DOI] [PubMed] [Google Scholar]
- 10.Petersen O.B., Vogel I., Ekelund C., Hyett J., Tabor A., Danish Fetal Medicine Study Group. Danish Clinical Genetics Study Group Potential diagnostic consequences of applying non-invasive prenatal testing: population-based study from a country with existing first-trimester screening. Ultrasound Obstet. Gynecol. 2014;43:265–271. doi: 10.1002/uog.13270. [DOI] [PubMed] [Google Scholar]
- 11.Jensen T.J., Dzakula Z., Deciu C., van den Boom D., Ehrich M. Detection of microdeletion 22q11.2 in a fetus by next-generation sequencing of maternal plasma. Clin. Chem. 2012;58:1148–1151. doi: 10.1373/clinchem.2011.180794. [DOI] [PubMed] [Google Scholar]
- 12.Peters D., Chu T., Yatsenko S.A., Hendrix N., Hogge W.A., Surti U., Bunce K., Dunkel M., Shaw P., Rajkovic A. Noninvasive prenatal diagnosis of a fetal microdeletion syndrome. N. Engl. J. Med. 2011;365:1847–1848. doi: 10.1056/NEJMc1106975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen S., Lau T.K., Zhang C., Xu C., Xu Z., Hu P., Xu J., Huang H., Pan L., Jiang F. A method for noninvasive detection of fetal large deletions/duplications by low coverage massively parallel sequencing. Prenat. Diagn. 2013;33:584–590. doi: 10.1002/pd.4110. [DOI] [PubMed] [Google Scholar]
- 14.Srinivasan A., Bianchi D.W., Huang H., Sehnert A.J., Rava R.P. Noninvasive detection of fetal subchromosome abnormalities via deep sequencing of maternal plasma. Am. J. Hum. Genet. 2013;92:167–176. doi: 10.1016/j.ajhg.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu S.C., Jiang P., Choy K.W., Chan K.C., Won H.S., Leung W.C., Lau E.T., Tang M.H., Leung T.Y., Lo Y.M., Chiu R.W. Noninvasive prenatal molecular karyotyping from maternal plasma. PLoS ONE. 2013;8:e60968. doi: 10.1371/journal.pone.0060968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wapner R.J., Babiarz J.E., Levy B., Stosic M., Zimmermann B., Sigurjonsson S., Wayham N., Ryan A., Banjevic M., Lacroute P. Expanding the scope of noninvasive prenatal testing: detection of fetal microdeletion syndromes. Am. J. Obstet. Gynecol. 2015;212 doi: 10.1016/j.ajog.2014.11.041. 332.e1–332.e9. [DOI] [PubMed] [Google Scholar]
- 17.Lo K.K., Boustred C., Chitty L.S., Plagnol V. RAPIDR: an analysis package for non-invasive prenatal testing of aneuploidy. Bioinformatics. 2014;30:2965–2967. doi: 10.1093/bioinformatics/btu419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan H.C., Quake S.R. Sensitivity of noninvasive prenatal detection of fetal aneuploidy from maternal plasma using shotgun sequencing is limited only by counting statistics. PLoS ONE. 2010;5:e10439. doi: 10.1371/journal.pone.0010439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krumm N., Sudmant P.H., Ko A., O’Roak B.J., Malig M., Coe B.P., Quinlan A.R., Nickerson D.A., Eichler E.E., NHLBI Exome Sequencing Project Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Killick R., Fearnhead P., Eckley I.A. Optimal Detection of Changepoints With a Linear Computational Cost. J. Am. Stat. Assoc. 2012;107:1590–1598. [Google Scholar]
- 21.R Development Core Team . R Foundation for Statistical Computing; 2014. R: A language and environment for statistical computing. [Google Scholar]
- 22.Rava R.P., Srinivasan A., Sehnert A.J., Bianchi D.W. Circulating fetal cell-free DNA fractions differ in autosomal aneuploidies and monosomy X. Clin. Chem. 2014;60:243–250. doi: 10.1373/clinchem.2013.207951. [DOI] [PubMed] [Google Scholar]
- 23.Warsof S.L., Larion S., Abuhamad A.Z. Overview of the impact of noninvasive prenatal testing on diagnostic procedures. Prenat. Diagn. 2015;35:972–979. doi: 10.1002/pd.4601. [DOI] [PubMed] [Google Scholar]
- 24.Lewis C., Hill M., Skirton H., Chitty L.S. Non-invasive prenatal diagnosis for fetal sex determination: benefits and disadvantages from the service users’ perspective. Eur. J. Hum. Genet. 2012;20:1127–1133. doi: 10.1038/ejhg.2012.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu S.C., Chan K.C., Zheng Y.W., Jiang P., Liao G.J., Sun H., Akolekar R., Leung T.Y., Go A.T., van Vugt J.M. Size-based molecular diagnostics using plasma DNA for noninvasive prenatal testing. Proc. Natl. Acad. Sci. USA. 2014;111:8583–8588. doi: 10.1073/pnas.1406103111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao C., Tynan J., Ehrich M., Hannum G., McCullough R., Saldivar J.S., Oeth P., van den Boom D., Deciu C. Detection of fetal subchromosomal abnormalities by sequencing circulating cell-free DNA from maternal plasma. Clin. Chem. 2015;61:608–616. doi: 10.1373/clinchem.2014.233312. [DOI] [PubMed] [Google Scholar]
- 27.Shaffer L.G., Bejjani B.A., Torchia B., Kirkpatrick S., Coppinger J., Ballif B.C. The identification of microdeletion syndromes and other chromosome abnormalities: cytogenetic methods of the past, new technologies for the future. Am. J. Med. Genet. C. Semin. Med. Genet. 2007;145C:335–345. doi: 10.1002/ajmg.c.30152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.