Abstract
Background
The present study aimed to measure miR-137 expression in patients with cutaneous melanoma (CM) and to estimate the correlation of miR-137 expression and the prognosis of CM patients.
Material/Methods
The expression level of miR-137 was assayed by quantitative real-time PCR (qRT-PCR) and presented as mean ±SD. Chi-square was used to evaluate the relationship between miR-137 expression and clinical characteristics. We used a Kaplan-Meier survival curve to determine the overall survival rate of CM patients. Moreover, the correlation between miR-137 expression and the prognosis of CM patients was confirmed by Cox regression analysis.
Results
The relative expression of miR-137 in CM tissue was 1.59±0.43, while that in paired normal tissue was 2.41±0.54, which was significantly higher. Chi-square analysis showed statistical significance between miR-137 expression and clinical characteristics such as TNM stage, ulcer, and occurrence site (P<0.05). However, no association was found between miR-137 expression and age, sex, or family history (P>0.05). According to the survival curve outcome, patients with low miR-137 expression showed relatively higher mortality (P=0.000) and multivariate analysis verified that low expression of miR-137 predicted poor prognosis of CM patients (HR=8.531, 95% CI=2.950–24.668, P=0.000).
Conclusions
Compared with paired normal tissues, miR-137 expression was lower in CM tissues. Patients with low miR-137 expression had higher mortality than those with high miR-137 expression, suggesting that low miR-137 expression indicated poor prognosis for CM patients.
MeSH Keywords: Administration, Cutaneous; MicroRNAs; Prognosis
Background
Skin cancer is one of the most common malignancies among people, whose incidence exceeds the sum of other malignant tumors [1–3]. Cutaneous melanoma (CM) is a member of skin cancers and is of high malignancy in clinical. It accounts for about 6.8–20% of skin cancers, with an increasing incidence of 3–8% every year [4,5]. CM is becoming the leading cause of skin cancer-related deaths. Therapy for CM is mainly surgery, and sometimes chemo- or radio- therapy is also used [6–8]. Because of its early metastasis and high mortality, it is important to diagnose and treat CM as early as possible [9]. In addition, most CM cases suffer from poor prognosis. Therefore, novel biomarkers are urgently needed for therapy and prognosis of CM patients.
MicroRNAs (miRNAs) are a kind of noncoding RNAs and consist of about 21–23 nucleotides, which can regulate the gene expression at post-transcriptional level by specific binding to the messenger ribonucleic acids (mRNAs) [10,11]. They also play important roles on some other aspects, such as differentiation and apoptosis of cells, biological development, and disease processes [12–14]. MiRNAs offer great potential as cancer biomarkers.
miR-137, located on chromosome 1p22 [15], is a brain-enriched micro-RNA with important roles during neurogenesis, including the proliferation and differentiation of neural stem cells, the regulation of dendritic morphogenesis, and synaptic plasticity [16]. Several existing reports confirmed that miR-137 participated in the development and processes of various cancers, such as gastric cancer, head and neck cancer, colorectal cancer, and brain tumors [17–20]. Previous studies have also demonstrated that miR-137 expression is associated with melanoma progression [21,22]. Bemis et al. first reported that miR-137 downregulated MITF, which is an important regulator in melanocyte development [23]. Recently, Chen et al. demonstrated that miR-137 inhibited melanoma cell proliferation by down-regulation of MITF and cyclin-dependent kinase 6 [24].
Although considerable evidence has confirmed that miR-137 can serve as a cancer suppressor or candidate site for cancer therapies, proof of a link between miR-137 and its clinical value in CM is still lacking. Therefore, additional studies are needed to elucidate the function of miR-137 in CM. In this study, we aimed to explore the miR-137 expression in CM tissues and paired normal tissues and evaluate its possibility as a prognostic biomarker for CM patients.
Material and Methods
Patients and samples
A total of 97 patients, who were diagnosed with malignant cutaneous melanoma at Guangdong General Hospital, were enrolled in this study. Among them, 55 were males and the others were females. Clinical characteristics of CM patients were collected based on the hospital records. Our study was approved by the Ethics Committee of Guangdong General Hospital. Patients selected in the study all signed consent forms. Fresh CM tissues and paired normal tissues were reserved to use immediately after resection.
A 5-year follow-up was performed with all CM patients. The data, including age, sex, family history, TNM stage, ulcer, and occurrence site, were recorded in a database. The information about follow-up was updated every 3 months by telephone or questionnaire.
