Abstract
Missense mutations in TP53 are common in human breast cancer, have been associated with worse prognosis, and may predict therapy effect. TP53 missense mutations are associated with aberrant accumulation of p53 protein in tumor cell nuclei. Previous studies have used relatively arbitrary cutoffs to characterize breast tumors as positive for p53 staining by immunohistochemical assays. This study aimed to objectively determine optimal thresholds for p53 positivity by manual and automated scoring methods utilizing whole tissue sections from the Carolina Breast Cancer Study. P53 immunostained slides were available for 564 breast tumors previously assayed for TP53 mutations. Average nuclear p53 staining intensity was manually scored as negative, borderline, weak, moderate, or strong and percentage of positive tumor cells was estimated. Automated p53 signal intensity was measured using the Aperio nuclear v9 algorithm combined with the Genie® histology pattern recognition tool and tuned to achieve optimal nuclear segmentation. ROC curve analysis was performed to determine optimal cutoffs for average staining intensity and percent cells positive to distinguish between tumors with and without a missense mutation. ROC curve analysis demonstrated a threshold of moderate average nuclear staining intensity as a good surrogate for TP53 missense mutations in both manual (AUC=0.87) and automated (AUC=0.84) scoring systems. Both manual and automated immunohistochemical scoring methods predicted missense mutations in breast carcinomas with high accuracy. Validation of the automated intensity scoring threshold suggests a role for such algorithms in detecting TP53 missense mutations in high throughput studies.
Keywords: TP53, missense mutation, breast cancer, immunohistochemistry, image analysis, automated scoring, algorithm
Introduction
The tumor suppressor gene TP53 is one of the most commonly mutated genes in human neoplasia in general and in breast cancer in particular. Overall, 23-30% of human breast carcinomas harbor TP53 mutations [1-4]. These mutations are readily detectable by various DNA sequencing techniques; however, those assays are rarely performed in routine clinical practice. Approximately 70-75% of TP53 mutations are missense mutations and lead to accumulation of abnormal p53 protein in the nuclei of tumor cells [5,6]. While levels of wild- type p53 are typically very low, accumulation of mutant p53 can be detected by immunohistochemistry (IHC), a technique that is extensively used in pathology laboratories. Increased levels of p53 as detected by IHC have been widely used as a surrogate for detection of an underlying TP53 mutation, although non-missense mutations are typically not detected by this method (false negatives) [7,8] and accumulation of wild-type p53 under certain circumstances can lead to false positive results.
P53 is thought to play an important role in the etiology of breast carcinomas owing to its importance in regulating the cell cycle and apoptosis [9,10]; and p53 expression has been studied as a biomarker in human mammary neoplasia where it has been found to be a prognostic factor and to predict response to radiation, endocrine and chemotherapy [11,12]. However, the literature on the clinical significance of p53 in breast cancer is not consistent. This may be due in part to the fact that p53 over expression has been variably defined, and the great majority of studies have used arbitrary cutoffs. The primary goal of our study was to determine a rational threshold for IHC assessment of p53 protein overexpression to act as a surrogate for a TP53 missense mutation.
A second aim of our study was to establish a cutoff value that would distinguish tumors with and without TP53 missense mutations utilizing automated image analysis of p53 immunostained slides. Immunohistochemical stains of smaller series of tumors lend themselves to manual scoring. However, an increasing number of clinical and especially epidemiological studies include very large numbers of cases, and many of them utilize tissue microarrays (TMAs). Consequently these studies involve thousands of tissue samples that need to be scored, often for a long list of biomarkers. Image analysis is a helpful tool in scoring large numbers of immunohistochemical stains. Rational thresholds for manual and automated scoring of p53 immunostains should prove useful for researchers studying the role of p53 in breast cancer and other epithelial malignancies.
Materials and Methods
Study Population
The Carolina Breast Cancer Study (CBCS) is a population-based, case-control study of genetic and environmental risk factors for breast cancer among African American and Caucasian women residing in North Carolina [13]. Eligible cases were women aged 20-74 years with a first primary diagnosis of invasive breast cancer between 1993 and 1996 (Phase 1 of the CBCS), and were identified by the North Carolina Central Cancer Registry via a rapid case ascertainment protocol [14]. The distributions of pathological features and biomarkers in our study population have been previously described [15]. African American cases and women diagnosed before age 50 were oversampled to ensure that they comprised approximately half of recruited case participants [14]. The CBCS conducted two recruitment phases subsequent to Phase 1, and TP53 mutational data for those cases was not collected. Detailed CBCS study methods have been previously described [13]. A total of 861 breast cancer cases were eligible for and consented to participate in Phase 1 of the CBCS [14]. Clinical data and information on tumor characteristics were obtained from medical records or direct pathological review of tumor tissue [14]. All study procedures were approved by the University of North Carolina School of Medicine Institutional Review Board.
