Summary
Maintenance of genomic integrity is critical during neurodevelopment, particularly in rapidly dividing cerebellar granule neuronal precursors that experience constitutive replication-associated DNA damage. As Dicer was recently recognized to have an unexpected function in the DNA damage response, we examined whether Dicer was important for preserving genomic integrity in the developing brain. We report that deletion of Dicer in the developing mouse cerebellum resulted in the accumulation of DNA damage leading to cerebellar progenitor degeneration, which was rescued with p53 deficiency; deletion of DGCR8 also resulted in similar DNA damage and cerebellar degeneration. Dicer deficiency also resulted in DNA damage and death in other rapidly dividing cells including embryonic stem cells and the malignant cerebellar progenitors in a mouse model of medulloblastoma. Together, these results identify an essential function of Dicer in resolving the spontaneous DNA damage that occurs during the rapid proliferation of developmental progenitors and malignant cells.
Keywords: dicer, replicative stress, medulloblastoma, apoptosis, p53, embryonic stem cells
Graphical Abstract
Rapidly proliferating cells undergo replication-associated DNA damage. Swahari et al. use the developing cerebellum and embryonic stem cells to show that Dicer is critical for resolving endogenous DNA damage and preventing cell death. Medulloblastomas also rely on Dicer for survival, suggesting Dicer inhibitors could be developed as a potential therapy.
Introduction
Dicer, a ribonuclease that processes small RNAs, has a well-established role in microRNA (miRNA) biogenesis (Bartel, 2004). As miRNAs can target hundreds to thousands of genes, deletion of Dicer is known to affect diverse physiological and pathological pathways including development, metabolism, proliferation, apoptosis, and cancer (Calin and Croce, 2006; He and Hannon, 2004). Indeed, studies that have investigated the consequences of Dicer deletion have focused primarily on linking the observed phenotype with dysregulation of miRNAs. Recently, however, an unexpected miRNA-independent function of Dicer was identified (Francia et al., 2012; Wei et al., 2012). Specifically, Dicer-mediated processing of small noncoding RNAs (ncRNAs) was shown to be required for the DNA damage response (DDR) in the presence of exogenous DNA damage. These ncRNAs, which correspond to the sites of DNA double strand breaks, are thought to act as templates for efficient DNA repair (Chowdhury et al., 2013; Sharma and Misteli, 2013; Tang and Ren, 2012). Dicer-deficient cells were incapable of generating DDR-associated ncRNAs (DDRNAs) and, as a consequence, were unable to promote effective repair of the damaged DNA.
The discovery of this function of Dicer opens the possibility that the embryonic lethality seen in Dicer-deficient mice (Bernstein et al., 2003) may not be entirely due to the consequences of disrupting the canonical miRNA pathway but could also be due to this critical function of Dicer in DDR. This is important because, during development, cells undergoing rapid proliferation are known to experience replication-associated DNA damage (McKinnon, 2013). Whether Dicer is required for the efficient repair of replication-associated DNA damage has not been previously examined.
Results and Discussion
To determine whether Dicer is important for resolving replication-associated DNA damage during development, we examined the developing cerebellum, which is associated with massive expansion of the cerebellar granule neuron precursors (CGNPs). CGNP proliferation, which peaks between postnatal days 5 and 8, is driven by the Sonic Hedgehog (Shh) signaling pathway (Hatten and Roussel, 2011). The proliferative region of the cerebellum is spatially distinct, as CGNPs proliferate in the external granular layer (EGL), exit the cell cycle and migrate to become terminally differentiated cerebellar granule neurons (CGNs) of the internal granular layer (IGL) (Hatten and Roussel, 2011). Importantly, rapid proliferation of CGNPs during cerebellar development is known to be associated with replicative stress (Hatten and Roussel, 2011; Lee et al., 2012a, 2012b; Murga et al., 2009).
We found that Dicer expression correlates with the period of rapid proliferation in the developing cerebellum. Dicer mRNA and protein were high in cerebellar lysates at P7 (when proliferation is active) but downregulated by P20 (when the proliferation period is over) (Figures 1Afig1 and S1A). Dicer levels were also high in the proliferating CGNPs in vitro (Figure 1B). At P7, the majority of cells expressing Dicer in the cerebellum were proliferating CGNPs in the EGL (Figures 1C, S1B, and S1C). As these cells differentiate into cerebellar neurons and migrate to the IGL, Dicer staining becomes markedly reduced by P20 (Figure 1D).
Figure 1. Dicer Is Highly Expressed in Proliferating CGNPs, and Its Deletion Leads to Cerebellar Progenitor Degeneration.
(A) Western blotting analysis of cerebellar lysates from P7 and P20 wild-type mice. β-actin served as a loading control.
(B) CGNPs isolated from P5 wild-type mice cultured with (Shh) or without (Veh) Sonic Hedgehog. Cyclin D2 served as a marker of proliferation, and tubulin was used as a loading control.
(C and D) Immunohistochemical staining of Dicer in wild-type cerebella (P7 and P20, respectively) counterstained with the hematoxylin nuclear stain. Lower panels show magnified images (20×) of the boxed area.
