Abstract
AIM: To explore the germline mutations of the two main DNA mismatch repair genes (hMSH2 and hMLH1) between patients with hereditary non-polyposis colorectal cancer (HNPCC) and suspected (atypical) HNPCC.
METHODS: Genomic DNA was extracted from the peripheral blood of the index patient of each family, and germline mutations of hMSH2 and hMLH1 genes were detected by PCR-single strand conformation polymorphism (PCR-SSCP) and DNA sequencing techniques.
RESULTS: For PCR-SSCP analysis, 67% (4/6) abnormal exons mobility in typical group and 33% (2/6) abnormal exons mobility in atypical group were recognized. In direct DNA sequencing, 50% (3/6) mutation of MMR genes in typical group and 33% (2/6) mutation of MMR genes in atypical group were found, and 4/6 (66.67%) and 1/6 (16.67%) mutations of hMSH2 and hMLH1 were identified in typical HNPCC and atypical HNPCC, respectively.
CONCLUSION: Mutation detection of the patients is of benefit to the analysis of HNPCC and, PCR-SSCP is an effective strategy to detect the mutations of HNPCC equivalent to direct DNA sequence. It seems that there exist more complicated genetic alterations in Chinese HNPCC patients than in Western countries.
INTRODUCTION
Hereditary nonpolyposis colorectal cancer (HNPCC) is characterized by early onset of colorectal cancer, location of tumors in the proximal colon, and an increased risk of neoplasms of extracolonic organs, including endometrium, stomach, urothelium, small intestine, and ovary[1-6]. The International Collaborative Group on HNPCC (ICG-HNPCC) proposed a set of clinical diagnostic criteria (Amsterdam Criteria I) for HNPCC in 1990 and has revised them recently (Amsterdam Criteria II) to provide a uniformity in collaborative studies[7,8]. According to these criteria, there should have at least three patients in two consecutive generations who had colorectal cancer or the other extracolonic malignancies including endometrial cancer, small bowel cancer, cancer of the ureter and cancer of the renal pelvis. One of them should be a first-degree relative of the other two, one cancer should be diagnosed before age 50, and familial adenomatous polyposis (FAP) should be excluded. In Asia, on the other hand, the Japan Research Society for Cancer of the Colon and Rectum developed the clinical criteria (Japanese criteria) for HNPCC in 1991[9]. According to these criteria, at least two relatives in at least two successive generations should have colorectal cancer, and one of them should be a first-degree relative of the other, also, we called them atypical HNPCC.
Nevertheless, according to those criteria above, all families that are diagnosed as having HNPCC are identified on the basis of a family history of colorectal or other certain malignancies. Sometimes the accuracy and reliability of the family history provided by patients themselves and their relatives or recorded by physicians are questionable, as there is much viability for the family history. Therefore, the requirements for establishing objective strategies to identify this disease are imperative. In the last decade, the progress in genetic study that DNA mismatch repair genes have been identified as being mutated in HNPCC including hMSH2, hMLHI, hPMS1, hPMS2, and hMSH6 makes it possible to identify the cohort of patients through genetic test. Totally, these genes are now believed to account for about 50%-70% of all families with HNPCC and over 90% of the identified mutations focused on the two genes, hMSH2 and hMLH1[10-18]. There are many studies about the procedures of genetic testing of HNPCC, such as microsatellite instability (MSI), PCR-SSCP, immunohistochemistry (IHC) and direct DNA sequencing[19-24]. However, much knowledge and a higher grade of technical development are required, before this strategy becomes applicable to the general population.
It has no doubt that there is a large population of HNPCC in China[25,26]. Unfortunately, there have been few reports about clinical and genetic characteristics of HNPCC[27]. In this study, six HNPCC families and six atypical HNPCC families were enrolled and, the germline mutations of hMSH2 and hMLH1 in the index patients from each family were investigated.
