Abstract
In Noonan Syndrome (NS) 30% to 50% of subjects show cognitive deficits of unknown etiology and with no known treatment. Here, we report that knock-in mice expressing either of two NS-associated Ptpn11 mutations show hippocampal-dependent spatial learning impairments and deficits in hippocampal long-term potentiation (LTP). In addition, viral overexpression of the PTPN11D61G in adult hippocampus results in increased baseline excitatory synaptic function, deficits in LTP and spatial learning, which can all be reversed by a MEK inhibitor. Furthermore, brief treatment with lovastatin reduces Ras-Erk activation in the brain, and normalizes the LTP and learning deficits in adult Ptpn11D61G/+ mice. Our results demonstrate that increased basal Erk activity and corresponding baseline increases in excitatory synaptic function are responsible for the LTP impairments and, consequently, the learning deficits in mouse models of NS. These data also suggest that lovastatin or MEK inhibitors may be useful for treating the cognitive deficits in NS.
Introduction
Noonan syndrome (NS) is an autosomal dominant genetic disorder with an incidence of ~1 in 2,500 live births characterized by facial abnormalities, short stature, motor delay and cardiac defects1, 2. Importantly, 30% to 50% of NS patients show cognitive deficits3–6. NS patients also show clumsiness, motor delay, hearing loss, deficits in spatial knowledge, planning, and social/emotional problems3, 4, 7. Recent studies showed that NS patients show impairments in hippocampal-dependent memory tasks4, 8, 9.
Germ line mutations in genes involved in Ras-Erk signaling such as PTPN11, SOS1, KRAS, NRAS, RAF1, BRAF, SHOC2, MEK1 and CBL have been reported to cause NS1, 10. Among those, mutations in the PTPN11 gene, which encodes the non-receptor protein tyrosine phosphatase SHP2, account for ~ 50% of NS cases1. SHP2 is a positive regulator for Ras-Erk signaling11 which is critically involved in many cellular processes including learning and memory12. The PTPN11 mutations found in NS patients result in gain-of-function alleles that up-regulate this signaling cascade11, 13–15. Cognitive problems, such as learning disabilities and memory impairments, are common in NS3, 5, 6. However, little is known about the role of PTPN11 in synaptic plasticity and learning and memory in the mammalian brain. Furthermore, there is no available treatment for cognitive deficits associated with this common genetic disorder.
Previous studies, that used NS mouse models derived by knocking-in mutations in the NS-associated Ptpn11 gene, demonstrated that the heterozygous knock-in mice show phenotypes similar to those found in NS patients. These include short stature, craniofacial abnormalities, myeloproliferative disease and multiple cardiac defects14, 16. In the present study, we first tested whether NS mutant mice have deficits in learning and memory and synaptic plasticity. Then, we asked whether increasing SHP2 activity in adult brain affects synaptic function, LTP and learning and memory. Finally, we examined whether it is possible to rescue the LTP and learning deficits of NS mutant mice in adults.
Results
NS mutant mice show deficits in spatial learning and memory
To investigate the underlying mechanism of the learning and memory deficits associated with NS, we studied two lines of heterozygous knock-in mice harboring gain-of-function mutations found in NS patients14, 16: Ptpn11D61G/+ and Ptpn11N308D/+. Previous studies showed that the Ptpn11D61G/+ mutation causes more severe phenotypes than the Ptpn11N308D/+ mutation14, 16. As NS patients show deficits in spatial function and in memory tasks dependent on the hippocampus4, 8, 9, we tested both Ptpn11 mutants in the hidden platform-version of the Morris water maze17. In this task, mice learn to use spatial cues around a pool to find an escape platform hidden beneath the water surface. Following training, memory is assessed in probe trials wherein the mice search for 60 seconds with the platform removed from the pool. Ptpn11N308D/+ mutants showed comparable latencies to find the hidden platform to their wild-type (WT) controls during training (Fig. 1a; Repeated measures ANOVA, F1, 18 = 2.078, P = 0.167) and showed normal swimming speeds in probe trials (WT, 17.33 ± 1.55 cm/s, n = 11 mice; Ptpn11N308D/+, 18.37 ± 0.82 cm/s, n = 9 mice; unpaired two-tailed t-test, t = 0.554, P = 0.586). However, in probe trials Ptpn11N308D/+ mutants spent significantly less time than WT mice in the target quadrant where the platform was located during training (Fig. 1b; WT, 57.87 ± 4.83 %; Ptpn11N308D/+, 41.85 ± 4.30 %; unpaired two-tailed t-test, t = 2.421, * P < 0.05). Also, the searches of WT mice during the probe trials were closer to the target platform than those of the mutants (WT, 32.53 ± 2.26 cm; Ptpn11N308D/+, 40.18 ± 2.05 cm; unpaired two-tailed t-test, t = 2.450, * P < 0.05). In contrast, Ptpn11N308D/+ mutants performed normally in the visible-platform version of the water maze (Supplementary Fig. 1), suggesting that the Ptpn11N308D/+ mutation does not impair either visuomotor function or motivation. After extended training, the Ptpn11N308D/+ mutants reached a level of performance comparable to WT mice in probe trials, demonstrating that they can acquire spatial information, albeit at a slower rate than WT mice (Supplementary Fig. 2). In addition, Ptpn11N308D/+ mutants also showed deficits in contextual fear conditioning, another hippocampus-dependent task (Supplementary Fig. 3).
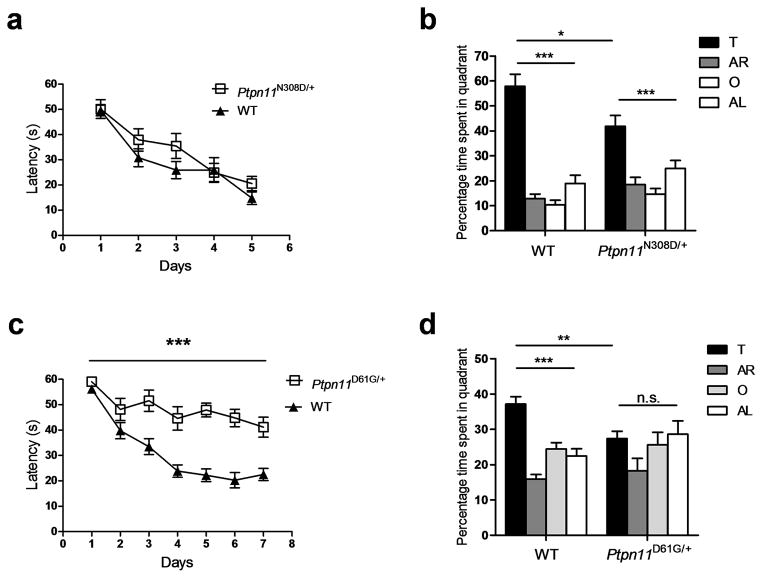
Figure 1. NS mice show spatial memory deficits.
a. Escape latencies of Ptpn11N308D/+ (n = 9) and WT littermates (n = 11) were not different in the hidden platform version of the water maze.
b. Ptpn11N308D/+ and WT littermates selectively searched in the target quadrant in a probe trial given after 3 days of training (Ptpn11N308D/+, n = 9 mice, One-way ANOVA, F3, 32 = 13.82, *** P < 0.001; WT, n = 11 mice, One-way ANOVA, F3, 40 = 48.48, *** P < 0.001). However, Ptpn11N308D/+ mice spent significantly less time in the target quadrant than WT mice. Two-way ANOVA for quadrant occupancy with genotype as between-subjects factor and pool quadrant as within-subjects factor, genotype x pool quadrant interaction: F3,54 = 4.091, * P < 0.05. Pool quadrants; target (T), adjacent right (AR), opposite (O), and adjacent left (AL) quadrant.
c. Ptpn11D61G/+ mutants (n = 10) showed significantly longer latency to the platform during training compared with WT controls (n = 15) in the hidden–platform version of the water maze.
d. Quadrant occupancy for the probe trial conducted after 3 days of training reveals that Ptpn11D61G/+ mice (n=10) did not show preference for the target quadrant, but their WT littermates (n=15) did. In addition, Ptpn11D61G/+ mice spent significantly less time in the target quadrant than did WT mice (Ptpn11D61G/+, 27.44 ± 2.04 %; WT, 37.14 ± 2.09, ** P < 0.01; unpaired two-tailed t-test). Two-way ANOVA for quadrant occupancy with genotype as between-subjects factor and pool quadrant as within-subjects factor, genotype x pool quadrant interaction: F3, 69 = 2.884, * P < 0.05. n.s., not significant (P > 0.05).
In agreement with the greater severity of phenotypes associated with the D61G mutation compared with the N308D mutation in both mutant mice and NS subjects6, 15, 16, 18, Ptpn11D61G/+ mice showed a more severe behavioral phenotype than Ptpn11N308D/+ mice. In probe trials, Ptpn11D61G/+ mice did not search selectively in the target quadrant (Fig. 1d; F3,36 = 2.029, P = 0.127 and F3,56 = 23.51, *** P < 0.0001 for Ptpn11D61G/+ and WT, respectively; one-way ANOVA) and spent more time searching further from the former platform location than did WT littermates (WT, 46.23 ± 1.29 cm, n = 15 mice; Ptpn11D61G/+, 52.43 ± 2.14 cm, n = 10 mice; unpaired two-tailed t-test, t = 3.178, * P < 0.05). Even with additional training, Ptpn11D61G/+ mice were unable to reach WT performance levels (Supplementary Fig. 2). Furthermore, Ptpn11D61G/+ mice took longer to reach the platform during training for both the hidden (Fig. 1c; Repeated measures ANOVA, F1, 23 = 38.54, *** P < 0.0001) and the visible-platform versions (Supplementary Fig. 1) of the Morris water maze, and showed slower swimming speeds (Ptpn11D61G/+, 11.98 ± 1.27 cm/s, n = 10; WT, 19.72 ± 0.46 cm/s, n = 15; unpaired two-tailed t-test, t = 6.618, *** P < 0.0001), which might have contributed to their longer latencies to reach the platform. Additional behavioral characterization in an open field test revealed that Ptpn11D61G/+ mice were hypoactive (Supplementary Fig. 1). These data demonstrate that the behavioral deficits of Ptpn11D61G/+ mice go beyond spatial learning and memory abnormalities. Importantly, the phenotype of NS patients also is not limited to cognitive deficits and can include other neurologic abnormalities, such as a higher rates of motor delay, clumsiness and poor coordination2.
