Abstract
Intellectual disability (ID) disorders are genetically and phenotypically extremely heterogeneous. Can this complexity be depicted in a comprehensive way as a means of facilitating the understanding of ID disorders and their underlying biology? We provide a curated database of 746 currently known genes, mutations in which cause ID (ID-associated genes [ID-AGs]), classified according to ID manifestation and associated clinical features. Using this integrated resource, we show that ID-AGs are substantially enriched with co-expression, protein-protein interactions, and specific biological functions. Systematic identification of highly enriched functional themes and phenotypes revealed typical phenotype combinations characterizing process-defined groups of ID disorders, such as chromatin-related disorders and deficiencies in DNA repair. Strikingly, phenotype classification efficiently breaks down ID-AGs into subsets with significantly elevated biological coherence and predictive power. Custom-made functional Drosophila datasets revealed further characteristic phenotypes among ID-AGs and specific clinical classes. Our study and resource provide systematic insights into the molecular and clinical landscape of ID disorders, represent a significant step toward overcoming current limitations in ID research, and prove the utility of systematic human and cross-species phenomics analyses in highly heterogeneous genetic disorders.
Introduction
Intellectual disability (ID) affects as much as 2% of our population and is characterized by significant limitations in intellectual functioning and adaptive behavior.1, 2 Because of its high frequency, limited treatability, and required lifelong care, ID is an important socioeconomic and health-care issue.
The clinical presentation of ID is highly heterogeneous. It can range from learning difficulties to profound cognitive impairment, and it can occur either non-specifically without further anomalies or in a more complex, syndromic context. A large proportion of ID is caused by mutations in single genes (ID-associated genes [ID-AGs]). Identification of these genes is still largely incomplete,3, 4 limiting our understanding of the underlying biology. This makes ID a major challenge in diagnostics and translational medicine.
Whereas studies on specific subgroups of ID disorders have indicated that convergent molecular pathways underlie common phenotypic aspects,5, 6, 7 ID-AGs on a more global scale (yet not systematic) have been argued to differ from genes implicated in autism in that they show poor biological convergence.8 A comprehensive picture of ID-AGs, ID-AG properties, gene-phenotype relations, and molecular modularity of ID is yet to be established.
Here, we provide a systematic, curated ID-AG catalog with associated core phenotypes and introduce a clinical classification system, accessible to other scientists in an interactive web-based database. Integration of custom-made and public data into this resource allowed resolving ID-AGs and ID disorders into biologically meaningful subgroups with significantly increased co-expression, protein interactions, and specific functions. Our analyses also identified typical phenotype combinations characterizing ID disorders that are linked to specific molecular processes, such as chromatin regulation and DNA repair, and found that public datasets contain patterns that provide insights into ID pathology and increase predictive power. Finally, we provide two large-scale functional ID-AG datasets generated in Drosophila and use these to define further predictive patterns that underlie ID.
Material and Methods
ID-Associated Gene Catalog
The ID-AG list was compiled from primary and secondary9 literature and OMIM. Reasons to exclude genes from the ID-AG catalog were as follows:
-
(1)
Low evidence, e.g., genetic data based on only one or two single individuals with de novo mutations or a single family with a missense mutation (from 2014 on); gene-disrupting translocations or deletions but no mutational confirmation of the candidate gene in other individuals; or clinical description of the disorder without genetic testing or confirmation.
-
(2)
Pure neurodegenerative manifestation, indicated by secondary onset of intellectual disability with regression of initial normal abilities or by a progressive disease course with deterioration of cognitive abilities.
-
(3)
Very early lethality, thus precluding proper assessment of psychomotor development.
-
(4)
Treatability, indicated by avoidance of decline in cognitive ability by substitution of certain factors, e.g., consequences of hypothyroidism, which can be avoided by thyroid hormone substitution.
-
(5)
Neurologic phenotype without a clear indication of cognitive impairment.
The excluded genes were compiled in an ID-AG candidate list and are available in the Systems Biology Approaches to ID (SysID) database (see Web Resources). Two versions of the ID-AG list were used in this study: (1) a set of 388 ID-AGs, published as of mid-2010, for the basis of the in-house Drosophila screens and (2) a larger set of 650 ID-AGs, published as of January 2014, for all non-Drosophila analyses.
Clinical Classification
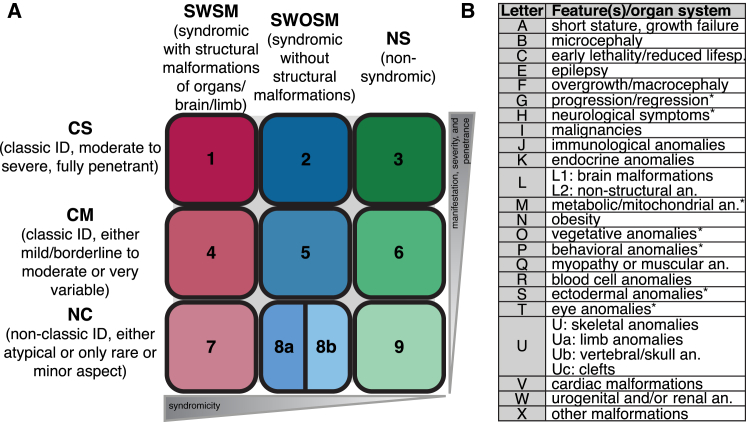
The large clinical heterogeneity of ID disorders comprises isolated (non-syndromic) ID, syndromic ID accompanied by various specific clinical phenotypes, and multisystemic disorders where ID constitutes only one of many aspects. Moreover, ID varies in severity and penetrance. In order to obtain a systematic and versatile yet manageable amount of phenotypic information, we designed a bipartite phenotype-based classification system for ID disorders. The classification consists of six higher-order super-classes comprising nine clinical classes according to the occurrence of non-syndromic or syndromic ID with or without congenital malformations (“syndromicity,” x axis in Figure 2A) and according to manifestation (e.g., atypical), severity, and penetrance of ID, the latter two of which correlate with each other (“manifestation, severity, and penetrance,” y axis in Figure 2A; see also Figure S2). In the case of genetically heterogeneous disorders, only gene-specific clinical information was used for the respective phenotype classification. A clinical expert annotated the phenotypic classification, and a second clinical expert revised the main classes independently. Discrepancies were discussed and jointly agreed on. In addition, ID-accompanying phenotypes for all ID disorders in the list were assembled. These comprise 27 additional features describing further symptoms and anomalies of various organ systems (Figure 2B and Figure S2). Letters A–X indicate the presence of specific clinical features and were added when the (estimated) reported frequency of the respective symptom was at least 20%–30%. A confidence criterion “limited number of affected individuals” was implemented in the database and indicates limited availability of clinical information.
Figure 2.
Bipartite Clinical ID-Classification System
(A) Main clinical classes. The “syndromicity” axis of ID entities is defined as follows: classes 1, 4, and 7 comprise disorders that are syndromic with structural malformations (SWSM); classes 2, 5, and 8 include disorders that are syndromic without structural malformations (SWOSM); and classes 3, 6, and 9 comprise non-syndromic (NS) ID disorders. The “manifestation, severity, and penetrance” axis of ID entities is defined as follows: classes 1–3 contain disorders with severe and fully penetrant manifestation of ID (CS), classes 4–6 include disorders with mild to moderate or very variable ID (CM), and classes 7–9 comprise disorders with ID in a rare (8a) or atypical (e.g., progressive, neurodegenerative features) (8b) manifestation (NC).
(B) ID-accompanying phenotypes: ID-accompanying clinical features that occur with an estimated frequency of >20% within the respective disorder. Abbreviations are as follows: lifesp, lifespan; and an, anomalies. Clinical features marked with an asterisk are explained as follows: progression/regression, progression of disease and regression of development; neurological symptoms, e.g., hypotonia, ataxia, and tremor; metabolic/mitochondrial an., e.g., enzymatic defects; vegetative anomalies, e.g., breathing anomalies and increased sweating; behavioral anomalies, e.g., autism and aggression; ectodermal anomalies, e.g., skin, hair, and nail anomalies; and eye anomalies, structural and functional. Figure S2 shows the numbers of genes per clinical class and ID-accompanying phenotype, a network view of the distribution of genes per clinical class, and a distribution of ID-accompanying phenotypes over main clinical classes.