MATERIALS AND METHODS
Subjects
The project was approved by the Institutional Review Board and informed consent was obtained from each participant before the procedures were carried out. Personal and family cancer history was obtained from the proband and participating relatives, and cancer diagnosis and deaths were confirmed by review of medical records, pathological reports or death certificates. Families were identified by the Amsterdam or Japanese criteria for HNPCC. Patients and families were classified as the HNPCC group according to the Amsterdam criteria, suspected HNPCC group according to the Japanese criteria and control group without any family history.
Genetic testing
Preparation of peripheral blood samples and DNA amplification Genetic analysis was performed on a blood specimen from the proband in each family. Specimens were collected and immediately frozen in liquid nitrogen. DNA was extracted from blood specimens using the Wizard genomic purification kit (Qiagen, Shanghai) according to the manufacturer’s instructions. Each of the exons from hMSH2 and hMLH1 genes was amplified by polymerase chain reaction (PCR). Next, the samples were heat-treated at 95 °C for 5 minutes to inactivate the enzyme, and used as the template DNA. All DNA amplification was performed in a 50 μl volume containing 100 ng template DNA, 10-3 M Tris-HCl (pH8.9), 50-3M KCl, 2.5 mom MgCl2, 0.2-3M of each dNTP,10 pmol each primer, and 1U Taq polymerase was subjected to 35 PCR cycles (5 mins at 94 °C, 40 s at 94 °C, 60 s at 55 °C, and 40 s at 72 °C). Oligonucliotide sequences were designed from the sequences published in Genebank.
PCR-SSCP analysis and direct sequencing
Single-strand conformation polymorphism (SSCP) The technique of single-strand conformational polymorphism (SSCP) was used to identify mutations in the mismatch repair genes[28,29]. Each exon of hMSH2 and hMLH1 was amplified specifically using PCR (details of oligonucleotide sequences and conditions are available from the authors). PCR products were electrophoresed on 10% non-denaturing polyacrylamide gels with an acrylamide: bisacrylaminde ratio of 30:0.8 at 70volt for 12 h at room temperature. DNA was detected by silver staining according to the methods described by others. Those single stranded DNAs that took up an altered conformation appeared as aberrantly migrating bands on the electrophoresis gel.
Direct sequencing
The nucleotide sequence of PCR products showing an abnormal mobility on SSCP was determined by direct sequencing. The PCR products were purified with 1.5% low melted point agrose, and then performed using automated DNA sequencer. Sequence alterations with an allele frequency of at least 5% were considered as normal variants (polymorphisms) and not reported.
Statistical analysis
The genetic differences of the typical HNPCC groups and the suspected HNPCC groups were analyzed for statistical significance using the chi square test. P < 0.05 was considered statistically significant.
RESULTS
Results of genetic testing
Characterization of the variants of PCR-SSCP and DNA sequencing analysis found in the 12 families/individuals in the study is presented in Table 1. A total of seven abnormal motilities were identified in PCR-SSCP including five in typical group and two in atypical group (part of PCR-SSCP analysis is shown in Figure 1). DNA sequencing found 6/7 (85.7%) abnormal motilities of PCR-SSCP, which were proven to be pathogenic germline mutations of hMSH2 and hMLH1. The other abnormal mobility, an “A” insertion in hMLH1 intron 10 occurred in the C10-1 patient, was proven to be polymorphism.
Table 1.