NS mutant mice show deficits in synaptic plasticity
Hippocampal long-term potentiation (LTP) in the Schaffer collateral synapses of CA1 cells has a key role in spatial learning and memory19. To identify the mechanism responsible for the learning and memory deficits caused by the Ptpn11 mutations, we examined CA1 Schaffer collateral LTP in Ptpn11N308D/+ and Ptpn11D61G/+ mice by performing extracellular field recordings in acute hippocampal slices. Ptpn11N308D/+ and WT slices showed no significant differences in basal synaptic transmission or paired-pulse facilitation (Supplementary Fig. 4). However, LTP induced with theta-burst stimulation (TBS; 2 or 5 theta bursts) was significantly reduced in Ptpn11N308D/+ mice (Fig. 2a; last 10 min of recording, WT, 159.5 ± 4.23 %, n=6 slices from 6 mice; Ptpn11N308D/+, 143.4 ± 4.81 %, n=6 slices from 6 mice; unpaired two-tailed t-test, t = 2.506, P < 0.05; Supplementary Fig. 5). Consistent with the hypothesis that these LTP deficits account for the learning impairments in Ptpn11 mutant mice, Ptpn11D61G/+ mice, with the bigger learning impairments, also showed more severe LTP deficits than those in Ptpn11N308D/+ mice (Fig. 2b; last 10 min of recording, WT, 139.2 ± 8.41 %,, n=7 slices from 7 mice; Ptpn11D61G/+, 110.8 ± 6.30 %, n=7 slices from 6 mice; unpaired two-tailed t-test, t = 2.698, P < 0.05). As in Ptpn11N308D/+ mice, basal synaptic transmission and paired-pulse facilitation were normal in Ptpn11D61G/+ mutants (Supplementary Fig. 4).
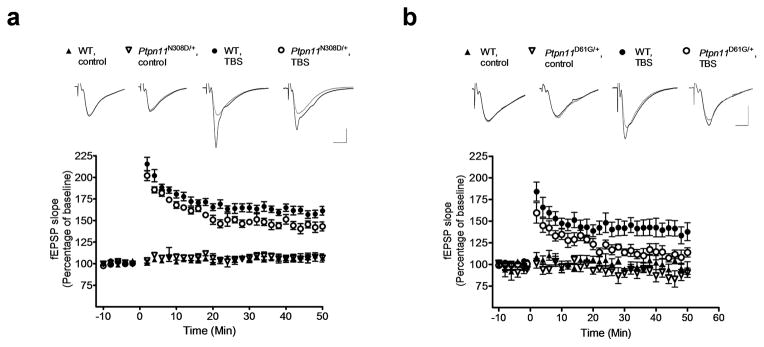
Figure 2. NS mice show LTP deficits.
a. LTP induced by a 5 TBS was reduced significantly in hippocampal slices from Ptpn11N308D/+ mice compared with their WT littermates (WT, n = 6 slices from 6 mice; Ptpn11N308D/+, n = 6 slices from 6 mice; Repeated-measures ANOVA: F1, 10 = 7.893, P < 0.05).
b. LTP induced by a 5 TBS protocol was reduced in hippocampal slices from Ptpn11D61G/+ mice compared with those from WT mice (WT, n = 7 slices from 7 mice; Ptpn11D61G/+, n = 7 slices from 6 mice; Repeated-measures ANOVA: F1,12 = 5.828, P < 0.05). fEPSP slopes normalized to the average baseline response before LTP induction (at time 0) are plotted in 2-min blocks. Sample traces show responses during baseline (gray) and the last 10 min (black) of the recording (average of ten recording traces). Scale: vertical bar, 0.5 mV; horizontal bar, 4 ms. Error bars represent s.e.m.
Adult expression of PTPN11D61G impairs LTP and memory
The mutations in Ptpn11 mice are present throughout development, affect the entire body and could disrupt the function of brain structures other than the hippocampus. Similarly, NS is a systemic developmental disorder, and it has been assumed that developmental defects are responsible for the cognitive deficits in these patients20. Viral vectors provide spatial and temporal regulation of gene expression critical for testing the specific role of Ptpn11 mutations in the adult brain. Moreover, NS alleles severely compromise the viability of mutant mice14, thus making it very difficult to obtain sufficient number of mutant mice for all studies envisioned (Supplementary table 1). To test whether altered Shp2 signaling in the adult hippocampus can cause LTP and, consequently, learning deficits, we overexpressed mutant PTPN11D61G using recombinant adeno-associated virus (AAV–PTPN11D61G) in the CA fields (CA1, CA2 and CA3) of the hippocampus of adult WT mice. PTPN11D61G overexpression in the hippocampus (Fig. 3a and Supplementary Fig. 6) resulted in increased Erk activation as assessed by immunoblotting p-Erk, confirming that AAV–expressed PTPN11D61G is functional (Fig. 3b; n = 5 hippocampi for each group, unpaired two-tailed t-test, t = 2.452, * P < 0.05). Consistently, AAV–PTPN11D61G expression impaired performance in probe trials of the water maze (Fig. 3c). AAV–PTPN11D61G–expressing mice spent significantly less time in the target quadrant than did AAV–GFP/vehicle-injected control mice (Fig. 3c; PTPN11D61G/veh, 25.89 ± 3.38 %, n=10; GFP/veh, 35.88 ± 2.95 %, n=13; unpaired two-tailed t-test, t = 2.231, * P < 0.05). Unlike the Ptpn11D61G/+ mutation in mice, AAV–PTPN11D61G expression did not affect swimming speed or other performance variables during the acquisition phase of the water maze (Supplementary Fig. 7), suggesting that the acute expression of PTPN11D61G in the hippocampus only affects learning and memory, whereas deregulation of Shp2–Erk signaling during development in the Ptpn11D61G/+ mice might affect other functions including motor coordination. Notably, overexpressing WT PTPN11 did not affect basal p-Erk levels or spatial learning and memory (Supplementary Fig. 8), demonstrating that the adverse impact on Erk signaling and learning and memory is specific to the NS-related PTPN11 mutation.
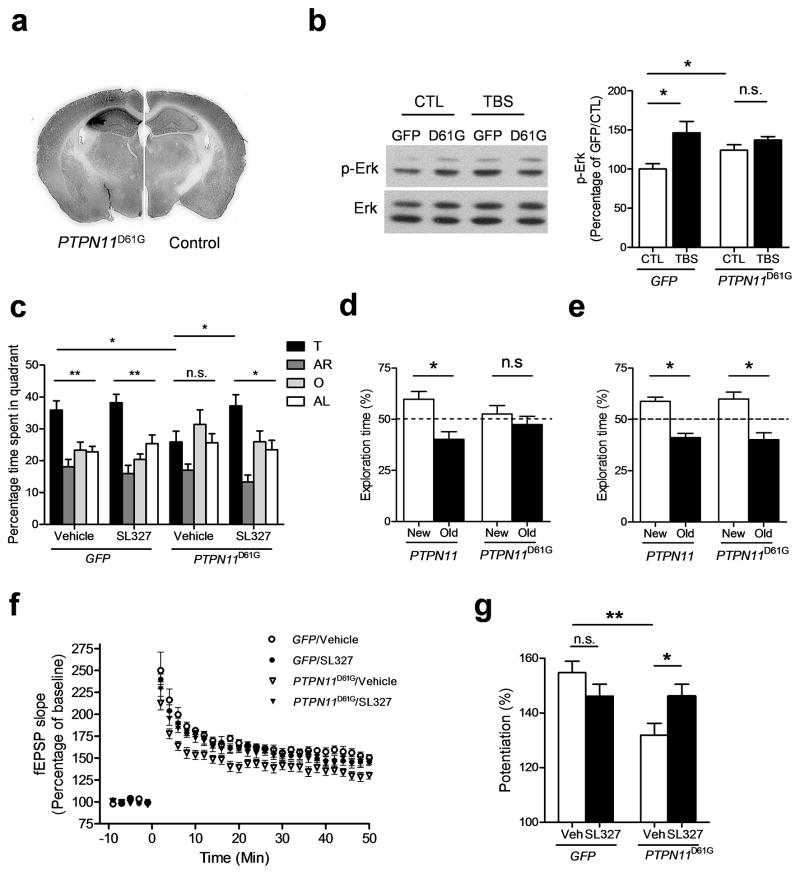
Figure 3. PTPN11D61G overexpression induces learning and memory and LTP deficits that can be reversed by MEK inhibition.
a. AAV–PTPN11D61G infection results in overexpression of SHP2D61G. Anti–SHP2 immunohistochemistry shows robust overexpression of SHP2 in the hippocampus of AAV–PTPN11D61G–infused brains (left) compared with AAV–GFP infused brains (right). Full-length blots/gels are presented in Supplementary Figure 11.