TP53 Mutation Screening
A screening algorithm incorporating single-strand conformational polymorphism (SSCP) analysis and manual DNA sequencing was employed to evaluate mutations in exons 4-8 of the TP53 gene [16,17]. Briefly, PCR amplification was carried out for exons 5-8, while exon 4 was amplified as two overlapping segments. Detailed mutation screening has been previously described [14].
Tumor Tissue Preparation and Histopathological Evaluation
Tumor tissue from formalin-fixed, paraffin-embedded tumor blocks was obtained for 798 of the 861 breast cancer cases (93%) and sectioned as previously described [18]. Tissue sections underwent standardized histopathological review by the study pathologist (JG). The invasive components of the tumors were identified by the study pathologist and selectively dissected for molecular analyses by a proteinase K extraction method as previously described [16,17]. We analyzed TP53 mutational status for 564 (71%) of the 798 cases for whom tissue was obtained. Of the 234 tumors not evaluated, 210 were determined by histopathological review to have insufficient tumor volume for molecular analysis and 24 tumors could not be assessed because of poor DNA quality. Four- micron paraffin sections were stained for p53 protein as previously described [19]. Briefly, sections underwent heat induced epitope retrieval in sub-boiling citrate buffer (pH 6.0) for 10 minutes. Mouse monoclonal p53 antibody clone DO7 (Dako, Glostrup, Denmark) was applied at a 1:100 dilution and incubated in a humidity chamber for 1 hour. The detection reaction utilized the Vector Labs Elite ABC Kit with Innovex Biosciences 3,3’-diaminobenzidine as chromogen and hematoxylin as counterstain. A board certified pathologist (JG) performed manual histopathological review of these tissue slides, recording percentage of positive cells and average nuclear staining intensity on a five-category scale: negative, borderline, weak, moderate, and strong. The reviewing pathologist assessed p53 staining across the entire tumor section and was blinded to other pathological and biomarker data, as well as TP53 mutational status. Additionally, a modified histoscore (H-score) [20] was calculated as the product of the manually scored average staining intensity score (0 for no staining, 0.5 for borderline, 1 for weak, 2 for moderate, 3 for strong) and manually estimated percentage of positive tumor cells (range 0-300). The borderline category was added because some tumors contained nuclei that were not completely negative but were very faintly stained, significantly less than weakly stained nuclei.
Aperio Automated Analysis
P53 stained slides were digitally imaged at 20× magnification using the Aperio ScanScope XT (Aperio Technologies, Vista, CA). Digital images were stored and analyzed within the Aperio Spectrum Database. P53 signal intensity was measured using the Aperio nuclear v9 algorithm (cell quantification) combined with the Genie® Histology Pattern Recognition tool. Genie® was used to exclude from the analysis artifacts related to the tissue processing (ink) and the sectioning and staining protocol (folds, debris). Algorithm parameters, including nuclear compactness, curvature and intensity thresholds, maximum and minimum nuclear size and hematoxylin optical density were tuned to achieve optimal nuclear segmentation. The Aperio v9 algorithm uses a 3-point scale to score average staining intensity, with a 1+ score indicating relative lack of staining intensity (corresponding to combined manual scores of negative, borderline, and weak), a 2+ score indicating relatively moderate staining intensity and a 3+ score corresponding to relatively strong staining intensity.
Statistical Analysis
Receiver operating characteristic curve (ROC) analysis was performed to assess the accuracy and optimal threshold of p53 staining (average intensity, percent positive tumor cells, modified H-score) to predict the presence of a missense mutation in TP53. TP53 mutational status was dichotomized, with a missense mutation representing the index category and all other mutational dispositions (no mutation, silent mutation, non-missense mutation) coded as referent. Unconditional logistic regression was also performed to evaluate the association between missense mutations in TP53 and average p53 protein IHC nuclear staining intensity. All statistical analyses were performed using SAS v9.3 software (SAS Institute, Cary, NC).
Results
Figure 1 illustrates the manually scored staining intensity categories. Figure 2 shows the distribution of p53 staining across the cohort of 564 breast carcinomas. Seventy-six tumors (13.5%) were completely devoid of nuclear staining, 336 (59.6%) showed borderline or weak nuclear staining, and 152 (27.0%) showed moderate or strong nuclear staining (Panel 2a). Among the 152 tumors with moderate or strong nuclear staining, 104 (68.4%) had >70% cells showing positivity for p53 (Panel 2b). Across the entire cohort, 374 tumors (66.3%) showed reactivity in less than 30% of tumor cells while 118 tumors (20.9%) had staining in over 70% of tumor cells (Panel 2c).