RNA extraction and qRT-PCR
Total RNA was isolated and purified from tissues by Trizol Reagent (Ambion, Foster City). Then the RNA was used to synthesize the cDNAs of miR-137 with M-MLV reverse transcriptase (Promega, Madison, WI) according to the manufacturer’s directions. The reverse transcriptive products were amplified and assayed by qRT-PCR. Each sample was determined in triplicate under optimal conditions. U6 was used as an endogenous standard. The relative expression of miR-137 was calculated with cycle threshold (CT) method and was normalized to U6. The data are presented as mean ± standard deviation (SD).
Statistical analysis
All computations were carried out with SPSS 18.0 software. The relationship of miR-137 expression and clinical characteristics was confirmed by chi-square testing. Kaplan-Meier survival analysis was performed to estimate the overall survival rate of CM patients, and the differences between the survival curves were tested by using the log-rank test. Correlation between miR-137 expression and prognosis of CM patients was evaluated by Cox regression analysis. Statistical difference was considered to be significant when P value was less than 0.05.
Results
Down-regulation of miR-137 in CM tissues
We used qRT-PCR to determine the expression of miR-137. The relative expression of miR-137 in CM tissues was 1.59±0.43 (mean ±SD), while that in normal tissues was 2.41±0.54, indicating that miR-137 expression was lower in CM tissues (Figure 1, P<0.05).
Figure 1.
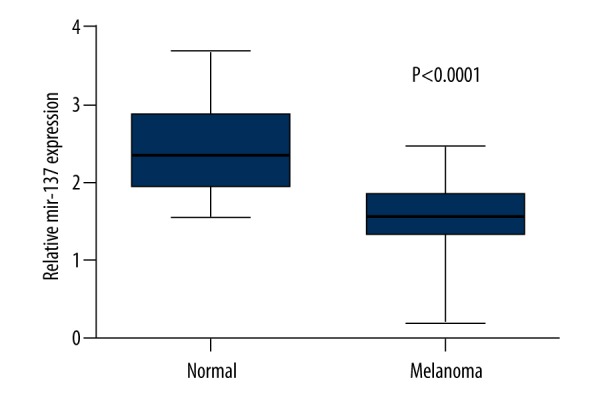
The expression levels of miR-137 both in CM tissues and paired normal tissues were measured by qRT-PCR. The expression levels of miR-137 were presented as mean ±SD. The result showed that miR-137 expression was lower in CM tissues by comparison with normal tissues.
Relationship between miR-137 expression and clinical characteristics
Further detection was conducted to explore the potential relationship of the miR-137 expression and clinical characteristics. We manually divided the CM tissues into 2 groups: the low expression group had a miR-137 expression level of less than 1.35, and others belonged to the high expression group. miR-137 expression was tightly associated with clinical characteristics, such as TNM stage, ulcer, and occurrence site (P<0.05). However, no significant relation was found between miR-137 expression and age, sex, and family history (Table 1, P>0.05).
Table 1.
The association between clinical characteristics and the miR-137 expression.
| Clinical characteristics | Case (n) | miR-137 expression | χ2 | P value | |
|---|---|---|---|---|---|
| Low | High | ||||
| Age (years) | 0.001 | 0.997 | |||
| ≤20 | 60 | 42 | 18 | ||
| >20 | 37 | 26 | 11 | ||
| Gender | 0.039 | 0.843 | |||
| Male | 55 | 39 | 16 | ||
| Female | 42 | 29 | 13 | ||
| Family history | 1.712 | 0.191 | |||
| Yes | 50 | 38 | 12 | ||
| No | 47 | 30 | 17 | ||
| TNM stage | 5.882 | 0.015 | |||
| I, II | 52 | 31 | 21 | ||
| III | 45 | 37 | 8 | ||
| Ulcer | 3.923 | 0.048 | |||
| Yes | 45 | 36 | 9 | ||
| No | 52 | 32 | 20 | ||
| Occurrence site | 5.432 | 0.020 | |||
| Trunk + head | 51 | 41 | 10 | ||
| Extremities | 46 | 27 | 19 | ||
Low expression of miR-137 presented poor prognosis in CM patients
A follow-up was conducted to estimate the overall survival rate of CM patients. The follow-up lasted for about 60 months and 53 patients died. Among the dead patients, 49 were in the low miR-137 expression group and the rest were in the high miR-137 expression group. Kaplan-Meier survival curve showed that the CM patients with low miR-137 expression presented significantly shorter survival time compared to those with high miR-137 expression (Figure 2, P=0.000). In addition, the correlation of miR-137 expression and prognosis of CM patients was evaluated by Cox regression analysis and the results revealed that low expression of miR-137 was an independent prognostic marker of CM patients (Table 2, HR=8.531, 95% CI=2.950–24.668, P=0.000).