b. PTPN11D61G overexpression increases basal Erk activity (phospho–Erk level) and prevents further Erk activation in response to TBS. Left, Representative immunoblot showing p–Erk (upper) and total Erk (lower) in PTPN11D61G–expressing slices and GFP–expressing slices. Slices were prepared 1 h after TBS. Right, Bar graph displays normalized p–Erk levels (mean ± s.e.m.). CTL, control without TBS.
c. MEK inhibitor SL327 reverses spatial memory deficits in PTPN11D61G–overexpressing mice in the Morris water maze. Quadrant occupancy analysis for the probe trial reveals that PTPN11D61G/veh mice showed no preference for the target quadrant (target vs. other quadrants, Dunnett’s Multiple Comparison Test after one-way ANOVA, P > 0.05). PTPN11D61G/veh mice also spent significantly less time in the target quadrant compared with GFP/veh mice. SL327 treatment significantly increased the time spent in the target quadrant in PTPN11D61G-expressing mice compared with vehicle-treated PTPN11D61G mice (PTPN11D61G/SL327, 37.25 ± 3.50 %, n=10, unpaired two-tailed t-test, t = 2.335, * P < 0.05).
d. PTPN11D61G overexpression in the hippocampus impairs memory in the object–place recognition test. Control mice expressing WT PTPN11 spent significantly more time exploring the object in the new place than exploring the object in the old place during the test session 24 h after training. However, PTPN11D61G–overexpressing mice did not show preference for the object in the new place.
e. MEK inhibitor SL327 rescues memory deficits in object–place recognition test caused by PTPN11D61G overexpression. When SL327 (32 mg/kg) was injected 30 min before training in the object-place recognition test, both PTPN11– and PTPN11D61G–expressing mice spent significantly more time exploring the object in the new place than exploring the object in the old place during the test session 24 h after training.
f. and g. MEK inhibitor SL327 reverses LTP deficits caused by PTPN11D61G overexpression. f. PTPN11D61G overexpression significantly impaired 5 TBS–induced LTP, and bath application of SL327 reversed the deficit (Repeated-measures ANOVA, F3, 72 = 140.2, P < 0.0001). SL327 (1 μM) was applied for 1 h before LTP induction, and then maintained in the bath throughout recording. g. Average % fEPSP changes (last 10 minutes of recording) shows a significant LTP deficit in the vehicle-treated PTPN11D61G group compared with the vehicle-treated GFP group (GFP/veh, 154.8 ± 4.18 %, n=7; PTPN11D61G/veh, 131.9 ± 4.38, n=10; unpaired two-tailed t-test, t = 3.625, ** P < 0.01) and significant reversal by SL327 treatment (PTPN11D61G/SL327, 146.1 ± 4.36 %, n=10; unpaired two-tailed t-test, t = 2.309, * P < 0.05). SL327 did not affect LTP in the GFP group (GFP/SL327, 146.2 ± 4.37 %, n=7; unpaired two-tailed t-test, t = 1.414, P = 0.183).
AAV–PTPN11D61G–expressing mice were also tested in another hippocampus-dependent task (object-place recognition) 24-hours after training; WT AAV–PTPN11–expressing mice were used as controls (Fig. 3d). Consistent with the water maze results, PTPN11D61G expression in the CA fields of the hippocampus caused memory deficits in this task: the control mice spent significantly more time exploring the object at the new location (Fig. 3d; n = 15 mice, 59.79 ± 3.72 % for new place, one-sample paired t-test compared to 50 %, t = 2.633, * P < 0.05), but the PTPN11D61G mice did not (Fig. 3d; n = 15 mice, 52.61 ± 4.10 % for new place, one-sample paired t-test compared to 50 %, t = 0.636, P = 0.535). Importantly, AAV–PTPN11D61G–expressing mice showed comparable total exploration time to that of WT AAV–PTPN11–expressing mice during training (WT AAV–PTPN11, 43.70 ± 3.98 s, n = 15; AAV–PTPN11D61G, 39.29 ± 4.94 s, n = 15; unpaired t-test, t = 0.695 P = 0.493). All together, these data show that expressing PTPN11D61G in the adult CA fields of the hippocampus is sufficient to disrupt memory, and demonstrate that PTPN11 plays a critical role in adult brain function, in addition to its effects on development20.
To test whether reducing Erk activity could reverse the memory deficits in AAV–PTPN11D61G–expressing mice, we treated these mice with the MEK inhibitor SL327 or vehicle daily, 30 min before training. SL327 treatment (32 mg/kg, intraperitoneal injection) decreased Erk activation in the hippocampus of control and AAV–PTPN11D61G mice (Supplementary Fig. 9). We choose a sub-threshold dose of the drug that does not impair spatial learning in WT mice and only decreases hippocampal Erk activation in WT mice by ~ 25% (Supplementary Fig. 9). Importantly, this SL327 treatment rescued the spatial learning deficits of the AAV–PTPN11D61G mice without affecting the performance of the AAV–GFP group (Fig. 3c; PTPN11D61G/SL327, one-way ANOVA, F3, 36 = 10.44, P < 0.001; target vs. other quadrants, Dunnett’s Multiple Comparison Test, * P < 0.05). Consistent with the water maze results, the same SL327 treatment also rescued the memory deficits in the object-place recognition task (Fig. 3e; WT PTPN11, n = 5, 58.83 ± 2.01 % for new place, two-tailed paired t-test compared to 50 %, t = 4.395, * P < 0.05; PTPN11D61G, n = 8, 59.90 ± 3.41 % for new place, two-tailed paired t-test compared to 50 %, t = 2.904, * P < 0.05). These results demonstrate that increased Ras–Erk signaling in adult CA fields of the hippocampus contribute to the memory deficits in AAV–PTPN11D61G–expressing mice. Remarkably, SL327 also reversed the memory deficits of the adult Ptpn11D61G/+ mutant mice in the Morris water maze, showing that normalizing Erk activity in adults can reverse the behavioral deficits even in mutant mice with germ line mutations (Supplementary Fig. 10).
Next, we asked whether AAV–PTPN11D61G expression in adults also impairs CA1 Schaffer collateral LTP. As in Ptpn11D61G/+ mutant mice, hippocampal slices from AAV–PTPN11D61G–transfected mice showed significantly reduced LTP in response to a TBS tetanus (Fig. 3f, g; GFP/veh, 154.8 ± 4.18 %, n = 7 slices from 7 mice; PTPN11D61G/veh, 131.9 ± 4.38 %, n = 10 slices from 10 mice; unpaired two-tailed t-test, t = 3.625, ** P < 0.01), demonstrating that manipulating Shp2 signaling specifically in the adult CA fields of the hippocampus is sufficient to impair LTP. In addition, TBS failed to further activate Erk in AAV–PTPN11D61G-transfected hippocampi (Fig. 3b; n=5 hippocampi for each group, unpaired two-tailed t-test, t = 1.580, P = 0.1527). SL327 treatment, which reversed their learning deficits, also normalized CA1 LTP in hippocampal slices from the AAV–PTPN11D61G-transfected mice (Fig. 3f, g; Two-way ANOVA, F1, 30 = 6.526, * P < 0.05; Bonferroni post-test reveals significant effect of SL327 treatment only on the PTPN11D61G group, * P < 0.05). It is noteworthy that basal synaptic transmission and paired-pulse facilitation were not affected either by AAV–PTPN11D61G expression or by the SL327 treatment we used (Supplementary Fig. 7). Taken together, these results indicate that deregulated Erk activity causes CA1 LTP deficits, and these deficits are responsible for the learning and memory impairments in mouse models of NS.