Figure 1. Examples of manually scored p53 staining intensity categories.
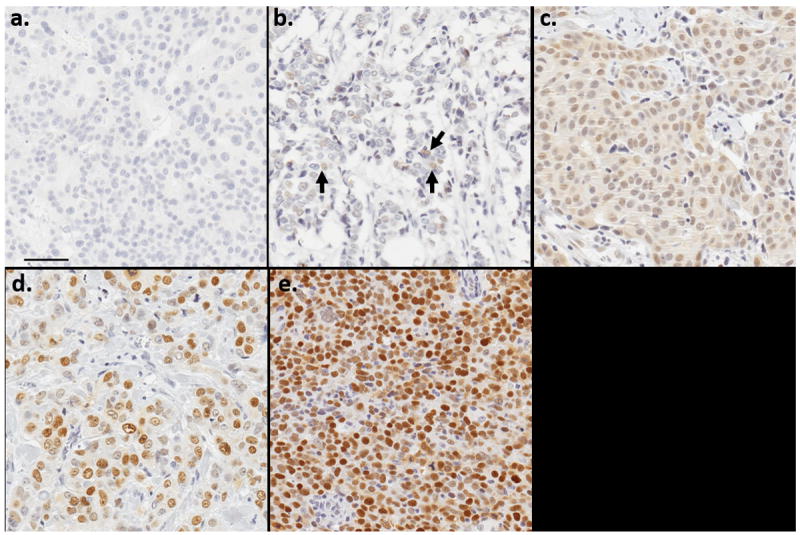
- No staining (0)
- Borderline staining (0.5)
- Weak staining (1)
- Moderate staining (2)
- Strong staining (3)
Figure 2. Distribution of manually scored p53 staining in breast carcinomas.
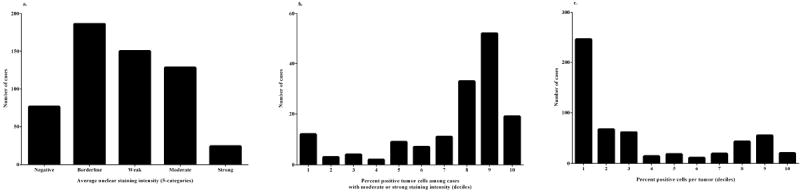
- Distribution of average nuclear staining intensity (5-category; 564 cases)
- Distribution of percentage positive tumor cells by decile (moderate and strong average nuclear staining only; 152 cases)
- Distribution of percentage positive tumor cells by decile (all tumors; 564 cases)
Examination of agreement between manual (3-category with combined negative, borderline, and weak serving as referent category) and automated systems for scoring average nuclear p53 staining intensity indicated good correlation (Spearman’s ρ=0.80). Using 3-category coding, agreement was 81.6% (Table 1). Dichotomizing the stains into negative (manual scores of negative, borderline, and weak combined; automated scores of 1) versus positive (manual scores of moderate and strong combined; automated scores of 2 and 3 combined) increased concordance to 91.5% (Table 1). ROC curve analysis revealed that a 3-category manual scoring system of average nuclear staining intensity (Figure 3b; AUC=0.87) was comparable to the previously used 5-category scale (Figure 3a; AUC=0.90). We noted that the AUC was slightly augmented by combining percentage cells positive and average nuclear staining intensity into a modified H-score with an optimal cut point of 60 (Figure 3c; AUC=0.92). The ROC curve for the automated 3-category scoring algorithm (Figure 3d; AUC=0.84) showed similar results to the corresponding 3-category manual scoring system. Both manual and automated scoring systems had an optimal threshold of at least moderate average nuclear staining.
Table 1.
Manual versus automated immunohistochemical scores for average nuclear p53 staining intensity
| Manual Score | Automated Score
|
|||
|---|---|---|---|---|
| 1 | 2 | 3 | Total | |
| Negative, Borderline, Weak | 405 (71.8%) | 4 (0.71%) | 3 (0.53%) | 412 (73.1%) |
|
| ||||
| Moderate | 41 (7.3%) | 33 (5.9%) | 54 (9.6%) | 128 (22.7%) |
|
| ||||
| Strong | 0 | 2 (0.35%) | 22 (3.9%) | 24 (4.3%) |
|
| ||||
| Total | 446 (79.1%) | 39 (6.9%) | 79 (14.0%) | 564 (100%) |
Figure 3. Receiver operating characteristic (ROC) curves for the association between TP53 mutational status and p53 protein staining intensity.