Figure 2.
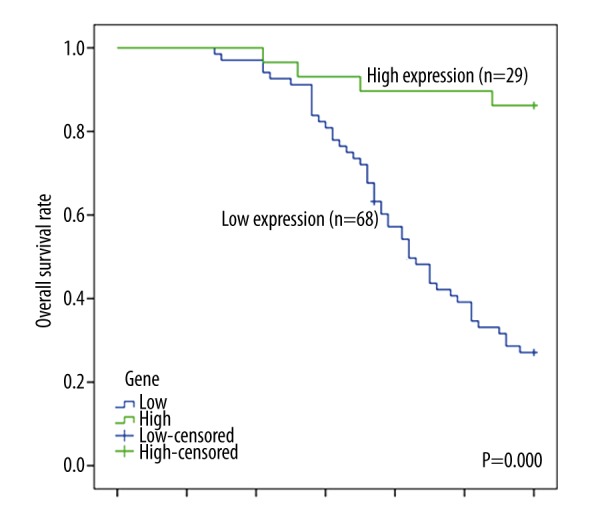
The overall survival rate of CM patients was evaluated by Kaplan-Meier survival analysis. Patients with low miR-137 expression showed higher mortality than those with high miR-137 expression.
Table 2.
The multivariate analysis of prognostic factors in CM.
| Clinical characteristics | P value | HR | 95% CI |
|---|---|---|---|
| TNM stage | 0.486 | 1.313 | 0.610–2.823 |
| Occurrence site | 0.889 | 1.052 | 0.518–2.133 |
| miR-137 expression | 0.000 | 8.531 | 2.950–24.668 |
Discussion
CM is a common skin tumor that derives from excessive hyperplasia of abnormal melanocytes. Clinical manifestations include bleeding, itching, tenderness, and ulcer [25]. Because of its high malignancy, incidence, and mortality, CM has drawn more and more attention. Previous studies have demonstrated that malignant melanoma patients have a very poor prognosis, with median survival of 6–10 months and a 5-year survival rate of <5% [26]. It is urgent to find new strategies and novel indicators to improve the diagnosis, treatment, and prognosis of CM patients. In recent years, miRNAs have been recognized as important factors in various cancers, including CM.
miRNAs are single-stranded fragments with about 21 nucleotides and they participate in many kinds of cell regulation. They have no open reading frame and encode no protein. Each miRNA may have several target genes or several miRNAs that co-regulate the same gene. These agents make miRNA tightly related to metabolism, cell cycle, cell differentiation, cell apoptosis, and development of individuals. The misregulation of miRNAs has been verified to be related with cancer initiation, promotion, and progression [27,28]. Functionally, miRNAs can serve as diagnostic and prognostic biomarkers for cancers. Since miR-137 was first found as a regulator via targeting MITF [23], there has been increasing interest in the role of miR-137 in the development and progression of different tumors. Many studies have reported that miR-137 presented significant down-regulation in various cancers [29,30].
In the present study, we investigated the miR-137 expression in CM tissues and evaluated the relationship between miR-137 expression and the prognosis of CM patients. The results showed that the miR-137 expression level in CM tissues was lower than in paired normal tissues, which was in accord with existing reports. The association of miR-137 expression and clinical characteristics was evaluated by chi-square. The result revealed that miR-137 expression was closely related with TNM stage, ulcer and occurrence site. miR-137 has been proved to be a tumor suppressor in several diseases. Zhang et al. verified that miR-137 acted as a tumor suppressor in non-small cell lung cancer [31]. Zheng et al. showed that miR-137 serves as a suppressor for gastric cancer [32]. Based on previous evidence, further investigations were conducted in this study. The survival curve showed that patients with low miR-137 expression had shorter survival time than those with high miR-137 expression. Moreover, the Cox regression analysis showed statistical significance between miR-137 expression and prognosis of CM patients, suggesting miR-137 was a target biomarker for CM. Thus, we presumed that miR-137 might act as a prognostic factor for CM patients.