Characterization of variants in PCR-SSCP and sequence detection
| Family | Abnormal mobility | DNA change | Mutation | Group |
| C4-1 | hMLH1 exon18 | T insert in 2 081 | Frameshift | Typical HNPCC |
| hMSH2 exon15 | C insert in 2 469 | Frameshift | ||
| G insert in 2 471 | Frameshift | |||
| C13-1 | hMSH2 exon11 | T insert in 1 760 | Frameshift | |
| A→C missense in 1 688 | Tyr563Ser | Typical HNPCC | ||
| C11-1 | hMSH2 exon13 | T to A missense in 2 091 | Cys697 | |
| Terminator Condon | Typical HNPCC | |||
| C1-1 | hMLH1 exon11 | A insert in 934 | Frameshift | Atypical HNPCC |
| C8-2 | hMLH1 exon12 | C to G missense in 1 198 | Leu400Val | |
| C to G missense in 1 261 | Val421Leu | |||
| C insert in 1 364 and 1 372 | Frameshift | Atypical HNPCC | ||
| C10-1 | hMLH1 intron10 | — | Polymorphism | Typical HNPCC |
Figure 1.

Results of PCR-SSCP in C13 family (hMSH2-exon11). Arrow shows the abnormal mobility.
Totally, five patients were found to harbor MMR gene mutations in the direct DNA sequencing. Three of six (50%) patients with HNPCC and 2/6 (33%) patients with suspected HNPCC were found to harbor mutations. However, we found the mutations were complicated as the total number of mutations was more than five and all the mutations we found were novel and of unknown pathogenicity (part of sequence analysis is shown in Figure 2).
Figure 2.
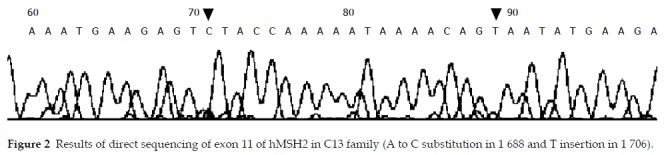
Results of direct sequencing of exon 11 of hMSH2 in C13 family (A to C substitution in 1 688 and T insertion in 1 706).
In typical HNPCC, the germline mutation of hMLH1 and hMSH2 was detected in 3 of 6 HNPCC families. Seven novel mutations were found in 3 exons of hMSH2 and 1 exon of hMLH1, two mutations were detected at 11 exons of hMSH2 in C13 family, one was the frame shift mutation resulted from an insertion of “T” at 1 760 so that a terminate cordon appeared ahead, the other was “A to C” missense mutation at 1 688 leading to Tyr-Ser substitution. A “T to A” substitution at 2 091 of exon 13 of hMSH2 in C11 family resulted in a terminate cordon at 697. The mutations in C4 family were worthy of specific comment, as the mutation occurred in hMLH1 as well as hMSH2, there was a frame shift mutation resulted from an insertion of T at 2 081 of exon 18 of hMLH1, there were “C, G” inserts at 2 469, 2 471 respectively. However, all families without germline hMSH2 and hMLH1 mutations were combined with extracolonic tumors.
In atypical HNPCC, only the germline mutation of hMLH1 was detected in 2 of 6 HNPCC families, one was the frame shift mutation in exon 11 resulted from an insertion of “A” at 934 in C13 family. In C8 family, the mutation occurred at four locations, one was “C to G” missense mutation at 1 198 leading to Leu400Val substitution, one was “C to G” missense mutation at 1 261 leading to Val421Leu substitution and the other two shift mutations occurred at 1 364 and 1 372 resulted from the A insertion at the two locations respectively. The detailed results of sequencing are shown in Table 2.
Table 2.
Comparison of two kinds of HNPCC with PCR-SSCP and sequencing
| Typical HNPCC | Atypical HNPCC | P | |
| (n = 6) | (n = 6) | ||
| PCR-SSCP | 4 (67%) | 2 (33%) | 0.269 |
| Sequencing | 3 (50%) | 2 (33%) | 0.575 |
DISCUSSION
To date, HNPCC is defined either by the so-called Amsterdam I+II criteria or by detection of a mutation in one of the mismatch repair genes. Once the positive mutation is identified, predictive testing of family members at risk is available. Screening recommendations for clinically identified families, mutation carriers, and their unaffected relatives at risk must be defined for clinical management[30].