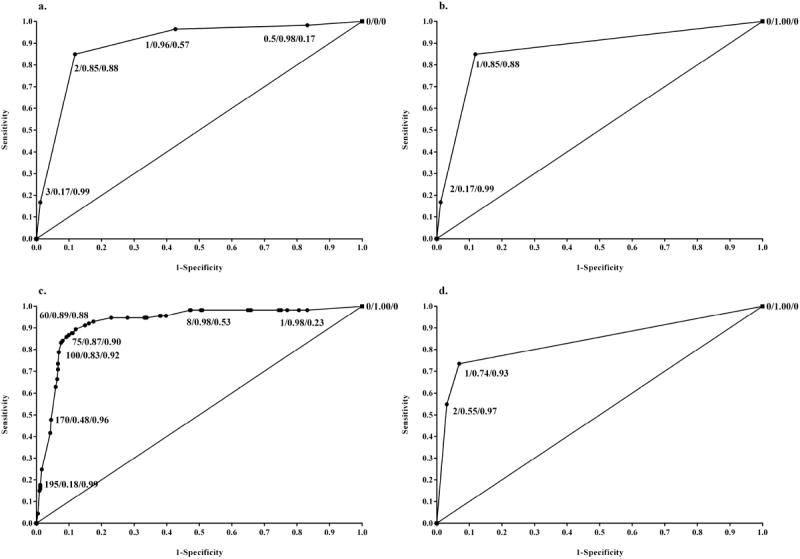
- ROC curve plot of 5-category manually scored IHC staining intensity predicting missense mutations in TP53 versus all other mutations or wild type (AUC=0.90). Labels contain staining intensity value/sensitivity/specificity. Staining intensity values are coded as: 0=negative, 0.5=borderline, 1=weak, 2=moderate, 3=strong.
- ROC curve plot of 3-category manually scored IHC staining intensity predicting missense mutations in TP53 versus all other mutations or wild type (AUC=0.87). Labels contain staining intensity value/sensitivity/specificity. Staining intensity values are coded as: 0=negative/borderline/weak, 1=moderate, 2=strong.
- ROC curve plot of modified H-score predicting a missense mutation in TP53 versus all other mutations or wild type (AUC=0.92). Labels contain modified H-score value/sensitivity/specificity.
- ROC curve plot of IHC staining intensity predicting a missense mutation in TP53 versus all other mutations or wild type using 3-category automated scoring (AUC=0.84). Labels contain staining intensity value/sensitivity/specificity. Staining intensity values are coded as: 0=negative/low, 1=moderate, 2=strong.
Manual and automated scores of average p53 nuclear staining intensity showed similar predictive capacity for TP53 missense mutations. Manual scoring correctly classified 96 of 113 missense mutations (sensitivity=0.85) and 372 of 422 tumors without such mutations (specificity=0.88). The automated scoring system correctly classified 83 of 113 missense mutations (sensitivity=0.73) and 393 of 422 tumors without such mutations (specificity=0.93) (Table 2).
Table 2.
Manual versus automated average immunohistochemical p53 protein staining intensity scores as predictors of TP53 missense mutations
| Missense Mutation | Accuracy | |||
|---|---|---|---|---|
| Present | Absent | |||
| Manual Score1 | + | 96 | 50 | 0.87 |
| - | 17 | 372 | ||
| Automated Score2 | + | 83 | 29 | 0.89 |
| - | 30 | 393 | ||
Positivity is defined as moderate or strong average nuclear staining intensity based on a 5-category scale (Negative-Strong)
Positivity is defined as 2 or 3 average staining intensity based on Aperio 3-category scale
When scored manually, an average nuclear staining intensity of at least moderate, ≥ 50% positive tumor cells, and a modified H-score of at least 60 had comparable sensitivities and specificities with regard to prediction of TP53 missense mutations; the automated scoring algorithm performed similarly to the manual system, although a lower sensitivity was noted while specificity was slightly higher (Table 3). Odds ratios (ORs) and 95% confidence intervals (CIs) estimating the association between manually scored average p53 nuclear staining intensity and presence of a missense mutation suggested robust prediction of a missense mutation when moderate to strong average nuclear staining intensity was observed (OR= 42.01; 95% CI: 23.19, 76.10) (Table 3). These findings were recapitulated using the Aperio v9 automated algorithm (OR=37.49; 95% CI: 21.36, 65.81) (Table 3).
Table 3.