SysID Database
ID-AG-related information includes a short gene description and the human gene info (Entrez ID, Ensembl ID, HUGO gene name, Human Protein Reference Database [HPRD] ID, synonyms, Gene Ontology-based terms, and chromosomal location). Gene-related disease information is provided per associated disease and includes OMIM disease numbers and mode of inheritance. Further clinical information is provided either by a non-vocabulary-controlled summary of characteristic symptoms or by either a PMID from GeneTest review entries or a primary reference (Table S1). Furthermore, Drosophila orthologs and identified phenotypes were uploaded into the database (CG number; FlyBase ID, gene name, and symbol; Vienna Drosophila Resource Center [VDRC] RNAi line identifiers; and phenotypes), as shown in Table S2. Candidate genes associated with autism were annotated according to the Simons Foundation’s SFARI database.10 Only genes of high-confidence categories S and 1–3 were considered. The URL for the SysID database is provided in the Web Resources.
Gene Ontology-Based Analysis
We used Golem v.2_111 to manually assemble 32 Gene Ontology-based groups from related Gene Ontology12 terms, and we downloaded associated genes and matched them to Entrez IDs. We used terms of biological process (BP), molecular function (MF), and cellular component (CC) according to the authors’ knowledge and research about processes relevant to ID (Table S3).
Protein-Protein-Interaction Network
For analysis of protein-protein interactions (PPIs), we created a human-specific PPI network containing physical interactions between proteins. Using Entrez IDs, we merged BioGrid 3.2.108 (release January 1, 2014)13 interactions based on physical associations (association, direct interaction, physical interaction, and co-localization with additional biochemical evidence, together ca. 97% of all BioGrid interactions) with PPIs from HPRD14 (release 9, April 13, 2010). After removal of duplicates and self-loops, this reference network contained 15,511 proteins and 138,029 connections, including 610 ID-AGs with 505 connections (Figure 1). We used this reference PPI network for all manually performed analyses of enrichment, PIE (physical interaction enrichment) scores, and connectivity.
Figure 1.
Systematic Analyses of Genes Implicated in ID Reveal Functional Groups and Molecular Modules
(A) Gene Ontology-based annotation of ID-AG function. Bar diagrams show enrichments of ID-AGs in each of the indicated Gene Ontology-based groups against the genome-wide background. The total number of genes per group is displayed in the respective bar. (Benjamini-Hochberg, ∗padj < 0.05, ∗∗padj < 0.01, ∗∗∗padj < 0.001.)
(B) Physical PPI network of ID-AG products. Circles indicate highly connected ID-AG communities; similar colors illustrate functional proximity (Figure S1). Genes directly connecting to communities are colored if they share Gene Ontology-based terms with the connected communities. Dark gray indicates nodes without associated Gene Ontology-based terms, and light gray indicates nodes with at least a first-degree connection to communities. ID-AGs without connections to other ID-AGs are not shown.
Community Clustering
We hierarchically clustered PPI communities obtained from the R package linkcomm15 on the basis of the number of shared nodes (after using the Jaccard coefficient to score the pairwise similarities; tree cutoff = 0.99) and visualized them as circles in Figure 1B and Figure S2.
Clustering ID-Accompanying Phenotypes
We determined the binary matrix of ID-accompanying phenotypes per gene on the basis of the computed row and column dendrograms and used gplots to visualize them as a heatmap.16
Network Visualization and Figures
Network visualization was carried out with Cytoscape (v.3.1.1)17 and Adobe Illustrator CS5.
Multiple-Testing Corrections
When performing multiple comparisons, we applied Benjamini-Hochberg corrections to control for the false-discovery rate. We determined Benjamini-Hochberg-adjusted p values (padj) with the R-stats package (v.2.16.0) and the number (n) of p values obtained within the test (i.e., n = 10 in a test of nine main clinical classes [1–8b] plus the group “all 650 ID-AGs,” or n = 28 in a test of 27 accompanying phenotypes [A–X] plus the group “all 650 ID-AGs genes”).
PIE Score
We calculated PIE scores and associated p values for all ID-AGs against direct PPIs from our reference network by using the PIE algorithm18 to account for biases in the number of reported interactions for disease-associated genes. Random protein groups were formed by number-matched sub-samplings selected from the 650 ID-AG set.
Enrichment Analyses
Enrichment for human and Drosophila datasets was calculated as follows: (a/b)/[(c − a)/(d − b)], where a is the number of genes that are in a class or ID-accompanying phenotype and have a specific feature, b is the number of genes in that class or ID-accompanying phenotype, c is the number of genes with that specific feature, and d is the total number of genes. For analyses of human datasets (phenotypes, Gene Ontology-based terms, and human postsynaptic density [hPSD]), we used the human genome with an estimated 20,500 genes or the 650 ID-AG set as a background, as indicated. For analyses of the generated phenotype groups for the Drosophila orthologs of the 388 ID-AG set, we used the background of all targeted fly orthologs. Uncorrected p values were determined with a two-sided Fisher’s exact test in R.
Co-expression Networks Based on BrainSpan and GTEx
We used the BrainSpan19 developmental transcriptome dataset (RNA sequencing with Gencode v.10) to examine the overrepresentation of highly co-expressed genes over all or within brain regions and time points, and we used GTEx20 data to determine overrepresentation of highly co-expressed genes over various tissues. To examine the overrepresentation of highly co-expressed genes among all ID-AGs, ID classes, and ID-accompanying phenotype groups in relation to the rest of the genome, we concatenated the expression coefficients per gene over all time points (embryonic stage to adulthood) and brain regions in the BrainSpan dataset. Additionally, we calculated co-expression enrichment for ID-AGs per brain region over all time points as well as enrichment for all ID-AGs per brain region at pre- and postnatal stages. Also, for the GTEx dataset, we combined expression values for all tissues. We calculated the co-expression correlation for gene pairs among 650 ID-AGs, per phenotype group (main classes 1–8b and ID-accompanying phenotypes A–X), and for random groups. For each phenotype group and the 650 ID-AGs, 10,000 random groups out of the entire gene-expression datasets met the following criteria: (1) same group size (e.g., main class 1 contains 65 genes, so 10,000 random groups contain 65 genes), (2) similar distribution of coding-sequence (CDS) length (as described in Honti et al.21), and (3) similar number of co-expressed genes (i.e., with a correlation coefficient > 0.3) in the complete network. We then calculated and compared the sum of co-expression coefficients for the real and random groups, and we again only included gene pairs with a correlation coefficient > 0.3. We calculated p values by comparing how many of the 10,000 co-expression coefficients of random groups were equal or higher than those of the corresponding test group, and we corrected for multiple testing. We calculated the enrichment score by dividing the sum of the co-expression coefficient per test group by the mean of the 10,000 random groups per group of interest.
For the co-expression of ID classes or phenotype groups, we compared the sum of co-expression values within ID classes or phenotype groups with the sum of co-expression values when the 650 ID-AGs were randomly distributed over the ID classes or phenotype groups, while considering group size.
Enrichment of Connectivity within ID Phenotype Classes
We calculated the overall connectivity of main clinical classes and ID-accompanying phenotypes among 650 ID-AGs by the median of (1) the number of PPIs based on our reference PPI network per clinical classe or ID-accompanying phenotype, (2) the co-expression levels per clinical class according to BrainSpan, and (3) the co-expression levels per ID-accompanying phenotype according to GTEx. For randomized classes, all 650 genes were shuffled between all classes controlled for class size, and the median of connectivity of all classes was calculated. This randomization was repeated 10,000 times. The enrichment (number of interactions divided by the median of the connectivity of random samples) was calculated, and p values (number of random samples with a connectivity equal to or higher than the real gene set) were obtained. This was done for both main clinical classes and ID-accompanying phenotypes.
Precision-Recall Analysis
To test whether the functional coherence of genes associated with the same ID class, ID super-class, or ID-accompanying phenotype manifests in their increased predictability, we performed leave-one-out cross-validations by taking advantage of an integrated phenotypic-linkage network described elsewhere.21 To assess the predictability of a class, we rank ordered all 17,011 genes in the integrated network by the sum of their link weights to the ten class genes most strongly linked to them, and the highest-ranking genes were predicted to be associated with the given class. The results of these analyses were represented by precision-recall curves, which show the proportion of true positives at different levels of coverage of known class genes (recall) in the predictions. To indicate the significance of the results, we evaluated expected precision values with randomly selected gene sets sub-sampled from among the 650 ID-AGs; the area they covered was color coded according to the corresponding p value. For evaluating all 650 ID-AGs, we used random genes controlled for both node degree and CDS length from the integrated network to calculate the expected precision values.