Recently, Watson et al[31] published their results about that carrier risk status changes resulted from mutation testing in hereditary non-polyposis colorectal cancer and hereditary breast-ovarian cancer. They concluded that mutation testing could raise the accuracy of carrier risk assessments, and lower the number of persons at high carrier risk. The most common risk assessment change resulted from DNA testing was a change from those at risk to non-carriers. To the extent that these persons were aware of their carrier risk and were obtaining heightened cancer surveillance test, this could be expected to lead to a reduced emotional toll and a reduced pressure on limited medical resources.
A better understanding of the molecular basis of hereditary colorectal cancer syndromes such as hereditary nonpolyposis colorectal cancer syndrome (HNPCC) and familial adenomatous polyposis (FAP) would have profound consequences for both the diagnosis and (prophylactic) treatment of (pre)malignant neoplastic lesions[32] Clinically, when we see a patient with colorectal cancer with a family history of suspected HNPCC, we need to work out our surgical strategies for this patient, the same as we do for the patients with HNPCC, in whom colonoscopy surveillance or prophylaxis surgeries such as subtotal colectomy are needed. One of the evidences of special surgical treatment has been found to be genetic detection[33,34]. Since HNPCC is resulted from the dysfunction of mismatch repair genes and some reports indicated that mutation of hMSH2 and hMLH 1 accounted for 40%-70% HNPCC[35-39], we decided to detect the germline mutation of hMSH2 and hMLH1 in this study. In PCR-SSCP analysis of genomic DNA of 12 probands, we identified seven abnormal mobilities, five in typical HNPCC and two in suspected HNPCC and six of them were testified to be mutations by DNA sequencing. When we compared the genetic investigation of two groups we studied, we also concluded that there were no statistical differences in the results of both PCR-SSCP and direct sequence.
We also found that the mutations occurred both in typical HNPCC group and in atypical HNPCC group. Beck et al[40] found germline HNPCC mutations in six families in which none fulfilled the Amsterdam criteria. They highlighted that if germline mutations of the mismatch repair genes were common in families with features of HNPCC, but not fulfilling the Amsterdam criteria, then it was very important that all such families were also referred to cancer clinics for assessment and possible genetic testing. It was suggested that many suspected HNPCC patients might not be recognized and might be excluded from genetic counseling[41,42].
Upon reviewing the literature and checking the variants registried in the ICG-HNPCC mutation database (http:// www.nfdht.nl), we found that all the mutations were novel. It is rarely reported that there exist multiple mutations in the same exons. However, it seemed to be common in our analysis, two mutations were found at 11 exons of hMSH2 in C13 family and C8 family. The mutation occurred at four locations, one was “C to G” missense mutation at 1 198 leading to Leu400Val substitution, one was “C to G” missense mutation at 1 261 leading to Val421Leu substitution and the other two shift mutations occurred at 1 364 and 1 372 resulted from the A insertion at the two locations, respectively. Furthermore, it is the first report that the mutation occurred in both hMLH1 and hMSH2 in C4 family.
Several techniques for genetic detection of HNPCC have been developed including MSI, immunohistochemistry, SSCP, denaturing gradient gel electrophoresis (DGGE) and sequencing. SSCP was considered inefficient in detecting mutations[43,44] and was expected to miss approximately 20% of point and frameshift mutations. Furthermore, it was unable to detect the whole exon deletions, which occured in some HNPCC families. We chose SSCP first because it was relatively simple, quick and cheap, and did not require special apparatus. In our study, 12 abnormal bands found in SSCP were proven to represent 11 mutations (91.7%). We found it was a practical procedure for the genetic screening of HNPCC.