Performance characteristics of manual and automated immunohistochemical scoring thresholds for the prediction of TP53 missense mutations
| Manual Scoring | AUC1 | Sensitivity | Specificity | OR (95% CI)2 |
|---|---|---|---|---|
| Intensity ≥ moderate (5-category)3 | 0.90 | 0.85 | 0.88 | 42.01 (23.19, 76.10) |
| Intensity ≥ moderate (3-category)4 | 0.87 | 0.85 | 0.88 | 42.01 (23.19, 76.10) |
| ≥ 50% cells positive | 0.93 | 0.90 | 0.88 | 46.23 (24.03, 88.93) |
| Modified H-score ≥ 605 | 0.92 | 0.89 | 0.88 | 61.23 (31.45, 119.20) |
| Automated Scoring | ||||
| Intensity ≥ 2 (3-category) | 0.84 | 0.73 | 0.93 | 37.49 (21.36, 65.81) |
Area under the receiver operating characteristic curve
Odds ratios and 95% confidence intervals for the association between scoring parameter and a missense mutation in TP53
Odds ratio calculated based on referent category combining negative, borderline, and weak staining intensities
All calculations based on a referent category combining negative, borderline, and weak staining intensities
A combination score incorporating percentage cells positive and average nuclear staining intensity
Finally, using a scoring threshold of at least moderate average nuclear staining, we examined the automated scoring system’s capacity to detect differential frequencies of TP53 missense mutations in 1) pre- versus post-menopausal breast carcinomas and 2) major breast cancer subtypes. As summarized in Table 4, moderate or strong average nuclear p53 staining intensity was more common in HER2-positive and premenopausal breast carcinomas, consistent with the TP53 mutational analysis.
Table 4.
Odds ratios (OR) and 95% confidence intervals (CI) for the associations between TP53 mutational status & automated nuclear p53 staining intensity and molecular breast cancer subtype & menopausal status in the Carolina Breast Cancer Study
| TP53 Missense Mutation1 | Automated Average p53 Staining Intensity2 | |
|---|---|---|
|
| ||
| Molecular Breast Cancer Subtype | ||
|
| ||
| Luminal A | 1.00 | 1.00 |
|
| ||
| Luminal B | 1.12 (0.56, 2.23) | 1.46 (0.74, 2.89) |
|
| ||
| HER2+/ER- | 1.65 (0.64, 4.24) | 2.73 (1.12, 6.64) |
|
| ||
| Basal-like | 1.26 (0.65, 2.44) | 1.33 (0.68, 2.63) |
|
| ||
| Unclassified | 0.81 (0.26, 2.50) | 1.63 (0.60, 4.38) |
|
| ||
| Menopausal Status3 | ||
|
| ||
| Postmenopausal | 1.00 | 1.00 |
|
| ||
| Premenopausal | 1.85 (1.19, 2.86) | 1.78 (1.17, 2.72) |
A missense mutation was coded as the index category with all other dispositions (no mutation, silent mutation, nonsense mutation) coded as the referent category
Automated staining intensity of ≥ 2 was coded as the index category with staining intensity of 1 coded as the referent category
For women under the age of 50, menopausal status was assigned to those who had undergone natural menopause, bilateral oopherectomy, or irradiation to the ovaries; in women aged 50 and over, menopausal status was assigned on the basis of cessation of menstruation
Discussion
It is well established that TP53 plays an important role in the pathogenesis of human breast cancer, owing to its importance in regulating both the cell cycle and apoptosis [9,10]. Most TP53 mutations are associated with decreased degradation of p53, leading to accumulation of the protein in tumor cells that is detectable as increased nuclear staining by IHC [5]. Mutant p53 is known to demonstrate gain-of-function oncogenic activity by binding and inactivating other regulatory proteins, such as p63 and p73 [21], and ovarian cancer patients carrying such gain-of-function missense mutations in TP53 showed greater resistance to platinum agents when compared to those without gain-of-function missense mutations [22]. Thus, identifying cancer patients carrying missense mutations in TP53 may be relevant to the selection of appropriate adjuvant therapy. Furthermore, TP53 mutations have been reported more often in basal-like and HER2+/ER- breast cancer subtypes compared to luminal breast cancers [23], and recent exome sequencing studies in TCGA have demonstrated that TP53 mutation type may differ by breast cancer molecular subtype [24]. Consequently, the utility of IHC to identify TP53 missense mutations in breast cancer may depend upon the tumor characteristics of a given population.
While mutational TP53 analysis is not routinely performed on human cancer specimens, IHC is a commonly used technique in pathology laboratories. IHC is frequently employed as a surrogate for TP53 mutational analysis, but suffers interpretability issues related to both false positive and false negative staining [25,26]. Despite these limitations, over expression of p53 has been shown to be a negative prognostic indicator in breast cancer [27,28] and has been associated with poor response to adjuvant endocrine [29,30], radiation [12], and chemotherapy [12]; additionally, p53 over expression has been associated with poor response to neoadjuvant chemotherapy [8,31,32]. However, rationally defined cut points for abnormally high p53 expression have been lacking, and few studies have utilized TP53 mutational data as a gold standard to calibrate p53 IHC scoring thresholds. The absence of objective cut points may partially explain why published data on p53 as a prognostic or predictive factor for human breast cancer have not been consistent [33,34].