Controlling for Node Degree and CDS Length
The node degree of a given gene was defined as the number of genes linked to this gene in the network. To control for both node degree and CDS length during the co-expression randomizations based on BrainSpan and GTEx and precision-recall calculation, we selected random genes that matched the node degree and CDS length of the studied genes. For each of the studied genes, we assigned a list of 100 genes with the same or most similar node degree and CDS length by using the longest CDS of each gene. We normalized the node degrees and CDS lengths and calculated the Euclidean distance between genes on the basis of these two measures. We used this Euclidean distance to form lists of the 100 genes most similar to each of the studied genes. We then assembled random gene sets by selecting one random gene from each of these lists.21
Fly Stocks and Breeding Conditions
For the neuronal screen, we used an elav-Gal4 promoter line from the Bloomington stock center (BL25750: P{w[+mW.hs] = GawB}elav[C155] w[1118]; P{w[+mC] = UAS-Dcr-2.D}2), and for the wing screen, we used the trp05/MS1096 promoter line from the Bloomington stock center (BL25706: w[1118] P{w[+mW.hs] = GawB}Bx[MS1096]; P{w[+mC] = UAS-Dcr-2.D}2). Stocks and crosses were cultured according to standard procedures and on standard fly food. Crosses for the neuronal screen were raised on 28°C and 60% humidity, and crosses for the wing-screen were raised on 25°C and 60% humidity, both at a 12/12 hr day/night cycle.
Drosophila Orthology and Genetic Manipulation
We mapped 388 human ID-AGs identified as of mid-2010 to their corresponding orthologs in Drosophila melanogaster by using Ensembl’s orthology classes (Ensemblv72_June2013),22 treefam annotations,23 and manual curation. One-to-one and one-to-many (fly-to-human) criteria identified 294 orthologs for 388 human ID-AGs. For conditional knockdown, we used the UAS-GAL4 system24 in combination with UAS-RNAi lines from the VDRC. Progenies from a cross of the Gal4 driver to the genetic backgrounds of the UAS-RNAi libraries (vdrc60000, vdrc60100)25 served as controls in all experiments and showed wild-type morphology and behavior.
Quality-Control Criteria of RNAi Lines
We used at least two independent constructs for each ID-AG (one from the GD [p-element-based transgenes] and one from the KK [phiC31-based transgenes] library) when available and selected RNAi lines with high s19 values, (0.98–1.00 in 97% of all cases; see the SysID database in the Web Resources), thereby exceeding the recommended threshold of 0.85 for ensuring high reproducibility.26 None of the described phenotypes was observed in the non-induced UAS-RNAi stocks.
Negative-Control Gene Set
We generated random lists of 35 conserved genes until we identified one that (1) contained no ID-AGs and (2) included genes that showed average expression in each of the three nervous system tissues (larval CNS, adult brain, and thoracic ganglion) in FlyAtlas.27 Like for ID-AGs, at least two independent RNAi-constructs against these genes were used if available.
Neuronal Screen with the Island Assay
The assay was performed as previously described28 with minor modifications. In brief, if gene silencing did not result in lethality, progenies of the appropriate genotype were collected in batches of 20 and either tested 2 days after collection (4–6 days old) or kept on standard food (changed to fresh food every other day) for later testing (14–16 days old). Phenotypes observed at the first testing point were annotated as “early,” and those at the second were annotated as “late.” A minimum of 10–20 flies were tested during the same time window of the day (10 a.m. to 3 p.m.). For the island assay, we used a soapy water bath with an elevated platform (“island,” 5 × 10 cm) in the middle. We evaluated locomotion defects by assessing the flies’ ability to immediately fly away after being dropped from their vial onto the platform from about 10 cm height. Fractions of the population flying away immediately (no phenotype) or remaining on the platform (phenotype) were scored. Behavior of the remaining flies was further evaluated (“walker” [flies walking on the platform], “sitter” [flies not moving on the platform], “jumper” [flies jumping on the platform], or a combination thereof). Whenever the independent RNAi lines tested for a particular gene did not all show the same phenotype, abnormal fly behavior or lethality was confirmed by two independent experiments blind to genotype. The phenotypes “progression” and “recovery” were assessed according to increasing phenotype frequency and/or tendency over age. RNAi-to-gene translation was done in a collective manner (all phenotypes observed among the tested RNAi lines were associated with the targeted Drosophila gene and its human ortholog) for the following phenotypes: developmental lethality, adult lethality, and all behavioral phenotypes (early or late walker, early or late sitter, and early or late jumper). Exceptions were the categories “phenotype progression,” “phenotype recovery,” and “no tendency,” for which only the strongest obtained phenotype was considered (whereby developmental lethality was stronger than behavioral phenotype). “Any phenotype” was annotated upon observation of at least one phenotype, and “no phenotype” was annotated if no phenotype was found in any of the tested RNAi lines per gene. See Table S2 for an overview of all observed phenotypes per gene within the neuronal screen (264 ID-AGs and 31 non-ID-AGs tested).
Wing Screen
For each genotype, we assessed the viability and the overall appearance of the wing before mounting the wings for closer phenotype evaluation. Of each genotype, three to five right wings of 8-day-old males were collected and dehydrated in a succession of three solutions (30/70 glycerol/isopropanol, 60/40 glycerol/isopropanol, and 90/10 glycerol/isopropanol, each for 10 min). Wings were mounted in 100% glycerol and stored at 4°C. The following phenotype categories were evaluated for the screen (Axio Imager Z1, magnification 5×, 10×, or 20×): wing shape (curled and cupped, size, and adhesion), posterior margin (notched or with missing hairs), wing fields (trichome polarity [missing, density, or disorganized], morphology, and other aspects [i.e., pigmented spots]), veins (missing and/or extra), and bristles (sensory organs). RNAi-to-gene translation for the wing screen was collective (see Neuronal Screen with the Island Assay above). See Table S2 for an overview and more detailed description of all observed phenotypes per human gene of the 261 ID-AGs and 31 non-ID-AGs tested.
Results
Mutations in More Than 650 Genes Cause ID
We assembled a systematic, manually curated catalog of 650 ID-AGs (as of January 2014; Table S1) according to criteria specified in the Material and Methods. Of the 650 ID-AGs, 101 (16%) are implicated in more than one specific ID disorder. Mutations in 400 of the genes (62%) follow autosomal-recessive inheritance, mutations in 139 genes (21%) are autosomal dominant (mostly de novo), and 103 ID-AGs (16%) are X-linked. Our SysID database (see Material and Methods) currently covers 777 ID-AGs (updated December 2015) with related phenotypic and functional data plus 389 ID-associated candidate genes.
Biological Functions of ID-Associated Genes
To characterize the functional coherence and connectivity among the complete set of 650 ID-AGs, we collected data from genome-wide resources of annotated gene function, physical PPIs, and gene co-expression. We used Gene Ontology-based annotations to ask, given the genome-wide frequency of genes in each of these processes, which of them are most enriched and thus most prone to bear ID-AGs. We found 560 ID-AGs (86%) to associate with at least one of the 32 Gene Ontology-based annotations shown in Figure 1A. Whereas the largest groups of ID-AGs were associated with metabolism, transporters, nervous system development, RNA metabolism, and transcription, the most enriched terms were hedgehog signaling, glutamate signaling, peroxisomes, glycosylation, and cilia (Figure 1A). Frequently discussed themes in ID, such as synaptic and chromatin-related processes,29, 30, 31, 32 although statistically significant enriched, were found to belong to neither the biggest nor the most enriched groups.
ID-Associated Genes Show High Connectivity and Significant Co-expression
We next asked whether ID-AGs and their products, ID-associated proteins, also show increased molecular connectivity, modularity, and co-expression. Constructing PPI networks from HPRD14 and BioGRID13 physical-interaction data, we found that nearly half (308 [47%]) of all ID-associated proteins physically interact with other ID-associated proteins. Of these, 66 are connected in small modules (pairs and tri- and quadromers), whereas 242 ID-associated proteins together form a single major network with 462 interactions (Figure 1B). Using the PIE approach to correct for inquisitional biases18 revealed that the 650 ID-AGs show more than a 30% increase in connectivity (PIE = 1.32, p < 0.0001) over randomly chosen proteins with the same number of known interactions.