Given the limitations of current techniques, direct PCR product sequencing analysis is more efficient for detecting mutations, but it is laborious, more expensive in term of reagents and data analysis time and not 100% efficient. It is much more acceptable clinically to take the screening techniques, such as SSCP, first and use sequencing to analyze the details of the mutation. Recently, some studies demonstrated that large genomic rearrangements accounted for 10%-20% of all hMSH2 mutations, and a lower proportion of all hMLH1 mutations. Nakagawa[45] used the multiplex ligation-dependent probe amplification (MLPA) method to screen hMSH2 and hMLH1 deletions in 70 patients whose colorectal or endometrial tumors were MSI positive, yet no mutation was found by genomic exon-by-exon sequencing of hMSH2, hMLH1, and hMSH6. They identified five candidates with four different hMSH2 deletions and one candidate with an hMLH1 deletion.
In summary, our study demonstrates that the Amsterdam criteria are important but inappropriate for the establishment of diagnosis, some atypical families not fulfilling all the Amsterdam criteria probably possess the similar genetic alterations. We conclude that the mutation detection of patients is of benefit to the analysis of HNPCC and, PCR-SSCP is as effective as direct DNA sequence in detecting the mutations of HNPCC. Furthermore, it seems that there exist more complicated genetic alterations in Chinese HNPCC patients than in Western countries.
However, there are also some cases without any mutation, further investigations should be carried out such as detection of other mismatch repair genes as well as detection of gene methylation of hMLH1.
Footnotes
Supported by the National Natural Science Foundation of China, No. 30170927
Edited by Ma JY and Wang XL
References
- 1.Lynch HT, Lynch JF. Hereditary nonpolyposis colorectal cancer. Semin Surg Oncol. 2000;18:305–313. doi: 10.1002/(sici)1098-2388(200006)18:4<305::aid-ssu5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 2.Vasen HF, Watson P, Mecklin JP, Jass JR, Green JS, Nomizu T, Müller H, Lynch HT. The epidemiology of endometrial cancer in hereditary nonpolyposis colorectal cancer. Anticancer Res. 1994;14(4B):1675–1678. [PubMed] [Google Scholar]
- 3.Jass JR. Pathology of hereditary nonpolyposis colorectal cancer. Ann N Y Acad Sci. 2000;910:62–73; discussion 73-74. doi: 10.1111/j.1749-6632.2000.tb06701.x. [DOI] [PubMed] [Google Scholar]
- 4.Vasen HF, Sanders EA, Taal BG, Nagengast FM, Griffioen G, Menko FH, Kleibeuker JH, Houwing-Duistermaat JJ, Meera Khan P. The risk of brain tumours in hereditary non-polyposis colorectal cancer (HNPCC) Int J Cancer. 1996;65:422–425. doi: 10.1002/(SICI)1097-0215(19960208)65:4<422::AID-IJC4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 5.Song YM, Zheng S. Analysis for phenotype of HNPCC in China. World J Gastroenterol. 2002;8:837–840. doi: 10.3748/wjg.v8.i5.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunlop MG, Farrington SM, Carothers AD, Wyllie AH, Sharp L, Burn J, Liu B, Kinzler KW, Vogelstein B. Cancer risk associated with germline DNA mismatch repair gene mutations. Hum Mol Genet. 1997;6:105–110. doi: 10.1093/hmg/6.1.105. [DOI] [PubMed] [Google Scholar]
- 7.Vasen HF, Mecklin JP, Khan PM, Lynch HT. The International Collaborative Group on HNPCC. Anticancer Res. 1994;14(4B):1661–1664. [PubMed] [Google Scholar]