Previous studies defined p53 over expression as: a certain minimum percentage of tumor cells demonstrating IHC staining, a certain minimum IHC staining intensity, or a combination of both. We tested all three models and found that cut points of 50% cells positive, moderate average nuclear staining intensity and a modified H-score of 60 all had comparable performance characteristics, as may be inferred from Figure 3 and Table 3. These results are consistent with previous studies using thresholds of 50% positive cells [35-37] or moderate staining intensity [38] as cutoffs for p53 over expression. Moreover, we found that IHC is a good surrogate for identifying TP53 missense mutations in exons 4-8, with accuracy approaching 90%. We did not examine mutations outside of exons 4-8, and it is possible that exclusion of potential missense mutations in these regions may have biased our results. However, previous reports have indicated a majority of missense mutations in TP53 are found within exons 4-8, with the promoter region, exons 1-3 and 9 containing few to none [39].
Our observation that a moderate or strong staining intensity is as informative as the percentage of positive cells or a certain H-score suggests great utility from an automated scoring perspective. Automated scoring algorithms have proved quite useful in measuring staining intensities, but have performed suboptimally in differentiating relevant (malignant) and irrelevant (benign) cells. Thus, scoring algorithms that include a percentage threshold typically do not lend themselves to automated analysis of human tumor samples stained for p53 or other immunohistochemical markers. Many contemporary clinical and epidemiological studies include over 1,000 samples, making manual scoring of IHC stains an onerous and lengthy task; utilizing automated algorithms to perform these high throughput tasks could improve efficiency and reduce variability. While pathologists often use a negative/weak/moderate/strong scale for manual scoring of immunohistochemical stains, these schemes inherently include an element of subjectivity; however, a strength of our study was that all cases were centrally scored by a single pathologist. A significant advantage of automated scoring algorithms is their ability to improve inter-observer concordance in the quantitation of IHC stains [40].
While sequencing approaches are becoming more cost-effective, they still rely on availability of sufficient DNA and are more costly than IHC assays. The efficiency of IHC in large epidemiologic studies ensures this approach will continue to be of value, despite potential misclassification rates. Better characterization of this misclassification will improve interpretation of IHC surrogates as predictors for TP53 mutational status. As may be inferred from Table 4, using an automated threshold of at least moderate average nuclear staining intensity demonstrated the higher prevalence of p53 over expression in HER2+/ER- and premenopausal breast carcinomas as reliably, if not better than, the DNA-based sequencing approach that shows higher frequencies of TP53 missense mutations in the same sub-groups.
To the best of our knowledge, our study is the first to calibrate and validate the Aperio p53 automated scoring algorithm in human breast carcinomas. The data we present here provide compelling evidence that a threshold of at least moderate average nuclear staining intensity is a good surrogate for detection of TP53 missense mutations, and this threshold translates well when applied to automated p53 staining intensity scoring algorithms.
Acknowledgments
We thank the staff and participants of the Carolina Breast Cancer Study for their invaluable contributions to this study. We also thank Jessica Tse for programming support. The authors also wish to acknowledge the staff of the UNC Translational Pathology Laboratory for performing all IHC assays. Finally, we appreciate the helpful comments on this manuscript by Drs. Melissa Troester and Lisa Carey of the Lineberger Comprehensive Cancer Center at the University of North Carolina, Chapel Hill.
Financial Support
This work was supported by the Specialized Program of Research Excellence (SPORE) in Breast Cancer at UNC, by the National Institutes of Health/National Cancer Institute [P50-CA58223 and 5R25CA147832-04], and by the University Cancer Research Fund (UCRF) (http://ucrf.unc.edu).
List of Abbreviations
- AUC
Area under the curve
- CBCS
Carolina Breast Cancer Study
- CI
Confidence interval
- IHC
Immunohistochemistry/immunohistochemical
- H-score
Histoscore
- JG
Joseph Geradts
- OR
Odds ratio
- ROC
Receiver operating characteristic
- SSCP
Single-strand conformational polymorphism
- TMA
Tissue microarray(s)
Footnotes
Author Contributions
JG and NJT conceived the study design and prepared the manuscript.
KCD, RCM and NJT contributed to the collection of data.
JG, RCM, NJT contributed to the analytic plan.
NNF, BM and NJT and performed the analyses of data.
KCD, NNF and BM contributed to manuscript preparation.