(E and F) H&E staining of P7 wild-type and DicerMath1-Cre cerebella (E); quantification of cell number in an equivalent unit area of the EGL is shown in (F).
(G and H) H&E staining of P20 wild-type and DicerMath1-Cre cerebella (G); quantification of cell number in an equivalent unit area of the IGL is shown in (H).
(I and J) Immunohistochemical staining of cleaved caspase-3 (cC3) in P4 wild-type and DicerMath1-Cre cerebella (I); quantification of cell number is shown in (J).
(K) Western blotting analysis of P4 wild-type, Dicerf/+; Math1-Cre, and DicerMath1-Cre. β-actin served as a loading control.
Scale bars, 300 μm (D, E, and G) or 100 μm (I) and 40 μm for magnified images in (C) and (D). **p < 0.01 (two-tailed, unpaired Student's t test). Error bars, means ± SEM. CB, cerebellum; IGL, internal granular layer; EGL, external granular layer; PL, Purkinje layer.
Dicer Deficiency Leads to Cerebellar Progenitor Degeneration
To investigate the role of Dicer in proliferating CGNPs in vivo, we generated mice in which Dicer could be deleted in the CGNPs using the Math1-Cre transgenic mice (Machold and Fishell, 2005) (Figures S1D–S1F). Dicerf/f; Math1-Cre (hereafter, DicerMath1-Cre) mice were viable at birth and were born at the expected Mendelian ratio. However, these mice developed ataxia and died at around P100, compared to wild-type (WT) and Dicer heterozygous mice (these control mice are hereafter referred to as DicerCtrl) (Figure S1G).
DicerMath1-Cre mice showed a striking phenotype with extensive loss of CGNPs during cerebellar development. Loss of CGNPs was detectable starting as early as P2 and resulted in the near complete absence of CGNs at P20 (Figures 1E–1H; data not shown). Interestingly, in some of these mice, the degeneration was restricted to the anterior half of the cerebellum. Immunohistochemistry of Dicer in the cerebella of these mice revealed that Dicer recombination occurred exclusively in the anterior half at this time point (Figure S1H). Indeed, previous studies have reported on the incidence of incomplete recombination in the posterior half in this Math1-Cre line (Lorenz et al., 2011; Machold and Fishell, 2005).
We investigated whether the reduced number of CGNPs at P7 was due to a decreased rate of proliferation or an increased rate of apoptosis. Cerebellar tissues from P4 mice were probed with phospho-histone H3 (pH3), Ki-67, and bromodeoxyuridine (BrdU) incorporation, markers of proliferation, and cleaved caspase-3, a marker of apoptosis. Minimal differences in rates of proliferation were detected between wild-type and Dicer-deficient cerebella (Figures S1I–S1N). In contrast, Dicer deficiency resulted in a marked increase in cleaved caspase-3 in Dicer-deficient CGNPs (Figures 1I–1K). Thus, the loss of CGNPs in the developing cerebellum in Dicer-deficient mice appeared to be a consequence of increased apoptosis in CGNPs.
Dicer Deletion Alone Results in the Accumulation of DNA Damage
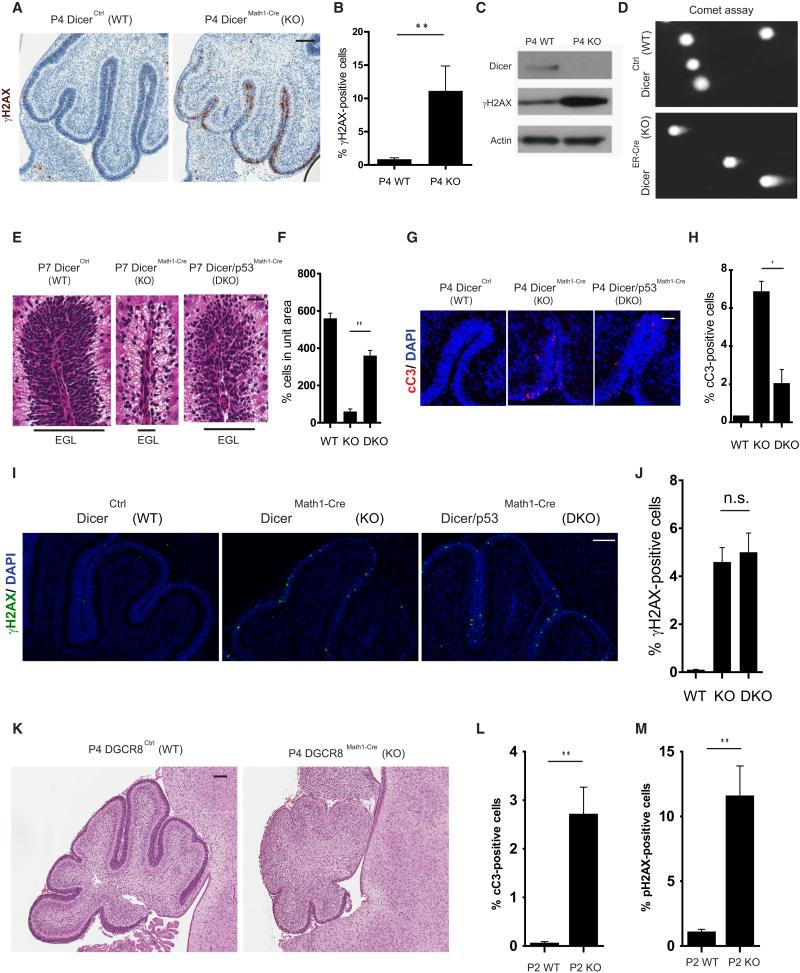
The recently described miRNA-independent function of Dicer in DDR (Francia et al., 2012; Wei et al., 2012) suggests that the cerebellar hypoplasia seen with Dicer deficiency could be a consequence of the inability of the CGNPs to repair the replication-associated DNA damage during this period of rapid proliferation. To specifically examine this possibility, we probed wild-type and Dicer-deficient cerebella for γH2AX (phosphorylation of the histone variant H2AX at serine 139) foci, a well-established marker for DNA damage (Rogakou et al., 1998). Indeed, DicerMath1-Cre CGNPs exhibited a marked increase in γH2AX foci as compared to the DicerCtrl CGNPs (Figures 2Afig2, 2B, S2A, and S2B). An increase in γH2AX levels was also detected in the cerebellar lysates from DicerMath1-Cre (Figure 2C). Consistent with the fact that replicating cells undergo DNA damage breaks with Dicer deficiency, the γH2AX staining is seen in the CGNPs that are positive for the proliferation marker PCNA (Figure S2C). Furthermore, 53BP1, another marker of DNA damage, also showed increased staining in the DicerMath1-Cre CGNPs (Figures S2D and S2E).