PTPN11D61G overexpression increases excitatory synaptic function
Next, we examined the electrophysiological mechanism underlying the LTP impairment in AAV–PTPN11D61G–transfected mice. Increases in Ras signaling are known to facilitate AMPA receptor trafficking to the surface membrane21. For example, expression of constitutively active Ras enhances AMPA receptor-mediated currents in hippocampal neurons and impairs LTP21. Hence, we asked whether the increases in activated Erk associated with PTPN11D61G expression enhanced AMPA currents. Whole-cell voltage clamp recordings revealed that the ratio of AMPA:NMDA currents was increased in AAV–PTPN11D61G-transfected hippocampi (Fig. 4a, b; AAV–PTPN11D61G, 1.51 ± 0.11, n = 10 cells from 5 mice; AAV–GFP, 0.96 ± 0.05, n = 10 cells from 5 mice, unpaired two-tailed t-test, t = 4.754, *** P < 0.001). Importantly, SL327 treatment normalized the AMPA:NMDA ratio (Fig. 4a, b; PTPN11D61G/SL327, 1.07 ± 0.11, n = 7 cells from 6 mice; GFP/SL327, 1.08 ± 0.07, n = 8 cells from 6 mice; PTPN11D61G/Veh vs. PTPN11D61G/SL327, unpaired two-tailed t-test, t = 2.832, * P <0.05). Although paired-pulse facilitation (PPF) ratio was unaffected by AAV–PTPN11D61G (Fig. 4c; AAV–PTPN11D61G, n = 12 cells from 5 mice; AAV–GFP, n= 11 cells from 5 mice; repeated-measures ANOVA, F1, 21 = 0.010, P = 0.921), mEPSC frequency (but not amplitude) was enhanced by this manipulation (Fig. 4d; GFP/Veh, 1.50 ± 0.53 Hz, n = 9 cells from 3 mice; PTPN11D61G/Veh, 4.64 ± 0.94 Hz, n = 9 cells from 3 mice; unpaired two-tailed t-test, t = 2.923, ** P < 0.01). The increased excitation in PTPN11D61G–transfected mice was reversed by SL327 treatment (Fig. 4d; PTPN11D61G/Veh, 4.64 ± 0.94 Hz, n = 9 cells from 3 mice; PTPN11D61G/SL327, 2.02 ± 0.32 Hz, n = 9 cells from 5 mice; unpaired two-tailed t-test, t = 2.645, * P < 0.05). Consistently, mEPSC frequency, but not amplitude, was significantly increased in pyramidal neurons of Ptpn11D61G/+ mice compared with WT (Fig. 4e; WT/Veh, 2.68 ± 0.55 Hz, n = 9 cells from 5 mice; Ptpn11D61G/+/Veh, 5.71 ± 0.56 Hz, n = 10 cells from 3 mice; unpaired two-tailed t-test, t = 3.858, ** P < 0.01). Moreover, mIPSC frequency and amplitude were unaffected in both AAV–PTPN11D61G-transfected mice and Ptpn11D61G/+ mutants (Fig. 4f; mIPSC frequency: GFP, 6.91 ± 0.87 Hz, n = 9 cells from 4 mice; PTPN11D61G, 6.93 ± 1.15 Hz, n = 7 cells from 4 mice, unpaired two-tailed t-test, t = 0.022, P = 0.983; mIPSC amplitude, GFP, 19.13 ± 1.19 pA, n = 9 cells from 4 mice; PTPN11D61G, 20.09 ± 1.62 pA, n = 7 cells from 4 mice, unpaired two-tailed t-test, t = 0.486, P = 0.634; Fig. 4g; mIPSC frequency: WT, 15.06 ± 2.08, n = 7 cells from 5 mice; Ptpn11D61G/+, 15.35 ± 3.50, n = 8 cells from 5 mice, unpaired two-tailed t-test, t = 0.0683, P = 0.947; mIPSC amplitude: WT, 32.91 ± 3.06 pA, n = 7 cells from 5 mice; Ptpn11D61G/+, 33.59 ± 2.32 pA, n = 8 cells from 5 mice, unpaired two-tailed t-test, t = 0.180, P = 0.860). Importantly, just as with AAV–PTPN11D61G mice, the increased excitation in Ptpn11D61G/+ mice was reversed by SL327 treatment (Fig. 4e; Ptpn11D61G/+/Veh, 5.71 ± 0.56 Hz, n = 10 cells from 3 mice; Ptpn11D61G/+/SL327, 2.87 ± 1.02 Hz, n = 9 cells from 3 mice; unpaired two-tailed t-test, t = 2.508, * P < 0.05), indicating that increased Ras-Erk signaling is responsible for the enhanced excitatory synaptic function associated with the Ptpn11D61G mutation.
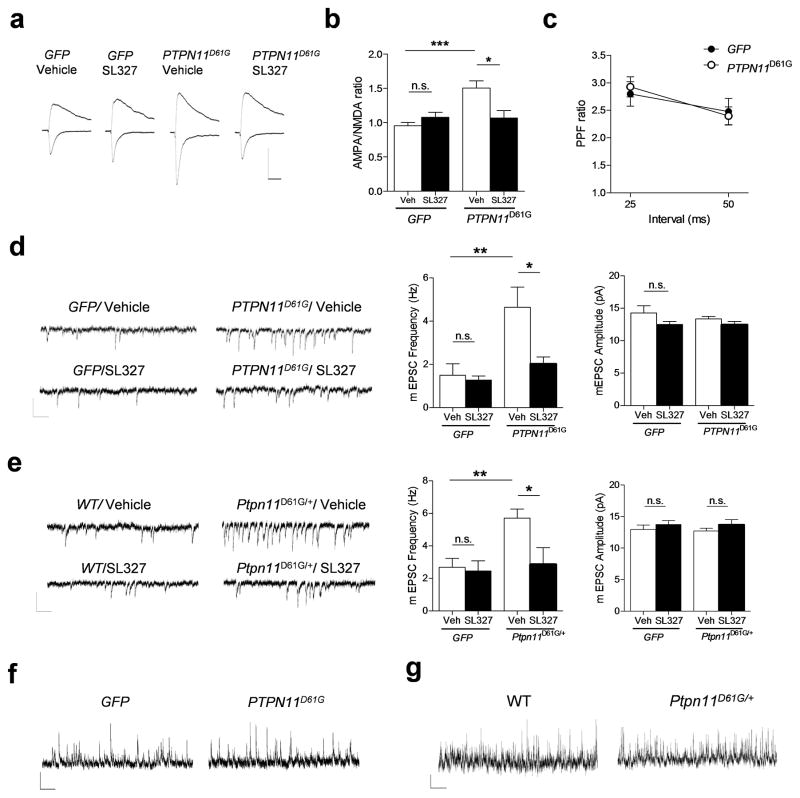
Figure 4. PTPN11D61G overexpression enhances excitatory synaptic function through increased Ras-Erk signaling.
a. AMPA receptor-mediated currents were measured at the peak of the currents at − 65 mV, and NMDA currents were measured 50 ms after onset at + 40 mV. The average of 15 traces is shown. Scale, 100 pA and 40 ms.
b. Group data showing the increased AMPA:NMDA current ratio in AAV–PTPN11D61G mice compared with AAV-GFP mice. SL327 treatment (1 μM, 1 h) significantly reversed the AMPA:NMDA current ratio in the PTPN11D61G group without affecting GFP–expressing mice. Two-way ANOVA, interaction between viral treatment and drug, F1, 31 = 10.53, ** P < 0.01. Bonferroni post-test reveals significant effect of SL327 treatment only on PTPN11D61G group (** P < 0.01).
c. Paired-pulse facilitation ratio is unaffected by PTPN11D61G. There was no significant difference at 25 ms or 50 ms intervals between the two groups.
d. PTPN11D61G overexpression increases excitatory synaptic function. Left, Representative traces of mEPSC recordings from GFP or PTPN11D61G–expressing hippocampus. Middle, mEPSC frequency was increased in AAV–PTPN11D61G–transfected mice compared with AAV–GFP mice, and was reversed by SL327 (1 μM) treatment without affecting on the AAV–GFP group. Two-way ANOVA with viral treatment as between-subjects factor, F1, 30 = 10.31, ** P < 0.01. Right, mEPSC amplitudes were not significantly different among groups. Two-way ANOVA with viral treatment as between-subjects factor, F1, 30 = 0.470, P = 0.498. Scale, 20 pA and 200 ms.
e. Excitatory synaptic function is increased in Ptpn11D61G/+ mutant mice and reversed by SL327 treatment. Left, Representative traces of mEPSC recordings from WT or Ptpn11D61G/+ mice. Middle, mEPSC frequency was increased in Ptpn11D61G/+ mice compared with WT littermates, and was reversed by SL327 (1 μM) treatment. Two-way ANOVA with genotype as between-subjects factor, F1, 33 = 5.914, * P < 0.05. Right, mEPSC amplitudes were not significantly different among groups. Two-way ANOVA with genotype as between-subjects factor, F1, 33 = 0.418, P = 0.839. Scale, 20 pA and 200 ms.
f. and g. mIPSC was not changed in either AAV–PTPN11D61G-transfected mice or Ptpn11D61G/+ mutants. f. Representative traces of mIPSC recordings from GFP or PTPN11D61G–expressing hippocampus. g. Representative traces of mIPSC recordings from Ptpn11D61G/+ mutant mice or WT littermates. Scale, 20 pA and 1 s.
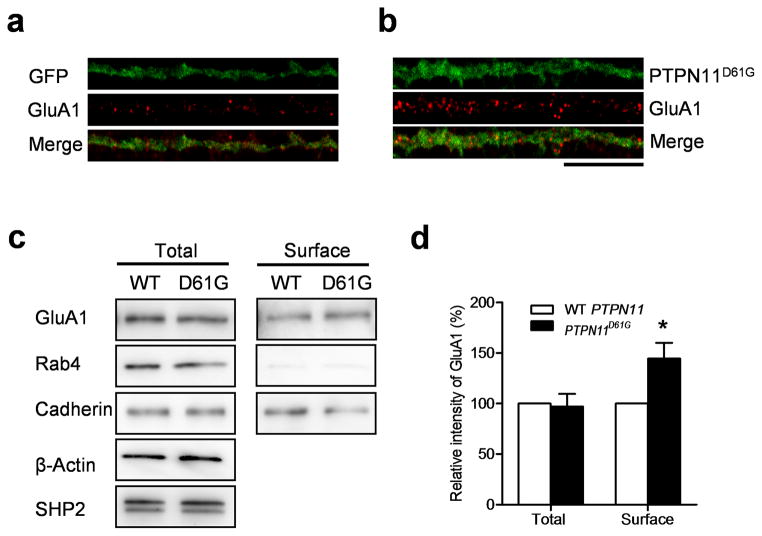
To test the hypothesis that the increase in excitation caused by the PTPN11D61G mutation is due to increases in the number of synapses with AMPA receptors, we transfected cultured hippocampal neurons (21 days in vitro, DIV) with PTPN11D61G and labeled surface GluA1 AMPA receptors (Fig. 5a, b). Indeed, the number of surface GluA1 receptor clusters was significantly increased in PTPN11D61G–transfected neurons compared with controls (Fig. 5a, b; GluA1 particle number per 10 μm: PTPN11D61G, 8.60 ± 0.59, n = 20 neurons, 1,432.6 μm of dendrites; GFP, 6.76 ± 0.34, n = 22 neurons, 1,759.6 μm of dendrites; unpaired two-tailed t-test, t = 2.763, ** P < 0.01), a result consistent with the increase in mEPSC frequency caused by PTPN11D61G. The size of GluA1 clusters, however, was not affected by PTPN11D61G expression (Fig. 5a, b; GluA1 particle size (μm2): PTPN11D61G, 0.19 ± 0.02, n=20 neurons, 1,432.6 μm of dendrites; GFP, 0.18 ± 0.02, n = 22 neurons, 1,759.6 μm of dendrites; unpaired two-tailed t-test, t = 0.319, P = 0.751), a result consistent with the finding of normal mEPSC amplitude. To quantitatively analyze the surface expression of GluA1, cultured neurons transfected with either WT PTPN11 or PTPN11D61G constructs were surface labeled with biotin and the biotinylated surface proteins were pulled-down and analyzed (Fig. 5c, d). While the total expression levels of GluA1 were similar in WT PTPN11 and PTPN11D61G–expressing neurons, the surface expression of GluA1 was significantly increased in PTPN11D61G expressing neurons compared with WT PTPN11–expressing neurons (Fig. 5c, d; Two-way ANOVA followed by Bonferroni post-test: interaction between fraction (total/surface) x virus (PTPN11/PTPN11D61G), F1, 12 = 5.704, * P < 0.05; total, PTPN11 vs PTPN11D61G, P > 0.05; surface, PTPN11 vs. PTPN11D61G, * P < 0.05). These data support the results from the immunocytochemistry experiments showing that PTPN11D61G expression facilitates the surface expression of GluA1. These results indicate that post-synaptic changes in AMPA receptor trafficking contribute to the increase in excitatory synaptic function caused by the PTPN11D61G mutation.