Although several targets of miR-137 have been identified in various cancers, the mechanism of miR-137 on CM remains unclear. Cheng et al. demonstrated that miR-137 functioned on gastric carcinoma by targeting PI3K/AKT signaling pathway [17]. Shin et al. [33] illustrated that miR-137 could regulate the p53 level in human keratinocytes. Besides, some research outcomes indicated that c-Met and YB1 were candidate targets of miR-137 in malignancy melanoma [34]. Interestingly, a recent study has shown that c-Met expression was decreased by transfection of miR-137 in the melanoma cells [24]. Therefore, we conjectured that miR-137 might affect CM by regulating the expression of its candidate target genes like MITF, c-Met and YB1, or by regulating some signaling pathways. This hypothesis needs to be validated by more studies, which might provide directions for future research.
Conclusions
There was a statistically significant relationship between miR-137 expression and clinical characteristics. Low expression of miR-137 predicted poor prognosis for CM patients. Thus, miR-137 could be regarded as a prognostic biomarker for CM patients.
Footnotes
Source of support: Departmental sources
References
- 1.Tierney P, de Gruil FR, Ibbotson S, Moseley H. Predicted increased risk of squamous cell skin cancer induction associated with sunbed exposure habits. Br J Dermatol. 2015;173(1):201–8. doi: 10.1111/bjd.13714. [DOI] [PubMed] [Google Scholar]
- 2.Kim Y, He YY. Ultraviolet radiation-induced non-melanoma skin cancer: Regulation of DNA damage repair and inflammation. Genes Dis. 2014;1:188–98. doi: 10.1016/j.gendis.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt SA, Schmidt M, Mehnert F, et al. Use of antihypertensive drugs and risk of skin cancer. J Eur Acad Dermatol Venereol. 2015;29(8):1545–54. doi: 10.1111/jdv.12921. [DOI] [PubMed] [Google Scholar]
- 4.Garay T, Kenessey I, Molnar E, et al. Prenylation inhibition-induced cell death in melanoma: Reduced sensitivity in BRAF mutant/PTEN wild-type melanoma cells. PLoS One. 2015;10:e0117021. doi: 10.1371/journal.pone.0117021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Junco JJ, Mancha-Ramirez A, Malik G, et al. Ursolic acid and resveratrol synergize with chloroquine to reduce melanoma cell viability. Melanoma Res. 2015;25(2):103–12. doi: 10.1097/CMR.0000000000000137. [DOI] [PubMed] [Google Scholar]
- 6.De Araujo ES, Kashiwabara AY, Achatz MI, et al. LINE-1 hypermethylation in peripheral blood of cutaneous melanoma patients is associated with metastasis. Melanoma Res. 2015;25(2):173–77. doi: 10.1097/CMR.0000000000000141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gallegos Hernandez JF, Nieweg OE. [Cutaneous Melanoma (CM): Current Diagnosis and Treatment]. Gac Med Mex. 2014;150(Suppl 2):175–82. [in Spanish] [PubMed] [Google Scholar]
- 8.Kaiser S, MacPherson MB, James TA, et al. Exploratory use of docetaxel loaded acid-prepared mesoporous spheres for the treatment of malignant melanoma. Cancer Nanotechnol. 2015;6:1. doi: 10.1186/s12645-015-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rajanna S, Rastogi I, Wojdyla L, et al. Current molecularly targeting therapies in NSCLC and melanoma. Anticancer Agents Med Chem. 2015;15(7):856–68. doi: 10.2174/1871520615666150202100130. [DOI] [PubMed] [Google Scholar]
- 10.Hudcova K, Trnkova L, Kejnovska I, et al. Novel biophysical determination of miRNAs related to prostate and head and neck cancers. Eur Biophys J. 2015;44(3):131–38. doi: 10.1007/s00249-015-1008-y. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Feng J, Tang L, et al. The Regulation and Function of miR-21-FOXO3a-miR-34b/c Signaling in Breast Cancer. Int J Mol Sci. 2015;16:3148–62. doi: 10.3390/ijms16023148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, Zeng Y, Wu S, et al. MiR-204, down-regulated in retinoblastoma, regulates proliferation and invasion of human retinoblastoma cells by targeting CyclinD2 and MMP-9. FEBS Lett. 2015;589(5):645–50. doi: 10.1016/j.febslet.2015.01.030. [DOI] [PubMed] [Google Scholar]
- 13.Ge S, Xie J, Liu F, He J. MicroRNA-19b reduces hepatic stellate cell proliferation by targeting GRB2 in hepatic fibrosis models in vivo and in vitro as part of the inhibitory effect of estradiol. J Cell Biochem. 2015;116(11):2455–64. doi: 10.1002/jcb.25116. [DOI] [PubMed] [Google Scholar]