- 8.Vasen HF, Watson P, Mecklin JP, Lynch HT. New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 1999;116:1453–1456. doi: 10.1016/s0016-5085(99)70510-x. [DOI] [PubMed] [Google Scholar]
- 9.Fujita S, Moriya Y, Sugihara K, Akasu T, Ushio K. Prognosis of hereditary nonpolyposis colorectal cancer (HNPCC) and the role of Japanese criteria for HNPCC. Jpn J Clin Oncol. 1996;26:351–355. doi: 10.1093/oxfordjournals.jjco.a023244. [DOI] [PubMed] [Google Scholar]
- 10.Lynch HT, Lynch JF. The Lynch Syndrome: Melding Natural History and Molecular Genetics to Genetic Counseling and Cancer Control. Cancer Control. 1996;3:13–19. doi: 10.1177/107327489600300101. [DOI] [PubMed] [Google Scholar]
- 11.Peltomäki P, Aaltonen LA, Sistonen P, Pylkkänen L, Mecklin JP, Järvinen H, Green JS, Jass JR, Weber JL, Leach FS. Genetic mapping of a locus predisposing to human colorectal cancer. Science. 1993;260:810–812. doi: 10.1126/science.8484120. [DOI] [PubMed] [Google Scholar]
- 12.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 13.Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 14.Nicolaides NC, Papadopoulos N, Liu B, Wei YF, Carter KC, Ruben SM, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM. Mutations of two PMS homologues in hereditary nonpolyposis colon cancer. Nature. 1994;371:75–80. doi: 10.1038/371075a0. [DOI] [PubMed] [Google Scholar]
- 15.Nyström-Lahti M, Parsons R, Sistonen P, Pylkkänen L, Aaltonen LA, Leach FS, Hamilton SR, Watson P, Bronson E, Fusaro R. Mismatch repair genes on chromosomes 2p and 3p account for a major share of hereditary nonpolyposis colorectal cancer families evaluable by linkage. Am J Hum Genet. 1994;55:659–665. [PMC free article] [PubMed] [Google Scholar]
- 16.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 17.Bronner CE, Baker SM, Morrison PT, Warren G, Smith LG, Lescoe MK, Kane M, Earabino C, Lipford J, Lindblom A. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 18.Peltomäki P, de la Chapelle A. Mutations predisposing to hereditary nonpolyposis colorectal cancer. Adv Cancer Res. 1997;71:93–119. doi: 10.1016/s0065-230x(08)60097-4. [DOI] [PubMed] [Google Scholar]
- 19.Wijnen J, Vasen H, Khan PM, Menko FH, van der Klift H, van Leeuwen C, van den Broek M, van Leeuwen-Cornelisse I, Nagengast F, Meijers-Heijboer A. Seven new mutations in hMSH2, an HNPCC gene, identified by denaturing gradient-gel electrophoresis. Am J Hum Genet. 1995;56:1060–1066. [PMC free article] [PubMed] [Google Scholar]
- 20.Ikenaga M, Tomita N, Sekimoto M, Ohue M, Yamamoto H, Miyake Y, Mishima H, Nishisho I, Kikkawa N, Monden M. Use of microsatellite analysis in young patients with colorectal cancer to identify those with hereditary nonpolyposis colorectal cancer. J Surg Oncol. 2002;79:157–165. doi: 10.1002/jso.10064. [DOI] [PubMed] [Google Scholar]
- 21.Merkelbach-Bruse S, Köse S, Losen I, Bosserhoff AK, Buettner R. High throughput genetic screening for the detection of hereditary non-polyposis colon cancer (HNPCC) using capillary electrophoresis. Comb Chem High Throughput Screen. 2000;3:519–524. doi: 10.2174/1386207003331409. [DOI] [PubMed] [Google Scholar]