Figure 5. PTPN11D61G overexpression increases surface AMPA receptor expression.
a. and b. Representative images of surface GluA1 staining in cultured neurons. GFP alone (a) or PTPN11D61G and GFP (b) were co–expressed using a bicistronic Sindbis viral vector in cultured hippocampal neurons (DIV21). Scale, 20μm.
c. Representative images of western blotting of total and biotinylated surface proteins. Cadherin and Rab-4 were used as markers for surface and cytosol expression, respectively. Full-length blots/gels are presented in Supplementary Figure 11.
d. Surface expression of GluA1 was significantly increased in PTPN11D61G expressing neurons compared to WT PTPN11 expressing neurons, while the total expression level of GluA1 did not differ between WT PTPN11 and PTPN11D61G transfected neurons.
Lovastatin treatment rescued LTP and learning deficits in Ptpn11D61G/+ mice
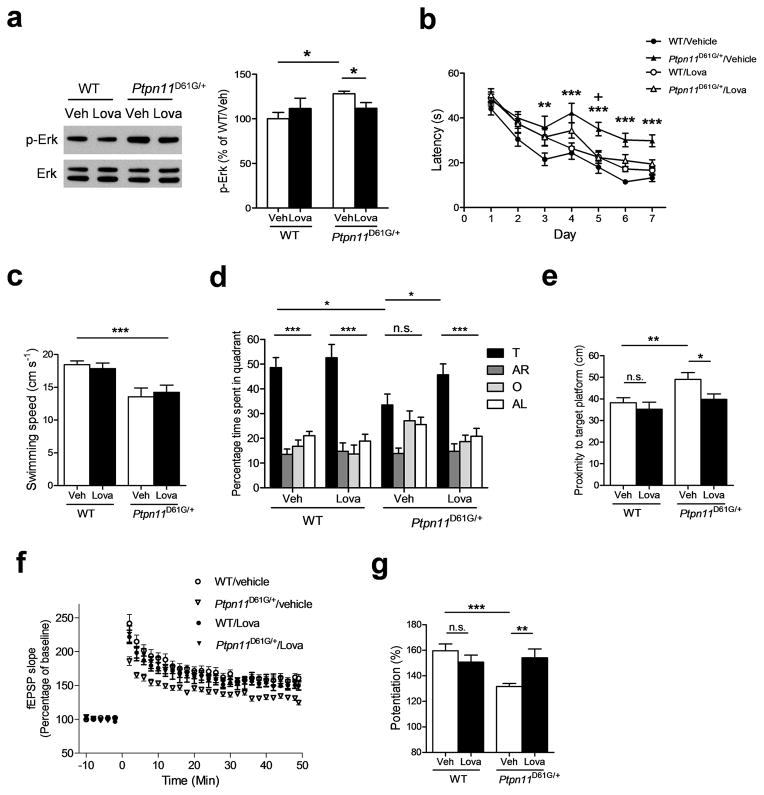
A MEK inhibitor SL327 rescued the spatial learning deficits in adult Ptpn11D61G/+ mice (Supplementary Fig. 10), suggesting that decreasing basal Erk activation can be a therapeutic strategy for learning deficits in NS. Previous studies also showed that lovastatin, a blood-brain-barrier-permeable member of a widely used class of FDA–approved drugs (statins), decreases the levels of isoprenyl groups required for Ras membrane localization and biological activity22, 23. As in AAV–PTPN11D61G–transfected mice, p-Erk levels were increased in Ptpn11D61G/+ hippocampi (Fig. 6a; WT/Veh, 100.0 ± 7.23 %, n = 8; Ptpn11D61G/+/Veh, 128.2 ± 2.87 %, n = 7; unpaired two-tailed t-test, t = 3.438, * P < 0.05). Lovastatin treatment normalized p-Erk levels in mutant hippocampi at concentrations that did not affect Erk activation in controls (Fig. 6a; Ptpn11D61G/+/Veh, 128.2 ± 2.87 %, n = 7; Ptpn11D61G/+/Lova, 111.8 ± 6.41 %, n = 8; unpaired two-tailed t-test, t = 2.231, * P <0.05). Importantly, lovastatin-treated Ptpn11D61G/+ mice showed better performance (e.g., faster times to reach the hidden platform of the Morris maze) than vehicle-treated Ptpn11D61G/+ mice (Fig. 6b; Two-way ANOVA followed by Bonferroni post-test, WT/Veh vs. Ptpn11D61G/+/Veh, ** P < 0.01, *** P < 0.001; Ptpn11D61G/+/Veh vs. Ptpn11D61G/+/Lova, + P < 0.05) although their swimming speeds were unchanged by the treatment (Fig. 6c; WT/veh, 18.5 ± 0.6 cm/s, n = 14 mice; Ptpn11D61G/+/veh, 13.6 ± 1.3 cm/s n = 11 mice; WT/lova, 17.9 ± 0.8 cm/s, n = 13 mice; Ptpn11D61G/+/lova, 14.2 ± 1.1 cm/s, n = 11 mice; two-way ANOVA with genotype as between-subjects factor and drug treatment as within-subjects factor, effect of genotype: F1, 45 = 19.79, *** P < 0.0001, interaction: F 1,45 = 0.4489, P = 0.506). These data suggest that the learning deficits in these animals are not due to their slower swimming speeds or other performance deficits. In probe trials, lovastatin-treated Ptpn11D61G/+ mice, unlike vehicle-treated Ptpn11D61G/+ mice, showed selective searching in the target quadrant. Also, during probe trials lovastatin-treated Ptpn11D61G/+ mice showed lower average proximity to the platform site (i.e., better performance) than vehicle-treated mutant mice, indicating that lovastatin treatment dramatically improved the performance of Ptpn11D61G/+ mice in probe trials (Fig. 6d, e; % time spent in target quadrant, Ptpn11D61G/+/veh, 33.48 ± 4.44 %, n = 11 mice; Ptpn11D61G/+/Lova, 45.70 ±4.43 %, n = 11 mice; unpaired two-tailed t-test, t = 1.947, * P < 0.05; proximity to target platform, WT/veh, 38.26 ± 2.33 cm, n = 14 mice; WT/Lova, 35.28 ± 3.26 cm, n = 13 mice; Ptpn11D61G/+/veh, 49.05 ± 3.15 cm, n = 11 mice; Ptpn11D61G/+/Lova, 39.82 ± 2.53 cm, n = 11 mice; unpaired two-tailed t-test, WT/veh vs. Ptpn11D61G/+/veh, t = 2.813, ** P < 0.01; Ptpn11D61G/+/veh vs. Ptpn11D61G/+/Lova, t = 2.284, * P < 0.05,). Importantly, the spatial learning performance of lovastatin–treated Ptpn11D61G/+ mice was indistinguishable from controls (Fig. 6d, e). Notably, at the concentration used, lovastatin had no effect on any measure of learning in WT animals (Fig. 6d, e).