To identify molecular units within the identified ID networks, we applied unsupervised community clustering on the protein interactions.15 This identified a molecular landscape of 21 highly intraconnected and partly interconnected ID modules (Figure 1B and Figure S1).
Furthermore, we found that ID-AGs, on average, show significantly enriched co-expression in two recently generated high-content gene-expression datasets, one specifically relevant to the brain (BrainSpan;19 E = 1.04, p = 0.0001) and one representing multiple organ systems (GTEx;20 E = 1.1, p < 0.0001). Interestingly, within the brain, the hippocampus, a primary region controlling learning and memory, shows the highest level of co-expression of ID-AGs (E = 1.21, padj < 0.0001; Table S4).
In summary, despite their extreme genetic heterogeneity, known ID-AGs show significantly elevated co-expression in the brain, particularly in a region relevant to cognitive processes. Moreover, the encoded proteins converge on a limited number of molecular networks and show considerable functional coherence.
An Expert-Curated, Phenotype-Based ID-Classification System
In order to obtain a systematic and versatile yet manageable amount of phenotypic information on clinically extremely heterogeneous ID disorders, we designed a bipartite phenotype-based classification system and annotated all ID disorders and genes accordingly. First, we defined ten main clinical classes relating to (1) manifestation, severity, and penetrance and (2) syndromicity of ID, and these are summarized in six super-classes (Figure 2A). The number of ID-AGs in the ten clinical classes varies from 19 (class 3) to 183 (class 5), with the exception of class 9, which harbors only one gene (Figures S2A and S2C). Second, 27 ID-accompanying phenotype categories, including structural malformations of various organ systems and functional or behavioral anomalies, were established (Figures 2B and S2B).
ID-Accompanying Phenotypes Are Characteristic of the Underlying Molecular Processes
We first performed hierarchical clustering of ID-AGs and their associated ID-accompanying phenotypes to map phenotypically similar groups of ID-AGs and to systematically unravel which ID-accompanying clinical features co-occur most frequently (Figure 3). Furthermore, Gene Ontology-based analysis revealed that one to several molecular processes were significantly overrepresented in ID-AGs associated with specific ID-accompanying phenotypes than among all ID-AGs. For example, short stature and ectodermal anomalies were much more co-morbid and enriched in genes operating in MAPK, growth factor signaling, and DNA repair than in all ID-AGs (Figure 3, right-hand side) and especially the whole genome (data not shown). Endocrine abnormalities and obesity were tightly linked with each other and co-occurred in a cluster of 18 ID-AGs and ID disorders dominated by genes with a function in cilia (Figure 3, red box). Epilepsy, neurological and metabolic abnormalities, myopathy, lethality, and non-structural MRI abnormalities co-occurred in a cluster of 20 genes enriched with mitochondrial function (blue box). Microcephaly and behavioral abnormalities were linked to two adjacent ID clusters comprising 20 genes enriched with chromatin-related function (yellow boxes). Twenty ID-AGs presenting merely with behavioral abnormalities were enriched with synaptic function (turquoise box). Of note, each of these clusters also contains genes that have not been previously associated with these molecular processes. Hence, this unsupervised analysis predicts a multitude of previously undescribed ID-AG functions (Figure 3 and Discussion).
Figure 3.
Relationships among Genes, Phenotypes, and Molecular Function in ID
Hierarchical clustering of ID-AGs and ID-accompanying phenotypes. Phenotypic similarity of (groups of) ID-AGs is indicated by the proximity of genes (x axis) and the proximity of ID-accompanying clinical features based on their co-occurrence in ID disorders (y axis). Gene Ontology-based terms that were significantly enriched after multiple-testing corrections in two or more adjacent ID-accompanying phenotypes are displayed on the right-hand side. Colored rectangles highlight randomly chosen clusters. These are highly enriched with cilia (red), chromatin (yellow), synapses (turquoise), and mitochondria (blue). Genes within the clusters are shown in the boxes below in the same color code. Those genes that are already associated with the respective Gene Ontology-based term are highlighted in bold. Abbreviations are as follow: an, anomalies; malf, malformation; non-struct, non-structural MRI anomalies; hedgehog, hedgehog signaling; Wnt, Wnt signaling; MAPK, MAPK signaling; and response to GF, response to growth factor.
In summary, phenotype-based cluster analyses systematically established gene-phenotype relationships in ID and revealed compromised molecular processes and machineries that underlie specific phenotype-defined subgroups of ID disorders.
Phenotype Delineation of Groups of Process-Defined ID Disorders
To define the typical phenotypic make-up of ID pathologies that are linked to specific biological processes, we calculated enrichments of ID-accompanying phenotypes among Gene Ontology-defined groups of ID-AGs in relation to their occurrence among all ID disorders (Figure 4). ID disorders linked to mitochondria, for example, were characterized by metabolic defects, myopathy, regression, neurological features, lethality, non-structural MRI brain defects, blood cell anomalies, and epilepsy, as commonly appreciated.33 These features were between 1.6- and 6.9-fold more enriched in ID-AGs linked to specific processes than in all ID-AGs (0.044 > padj > 2.45 × 10−29).
Figure 4.
Phenotype Delineation of Groups of Process-Defined ID Disorders
The typical phenotype combinations characterizing ID disorders associated with a specific molecular process or system were defined according to the Gene Ontology-based groups shown in Figure 1A. The volcano plots show relative enrichments (x axes in log10 scale) of ID-accompanying phenotypes (A–X) among the indicated molecular process or system in relation to their occurrence among all 650 ID-AGs, plotted against the corresponding p values (y axes in −log10 scale). Letters (A–X) refer to ID-accompanying phenotypes as listed in Figure 2B. ID-accompanying phenotypes highlighted in red show significant specificity (Benjamini Hochberg, padj < 0.05), thus identifying clinical features that are characteristic of the respective molecular-process-defined ID disorder group.
Among cilia-associated ID disorders, we found strong enrichment of obesity and urogenital, renal, skeletal, eye, and brain malformations, which are widely recognized features of ciliopathies34 (4.4 < E < 13.1; 1.8 × 10−4 > padj > 7.6 × 10−12), but also endocrine defects (E = 5.2; padj = 2.3 × 10−4) and behavioral anomalies (E = 3.4; padj = 1.6 × 10−3). ID-associated deficiencies in DNA repair were defined by malignancies, ectodermal anomalies, short stature, and microcephaly (4.1 < E < 8.8; 3.2 × 10−4 > padj > 1.6 × 10−4). Chromatin-related ID disorders can be identified by clefts, cardiac problems, other malformations, limb anomalies, and short stature (2.6 < E < 3.2; 1.1 × 10−2 > padj > 9.7 × 10−4). Clinical features reached even higher enrichment among ID disorders linked to specific signaling pathways, such as MAPK signaling (top features: ectodermal anomalies and cardiac malformations; E = 3.0), Wnt signaling (top feature: other malformations; E = 7.0), hedgehog signaling (top feature: limb anomalies; E = 9.7), and BMP signaling (top feature: vertebral and skull anomalies; E = 103.3) (Figure 4).
In summary, systematic analyses of ID-AGs permitted clinical delineation of groups of process-defined ID disorders.
Clinical Classification of ID Disorders Disentangles ID-Associated Genes into Biologically Meaningful Modules
To reveal the extent to which human phenotypes can be used for disentangling the large network of ID-AGs into biologically meaningful, physically interacting modules, we determined PIE scores for the clinical classes and for the ID-accompanying phenotype categories and asked whether these show a higher degree of connectivity than the complete group of all ID-AGs (PIE = 1.32, see above). Most clinical classes (2–5, 7, and 8b) had significantly higher PIE scores (1.7 ≤ PIE ≤ 10.8, padj < 0.05; Figure 5A and Figure S3). Comparing the total number of “within-class” PPIs with the number of interactions in randomly scrambled classes demonstrated that the disease-based classification successfully captures the molecular modularity of ID (E = 1.68, p < 0.0001). The same was true for the ID-accompanying phenotype categories (24 of 27 had PIE scores between 1.4 and 10.0 [Figure S3] and were significantly enriched in “within accompanying phenotype category” connectivity [E = 1.96; p < 0.0001]).