Conflict of interest statement: None declared
References
- 1.Forbes SA, Bindal N, Bamford S, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic acids research. 2011;39:D945–950. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coles C, Condie A, Chetty U, et al. p53 mutations in breast cancer. Cancer research. 1992;52:5291–5298. [PubMed] [Google Scholar]
- 3.Olivier M, Langerod A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 4.Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harbor perspectives in biology. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petitjean A, Mathe E, Kato S, et al. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Human mutation. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 6.Petitjean A, Achatz MI, Borresen-Dale AL, et al. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–2165. doi: 10.1038/sj.onc.1210302. [DOI] [PubMed] [Google Scholar]
- 7.Robles AI, Harris CC. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harbor perspectives in biology. 2010;2:a001016. doi: 10.1101/cshperspect.a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alsner J, Jensen V, Kyndi M, et al. A comparison between p53 accumulation determined by immunohistochemistry and TP53 mutations as prognostic variables in tumours from breast cancer patients. Acta oncologica. 2008;47:600–607. doi: 10.1080/02841860802047411. [DOI] [PubMed] [Google Scholar]
- 9.Gasco M, Shami S, Crook T. The p53 pathway in breast cancer. Breast cancer research : BCR. 2002;4:70–76. doi: 10.1186/bcr426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walerych D, Napoli M, Collavin L, et al. The rebel angel: mutant p53 as the driving oncogene in breast cancer. Carcinogenesis. 2012;33:2007–2017. doi: 10.1093/carcin/bgs232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Connor PM, Jackman J, Bae I, et al. Characterization of the p53 tumor suppressor pathway in cell lines of the National Cancer Institute anticancer drug screen and correlations with the growth-inhibitory potency of 123 anticancer agents. Cancer research. 1997;57:4285–4300. [PubMed] [Google Scholar]
- 12.Lowe SW, Bodis S, McClatchey A, et al. p53 status and the efficacy of cancer therapy in vivo. Science. 1994;266:807–810. doi: 10.1126/science.7973635. [DOI] [PubMed] [Google Scholar]
- 13.Newman B, Moorman PG, Millikan R, et al. The Carolina Breast Cancer Study: integrating population-based epidemiology and molecular biology. Breast cancer research and treatment. 1995;35:51–60. doi: 10.1007/BF00694745. [DOI] [PubMed] [Google Scholar]
- 14.Conway K, Edmiston SN, Cui L, et al. Prevalence and spectrum of p53 mutations associated with smoking in breast cancer. Cancer research. 2002;62:1987–1995. [PubMed] [Google Scholar]
- 15.Furberg H, Millikan R, Dressler L, et al. Tumor characteristics in African American and white women. Breast cancer research and treatment. 2001;68:33–43. doi: 10.1023/a:1017994726207. [DOI] [PubMed] [Google Scholar]
- 16.Li Y, Millikan RC, Carozza S, et al. p53 mutations in malignant gliomas. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 1998;7:303–308. [PubMed] [Google Scholar]
- 17.Olshan AF, Weissler MC, Pei H, et al. Alterations of the p16 gene in head and neck cancer: frequency and association with p53, PRAD-1 and HPV. Oncogene. 1997;14:811–818. doi: 10.1038/sj.onc.1200892. [DOI] [PubMed] [Google Scholar]
- 18.Dressler LG, Geradts J, Burroughs M, et al. Policy guidelines for the utilization of formalin-fixed, paraffin-embedded tissue sections: the UNC SPORE experience. University of North Carolina Specialized Program of Research Excellence. Breast cancer research and treatment. 1999;58:31–39. doi: 10.1023/a:1006354627669. [DOI] [PubMed] [Google Scholar]
- 19.Furberg H, Millikan RC, Geradts J, et al. Environmental factors in relation to breast cancer characterized by p53 protein expression. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2002;11:829–835. [PubMed] [Google Scholar]
- 20.Hatanaka Y, Hashizume K, Nitta K, et al. Cytometrical image analysis for immunohistochemical hormone receptor status in breast carcinomas. Pathology international. 2003;53:693–699. doi: 10.1046/j.1440-1827.2003.01547.x. [DOI] [PubMed] [Google Scholar]