Figure 6. Lovastatin treatment reverses spatial learning and memory and LTP deficits in Ptpn11D61G/+ mice.
a. Lovastatin treatment reverses increased Erk activation in hippocampi from Ptpn11D61G/+ mice. Left, Representative immunoblot showing p-Erk (upper) and total Erk (lower) levels in WT and Ptpn11D61G/+ mutant mice. Hippocampi were dissected 6 h after the 4th day of lovastatin injection (subcutaneous (s.c.) injections, 10 mg/kg). Full-length blots/gels are presented in Supplementary Figure 11. Right, Bar graph displaying normalized p-Erk levels (mean ± s.e.m.).
b. Vehicle-treated Ptpn11D61G/+ mutant mice showed significantly longer latency to the hidden platform during training sessions compared with vehicle-treated WT mice. Lovastatin-treated Ptpn11D61G/+ mice showed comparable latency to WT mice.
c. Lovastatin treatment did not improve swimming speed.
d and e. Lovastatin treatment (10 mg/kg) reverses spatial memory deficits in Ptpn11D61G/+ mice at a concentration that does not affect WT controls. d. Quadrant occupancy analysis for the probe trail reveals that Ptpn11D61G/+ mice with vehicle treatment (Ptpn11D61G/+/veh) showed no preference for the target quadrant (target vs. other quadrants, Dunnett’s Multiple Comparison Test after one-way ANOVA, P > 0.05). By contrast, the Ptpn11D61G/+/Lova group selectively searched for the target quadrant, suggesting that lovastatin treatment reversed the spatial memory deficit in Ptpn11D61G/+ mice (target vs. other quadrants, Dunnett’s Multiple Comparison Test after one-way ANOVA, *** P < 0.0001). The Ptpn11D61G/+/Lova group also spent significantly more time in the target quadrant compared with Ptpn11D61G/+/veh mice. e. Proximity analysis reveals that the spatial memory deficit in Ptpn11D61G/+ mice can be reversed by lovastatin treatment.
f ang g. Lovastatin treatment reverses LTP deficits in Ptpn11D61G/+ mice at concentrations that do not affect WT littermates. f. Ptpn11D61G/+ mice showed a deficit in 5 TBS-induced LTP that was reversed by systemic administration of lovastatin (Repeated-measures ANOVA, F3, 96 = 14.38, P < 0.0001). g. Average % fEPSP changes (last 10 minutes of recordings) show that lovastatin treatment significantly rescued the LTP deficit in Ptpn11D61G/+ mice (WT/veh, 159.6 ± 5.33 %, n=7; WT/Lova, 150.7 ± 5.49 %, n=6; Ptpn11D61G/+/veh, 131.7 ± 2.31 %, n=9; Ptpn11D61G/+/Lova, 154.2 ± 6.88 %, n=7; unpaired two-tailed t-test, ** P < 0.01, *** P < 0.001).
Consistent with the hypothesis that increased Ras–Erk activity leads to the LTP deficits responsible for spatial learning impairment in Ptpn11 mutant mice, the levels of 5 TBS-induced LTP in lovastatin–treated Ptpn11D61G/+ mice were significantly higher than those in the vehicle-treated mutants and indistinguishable from those in WT control animals (Fig. 6f, g; two-way ANOVA with genotype as between-subjects factor, F1, 25 = 5.936, * P < 0.05, Bonferroni post-test reveals a significant effect of lovastatin treatment only on Ptpn11D61G/+ group, ** P < 0.01). By contrast, lovastatin treatment had no effect on LTP in hippocampal slices from WT mice (Fig. 6f, g). Thus, lovastatin treatment can normalize LTP deficits and spatial learning impairments even in adult Ptpn11D61G/+ mice. Although we cannot exclude the possibility that lovastatin may affect other biological processes24, our data suggest that lovastatin reverses the spatial learning deficits of Ptpn11D61G/+ mice, by reducing Erk activation and consequently correcting LTP deficits.
Discussion
Our study provides compelling evidence that the spatial learning and memory deficits in mouse models of NS are caused by enhanced Ras-Erk activation, which disrupts the balance between excitation and inhibition (E/I) and impairs hippocampal long-term potentiation. Furthermore, our experiments with viral vectors demonstrate that Ptpn11 plays critical roles not only in regulating development20, 25, but also in adult brain functions. Consistent with our findings, expression of the fly ortholog of SHP2 (Csw) bearing gain-of-function mutations impaired long-term memory in Drosophila26.
In the present study, we used two knock-in mutant mice harboring a D61G or a N308D mutation in PTPN11. The D61G mutation is associated with both NS and leukemia and shows higher enzymatic activity than N308D, which is only associated with NS15. Consistently, Ptpn11D61G/+ mice showed more severe deficits in LTP and learning than Ptpn11N308D/+ mice. Although basal-level of p-Erk was significantly higher in the hippocampus of Ptpn11D61G/+ mice compared to WT littermates (Fig. 6), we could not detect significant increases in basal p-Erk levels in the hippocampus of Ptpn11N308D/+ mice, perhaps because these mice showed an overall milder phenotype (Supplementary Fig. 9c).
The activation of Ras-Erk signaling facilitates AMPA receptor trafficking during LTP21 and abnormal hyperactivation of postsynaptic Erk signaling impairs hippocampal LTP and learning27, 28. Our findings suggest that the PTPN11D61G mutation increases the number of synapses with postsynaptic AMPA receptors, thus occluding LTP and therefore impairing learning. In agreement with the hypothesis that there are more synapses with AMPA receptors, we found that PTPN11D61G expression increases mEPSC frequency, but does not affect PPF ratio (Fig. 4c), a form of plasticity very sensitive to changes in pre-synaptic function. Additionally, PTPN11D61G expression increased the evoked AMPA:NMDA ratios (Fig. 4a, b), another observation consistent with the hypothesis that the PTPN11D61G expression resulted in more synapses with AMPA receptors. Importantly, these observations were reproduced in both AAV–PTPN11D61G–transfected mice and in the germ line mutants. Importantly, PTPN11D61G expression enhanced the surface expression of GluA1 and increased the number of surface GluA1 clusters in cultured hippocampal neurons, a finding consistent with the hypothesis that the enhancement in excitatory synaptic function driven by PTPN11D61G expression is caused by postsynaptic mechanisms. Interestingly, deletion of a Ras–Erk regulator (SynGAP) was reported to increase ERK signaling, enhance the levels of AMPA receptors, increase mEPSC frequency and impair LTP27.
Deregulation of Ras–Erk signaling has been associated with other genetic disorders including neurofibromatosis type I (NF1), Costello syndrome, LEOPARD syndrome, CFC syndrome, and Legius syndrome10, 29. Among these, studies with the Nf1+/− mutant mouse, which is a model of NF1, demonstrated that increased Ras signaling results in increased GABA release (excitation is normal in these mice) that leads to deficits in LTP and, consequently, learning and memory impairments30–33. Altogether these findings demonstrate that similar behavioral (e.g., spatial learning deficits) and even electrophysiological phenotypes (i.e., LTP deficits) can be caused by different cellular mechanisms: increases in AMPARs in NS mice and increases in GABA release in NF1 mice. Homozygous deletion of the NF1 gene in mouse post-natal excitatory neurons does not affect either synaptic transmission or learning32, whereas expression of the NS–mutation PTPN11D61G in post-natal excitatory neurons does disrupt both synaptic transmission and learning, a direct demonstration of the distinct roles of these two Ras signaling modulators.
In this study, we show that postnatal treatment with an FDA–approved drug, lovastatin, can reverse learning and memory as well as LTP deficits in an adult NS mouse model. A previous study showed that lovastatin treatment can rescue spatial learning problems, attention deficits and pre-pulse inhibition deficits in Nf1+/− mutant mice22. Thus, our studies suggest that this FDA–approved drug with a strong safety profile may also be useful for treatment of cognitive deficits associated with NS.
ONLINE METHODS
Methods
Mice
Ptpn11D61G/+ mice were backcrossed to 129S6/SvEv and Ptpn11N308D/+ mice were backcrossed to C57Bl/6J mice at least 6 times before experiments. Three to six month–old male and female mice were used. For AAV experiments, 3 – 4 month-old male C57Bl/6J mice (Jackson Laboratory) were used. Mice were randomly assigned to treatment and experimental condition. All experiments used littermates as controls and were carried and analyzed with the experimenters blinded to genotype and treatment. Animals were group housed (2 – 4) on a 12 h light/dark cycle in vivarium at UCLA and CAU. All studies were approved by the Animal Research Committee at UCLA and CAU.
Drugs
SL327 (Tocris) was dissolved in DMSO (16 mg/ml) and was injected intraperitoneally once daily, 30 min before the water maze experiment at a dose of 32mg/kg. The volume of a single injection was under 80 μl. Lovastatin (Mevinolin, Sigma) was prepared as previously described22. Briefly, lovastatin was dissolved in ethanol (final concentration of 8%) and 1N NaOH was added to convert mevinolin to the sodium salt. The pH of the final solution (4 mg/ml) was adjusted to 7.5 with HCl. The Vehicle solution was prepared with the same procedure. Lovastatin was administered daily (subcutaneous injection, 10 mg/kg) for 3 days before the first training day of the water maze and 6 h before training every day thereafter.
AAV
The coding sequence of human PTPN11 with or without the D61G mutation was subcloned into the HindIII – NsiI site of the AAV expression vector pSOFF. The resultant vector expresses mutant PTPN11 under the synthetic CBA promoter (CMV enhancer and chicken beta-actin promoter). Recombinant virus (rAAV5) was purified as previously described34. Briefly, an iodixanol gradient purification was performed followed by an ion exchange chromatography step which results in a 99 % pure vector preparation as judged by silver stained-SDS acrylamide gel fractionation. After the chromatography, the buffer was exchanged and the virus was concentrated in Ringer’s solution using a Biomax 100 K concentrator (Millipore). Vector titers were determined by Real Time PCR. Typical titers were 3.09 × 1012 genome copies/ml. rAAV5-GFP expressing only GFP was used as a control. Virus was infused into two sites per hemisphere (1 μl per injection, AP=−2.5, Lat=+/−2, DV=−1.7; AP=−1.8, Lat=+/−1, DV=−1.6) over 5 min through a 30-gauge Hamilton microsyringe. Viruses (GFP, WT PTPN11 or PTPN11D61G) were randomly assigned for infusion. After completion of infusion, the syringe was left in place for an additional 5 min. All the experiments were done three weeks after the infusion.