Figure 5.
Genomic, Proteomic, and Phenotype Datasets Define Predictive Patterns in ID
(A–E) 650 ID-AGs and clinical subsets were matched to public datasets and show patterns relating to clinical classes. (A) PPIs, (B) co-expression in BrainSpan, (C) co-expression in GTEx, (D) hPSD, and (E) autism candidate genes. Enrichment scores are provided for nine main classes (1–8b) and the total set of 650 ID-AGs (outer frame). (Benjamini-Hochberg, ∗padj < 0.05; ∗∗padj < 0.01; ∗∗∗padj < 0.001.) Note that class 8b, belonging to the SWOSM super-class column, is depicted in the third column because of symmetry reasons and because class 9 contains only a single gene.
(F) The predictive power of 650 ID-AGs to identify ID-AGs in leave-one-out analysis on the basis of proximity in the reference gene network is illustrated by standard precision-recall analysis. Precision is defined as the number of correctly predicted ID-AGs as a proportion of all genes predicted for a given recall. Recall is the proportion of all ID-AGs that are recovered. The significance of these predictions was determined by comparison with precision-recall curves obtained with number-matched random genes. These are represented by gray areas shaded to indicate the p values as shown in the legend and reveal the highly significant power of the 650 ID-AGs to predict each other from the genome-wide background.
(G) Examples of precision-recall for individual ID clinical classes and ID-accompanying phenotype categories, notably from the 650 ID-AG background. Thus, deconvoluting ID-AGs according to phenotypes results in added predictive value (compared to that of random IDA-Gs).
We next asked whether discernible patterns that validate biological coherence of clinically defined ID classes also exist in other genome-wide data. In BrainSpan gene-expression data, most clinical classes showed an elevated level of co-expression in brain when they were compared to the genomic background and all ID-AGs (Figure 5B), and the level of co-expression of ID-AGs within ID classes was significantly higher than for ID-AGs that were randomly distributed over the ID classes (E = 1.07; p = 0.012). Likewise, co-expression levels across human tissues (GTEx) were elevated among most ID classes (Figure 5C) and among 24 of 27 ID-accompanying phenotype categories (padj < 0.05 [except F, I, Ub, and E = 1.13], p < 0.0001 for ID-accompanying phenotype classes versus randomly distributed ID-AGs).
In conclusion, annotation of ID-AGs to clinical classes and ID-accompanying phenotypes demonstrated that phenotype classification can deconvolute the large group of ID-AGs into modules with elevated biological coherence.
Added Value of Human Phenotype Classification for Prediction of Disease-Associated Genes
We wondered whether publically available functional or phenotype datasets would be enriched with specific ID classes. Given that synapse biology has been proposed to play a central role in ID,29, 32 we first determined the distribution of 1,458 previously reported hPSD proteins.35 Although genes encoding hPSD proteins were highly represented among all ID-AGs (105 proteins, E = 2.37, p < 0.0001), they were particularly enriched in ID classes 1, 3, and 6 while being strikingly underrepresented in classes 4 and 7 (Figure 5D). A similar distribution (highest enrichment in non-syndromic classes 3 and 6 and striking underrepresentation in class 7) was found when we matched genes associated with co-morbid autism spectrum disorder phenotypes (SFARI database)10 to ID classes (Figure 5E). We then performed precision-recall analysis by using a recently established phenotypic-linkage network21 to determine the ability of ID-AGs to predict each other on the basis of their increased functional similarity (compared to that among random genes) (Figure 5F). The resulting precision-recall curve was highly significant (p < 0.001), reinforcing our findings that known ID-AGs show considerable functional coherence to an extent that can be exploited for prioritization and prediction of disease-associated genes.
Strikingly, testing the added predictive value of individual ID clinical classes and categories (in comparison to that of randomly selected ID-AGs) validated that particular classes of ID-AGs form sub-clusters of significantly increased coherence and power. This was true for individual main clinical classes (1, 4, and 8b), super-classes (syndromic ID with structural malformations), and ID-accompanying phenotypes (e.g., ID disorders with brain malformations, obesity, behavioral abnormalities, and limb malformations; Figure 5G and data not shown).
Together, our findings prove the importance of phenotype consideration in ID-AG prediction and demonstrate the added value of our human-phenotype-based classification system to the predictive power.
Patterns in ID Are Revealed by Custom-Made Drosophila Phenotype Data
Lastly, because human phenotypic information is often limited, we also aimed to provide proof of concept that relevant functional and phenotypic information can be generated in a customized manner. We used Drosophila melanogaster, an established model for ID genetics and pathology,36 to generate two large-scale functional, multiparametric datasets for ID-AGs annotated in an earlier version of the SysID database (388 ID-AGs) by RNAi-mediated knockdown of their fly orthologs. We chose two assays covering different functional domains: behavior and morphology. The RNAi approach is a suitable global approximation to model the human disease conditions because (partial) loss of gene function is thought to be the causative mechanism for the vast majority of these ID-AGs.37 We used a total of 570 RNAi lines, including two independent RNAi constructs per gene whenever available (Table S2), and characterized ID knockdown models (1) in a behavioral assay upon neuronal knockdown at two different time points to distinguish early- and late-onset phenotypes (Figure 6A) and (2) in a morphological assay using the wing as a model (Figures 6A and 6B and Tables S2.1–S2.6).
Figure 6.
Custom-Made Functional Datasets in Drosophila Reveal Additional Patterns
(A) Schematic representation of the neuronal screen and assessed phenotypes. Viable pan-neuronal knockdown ID models were tested at two different time points for their ability to escape from a platform.
(B) Phenotypes evaluated in the wing screen. Examples of genes (human orthologs) and associated phenotypes are shown.
(C) Phenotypes and their frequencies upon neuronal knockdown. Note that knockdown of ID-AGs (green) tended to cause early phenotypes, whereas knockdown of non-ID-AGs (gray) caused significantly more late phenotypes.
(D) Phenotypes and their frequencies upon knockdown in the wing. ID-AGs are highly enriched with lethal, posterior-margin, and wing-field phenotypes. Broad morphological phenotypes are evenly represented among ID-AGs and non-ID-AGs.
Bar graphs in (C) and (D) show genes in each phenotype group as a percentage of all genes in each dataset (264 ID-AGs in the neuronal dataset, 261 ID-AGs in the wing dataset, and 31 non-ID-AGs in both assays). The p values were determined with Fisher’s exact test and corrected for multiple testing (Benjamini-Hochberg, ∗padj < 0.05; ∗∗padj < 0.01; ∗∗∗padj < 0.001). Note that each gene can be associated with more than one phenotype.
(E and F) Enrichment of early behavioral (early walker and early sitter, E) and wing morphological phenotypes (trichome density and missing veins, F), resolved according to ID clinical classes, shows that the increased abundance of the phenotypes among ID-AGs (Figure 6C) arises from enrichment of phenotypes in specific clinical classes. (E′ and F′) Precision-recall analysis (see Figure 5 for details) shows the significant predictive power of the custom-made phenotypes to identify other ID-AG orthologs associated with the same phenotype. p value curves from number-matched, randomly sub-sampled ID-AG sets are indicated.
We found that specific Drosophila phenotypes within both screens were significantly more or less abundant among ID-AGs than among non-ID-associated control genes (Material and Methods). Upon neuronal ablation, ID-AGs showed a consistent tendency for early-onset phenotypes (developmental lethality and all early behavioral phenotypes), but not late-onset phenotypes (Figure 6C). Progressive phenotypes (which become more frequent at late testing time points than at early time points) could be observed in more than 70% of controls but in only 40% of the ID-AG set, which was probably at least partially attributable to increased developmental lethality in the latter. In general, the congenital or early-onset phenotype of ID as a (neuro)developmental disorder appears to be reflected in corresponding fly phenotypes such as developmental lethality and early-onset behavioral phenotypes. In the second screen, ID-AG ablation in the wing revealed that phenotype rates associated with ID-AGs were higher than those associated with the control gene set. In particular, specific morphological phenotypes such as trichome and vein defects were highly enriched (Figure 6D).