Figure 2. Dicer Deletion Results in the Accumulation of DNA Damage in CGNPs.
(A and B) Immunohistochemical staining of γH2AX in P4 wild-type and DicerMath1-Cre cerebella (A); quantification of γH2AX-positive cells is shown in (B).
(C) Western blotting analysis for Dicer and γH2AX of P4 wild-type and DicerMath1-Cre cerebellar lysates. β-actin served as a loading control.
(D) Representative images of the comet assay (single-cell gel electrophoresis) performed in tamoxifen-treated CGNPs isolated from wild-type and DicerER-Cre mice.
(E and F) H&E staining of P7 WT, DicerMath1-Cre, and Dicer/p53Math1-Cre cerebella (E); quantification of cell number in the EGL of the cerebellum is shown in (F).
(G and H) Immunohistochemical staining of cleaved caspase-3 in DicerCtrl (WT), DicerMath-1Cre (KO), and Dicer/p53Math1-Cre (DKO) cerebella at P4 (G); quantification of cC3-positive cells is shown in (H).
(I and J) Immunohistochemical staining of γH2AX in DicerCtrl (WT), DicerMath-1Cre (KO), and Dicer/p53Math1-Cre (DKO) cerebella at P4 (I); quantification of γH2AX-positive cells is shown in (J).
(K–M) H&E staining of P4 WT and DGCR8Math1-Cre cerebella (K); quantification of γH2AX- and cC3-positive cells in P2 WT and DGCR8Math-1Cre cerebella are shown in (L) and (M), respectively.
Scale bars, 100 μm (A, I, and K) and 30 μm (E and G). **p < 0.01 (two-tailed, unpaired Student's t test). Error bars, means ± SEM.
Additionally, we used the comet assay (Singh et al., 1988) to directly detect the DNA damage caused by deletion of Dicer. To enable the efficient deletion of Dicer in CGNPs in culture, we first generated a tamoxifen-inducible Cre mouse model of Dicer deletion (DicerER-Cre) by crossing Dicer floxed mice with CAGG-Cre-ER mice. CGNPs isolated from P5 DicerER-Cre mice were treated with 4OH-Tamoxifen (1 μM) for 48 hr to induce recombination at the Dicer locus (Figure S2F). These Dicer-deleted CGNPs exhibited features of DNA damage as evidenced by increased comet tail moments (Figure 2D). This result confirms not only that deletion of Dicer alone results in DNA damage, but also that this process is cell autonomous.
Consistent with the accumulation of DNA damage, CGNPs in the EGL of DicerMath1-Cre mice also showed increased p53 staining (Figures S2G and S2H). To examine whether the cerebellar progenitor degeneration seen with Dicer deficiency was a consequence of DNA damage activating a p53-mediated apoptotic pathway, we crossed the DicerMath1-Cre mice with p53-deficient mice. Our results show that the apoptotic degeneration of CGNPs with Dicer deficiency could be rescued with co-deletion of p53 (Figures 2E–2H). The inability of p53 deficiency to completely rescue this DNA damage phenotype is consistent with previous observations that p53 deficiency only partially rescues the DNA damage phenotype seen with ATR deficiency in the developing cerebellum (Lee et al., 2012b). Importantly, γH2AX staining was still evident in these Dicer and p53 co-deleted mice, indicating that the DNA damage precedes cell death, and that the increase in γH2AX seen with Dicer deficiency is not simply a consequence of cell death (Figures 2I and 2J). Together, these results suggest that the cerebellar progenitor degeneration seen with Dicer deficiency is a consequence of endogenous DNA damage inducing cell death in the rapidly proliferating CGNPs.