Behavior
Behavioral experiments were performed during the light cycle. In the hidden platform-version of Morris water maze, mice were trained with two blocks of 2 trials (ITI = 1min) spaced about 45 min apart. In each training trial, mice were released from a different starting position and then were allowed to search for the escape platform for 60 s. The platform was submerged 1 cm under the surface of the water. Once a mouse found the platform, it was left there for 15 s. If a mouse did not find the platform within 60 s, it was guided to the platform and remained on the platform for 15 s before being removed from the pool. Mice were trained for 5 – 7 consecutive days. Memory was assessed in probe trials that were given after completion of training as described in the main text. During the probe trials, the platform was removed and the mice were allowed to search for it for 60 s. One mouse was excluded from further analysis because of floating (no voluntary movement for more than 10 s in more than 2 trials). The same group of mice was tested in the visible platform–version of Morris water maze. Data were acquired and analyzed using WaterMaze software (Actimetrics).
The object-place recognition task included a training and a test session. Before training, mice were handled 5 min per day for 4 days, and then habituated in a square box (27.5 cm × 27.5 cm × 25 cm) for 15 min for another 2 days. One side of the experimental box included a prominent cue. During the 10–min training session, mice were placed in the box, exposed to two identical objects and allowed to explore these objects. During the test session (24h after training), mice were placed back into the experimental box with the same two objects for 5 min: one object (Old location) stayed in the same location as during training, while the other object (New location) was moved to a new location. For the rescue experiment, SL327 (32 mg/kg, i.p.) was injected 30 min before the training session. The objects changed during the test sessions were randomly counterbalanced between mice. Experiments were videotaped and the exploration times were manually analyzed.
Electrophysiology
For extracellular recordings of field excitatory postsynaptic potentials (fEPSP), sagittal slices (400 μm) were prepared with a vibratome (VT1000S, Leica) in ice-cold artificial cerebrospinal fluid (ACSF). Slices recovered at room temperature for at least 90 min before recording in ACSF saturated with 95 % O2 and 5 % CO2 containing the following (in mM): 120 NaCl, 3.5 KCl, 2.5 CaCl2, 1.3 MgSO4, 1.25 NaH2PO4, 20 NaHCO3 and 10 D-glucose. Recording was performed in a submerged chamber perfused with ACSF (32 °C). fEPSPs were recorded with platinum-iridium electrodes placed in the CA1 stratum radiatum. Bipolar platinum stimulating electrodes were placed in Schaffer collaterals. Baseline responses were measured with stimulation (0.017 Hz, 0.1 ms pulse duration) at an intensity (typically 20 – 30 μA) that evoked a response that was approximately one third of the maximum evoked response. LTP was induced with theta-burst stimulation (2 or 5 bursts, each burst consisting of four pulses at 100 Hz with a 200 ms inter–burst interval). Initial fEPSP slopes were measured and normalized to the average of baseline (with Clampfit 10.2).
Whole-cell voltage clamp recordings were done with an Axopatch 200B amplifier (Axon Instrument) as previously described31, 32. Coronal slices (350 μm) were prepared in ice-cold slice cutting solution containing the following (in mM), 140 2-hydroxy-N,N,N-trimethylethanaminium chloride (Choline Chloride), 3 Na-Pyruvate, 2.5, KCl, 1 CaCl2, 7 MgSO4, 26 NaHCO3, 30 D-glucose, 1 kynurenic acid, 1.3 Na-ascorbate. Patch electrodes (3–6 MΩ when filled) were filled with a solution containing the following (in mM): 140 Cs-methanesulfonate, 7 NaCl, 10 HEPES, 0.2 EGTA, 4 Mg-ATP, 0.3 Na-GTP, 5 QX-314. For mEPSC recordings, voltage clamp recordings were performed at -60 mV in the presence of 100 μM picrotoxin and 1 μM TTX. mIPSCs were measured at + 10 mV in the presence of 1 mM kynurenic acid and 1 μM TTX. Only recordings during which series resistance changed less than 20 % throughout the experiment were analyzed. mPSCs were analyzed with an in-house analysis software (EVAN)35. For AMPA/NMDA currents ratio experiments, recordings were performed in ACSF containing 100 μM picrotoxin. Pyramidal neurons in CA1 were voltage-clamped at − 65 mV, and AMPA–mediated EPSCs were evoked by stimulating with a bipolar platinum stimulating electrode at 0.1 Hz. After recording 15 responses, the holding potential was manually changed to + 40 mV to record NMDA receptor–mediated EPSCs. The AMPA/NMDA ratio was calculated by dividing the mean value of 15 AMPA–mediated EPSC peak amplitudes by the mean value of 15 NMDA receptor-mediated EPSC amplitudes measured at 50 ms after the onset of stimulation (Clampfit 10.2).
Western blot and immunohistochemistry
Dissected hippocampi were homogenized in protein lysis buffer (10mM Tris-Cl pH 6.8, 1.6 % SDS) containing protease and phosphatase inhibitor cocktails (Sigma). Supernatants were collected after centrifugation and the protein concentration was determined using a BCA assay kit (Thermo). Equal amounts of proteins (5 μg) were separated by electrophoresis on a 4 % – 12 % SDS–PAGE (Invitrogen), and then transferred to nitrocellulose membranes. After blocking with 5% BSA in TBS-T (Tris–buffer saline containing 0.1% Tween-20) for 1 hr at room temperature, membranes were hybridized with a primary antibody overnight at 4°C. After washing with TBS–T, membranes were incubated with a secondary antibody in 5% non-fat milk/TBS–T for 1 hr at room temperature. Signals were visualized by ECL (Thermo) and exposure time was adjusted so that the signals measured were in a linear range. After detecting phospho-Erk, the membranes were stripped and re-probed with a total Erk antibody. The total Erk levels were used to normalize each sample. The following primary antibodies were used: anti-phospho-Erk (#9101S, Cell Signaling, 1:6000), anti-total Erk (#9102S, Cell Signaling, 1:5000) and anti-SHP2 (sc-280, Santa Cruz, 1:3000).
For immunohistochemistry of SHP2, rAAV5-PTPN11D61G– or rAAV5–GFP–injected mice were perfused with ice-cold 4 % paraformaldehyde and the brains were removed, followed by post-fixation in 4 % paraformaldehyde overnight at 4 °C. Coronal brain sections (30 μm thick) were mounted onto slide glasses and were treated with 0.3 % H2O2 in methanol for 30 min to quench endogenous peroxidase activity. After blocking with 5 % normal goat serum in TBS–T (0.1 % Triton X–100), sections were incubated with anti-SHP2 antibody (1:100; Sc-280, Santa Cruz Biotechnology) for 48 hrs at 4°C. A biotinylated anti-rabbit antibody (1:50, 1 h at room temperature; Vector laboratories) was used as a secondary, which was followed by avidin-biotin-peroxidase complex (Vector Laboratories) formation for 30 min. Signals were visualized by incubating sections in DAB substrate solution (Vector Laboratories). For fluorescent immunohistochemistry of SHP2 and Gad67, anti-SHP2 antibody (1:100, Santa Cruz Biotechnology) and anti-Gad67 antibody (1:500, Millipore, MAB5406) were used as primary antibodies, anti-rabbit Alexa-568 (1:250) and anti-mouse Alexa-647 (1:250) were used as secondary antibodies. Images were acquired by using a confocal microscope (Olympus).
Sindbis viral vector construction and immunocytochemistry
The coding sequence of human PTPN11 with or without the D61G mutation was subcloned into a Sindbis viral expression vector (pSinRep5; Invitrogen) and GFP was inserted into the 3′ region of the coding sequence along with an additional subgenomic promoter for bicistronic expression. Sindbis viruses were produced according to the manufacturer’s protocol (Invitrogen) and directly added to the medium of cultured rat hippocampal neurons (DIV21). Twelve hours after infection, immunocytochemistry was performed with or without permeabilization by using anti-GluA1-N (#AGC-004, Alomone labs) antibody and Cy3–conjugated anti-rabbit IgG antibody (Jackson ImmunoResearch Lab). Images were acquired by using confocal microscope (Zeiss LSM 710) and analyzed by using ImageJ (ver. 1.42q).
Biotinylation of surface proteins
Rat cortical neurons (16–18 DIV) were transfected with the Sindbis virus encoding wild type or mutant (D61G) PTPN11 and allowed to be expressed for 12 h. The cultures were incubated with sulfo-NHS-SS-biotin (1 mg/ml, Thermo Scientific) in ice–cold PBS for 30 min at 4°C, followed by a 10 min incubation in ice-cold Tris buffer (100 mM, pH 8.0), and subsequently lysed with a lysis buffer [50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1 % Triton X-100, 0.1 % Sodium deoxycholate, 1X protease inhibitor (Roche)]. Biotinylated surface proteins were precipitated with Streptavidin agarose (Thermo Scientific) through overnight incubation. The precipitated beads were washed and used in Western blotting analysis. Antibodies were as follows: anti-GluA1-N (1:1000, Alomone labs), anti-Rab4 (1:2000, #6100889, BD Transduction Laboratories), anti-Cadherine (1:4000, sc-59876, Santa Cruz Biotechnology), anti-β-Actin (1:4000, A5316, Sigma-Aldrich), anti-SHP2 (1:2000, Santa Cruz Biotechnology).
Statistics
For water maze data, we used ANOVAs to analyze quadrant occupancy (% time spent in quadrant). After initial ANOVA analyses, searching specificity for each genotype was determined by comparing target quadrant to other quadrants using Dunnett’s Multiple Comparison Test. We also used two-way ANOVAs to analyze the interaction between genotypes and pool quadrants. In addition, we compared target quadrant occupancy among different groups by using the unpaired two-tailed t-test. Proximity measures between two genotypes also were analyzed by the unpaired two-tailed t-test. Effects of drug treatments on different genotypes were analyzed by using two-way ANOVA followed by appropriate post-hoc tests. LTP data were analyzed by using repeated-measures ANOVA followed by Bonferroni test on the responses after LTP induction and unpaired two-tailed t-test on the average of the last 10 min of recording. For other experiments, we used Student’s t-test to compare two groups and ANOVA to compare three or more groups. We did not use statistical methods to predetermine the sample sizes, but our sample sizes are similar to those reported in previously published papers31–33. Data distribution was assumed to be normal but this was not formally tested. All the data are represented as mean ± s.e.m.