- 21.Oren M, Rotter V. Mutant p53 gain-of-function in cancer. Cold Spring Harbor perspectives in biology. 2010;2:a001107. doi: 10.1101/cshperspect.a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang HJ, Chun SM, Kim KR, et al. Clinical relevance of gain-of-function mutations of p53 in high-grade serous ovarian carcinoma. PloS one. 2013;8:e72609. doi: 10.1371/journal.pone.0072609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina Breast Cancer Study. JAMA : the journal of the American Medical Association. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 24.Cancer Genome Atlas N. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmann D, Ruhri C, Schmitt M, et al. Accumulation of p53 protein as an indicator for p53 gene mutation in breast cancer. Occurrence of false-positives and false-negatives. Diagnostic molecular pathology : the American journal of surgical pathology, part B. 1993;2:36–41. [PubMed] [Google Scholar]
- 26.Sjogren S, Inganas M, Norberg T, et al. The p53 gene in breast cancer: prognostic value of complementary DNA sequencing versus immunohistochemistry. Journal of the National Cancer Institute. 1996;88:173–182. doi: 10.1093/jnci/88.3-4.173. [DOI] [PubMed] [Google Scholar]
- 27.Thorlacius S, Thorgilsson B, Bjornsson J, et al. TP53 mutations and abnormal p53 protein staining in breast carcinomas related to prognosis. European journal of cancer. 1995;31A:1856–1861. doi: 10.1016/0959-8049(95)00399-4. [DOI] [PubMed] [Google Scholar]
- 28.Overgaard J, Yilmaz M, Guldberg P, et al. TP53 mutation is an independent prognostic marker for poor outcome in both node-negative and node-positive breast cancer. Acta oncologica. 2000;39:327–333. doi: 10.1080/028418600750013096. [DOI] [PubMed] [Google Scholar]
- 29.Yamashita H, Toyama T, Nishio M, et al. p53 protein accumulation predicts resistance to endocrine therapy and decreased post-relapse survival in metastatic breast cancer. Breast cancer research : BCR. 2006;8:R48. doi: 10.1186/bcr1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garimella V, Hussain T, Agarwal V, et al. Clinical response to primary letrozole therapy in elderly patients with early breast cancer: possible role for p53 as a biomarker. International journal of surgery. 2014;12:821–826. doi: 10.1016/j.ijsu.2014.06.009. [DOI] [PubMed] [Google Scholar]
- 31.Bottini A, Berruti A, Bersiga A, et al. p53 but not bcl-2 immunostaining is predictive of poor clinical complete response to primary chemotherapy in breast cancer patients. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6:2751–2758. [PubMed] [Google Scholar]
- 32.Chen MB, Zhu YQ, Xu JY, et al. Value of TP53 status for predicting response to neoadjuvant chemotherapy in breast cancer: a meta-analysis. PloS one. 2012;7:e39655. doi: 10.1371/journal.pone.0039655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bertheau P, Espie M, Turpin E, et al. TP53 status and response to chemotherapy in breast cancer. Pathobiology : journal of immunopathology, molecular and cellular biology. 2008;75:132–139. doi: 10.1159/000123851. [DOI] [PubMed] [Google Scholar]
- 34.Varna M, Bousquet G, Plassa LF, et al. TP53 status and response to treatment in breast cancers. Journal of biomedicine & biotechnology. 2011;2011 doi: 10.1155/2011/284584. 284584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alkushi A, Lim P, Coldman A, et al. Interpretation of p53 immunoreactivity in endometrial carcinoma: establishing a clinically relevant cut-off level. International journal of gynecological pathology : official journal of the International Society of Gynecological Pathologists. 2004;23:129–137. doi: 10.1097/00004347-200404000-00007. [DOI] [PubMed] [Google Scholar]
- 36.Yemelyanova A, Vang R, Kshirsagar M, et al. Immunohistochemical staining patterns of p53 can serve as a surrogate marker for TP53 mutations in ovarian carcinoma: an immunohistochemical and nucleotide sequencing analysis. Modern pathology : an official journal of the United States and Canadian Academy of Pathology, Inc. 2011;24:1248–1253. doi: 10.1038/modpathol.2011.85. [DOI] [PubMed] [Google Scholar]
- 37.Kikuchi S, Nishimura R, Osako T, et al. Definition of p53 overexpression and its association with the clinicopathological features in luminal/HER2-negative breast cancer. Anticancer research. 2013;33:3891–3897. [PubMed] [Google Scholar]
- 38.Chang MC, Cibas ES, Crum CP, et al. High-grade and low-grade pelvic serous neoplasms demonstrate differential p53 immunoreactivity in peritoneal washings. Acta cytologica. 2011;55:79–84. doi: 10.1159/000320862. [DOI] [PubMed] [Google Scholar]
- 39.Hartmann A, Blaszyk H, McGovern RM, et al. p53 gene mutations inside and outside of exons 5-8: the patterns differ in breast and other cancers. Oncogene. 1995;10:681–688. [PubMed] [Google Scholar]
- 40.Bloom K, Harrington D. Enhanced accuracy and reliability of HER-2/neu immunohistochemical scoring using digital microscopy. American journal of clinical pathology. 2004;121:620–630. doi: 10.1309/Y73U-8X72-B68T-MGH5. [DOI] [PubMed] [Google Scholar]