Previous studies have shown that, in addition to Dicer, Drosha is also important for efficient DDR (Francia et al., 2012). In mammalian cells, Drosha and DGCR8 together form the microprocessor complex that processes small RNAs (Macias et al., 2013). Thus, we examined whether DGCR8 was also important for resolving DNA damage during cerebellar development. Specifically, we crossed the DGCR8 floxed mice with Math1-Cre mice to generate the DGCR8Math1-Cre mice to conditionally delete DGCR8 in the CGNPs. These mice also exhibited marked accumulation of DNA damage and cerebellar degeneration during development just as seen with Dicer deficiency (Figures 2K–2M).
DNA Damage with Dicer Deficiency in Other Rapidly Proliferating Cells
To examine whether Dicer deficiency also induces replication-associated DNA damage in other proliferative regions of the brain, we generated a Dicer hGFAP-Cre mouse where recombination occurs in various parts of the brain including primitive neural precursors, the dentate gurus of the hippocampus and cerebellum (Zhuo et al., 2001). Dicerf/f; hGFAP-Cre (hereafter, DicerhGFAP-Cre) mice were viable but exhibited marked ataxia and growth defects and died at around P20 (data not shown). Consistent with our results with DicerMath1-Cre mice, we detected cerebellar progenitor degeneration with increased γH2AX staining in DicerhGFAP-Cre mice (Figures S3A and S3B). Strikingly, Dicer deletion also resulted in increased γH2AX foci and degeneration of the dentate gyrus, which corresponds to the area undergoing proliferation during postnatal development (Figures S3C and S3D).
To investigate the importance of this function of Dicer in non-neuronal cells, we examined mouse embryonic stem cells (mESCs), which are known to proliferate rapidly, undergo replicative stress (Tichy and Stambrook, 2008), and have higher baseline levels of γH2AX as compared to other less rapidly dividing cells (Banáth et al., 2009). We found Dicer levels to also be higher in mESCs as compared to mouse embryonic fibroblasts (Figure S3E). To determine whether Dicer was important for DDR in mESCs, we examined the outcome of knocking down Dicer in these cells. Our results show that knockdown of Dicer markedly increased γH2AX foci and cell death in mESCs (Figures 3A–3Ffig3). To examine whether Dicer was specifically important for resolving replicative stress-induced DNA damage, we treated mESCs with hydroxyurea, a known inducer of replicative stress. Our results show that Dicer knockdown sensitized mESCs to a low dose of hydroxyurea (1 μM) (Figures S3F and S3G). Importantly, consistent with our results in the CGNP model, cell death seen with Dicer inhibition in mESCs was also p53 dependent, as knockdown of p53 reduced the Dicer-deficiency-induced mESC death (Figures 3E and 3F).
Figure 3. Dicer Knockdown in mESCs Results in Increased DNA Damage and Cell Death.
(A and B) Western blotting analysis of control- and siDicer-transfected mESCs (A). Tubulin served as a loading control. Quantification of γH2AX levels is shown in (B).
(C and D) γH2AX staining (green) of control- and siDicer-transfected mESCs (C). DAPI (blue) stains the nucleus. Quantification of the number of mESCs with γH2AX foci is shown in (D).
(E and F) Propidium iodide (PI) staining in control-, siDicer-, and siDicer/p53-transfected mESCs (E). Quantification of the fraction of PI-positive cells is shown in (F).
Scale bars, 20 μm (C) or 50 μm (E). *p < 0.05 (two-tailed, unpaired Student's t test). Error bars, means ± SEM.
Dicer Deletion Alone Increases Spontaneous DNA Damage in Medulloblastoma and Reduces Tumor Growth
To determine whether the function of Dicer in DDR was also important for resolving replication-associated DNA damage in rapidly proliferating cancers, we utilized the SmoM2 medulloblastoma tumor model. SmoM2 mice express a Smoothened mutation that constitutively activates the Shh pathway in CGNPs, with all mice developing aggressive tumors by P20 (Mao et al., 2006). To assess the function of Dicer in medulloblastoma, we generated Dicerf/f Math1-Cre; SmoM2 (Dicer-deficient SmoM2) and Dicer+/+ Math1-Cre; SmoM2 (wild-type SmoM2) mice. Dicer-deficient SmoM2 mice developed markedly smaller tumors compared to wild-type SmoM2 mice (Figures 4A and 4Bfig4). Interestingly, the reduced tumor volume was not a consequence of reduced proliferation, as no differences were found in pH3 staining between Dicer-deficient and wild-type SmoM2 cerebella at P4 (Figures 4C and 4D). Instead, the Dicer-deficient SmoM2 cerebella at P4 exhibited a marked increase in DNA damage and apoptosis as detected by γH2AX and cleaved caspase-3 staining (Figures 4E and 4H).