A supplementary methods checklist is available.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Istvan Mody, Dr. Thomas O’Dell, Dr. Peyman Golshani and Silva lab members for their comments on the manuscript and for valuable discussions, Ryan Jones and Dr. Yu Zhou for helping with electrophysiological analysis, Dr. Denise Y. Cai for statistical advice, Aida Amin, Hwang Shan and Ryan Knier for technical support.
This work was supported by MH084315 to A.J.S. NRF–2013R1A1A1006766 and NRF–2013R1A3A1072570 to Y.–S.L, R37 CA49132 to B.G.N, MEST–2012–0005751 to H.K.K. B.G.N. is also a Canada Research Chair, Tier 1, and work in his lab is partially supported by the Ontario Ministry of Health and Long Term Care and the Princess Margaret Cancer Foundation.
Footnotes
Contributions
Y.–S.L. D.E. and A.J.S. conceptualized the research, designed the experiments and wrote the manuscript; Y. –S.L., D.E., M.Z., M.K., H.R., C.K. C.I.N. and Y.C. performed behavioral experiments; Y.-S.L performed whole-cell patch clamp recordings; Y. –S.L., M.Z. and Y.S. performed LTP recording and biochemical analyses; J–Y.O and H.K.K performed immunocytochemistry and biotinylation experiments; T.A. and B.G.N. provided Ptpn11D61G/+ and Ptpn11N308D/+ founders, discussed the results and edited the manuscript; D.B. and C.B. packaged viral vectors; Y. –S.L., D.E., M.Z. J.B., H.K.K, B.-K.K. analyzed the data and discussed the results.
References
- 1.Tartaglia M, Gelb BD. Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet. 2005;6:45–68. doi: 10.1146/annurev.genom.6.080604.162305. [DOI] [PubMed] [Google Scholar]
- 2.Romano AA, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics. 2010;126:746–759. doi: 10.1542/peds.2009-3207. [DOI] [PubMed] [Google Scholar]
- 3.Lee DA, Portnoy S, Hill P, Gillberg C, Patton MA. Psychological profile of children with Noonan syndrome. Dev Med Child Neurol. 2005;47:35–38. doi: 10.1017/s001216220500006x. [DOI] [PubMed] [Google Scholar]
- 4.van der Burgt I, et al. Patterns of cognitive functioning in school-aged children with Noonan syndrome associated with variability in phenotypic expression. J Pediatr. 1999;135:707–713. doi: 10.1016/s0022-3476(99)70089-2. [DOI] [PubMed] [Google Scholar]
- 5.Cesarini L, et al. Cognitive profile of disorders associated with dysregulation of the RAS/MAPK signaling cascade. Am J Med Genet A. 2009;149A:140–146. doi: 10.1002/ajmg.a.32488. [DOI] [PubMed] [Google Scholar]
- 6.Pierpont EI, et al. Genotype differences in cognitive functioning in Noonan syndrome. Genes Brain Behav. 2009;8:275–282. doi: 10.1111/j.1601-183X.2008.00469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verhoeven W, Wingbermuhle E, Egger J, Van der Burgt I, Tuinier S. Noonan syndrome: psychological and psychiatric aspects. Am J Med Genet A. 2008;146A:191–196. doi: 10.1002/ajmg.a.32115. [DOI] [PubMed] [Google Scholar]
- 8.Alfieri P, et al. Long term memory profile of disorders associated with dysregulation of the RAS-MAPK signaling cascade. Behav Genet. 2011;41:423–429. doi: 10.1007/s10519-011-9446-5. [DOI] [PubMed] [Google Scholar]
- 9.Pierpont EI, Tworog-Dube E, Roberts AE. Learning and memory in children with Noonan syndrome. Am J Med Genet A. 2013;161:2250–2257. doi: 10.1002/ajmg.a.36075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zenker M. Clinical manifestations of mutations in RAS and related intracellular signal transduction factors. Curr Opin Pediatr. 2011;23:443–451. doi: 10.1097/MOP.0b013e32834881dd. [DOI] [PubMed] [Google Scholar]
- 11.Neel BG, Gu H, Pao L. The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci. 2003;28:284–293. doi: 10.1016/S0968-0004(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 12.Sweatt JD. The neuronal MAP kinase cascade: a biochemical signal integration system subserving synaptic plasticity and memory. Journal of neurochemistry. 2001;76:1–10. doi: 10.1046/j.1471-4159.2001.00054.x. [DOI] [PubMed] [Google Scholar]
- 13.Fragale A, Tartaglia M, Wu J, Gelb BD. Noonan syndrome-associated SHP2/PTPN11 mutants cause EGF-dependent prolonged GAB1 binding and sustained ERK2/MAPK1 activation. Hum Mutat. 2004;23:267–277. doi: 10.1002/humu.20005. [DOI] [PubMed] [Google Scholar]
- 14.Araki T, et al. Mouse model of Noonan syndrome reveals cell type- and gene dosage-dependent effects of Ptpn11 mutation. Nat Med. 2004;10:849–857. doi: 10.1038/nm1084. [DOI] [PubMed] [Google Scholar]
- 15.Keilhack H, David FS, McGregor M, Cantley LC, Neel BG. Diverse biochemical properties of Shp2 mutants. Implications for disease phenotypes. The Journal of biological chemistry. 2005;280:30984–30993. doi: 10.1074/jbc.M504699200. [DOI] [PubMed] [Google Scholar]
- 16.Araki T, et al. Noonan syndrome cardiac defects are caused by PTPN11 acting in endocardium to enhance endocardial-mesenchymal transformation. Proc Natl Acad Sci U S A. 2009;106:4736–4741. doi: 10.1073/pnas.0810053106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris RG, Garrud P, Rawlins JN, O’Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- 18.Tartaglia M, et al. Diversity and functional consequences of germline and somatic PTPN11 mutations in human disease. American journal of human genetics. 2006;78:279–290. doi: 10.1086/499925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YS, Silva AJ. The molecular and cellular biology of enhanced cognition. Nat Rev Neurosci. 2009;10:126–140. doi: 10.1038/nrn2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier AS, et al. Control of CNS cell-fate decisions by SHP-2 and its dysregulation in Noonan syndrome. Neuron. 2007;54:245–262. doi: 10.1016/j.neuron.2007.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu JJ, Qin Y, Zhao M, Van Aelst L, Malinow R. Ras and Rap control AMPA receptor trafficking during synaptic plasticity. Cell. 2002;110:443–455. doi: 10.1016/s0092-8674(02)00897-8. [DOI] [PubMed] [Google Scholar]
- 22.Li W, et al. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- 23.Sebti SM, Tkalcevic GT, Jani JP. Lovastatin, a cholesterol biosynthesis inhibitor, inhibits the growth of human H-ras oncogene transformed cells in nude mice. Cancer Commun. 1991;3:141–147. doi: 10.3727/095535491820873371. [DOI] [PubMed] [Google Scholar]
- 24.Mailman T, Hariharan M, Karten B. Inhibition of neuronal cholesterol biosynthesis with lovastatin leads to impaired synaptic vesicle release even in the presence of lipoproteins or geranylgeraniol. Journal of neurochemistry. 2011;119:1002–1015. doi: 10.1111/j.1471-4159.2011.07474.x. [DOI] [PubMed] [Google Scholar]
- 25.Lee SH, et al. Synapses are regulated by the cytoplasmic tyrosine kinase Fer in a pathway mediated by p120catenin, Fer, SHP-2, and beta-catenin. J Cell Biol. 2008;183:893–908. doi: 10.1083/jcb.200807188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pagani MR, Oishi K, Gelb BD, Zhong Y. The phosphatase SHP2 regulates the spacing effect for long-term memory induction. Cell. 2009;139:186–198. doi: 10.1016/j.cell.2009.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc Natl Acad Sci U S A. 2006;103:4344–4351. doi: 10.1073/pnas.0600084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stornetta RL, Zhu JJ. Ras and Rap signaling in synaptic plasticity and mental disorders. The Neuroscientist: a review journal bringing neurobiology, neurology and psychiatry. 2011;17:54–78. doi: 10.1177/1073858410365562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tidyman WE, Rauen KA. The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev. 2009;19:230–236. doi: 10.1016/j.gde.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shilyansky C, Lee YS, Silva AJ. Molecular and cellular mechanisms of learning disabilities: a focus on NF1. Annu Rev Neurosci. 2010;33:221–243. doi: 10.1146/annurev-neuro-060909-153215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shilyansky C, et al. Neurofibromin regulates corticostriatal inhibitory networks during working memory performance. Proc Natl Acad Sci U S A. 2010;107:13141–13146. doi: 10.1073/pnas.1004829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui Y, et al. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costa RM, et al. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- 34.Zolotukhin S, et al. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods. 2002;28:158–167. doi: 10.1016/s1046-2023(02)00220-7. [DOI] [PubMed] [Google Scholar]
- 35.Hajos N, Nusser Z, Rancz EA, Freund TF, Mody I. Cell type- and synapse-specific variability in synaptic GABAA receptor occupancy. Eur J Neurosci. 2000;12:810–818. doi: 10.1046/j.1460-9568.2000.00964.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.