- 14.Qian J, Li R, Wang YY, et al. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403(1–2):33–41. doi: 10.1007/s11010-015-2334-1. [DOI] [PubMed] [Google Scholar]
- 15.Bemis LT, Chen R, Amato CM, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–68. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 16.Ma G, Yin J, Fu J, et al. Association of a miRNA-137 polymorphism with schizophrenia in a Southern Chinese Han population. Biomed Res Int. 2014;2014:751267. doi: 10.1155/2014/751267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng Y, Li Y, Liu D, et al. miR-137 effects on gastric carcinogenesis are mediated by targeting Cox-2-activated PI3K/AKT signaling pathway. FEBS Lett. 2014;588:3274–81. doi: 10.1016/j.febslet.2014.07.012. [DOI] [PubMed] [Google Scholar]
- 18.Liu M, Lang N, Qiu M, et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128:1269–79. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- 19.Langevin SM, Stone RA, Bunker CH, et al. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer. 2011;117:1454–62. doi: 10.1002/cncr.25689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silber J, Lim DA, Petritsch C, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller DW, Rehli M, Bosserhoff AK. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J Invest Dermatol. 2009;129:1740–51. doi: 10.1038/jid.2008.452. [DOI] [PubMed] [Google Scholar]
- 22.Philippidou D, Schmitt M, Moser D, et al. Signatures of microRNAs and selected microRNA target genes in human melanoma. Cancer Res. 2010;70:4163–73. doi: 10.1158/0008-5472.CAN-09-4512. [DOI] [PubMed] [Google Scholar]
- 23.Bemis LT, Chen R, Amato CM, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–68. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- 24.Chen X, Wang J, Shen H, et al. Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci. 2011;52:1193–99. doi: 10.1167/iovs.10-5272. [DOI] [PubMed] [Google Scholar]
- 25.Luk NM, Ho LC, Choi CL, et al. Clinicopathological features and prognostic factors of cutaneous melanoma among Hong Kong Chinese. Clin Exp Dermatol. 2004;29:600–4. doi: 10.1111/j.1365-2230.2004.01644.x. [DOI] [PubMed] [Google Scholar]
- 26.Tsao H, Atkins MB, Sober AJ. Management of cutaneous melanoma. N Engl J Med. 2004;351:998–1012. doi: 10.1056/NEJMra041245. [DOI] [PubMed] [Google Scholar]
- 27.Yan SY, Chen MM, Li GM, et al. MiR-32 induces cell proliferation, migration, and invasion in hepatocellular carcinoma by targeting PTEN. Tumour Biol. 2015;36(6):4747–55. doi: 10.1007/s13277-015-3124-9. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Tao W, Ni S, et al. Tumor-suppressive microRNA-145 induces growth arrest by targeting SENP1 in human prostate cancer cells. Cancer Sci. 2015;106(4):375–82. doi: 10.1111/cas.12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bier A, Giladi N, Kronfeld N, et al. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget. 2013;4:665–76. doi: 10.18632/oncotarget.928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li KK, Yang L, Pang JC, et al. MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol. 2013;23:426–39. doi: 10.1111/bpa.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang B, Liu T, Wu T, et al. microRNA-137 functions as a tumor suppressor in human non-small cell lung cancer by targeting SLC22A18. Int J Biol Macromol. 2014;74C:111–18. doi: 10.1016/j.ijbiomac.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Zheng X, Dong J, Gong T, et al. MicroRNA library-based functional screening identified miR-137 as a suppresser of gastric cancer cell proliferation. J Cancer Res Clin Oncol. 2015;141(5):785–95. doi: 10.1007/s00432-014-1847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin KH, Pucar A, Kim RH, et al. Identification of senescence-inducing microRNAs in normal human keratinocytes. Int J Oncol. 2011;39:1205–11. doi: 10.3892/ijo.2011.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo C, Tetteh PW, Merz PR, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol. 2013;133:768–75. doi: 10.1038/jid.2012.357. [DOI] [PubMed] [Google Scholar]