Figure 4. Dicer Deletion Leads to Reduced Tumor Volume in the SmoM2 Mouse Model of Medulloblastoma.
(A) H&E analysis of P20 WT (WT SmoM2) and Dicer-deleted medulloblastoma (KO SmoM2).
(B) Quantification of tumor load in (A).
(C and D) Phospho-histone H3 (pH3) staining of P4 WT SmoM2 and Dicer KO SmoM2 (C); quantification of pH3-positive cells is shown in (D).
(E and F) γH2AX staining of P4 WT SmoM2 and Dicer KO SmoM2 (E); quantification of γH2AX-positive cells is shown in (F).
(G and H) Cleaved caspase-3 (cC3) staining of P4 WT SmoM2 and Dicer KO SmoM2 (G); quantification of cC3-positive cells is shown in (H).
(I) Immunohistochemical staining for cC3 in P18 WT and Dicer KO SmoM2 mice that were injected with etoposide (5 mg/kg). Uninjected mice were used as controls.
(J) Quantification of cC3-positive cells is shown in (I).
Scale bars, 500 μm (A) or 300 μm (C, E, and G). *p < 0.05, **p < 0.01 (two-tailed, unpaired Student's t test). Error bars, means ± SEM.
Interestingly, once the tumors emerged, the background rates of apoptosis in wild-type and Dicer-deficient tumors at P18 was comparable (Figures 4I and 4J). We examined whether these Dicer-deficient medulloblastoma tumors were more sensitive to chemotherapy. Wild-type and Dicer-deficient SmoM2 mice were injected with etoposide at P18 and analyzed for cleaved caspase-3 24 hr later. Dicer-deficient tumors were indeed more sensitive to etoposide and exhibited increased cell death compared to wild-type tumors (Figures 4I and 4J). Together, these results show that the role of Dicer in DDR in the developing brain also extends to the context of rapidly proliferating tumor cells in medulloblastoma.
While the function of Dicer in miRNA biogenesis is well known, the emerging evidence indicating that Dicer is also important for processing other small RNAs (Johanson et al., 2013) opens the possibility that some of the phenotypes seen with Dicer deletion could be independent of miRNAs. In particular, studies in Arabidopsis and mammalian cell culture models, where DNA damage was induced either by radiation or by engineering site-specific breaks, identified Dicer-processed ncRNAs corresponding to the sites of DNA damage that were important for DDR (Francia et al., 2012; Wei et al., 2012). Our results show that this function of Dicer in DDR may be particularly important in development where rapidly proliferating cells have to cope with endogenous DNA damage generated as a result of replicative stress. Loss of key DNA damage signaling and repair proteins including ATR, TopBP1, DNA ligase IV, Xrcc2, and NBS1 is known to be sufficient to trigger degeneration of the cerebellum and other neural progenitors (Barnes et al., 1998; Deans et al., 2000; Frappart et al., 2005; McKinnon, 2013). Loss of Dicer also appears to trigger a similar response with increased DNA damage and degeneration of the cerebellum, which is rescued with p53 deficiency. These results suggest that the primary cause of cell death with Dicer deficiency may not be the global disruption of miRNA biogenesis but rather a more direct consequence of DNA damage. Consistent with this, we did not observe any marked changes in the expression of key DNA damage response genes in the Dicer-deficient brain (Figure S4). However, we cannot completely rule out the possibility that the DNA damage phenotype could be caused by the deficiency of a few miRNA that are specifically important for DNA damage repair. Indeed, it is challenging to precisely discern the miRNA-dependent and -independent functions of Dicer particularly in the context of replication-associated DNA damage. We also performed small RNA sequencing in proliferating wild-type cerebellum. Although we could not detect DDRNAs or double-strand break-induced small RNAs (diRNAs) that corresponded to any sites of DNA damage (data not shown), it is indeed very challenging to detect such low-frequency small RNAs as DNA damage during development likely occurs at very low levels and is spread throughout the genome. More detailed studies are needed in the future to functionally examine the presence of DDRNAs in proliferating cerebellum and medulloblastoma. Importantly, our results identify a previously unappreciated essential function of Dicer and DGCR8 in maintaining genomic integrity during development.
Previous studies that generated mice with conditional deletions of Dicer in the developing brain have also reported striking cellular degeneration phenotypes. For example, deletion of Dicer in the neural progenitors of the developing cortex with Emx1-Cre (De Pietri Tonelli et al., 2008; Kawase-Koga et al., 2009), Nestin-Cre (Kawase-Koga et al., 2009; McLoughlin et al., 2012; Zindy et al., 2015), hGFAP-Cre (Nigro et al., 2012), or Foxg1-Cre (Makeyev et al., 2007; Nowakowski et al., 2011) induces cell death resulting in cortical and forebrain thinning. In contrast, deletion of Dicer in postmitotic neurons with CaMKII-Cre (Davis et al., 2008; Hébert et al., 2010; Konopka et al., 2010), Nex-Cre (Hong et al., 2013; Volvert et al., 2014), and DR-1-Cre (Cuellar et al., 2008) affects neuronal functions but has a relatively modest effect on cell survival. The different outcomes of Dicer deletion in rapidly dividing neural progenitors versus postmitotic neurons are also consistent with our results that point to an essential function of Dicer in resolving replication-associated DNA damage.