Resolving ID-characteristic Drosophila phenotypes such as early behavioral phenotypes and wing morphological phenotypes according to our human-ID-classification system, we found that the identified ID phenotype patterns did not arise from an overall enrichment of the fly phenotype among ID models but rather derived from specific clinical ID classes. For example, in the neuronal screen, the phenotypes “early walker” and “early sitter” derived from a high enrichment of phenotypes associated with ID-AG orthologs of classes 4 and 7 and 1, 4, and 7, respectively (Figure 6E). In the wing screen, classes 4 and 7 were highly enriched with Drosophila phenotypes “wing trichome density,” and classes 3 and 7 were enriched with “wing veins missing,” suggesting that these can be considered phenologs38 of these human phenotype classes (Figure 6F). Precision-recall analysis21 of the fly phenotypes “early sitter” and “wing veins missing” (Figures 6D and 6E), analogous to analysis of the human phenotypes (Figures 5F and 5G), revealed their predictive value (p < 0.01) (Figures 6E′ and 6F′). No striking patterns of enrichment were observed for ID-atypical Drosophila late behavioral and gross wing-growth phenotypes or for a number of other phenotypes.
Together, the data generated in Drosophila provide experimental support that ID-AGs exert important functions during development, in agreement with the strong developmental origin of the human pathologies. Moreover, relations can be established between distinct phenotypes in Drosophila and humans, providing evidence that cross-species phenotyping can contribute to ID-AG prediction and identification.
Discussion
To demonstrate that highly heterogeneous ID disorders can be systematically broken down into biologically coherent modules, we set up a curated inventory of currently known ID-AGs and their associated phenotypes, classified in a number of clinical categories and linked to various publically available and previously undescribed functional data. We provide an easily exploitable database (SysID database, see Web Resources) representing a comprehensive resource of ID-AGs, their properties, functional connectivity, and gene-phenotype relations. These aspects are fundamental to a better understanding of the molecular processes underlying cognitive (dys)function for furthering genetic diagnostics and developing treatment strategies that aim to target shared pathways and processes rather than single genes.
Apart from comprehensiveness, the main achievements of our work in comparison to those of previous studies reporting ID-AG lists8, 9, 39, 40, 41 include (1) manual curation, (2) a conservative annotation of ID-AGs only when independent evidence from several individuals exists, and (3) a strategy that integrates genes and phenotypes. In order to reduce phenotypic complexity and to create a manageable amount of clinical data, we applied a bipartite ID-classification system based on (1) the manifestation, severity, penetrance, and ‘syndromicity’ of ID and (2) recurrently reported ID-accompanying phenotypes. Because inconsistent terminology, incomplete phenotype annotation or functional knowledge, diagnostic biases in published reports, and disorders with only a few affected individuals limit reliability in systematic phenotyping and are most likely more pronounced in complex, high-resolution approaches (such as that established by the Human Phenotype Ontology42, 43), we used a limited amount of 27 ID-accompanying phenotypes covering main organ systems and features. Growing clinical data and ongoing attempts to improve annotation and curation of phenotyping and phenotype ontologies43, 44, 45 still need investment,44, 46 but they hold potential for phenomics approaches with eventually higher resolution.
This study systematically revealed quantitative overrepresentation of biological processes and molecular modules in ID and used phenotypic information to distinguish various biologically meaningful subgroups of ID-AGs. The differential representation of genes encoding synaptic (hPSD) proteins35 among clinical classes (Figure 5D), for example, is striking. Differently selected groups of affected individuals are thus likely to account for the reported discrepancies in the contribution of genes with synaptic function to ID.30, 47 Furthermore, genes associated with autism spectrum disorders show a similar pattern, in agreement with the notion that synapse biology is a major theme in these disorders.32, 48, 49
Phenotypic and Molecular Coherence in ID and Related Disorders
Our findings of phenotype-based functional modules add to widely accumulated evidence that similar clinical phenotypes are caused by genetic defects in common pathways and processes.50 However, phenotype-based systematic deconvolution of highly heterogeneous disorders has not been accomplished previously.
Interestingly, a recent study reported significant connectivity and anatomical specificity of ASD, but not ID, on the basis of analysis of a single brain region, the cerebral cortex, during a limited time window.8 Applying our analysis to the published list of ASD-associated genes (118 genes indicated as ASD, but not ID; 11 of these genes are, however, convincingly implicated in syndromic ID disorders) surprisingly revealed an overall lower molecular connectivity of ASD-associated genes (PIE = 0.89) than of random genes (PIE = 1) and our ID-AG catalog (PIE = 1.32). Likewise, co-expression in the cerebral cortex across all stages was 2.4-fold higher for all 650 ID-AGs than for the indicated ASD-associated genes and even up to more than 4-fold higher in clinical classes 1 and 4. This illustrates the extensive nature of biological coherence in ID. Given that the observed co-expression of ID-AGs can primarily be attributed to their increased co-expression in the embryonic brain (E = 1.05 [p < 0.0001] versus postnatal E = 1.02 [p = 0.06]), it will be interesting to further map ID modules onto specific embryonic stages and brain regions such as the cortex and hippocampus.
Fundamental and Translational Potential of Human and Drosophila Phenotypes
By establishing the relationships among genes, phenotypes, and molecular function, we have generated knowledge and predictive value. First, clustering ID-AGs according to their accompanying clinical features resulted in a landscape of human phenotypes (Figure 3) and suggested previously undescribed biological functions for many of them, given that genes close to one another are likely to share function. Five of 18 ID-AGs in our highlighted, red cluster in Figure 3 have not yet been annotated with cilia-related Gene Ontology terms. Strikingly, a recent study demonstrated that one of these, the GNAS (MIM: 139320)-encoded protein Gαs, is highly enriched at the primary cilium of granule neuron precursors and regulates ciliary trafficking of hedgehog-pathway components.51 HDAC8 might function in analogy to HDAC6 and regulate cilia disassembly via acetylation of tubulin.52 MAGEL2 was shown to co-immunoprecipitate with BBS4,53 a ciliary protein present in the same cluster. Second, this phenotype-based approach can also identify indirect gene functions. The turquoise-labeled cluster, enriched with genes encoding synaptic proteins, also contains transcription factors DEAF1 and FOXP1, which have already been implicated in memory and synaptic function, respectively.54, 55 Third, a specific combination of phenotypes, biological processes, and gene function allowed the recognition of typical phenotype combinations characterizing groups of process-defined ID disorders, such as chromatin-related ID disorders, ciliopathies, and ID-associated deficiencies in DNA repair. Fourth, precision-recall analyses of phenotype-defined groups of ID-AGs unambiguously demonstrated the added predictive value of phenotypes and the phenotype-based classification systems.
How can further informative phenotype data be generated in a fast, customized, and large-scale approach? Cross-species phenotyping using efficient genetic models has emerged as a promising approach with translational potential.26, 37, 56, 57 We generated two ID datasets in Drosophila. Both highlight ID-AGs with important roles in development, in perfect agreement with their association with clinical classes 1, 4, and 7 (Figures 6E and 6F) and thus with the super-class of syndromic ID with structural malformations. It is conceivable that other experimental readouts in Drosophila would produce enrichments among different clinical classes. Synapse or learning and memory phenotypes, for example, might support non-syndromic classes 3 and 6. Such patterns of a particular fly phenotype similar to a specific human phenotype could be exploited to support the causative nature of mutations in ID-associated candidate genes by rapid, custom-made assays in Drosophila.
In addition to providing insights into the biology and modularity of ID, our study has immediate translational benefit, including the broad applicability of our database, which now contains 746 high-confidence ID-AGs and associated data. The ID-AG catalog can serve as a basis for either targeted sequencing of diagnostic gene panels or evaluating data from exome or genome sequencing, as already implemented in our diagnostic centers. Furthermore, we propose exploiting the patterns identified in our study to not only contribute to the evaluation of novel ID-AG candidates but also pursue the systematic characterization of the underlying biological mechanisms.58 Because our experiments provide proof of principle that functional data with translational value can be generated in Drosophila on demand, application of efficient disease models in diagnostic settings should be encouraged. In conclusion, our gene catalog, human and cross-species phenotype annotations, integrated analyses, and flexible database provide a significant step toward overcoming current limitations in ID research and diagnostics and the basis for objective application of human phenotype and functional annotations.