- 22.Holinski-Feder E, Muller-Koch Y, Friedl W, Moeslein G, Keller G, Plaschke J, Ballhausen W, Gross M, Baldwin-Jedele K, Jungck M, et al. DHPLC mutation analysis of the hereditary nonpolyposis colon cancer (HNPCC) genes hMLH1 and hMSH2. J Biochem Biophys Methods. 2001;47:21–32. doi: 10.1016/s0165-022x(00)00148-2. [DOI] [PubMed] [Google Scholar]
- 23.Kurzawski G, Safranow K, Suchy J, Chlubek D, Scott RJ, Lubiński J. Mutation analysis of MLH1 and MSH2 genes performed by denaturing high-performance liquid chromatography. J Biochem Biophys Methods. 2002;51:89–100. doi: 10.1016/s0165-022x(02)00003-9. [DOI] [PubMed] [Google Scholar]
- 24.Wahlberg SS, Schmeits J, Thomas G, Loda M, Garber J, Syngal S, Kolodner RD, Fox E. Evaluation of microsatellite instability and immunohistochemistry for the prediction of germ-line MSH2 and MLH1 mutations in hereditary nonpolyposis colon cancer families. Cancer Res. 2002;62:3485–3492. [PubMed] [Google Scholar]
- 25.Cai SJ, Xu Y, Cai GX, Lian P, Guan ZQ, Mo SJ, Sun MH, Cai Q, Shi DR. Clinical characteristics and diagnosis of patients with hereditary nonpolyposis colorectal cancer. World J Gastroenterol. 2003;9:284–287. doi: 10.3748/wjg.v9.i2.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai Q, Sun MH, Lu HF, Zhang TM, Mo SJ, Xu Y, Cai SJ, Zhu XZ, Shi DR. Clinicopathological and molecular genetic analysis of 4 typical Chinese HNPCC families. World J Gastroenterol. 2001;7:805–810. doi: 10.3748/wjg.v7.i6.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beck NE, Tomlinson IP, Homfray T, Frayling I, Hodgson SV, Harocopos C, Bodmer WF. Use of SSCP analysis to identify germline mutations in HNPCC families fulfilling the Amsterdam criteria. Hum Genet. 1997;99:219–224. doi: 10.1007/s004390050343. [DOI] [PubMed] [Google Scholar]
- 28.Viel A, Genuardi M, Capozzi E, Leonardi F, Bellacosa A, Paravatou-Petsotas M, Pomponi MG, Fornasarig M, Percesepe A, Roncucci L, et al. Character-ization of MSH2 and MLH1 mutations in Italian families with hereditary nonpolyposis colorectal cancer. Genes Chromosomes Cancer. 1997;18:8–18. doi: 10.1002/(sici)1098-2264(199701)18:1<8::aid-gcc2>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 29.Möslein G. Clinical implications of molecular diagnosis in hereditary nonpolyposis colorectal cancer. Recent Results Cancer Res. 2003;162:73–78. doi: 10.1007/978-3-642-59349-9_6. [DOI] [PubMed] [Google Scholar]
- 30.Watson P, Narod SA, Fodde R, Wagner A, Lynch JF, Tinley ST, Snyder CL, Coronel SA, Riley B, Kinarsky Y, et al. Carrier risk status changes resulting from mutation testing in hereditary non-polyposis colorectal cancer and hereditary breast-ovarian cancer. J Med Genet. 2003;40:591–596. doi: 10.1136/jmg.40.8.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Church JM. Prophylactic colectomy in patients with hereditary nonpolyposis colorectal cancer. Ann Med. 1996;28:479–482. doi: 10.3109/07853899608999111. [DOI] [PubMed] [Google Scholar]
- 32.Hanna NN, Mentzer RM. Molecular genetics and management strategies in hereditary cancer syndromes. J Ky Med Assoc. 2003;101:100–107. [PubMed] [Google Scholar]
- 33.Lynch HT, Watson P, Shaw TG, Lynch JF, Harty AE, Franklin BA, Kapler CR, Tinley ST, Liu B, Lerman C. Clinical impact of molecu-lar genetic diagnosis, genetic counseling, and management of he-reditary cancer. Part II: Hereditary nonpolyposis colorectal carci-noma as a model. Cancer. 1999;86(11 Suppl):2457–2463. doi: 10.1002/(sici)1097-0142(19991201)86:11+<2457::aid-cncr2>3.3.co;2-9. [DOI] [PubMed] [Google Scholar]