A pathological context in which rapidly proliferating cells are known to undergo replicative stress is tumors (Burrell et al., 2013). Previous studies that have deleted Dicer in primary tumor models have reported that Dicer deficiency is incompatible with tumor growth (Kumar et al., 2009). In contrast, deletion of one copy of Dicer accelerates tumor growth in multiple models, including in medulloblastomas (Lambertz et al., 2010; Zhang et al., 2013; Zindy et al., 2015). Likewise, while biallelic mutations that result in complete loss of Dicer function are very rare, mutations in one Dicer allele have been associated with cancers in humans (Foulkes et al., 2014). From the perspective of Dicer function in DDR, one reason why partial, but not complete, loss of Dicer is associated with cancers could be that reduced Dicer function results in an increased but sublethal rate of DNA damage that is tumorigenic. Complete loss of Dicer would result in the marked increase in DNA damage and cell death, as shown in our medulloblastoma model.
Together, our findings highlight the emerging importance of Dicer in DDR. As shown here, the function of Dicer in resolving endogenous DNA damage is particularly critical in rapidly proliferating cells during development, a task that also appears to be co-opted in tumors.
Experimental Procedures
Mice
Dicerf/f mice were kindly provided by Dr. Gregory Hannon (Cold Spring Harbor Laboratory). Math1-Cre mice were generously shared by David Rowitch (University of California, San Francisco) and Robert Wechsler-Reya (Sanford-Burnham Medical Research Institute, La Jolla, CA). hGFAP-Cre mice were generously provided by Eva Anton (University of North Carolina; UNC). ER-Cre (CAG-Cre/Esr1; strain: 004453), DGCR8f/f, p53f/f, and SmoM2 mice (strain: 5130) were obtained from The Jackson Laboratory. All animal handling and protocols were carried out in accordance with the Animal Care and Use Committee of UNC.
Comet Assay
Comet Assay (single-cell gel electrophoresis) was performed according to the manufacturer's instructions (Trevigen). In brief, CGNPs from DicerCtrl and DicerER-Cre were cultured in the presence of 2 μM 4-OH Tamoxifen for 24–48 hr. The cells were scraped and washed once with ice-cold 1 × PBS (Gibco). Cells were resuspended at 105 cells/ml in PBS and mixed with molten LMAgarose (at 37°C) at a ratio of 1:10. 50 μl of the mixture was immediately pipetted onto the CometSlide. The slide was immersed first in lysis solution and then in alkaline unwinding solution. The electrophoresis was performed using the alkaline electrophoresis solution at 21 V for 30 min. The slides were dried and immersed in SYBR Gold solution and were visualized on a DMIRE2 inverted fluorescence microscope (Leica). The experiments were done at least in triplicate.
Immunohistochemistry
IHC was carried out in the Bond stainer (Leica). In brief, slides were dewaxed in Bond Dewax solution) and hydrated in bond wash solution. Antigen retrieval for antibodies was performed for 30 min at 100°C in bond-epitope retrieval solution 1 (pH 6.0). Slides were incubated with primary antibody for 1 hr. Primary antibodies used were Dicer 13D6 (Abcam), cleaved caspase-3 (Cell Signaling Technology), γH2AX (Cell Signaling Technology), p27-Kip1 (Dako), PCNA (Cell Signaling Technology), BrdU (AbD Serotec), Ki-67 (Leica), phospho-histone H3 (Cell Signaling Technology), and NeuN (Millipore). Nuclei were counterstained with hematoxylin or DAPI. Antibody detection was performed using the Bond Polymer Refine Detection System (DS9800). Stained slides were dehydrated and coverslipped. Stained slides were digitally imaged at 20× magnification using the Aperio ScanScope XT (Aperio Technologies), and digital images were stored in the Aperio eSlide Manager Database at Translation Pathology Laboratory (TPL).
Cell Counts
The expression of all markers was measured in manually annotated regions using the Nuclear v9 algorithm (Aperio Technologies), with minor adjustments for stain optical density and nuclear shape. The intensity score (1+ = weak positive, 2+ = moderately positive, and 3+ = strong positive), and the percentage of positive cells for each score was used to calculate the H-score using the formula
Supplementary Material
Highlights.