Acknowledgments
We thank H.G. Brunner and A. Hoischen for critical reading of the manuscript, J. de Gooyert for input into database design and excellent technical assistance, and I. Küçükkalıpçı and K. Narcı for help with the Drosophila screens. We acknowledge the Vienna Drosophila Resource Center and Bloomington Drosophila Stock Center (NIH P40OD018537) for providing Drosophila strains. This research was supported in part by the European Union’s FP7 large-scale integrated network Gencodys (HEALTH-241995 to C.W., M.A.H., and A.S.), by the IZKF (Interdisziplinäres Zentrum für Klinische Forschung) Erlangen and grants (ZW184/1-1 and ZW184/1-2) from the DFG (Deutsche Forschungsgemeinschaft) to C.Z., by a ZonMw grant (907-00-365) from the NWO (Netherlands Organization for Scientific Research) to T.K., by a grant from the BMBF (German Federal Ministry of Education and Research; 01GS08160, funding the German Intellectual Disability Network [MRNET]), and by Vidi and TOP grants (917-96-346 and 912-12-109) from ZonMw and NWO to A.S.
Published: January 7, 2016
Footnotes
Supplemental Data include three figures and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.ajhg.2015.11.024.
Contributor Information
Martijn A. Huynen, Email: martijn.huijnen@radboudumc.nl.
Annette Schenck, Email: annette.schenck@radboudumc.nl.
Web Resources
The URLs for data presented herein are as follows:
Ensembl, http://www.ensembl.org/index.html
FlyAtlas, http://flyatlas.org/atlas.cgi
FlyBase, http://flybase.org/
Gene Ontology, http://geneontology.org/
GeneReviews, http://www.ncbi.nlm.nih.gov/books/NBK1116/
HUGO, http://www.genenames.org
Human Protein Reference Database (HPRD), http://www.hprd.org
NCBI Gene, http://www.ncbi.nlm.nih.gov/gene
Simons Foundation SFARI database, http://sfari.org/
SysID database, http://sysid.cmbi.umcn.nl/
Vienna Drosophila Resource Center (VDRC), http://stockcenter.vdrc.at/control/main
Supplemental Data
This table, exported from the SysID database, comprises main information on the IDAG catalog used throughout the paper: gene name, Entrez ID, OMIM gene number, Ensembl ID, gene group (388_ID version from mid 2010, 650_ID version from beginning of 2014), main classes, ID accompanying phenotypes, confidence criteria, disease types, disease subtypes, OMIM disease numbers and information whether the gene was screened in in-house Drosophila assays (0 = no, 1 = yes). Gene associated information was obtained as described in Material and Methods.
(2.1) Data table Drosophila neuronal screen: phenotype (column B), number of associated ID-AGs (column C), number of associated non-ID/control genes (column D), and description of phenotype (column E). The lower table provides the numbers of the tested Drosophila genes associated with the human phenotype class (main clinical classes & ID accompanying phenotypes). (2.2–2.3) Gene-phenotype matrix for neuronal screen for associated ID-AGs (2.2), and non-ID controls (2.3): human gene identifiers (column A–C), identifiers of fly orthologs and tested RNAi lines (columns D–H), and phenotype information (columns I–X). (2.4–2.6) Information on wing-screen analogous to 2.1–2.2. Phenotype information in all tables: 0 = false and 1 = true.
List of co-expression scores for 650 ID-AGs in different brain regions (BrainSpan).
References
- 1.Schalock R.L., Borthwick-Duffy S.A., Bradley V.J., Buntinx W.H.E., Coulter D.L., Craig E.M., Gomez S.C., Lachapelle Y., Luckasson R., Reeve A. American Association on Intellectual and Developmental Disabilities; 2010. Intellectual Disability: Definition, Classification, and Systems of Supports. [Google Scholar]
- 2.Ropers H.H. Genetics of early onset cognitive impairment. Annu. Rev. Genomics Hum. Genet. 2010;11:161–187. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 3.Grozeva D., Carss K., Spasic-Boskovic O., Tejada M.I., Gecz J., Shaw M., Corbett M., Haan E., Thompson E., Friend K., Italian X-linked Mental Retardation Project. UK10K Consortium. GOLD Consortium Targeted Next-Generation Sequencing Analysis of 1,000 Individuals with Intellectual Disability. Hum. Mutat. 2015;36:1197–1204. doi: 10.1002/humu.22901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Redin C., Gérard B., Lauer J., Herenger Y., Muller J., Quartier A., Masurel-Paulet A., Willems M., Lesca G., El-Chehadeh S. Efficient strategy for the molecular diagnosis of intellectual disability using targeted high-throughput sequencing. J. Med. Genet. 2014;51:724–736. doi: 10.1136/jmedgenet-2014-102554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bahi-Buisson N., Poirier K., Fourniol F., Saillour Y., Valence S., Lebrun N., Hully M., Bianco C.F., Boddaert N., Elie C., LIS-Tubulinopathies Consortium The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain. 2014;137:1676–1700. doi: 10.1093/brain/awu082. [DOI] [PubMed] [Google Scholar]
- 6.Zenker M. Clinical manifestations of mutations in RAS and related intracellular signal transduction factors. Curr. Opin. Pediatr. 2011;23:443–451. doi: 10.1097/MOP.0b013e32834881dd. [DOI] [PubMed] [Google Scholar]
- 7.Zaghloul N.A., Katsanis N. Functional modules, mutational load and human genetic disease. Trends Genet. 2010;26:168–176. doi: 10.1016/j.tig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parikshak N.N., Luo R., Zhang A., Won H., Lowe J.K., Chandran V., Horvath S., Geschwind D.H. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155:1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inlow J.K., Restifo L.L. Molecular and comparative genetics of mental retardation. Genetics. 2004;166:835–881. doi: 10.1093/genetics/166.2.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Basu S.N., Kollu R., Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sealfon R.S., Hibbs M.A., Huttenhower C., Myers C.L., Troyanskaya O.G. GOLEM: an interactive graph-based gene-ontology navigation and analysis tool. BMC Bioinformatics. 2006;7:443. doi: 10.1186/1471-2105-7-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., The Gene Ontology Consortium Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stark C., Breitkreutz B.J., Reguly T., Boucher L., Breitkreutz A., Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keshava Prasad T.S., Goel R., Kandasamy K., Keerthikumar S., Kumar S., Mathivanan S., Telikicherla D., Raju R., Shafreen B., Venugopal A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kalinka A.T., Tomancak P. linkcomm: an R package for the generation, visualization, and analysis of link communities in networks of arbitrary size and type. Bioinformatics. 2011;27:2011–2012. doi: 10.1093/bioinformatics/btr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Warnes, G.R., Bolker, B., Bonebakker, L., Gentleman, R., Huber, W., Liaw, A., Lumley, T., Maechler, M., Magnusson, A., Moeller, S., et al. (2014). gplots: Various R programming tools for plotting. https://cran.r-project.org/web/packages/gplots/index.html.
- 17.Cline M.S., Smoot M., Cerami E., Kuchinsky A., Landys N., Workman C., Christmas R., Avila-Campilo I., Creech M., Gross B. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sama I.E., Huynen M.A. Measuring the physical cohesiveness of proteins using physical interaction enrichment. Bioinformatics. 2010;26:2737–2743. doi: 10.1093/bioinformatics/btq474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.BrainSpan (2013). Atlas of the Developing Human Brain. http://www.brainspan.org.