- 34.Boland CR. Molecular genetics of hereditary nonpolyposis colorectal cancer. Ann N Y Acad Sci. 2000;910:50–59; discussion 59-61. doi: 10.1111/j.1749-6632.2000.tb06700.x. [DOI] [PubMed] [Google Scholar]
- 35.Jacob S, Praz F. DNA mismatch repair defects: role in colorectal carcinogenesis. Biochimie. 2002;84:27–47. doi: 10.1016/s0300-9084(01)01362-1. [DOI] [PubMed] [Google Scholar]
- 36.Bocker T, Rüschoff J, Fishel R. Molecular diagnostics of cancer predisposition: hereditary non-polyposis colorectal carcinoma and mismatch repair defects. Biochim Biophys Acta. 1999;1423:O1–O10. doi: 10.1016/s0304-419x(99)00008-6. [DOI] [PubMed] [Google Scholar]
- 37.Planck M, Koul A, Fernebro E, Borg A, Kristoffersson U, Olsson H, Wenngren E, Mangell P, Nilbert M. hMLH1, hMSH2 and hMSH6 mutations in hereditary non-polyposis colorectal cancer families from southern Sweden. Int J Cancer. 1999;83:197–202. doi: 10.1002/(sici)1097-0215(19991008)83:2<197::aid-ijc9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 38.Krüger S, Plaschke J, Jeske B, Görgens H, Pistorius SR, Bier A, Kreuz FR, Theissig F, Aust DE, Saeger HD, et al. Identification of six novel MSH2 and MLH1 germline mutations in HNPCC. Hum Mutat. 2003;21:445–446. doi: 10.1002/humu.9121. [DOI] [PubMed] [Google Scholar]
- 39.Beck NE, Tomlinson IP, Homfray T, Hodgson SV, Harocopos CJ, Bodmer WF. Genetic testing is important in families with a history suggestive of hereditary non-polyposis colorectal cancer even if the Amsterdam criteria are not fulfilled. Br J Surg. 1997;84:233–237. [PubMed] [Google Scholar]
- 40.Wang Q, Lasset C, Desseigne F, Saurin JC, Maugard C, Navarro C, Ruano E, Descos L, Trillet-Lenoir V, Bosset JF, et al. Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum Genet. 1999;105:79–85. doi: 10.1007/s004399900064. [DOI] [PubMed] [Google Scholar]
- 41.Wagner A, Hendriks Y, Meijers-Heijboer EJ, de Leeuw WJ, Morreau H, Hofstra R, Tops C, Bik E, Bröcker-Vriends AH, van Der Meer C, et al. Atypical HNPCC owing to MSH6 germline mutations: analysis of a large Dutch pedigree. J Med Genet. 2001;38:318–322. doi: 10.1136/jmg.38.5.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Möslein G, Pistorius S, Saeger HD, Schackert HK. Preventive surgery for colon cancer in familial adenomatous polyposis and hereditary nonpolyposis colorectal cancer syndrome. Langenbecks Arch Surg. 2003;388:9–16. doi: 10.1007/s00423-003-0364-8. [DOI] [PubMed] [Google Scholar]
- 43.Syngal S, Fox EA, Eng C, Kolodner RD, Garber JE. Sensitivity and specificity of clinical criteria for hereditary non-polyposis colorectal cancer associated mutations in MSH2 and MLH1. J Med Genet. 2000;37:641–645. doi: 10.1136/jmg.37.9.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu B, Parsons R, Papadopoulos N, Nicolaides NC, Lynch HT, Watson P, Jass JR, Dunlop M, Wyllie A, Peltomäki P, et al. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med. 1996;2:169–174. doi: 10.1038/nm0296-169. [DOI] [PubMed] [Google Scholar]
- 45.Nakagawa H, Hampel H, de la Chapelle A. Identification and characterization of genomic rearrangements of MSH2 and MLH1 in Lynch syndrome (HNPCC) by novel techniques. Hum Mutat. 2003;22:258. doi: 10.1002/humu.9171. [DOI] [PubMed] [Google Scholar]