Dicer is highly expressed in rapidly proliferating cells (e.g., CGNPs and ESCs)
Dicer is essential for resolving replication-associated DNA damage in these cells
Degeneration seen with Dicer deficiency can be partially rescued by deletion of p53
Dicer deficiency induces DNA damage and reduces tumor burden in medulloblastomas
Acknowledgments
We thank the members of M.D.'s lab for critical review of this manuscript and insightful discussions. We thank Stephanie Cohen, Mervi Eeva, Mark Vincent Olorvida, Ying Li, Albert Wigus, and Bentley Midkiff at the UNC TPL for expert technical assistance. We also appreciate the technical assistance provided by Janice Weaver and Carolyn Suitt at the UNC Animal Histopathology and the Center for Gastrointestinal Biology and Disease, respectively. The UNC TPL is supported in part by grants from the National Cancer Institute (3P30CA016086) and the UNC University Cancer Research Fund. This work was supported by NIH grants NS042197 and NS095650 to M.D. and DK091318 to P.S.
Footnotes
Supplemental Information
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at *bxs.
Supplemental Information
Document S1. Supplemental Experimental Procedures and Figures S1–S4
Document S2. Article plus Supplemental Information
References
- Banáth JP, Bañuelos CA, Klokov D, MacPhail SM, Lansdorp PM, Olive PL. Explanation for excessive DNA single-strand breaks and endogenous repair foci in pluripotent mouse embryonic stem cells. Exp. Cell Res. 2009;315:1505–1520. doi: 10.1016/j.yexcr.2008.12.007. [DOI] [PubMed] [Google Scholar]
- Barnes DE, Stamp G, Rosewell I, Denzel A, Lindahl T. Targeted disruption of the gene encoding DNA ligase IV leads to lethality in embryonic mice. Curr. Biol. 1998;8:1395–1398. doi: 10.1016/s0960-9822(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller M-C, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. Replication stress links structural and numerical cancer chromosomal instability. Nature. 2013;494:492–496. doi: 10.1038/nature11935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Choi YE, Brault ME. Charity begins at home: non-coding RNA functions in DNA repair. Nat. Rev. Mol. Cell Biol. 2013;14:181–189. doi: 10.1038/nrm3523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuellar TL, Davis TH, Nelson PT, Loeb GB, Harfe BD, Ullian E, McManus MT. Dicer loss in striatal neurons produces behavioral and neuroanatomical phenotypes in the absence of neurodegeneration. Proc. Natl. Acad. Sci. USA. 2008;105:5614–5619. doi: 10.1073/pnas.0801689105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TH, Cuellar TL, Koch SM, Barker AJ, Harfe BD, McManus MT, Ullian EM. Conditional loss of Dicer disrupts cellular and tissue morphogenesis in the cortex and hippocampus. J. Neurosci. 2008;28:4322–4330. doi: 10.1523/JNEUROSCI.4815-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers JN, Haffner C, Murchison EP, Hannon GJ, Huttner WB. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. Development. 2008;135:3911–3921. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans B, Griffin CS, Maconochie M, Thacker J. Xrcc2 is required for genetic stability, embryonic neurogenesis and viability in mice. EMBO J. 2000;19:6675–6685. doi: 10.1093/emboj/19.24.6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Priest JR, Duchaine TF. DICER1: mutations, microRNAs and mechanisms. Nat. Rev. Cancer. 2014;14:662–672. doi: 10.1038/nrc3802. [DOI] [PubMed] [Google Scholar]
- Francia S, Michelini F, Saxena A, Tang D, de Hoon M, Anelli V, Mione M, Carninci P, d'Adda di Fagagna F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature. 2012;488:231–235. doi: 10.1038/nature11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frappart P-O, Tong W-M, Demuth I, Radovanovic I, Herceg Z, Aguzzi A, Digweed M, Wang Z-Q. An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat. Med. 2005;11:538–544. doi: 10.1038/nm1228. [DOI] [PubMed] [Google Scholar]
- Hatten ME, Roussel MF. Development and cancer of the cerebellum. Trends Neurosci. 2011;34:134–142. doi: 10.1016/j.tins.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hébert SS, Papadopoulou AS, Smith P, Galas M-C, Planel E, Silahtaroglu AN, Sergeant N, Buée L, De Strooper B. Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet. 2010;19:3959–3969. doi: 10.1093/hmg/ddq311. [DOI] [PubMed] [Google Scholar]