- 20.GTEx Consortium The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013;45:580–585. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honti F., Meader S., Webber C. Unbiased functional clustering of gene variants with a phenotypic-linkage network. PLoS Comput. Biol. 2014;10:e1003815. doi: 10.1371/journal.pcbi.1003815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flicek P., Ahmed I., Amode M.R., Barrell D., Beal K., Brent S., Carvalho-Silva D., Clapham P., Coates G., Fairley S. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruan J., Li H., Chen Z., Coghlan A., Coin L.J., Guo Y., Hériché J.K., Hu Y., Kristiansen K., Li R. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 25.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–156. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 26.Neumüller R.A., Richter C., Fischer A., Novatchkova M., Neumüller K.G., Knoblich J.A. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chintapalli V.R., Wang J., Dow J.A. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2007;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 28.Schmidt I., Thomas S., Kain P., Risse B., Naffin E., Klämbt C. Kinesin heavy chain function in Drosophila glial cells controls neuronal activity. J. Neurosci. 2012;32:7466–7476. doi: 10.1523/JNEUROSCI.0349-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grant S.G. Synaptopathies: diseases of the synaptome. Curr. Opin. Neurobiol. 2012;22:522–529. doi: 10.1016/j.conb.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 30.Pavlowsky A., Chelly J., Billuart P. Emerging major synaptic signaling pathways involved in intellectual disability. Mol. Psychiatry. 2012;17:682–693. doi: 10.1038/mp.2011.139. [DOI] [PubMed] [Google Scholar]
- 31.Kleefstra T., Schenck A., Kramer J.M., van Bokhoven H. The genetics of cognitive epigenetics. Neuropharmacology. 2014;80:83–94. doi: 10.1016/j.neuropharm.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 32.Krumm N., O’Roak B.J., Shendure J., Eichler E.E. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37:95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morava E., van den Heuvel L., Hol F., de Vries M.C., Hogeveen M., Rodenburg R.J., Smeitink J.A. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823–1826. doi: 10.1212/01.wnl.0000244435.27645.54. [DOI] [PubMed] [Google Scholar]
- 34.Valente E.M., Rosti R.O., Gibbs E., Gleeson J.G. Primary cilia in neurodevelopmental disorders. Nat. Rev. Neurol. 2014;10:27–36. doi: 10.1038/nrneurol.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayés A., van de Lagemaat L.N., Collins M.O., Croning M.D., Whittle I.R., Choudhary J.S., Grant S.G. Characterization of the proteome, diseases and evolution of the human postsynaptic density. Nat. Neurosci. 2011;14:19–21. doi: 10.1038/nn.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Voet M., Nijhof B., Oortveld M.A., Schenck A. Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci. Biobehav. Rev. 2014;46:326–342. doi: 10.1016/j.neubiorev.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 37.Oortveld M.A., Keerthikumar S., Oti M., Nijhof B., Fernandes A.C., Kochinke K., Castells-Nobau A., van Engelen E., Ellenkamp T., Eshuis L. Human intellectual disability genes form conserved functional modules in Drosophila. PLoS Genet. 2013;9:e1003911. doi: 10.1371/journal.pgen.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGary K.L., Park T.J., Woods J.O., Cha H.J., Wallingford J.B., Marcotte E.M. Systematic discovery of nonobvious human disease models through orthologous phenotypes. Proc. Natl. Acad. Sci. USA. 2010;107:6544–6549. doi: 10.1073/pnas.0910200107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Betancur C. Etiological heterogeneity in autism spectrum disorders: more than 100 genetic and genomic disorders and still counting. Brain Res. 2011;1380:42–77. doi: 10.1016/j.brainres.2010.11.078. [DOI] [PubMed] [Google Scholar]
- 40.Kou Y., Betancur C., Xu H., Buxbaum J.D., Ma’ayan A. Network- and attribute-based classifiers can prioritize genes and pathways for autism spectrum disorders and intellectual disability. Am. J. Med. Genet. C. Semin. Med. Genet. 2012;160C:130–142. doi: 10.1002/ajmg.c.31330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilissen C., Hehir-Kwa J.Y., Thung D.T., van de Vorst M., van Bon B.W., Willemsen M.H., Kwint M., Janssen I.M., Hoischen A., Schenck A. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–347. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 42.Köhler S., Doelken S.C., Mungall C.J., Bauer S., Firth H.V., Bailleul-Forestier I., Black G.C., Brown D.L., Brudno M., Campbell J. The Human Phenotype Ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oti M., Huynen M.A., Brunner H.G. The biological coherence of human phenome databases. Am. J. Hum. Genet. 2009;85:801–808. doi: 10.1016/j.ajhg.2009.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deans A.R., Lewis S.E., Huala E., Anzaldo S.S., Ashburner M., Balhoff J.P., Blackburn D.C., Blake J.A., Burleigh J.G., Chanet B. Finding our way through phenotypes. PLoS Biol. 2015;13:e1002033. doi: 10.1371/journal.pbio.1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freimer N., Sabatti C. The human phenome project. Nat. Genet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- 46.Houle D., Govindaraju D.R., Omholt S. Phenomics: the next challenge. Nat. Rev. Genet. 2010;11:855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 47.Najmabadi H., Hu H., Garshasbi M., Zemojtel T., Abedini S.S., Chen W., Hosseini M., Behjati F., Haas S., Jamali P. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63. doi: 10.1038/nature10423. [DOI] [PubMed] [Google Scholar]
- 48.Bourgeron T. From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat. Rev. Neurosci. 2015;16:551–563. doi: 10.1038/nrn3992. [DOI] [PubMed] [Google Scholar]
- 49.Pinto D., Delaby E., Merico D., Barbosa M., Merikangas A., Klei L., Thiruvahindrapuram B., Xu X., Ziman R., Wang Z. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am. J. Hum. Genet. 2014;94:677–694. doi: 10.1016/j.ajhg.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oti M., Brunner H.G. The modular nature of genetic diseases. Clin. Genet. 2007;71:1–11. doi: 10.1111/j.1399-0004.2006.00708.x. [DOI] [PubMed] [Google Scholar]
- 51.He X., Zhang L., Chen Y., Remke M., Shih D., Lu F., Wang H., Deng Y., Yu Y., Xia Y. The G protein α subunit Gαs is a tumor suppressor in Sonic hedgehog-driven medulloblastoma. Nat. Med. 2014;20:1035–1042. doi: 10.1038/nm.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pugacheva E.N., Jablonski S.A., Hartman T.R., Henske E.P., Golemis E.A. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129:1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee S., Walker C.L., Karten B., Kuny S.L., Tennese A.A., O’Neill M.A., Wevrick R. Essential role for the Prader-Willi syndrome protein necdin in axonal outgrowth. Hum. Mol. Genet. 2005;14:627–637. doi: 10.1093/hmg/ddi059. [DOI] [PubMed] [Google Scholar]
- 54.Vulto-van Silfhout A.T., Rajamanickam S., Jensik P.J., Vergult S., de Rocker N., Newhall K.J., Raghavan R., Reardon S.N., Jarrett K., McIntyre T. Mutations affecting the SAND domain of DEAF1 cause intellectual disability with severe speech impairment and behavioral problems. Am. J. Hum. Genet. 2014;94:649–661. doi: 10.1016/j.ajhg.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bacon C., Schneider M., Le Magueresse C., Froehlich H., Sticht C., Gluch C., Monyer H., Rappold G.A. Brain-specific Foxp1 deletion impairs neuronal development and causes autistic-like behaviour. Mol. Psychiatry. 2015;20:632–639. doi: 10.1038/mp.2014.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto S., Jaiswal M., Charng W.L., Gambin T., Karaca E., Mirzaa G., Wiszniewski W., Sandoval H., Haelterman N.A., Xiong B. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnorrer F., Schönbauer C., Langer C.C., Dietzl G., Novatchkova M., Schernhuber K., Fellner M., Azaryan A., Radolf M., Stark A. Systematic genetic analysis of muscle morphogenesis and function in Drosophila. Nature. 2010;464:287–291. doi: 10.1038/nature08799. [DOI] [PubMed] [Google Scholar]
- 58.Shendure J. Life after genetics. Genome Med. 2014;6:86. doi: 10.1186/s13073-014-0086-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
This table, exported from the SysID database, comprises main information on the IDAG catalog used throughout the paper: gene name, Entrez ID, OMIM gene number, Ensembl ID, gene group (388_ID version from mid 2010, 650_ID version from beginning of 2014), main classes, ID accompanying phenotypes, confidence criteria, disease types, disease subtypes, OMIM disease numbers and information whether the gene was screened in in-house Drosophila assays (0 = no, 1 = yes). Gene associated information was obtained as described in Material and Methods.
(2.1) Data table Drosophila neuronal screen: phenotype (column B), number of associated ID-AGs (column C), number of associated non-ID/control genes (column D), and description of phenotype (column E). The lower table provides the numbers of the tested Drosophila genes associated with the human phenotype class (main clinical classes & ID accompanying phenotypes). (2.2–2.3) Gene-phenotype matrix for neuronal screen for associated ID-AGs (2.2), and non-ID controls (2.3): human gene identifiers (column A–C), identifiers of fly orthologs and tested RNAi lines (columns D–H), and phenotype information (columns I–X). (2.4–2.6) Information on wing-screen analogous to 2.1–2.2. Phenotype information in all tables: 0 = false and 1 = true.
List of co-expression scores for 650 ID-AGs in different brain regions (BrainSpan).