- Hong J, Zhang H, Kawase-Koga Y, Sun T. MicroRNA function is required for neurite outgrowth of mature neurons in the mouse postnatal cerebral cortex. Front. Cell. Neurosci. 2013;7:151. doi: 10.3389/fncel.2013.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanson TM, Lew AM, Chong MMW. MicroRNA-independent roles of the RNase III enzymes Drosha and Dicer. Open Biol. 2013;3:130144. doi: 10.1098/rsob.130144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase-Koga Y, Otaegi G, Sun T. Different timings of Dicer deletion affect neurogenesis and gliogenesis in the developing mouse central nervous system. Dev. Dyn. 2009;238:2800–2812. doi: 10.1002/dvdy.22109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka W, Kiryk A, Novak M, Herwerth M, Parkitna JR, Wawrzyniak M, Kowarsch A, Michaluk P, Dzwonek J, Arnsperger T, et al. MicroRNA loss enhances learning and memory in mice. J. Neurosci. 2010;30:14835–14842. doi: 10.1523/JNEUROSCI.3030-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, Kirsch DG, Golub TR, Jacks T. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23:2700–2704. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambertz I, Nittner D, Mestdagh P, Denecker G, Vandesompele J, Dyer MA, Marine JC. Monoallelic but not biallelic loss of Dicer1 promotes tumorigenesis in vivo. Cell Death Differ. 2010;17:633–641. doi: 10.1038/cdd.2009.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Katyal S, Downing SM, Zhao J, Russell HR, McKinnon PJ. Neurogenesis requires TopBP1 to prevent catastrophic replicative DNA damage in early progenitors. Nat. Neurosci. 2012a;15:819–826. doi: 10.1038/nn.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Shull ER, Frappart PO, Katyal S, Enriquez-Rios V, Zhao J, Russell HR, Brown EJ, McKinnon PJ. ATR maintains select progenitors during nervous system development. EMBO J. 2012b;31:1177–1189. doi: 10.1038/emboj.2011.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Deutschmann M, Ahlfeld J, Prix C, Koch A, Smits R, Fodde R, Kretzschmar HA, Schüller U. Severe alterations of cerebellar cortical development after constitutive activation of Wnt signaling in granule neuron precursors. Mol. Cell. Biol. 2011;31:3326–3338. doi: 10.1128/MCB.05718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machold R, Fishell G. Math1 is expressed in temporally discrete pools of cerebellar rhombic-lip neural progenitors. Neuron. 2005;48:17–24. doi: 10.1016/j.neuron.2005.08.028. [DOI] [PubMed] [Google Scholar]
- Macias S, Cordiner RA, Cáceres JF. Cellular functions of the microprocessor. Biochem. Soc. Trans. 2013;41:838–843. doi: 10.1042/BST20130011. [DOI] [PubMed] [Google Scholar]
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell. 2007;27:435–448. doi: 10.1016/j.molcel.2007.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, McMahon AP. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon PJ. Maintaining genome stability in the nervous system. Nat. Neurosci. 2013;16:1523–1529. doi: 10.1038/nn.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoughlin HS, Fineberg SK, Ghosh LL, Tecedor L, Davidson BL. Dicer is required for proliferation, viability, migration and differentiation in corticoneurogenesis. Neuroscience. 2012;223:285–295. doi: 10.1016/j.neuroscience.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murga M, Bunting S, Montaña MF, Soria R, Mulero F, Cañamero M, Lee Y, McKinnon PJ, Nussenzweig A, Fernandez-Capetillo O. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat. Genet. 2009;41:891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro A, Menon R, Bergamaschi A, Clovis YM, Baldi A, Ehrmann M, Comi G, De Pietri Tonelli D, Farina C, Martino G, Muzio L. MiR-30e and miR-181d control radial glia cell proliferation via HtrA1 modulation. Cell Death Dis. 2012;3:e360. doi: 10.1038/cddis.2012.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowakowski TJ, Mysiak KS, Pratt T, Price DJ. Functional dicer is necessary for appropriate specification of radial glia during early development of mouse telencephalon. PLoS ONE. 2011;6:e23013. doi: 10.1371/journal.pone.0023013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Sharma V, Misteli T. Non-coding RNAs in DNA damage and repair. FEBS Lett. 2013;587:1832–1839. doi: 10.1016/j.febslet.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- Tang K-F, Ren H. The role of dicer in DNA damage repair. Int. J. Mol. Sci. 2012;13:16769–16778. doi: 10.3390/ijms131216769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Stambrook PJ. DNA repair in murine embryonic stem cells and differentiated cells. Exp. Cell Res. 2008;314:1929–1936. doi: 10.1016/j.yexcr.2008.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volvert M-L, Prévot P-P, Close P, Laguesse S, Pirotte S, Hemphill J, Rogister F, Kruzy N, Sacheli R, Moonen G, et al. MicroRNA targeting of CoREST controls polarization of migrating cortical neurons. Cell Rep. 2014;7:1168–1183. doi: 10.1016/j.celrep.2014.03.075. [DOI] [PubMed] [Google Scholar]
- Wei W, Ba Z, Gao M, Wu Y, Ma Y, Amiard S, White CI, Rendtlew Danielsen JM, Yang YG, Qi Y. A role for small RNAs in DNA double-strand break repair. Cell. 2012;149:101–112. doi: 10.1016/j.cell.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Zhang B, Chen H, Zhang L, Dakhova O, Zhang Y, Lewis MT, Creighton CJ, Ittmann MM, Xin L. A dosage-dependent pleiotropic role of Dicer in prostate cancer growth and metastasis. Oncogene. 2013 doi: 10.1038/onc.2013.281. 10.1038/onc.2013.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]
- Zindy F, Lee Y, Kawauchi D, Ayrault O, Merzoug LB, Li Y, McKinnon PJ, Roussel MF. Dicer Is Required for Normal Cerebellar Development and to Restrain Medulloblastoma Formation. PLoS ONE. 2015;10:e0129642. doi: 10.1371/journal.pone.0129642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.