Abstract
Purpose
To describe the long noncoding RNA (lncRNA) profiles in cumulus cells isolated from polycystic ovary syndrome (PCOS) patients by employing a microarray and in-depth bioinformatics analysis. This information will help us understand the occurrence and development of PCOS.
Methods
In this study, we used a microarray to describe lncRNA profiles in cumulus cells isolated from ten patients (five PCOS and five normal women). Several differentially expressed lncRNAs were chosen to validate the microarray results by quantitative RT-PCR (qRT-PCR). Then, the differentially expressed lncRNAs were classified into three subgroups (HOX loci lncRNA, enhancer-like lncRNA, and lincRNA) to deduce their potential features. Furthermore, a lncRNA/mRNA co-expression network was constructed by using the Cytoscape software (V2.8.3, http://www.cytoscape.org/).
Results
We observed that 623 lncRNAs and 260 messenger RNAs (mRNAs) were significantly up- or down-regulated (≥2-fold change), and these differences could be used to discriminate cumulus cells of PCOS from those of normal patients. Five differentially expressed lncRNAs (XLOC_011402, ENST00000454271, ENST00000433673, ENST00000450294, and ENST00000432431) were selected to validate the microarray results using quantitative RT-PCR (qRT-PCR). The qRT-PCR results were consistent with the microarray data. Further analysis indicated that many differentially expressed lncRNAs were transcribed from chromosome 2 and may act as enhancers to regulate their neighboring protein-coding genes. Forty-three lncRNAs and 29 mRNAs were used to construct the coding-non-coding gene co-expression network. Most pairs positively correlated, and one mRNA correlated with one or more lncRNAs.
Conclusions
Our study is the first to determine genome-wide lncRNA expression patterns in cumulus cells isolated from PCOS patients by microarray. The results show that clusters of lncRNAs were aberrantly expressed in cumulus cells of PCOS patients compared with those of normal women, which revealed that lncRNAs differentially expressed in PCOS and normal women may contribute to the occurrence of PCOS and affect oocyte development.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-015-0630-z) contains supplementary material, which is available to authorized users.
Keywords: Polycystic ovary syndrome (PCOS), Long noncoding RNA (lncRNA), Cumulus cells, Microarray
Introduction
Polycystic ovary syndrome (PCOS) is one of the most common endocrinopathies in women of reproductive age [1] and accounts for approximately 75 % of anovulatory infertility [2]. The phenotype of PCOS is variable and includes hyperandrogenism, menstrual irregularity, and polycystic ovarian morphology [3]. Patients suffering from PCOS are also characterized by obesity, hirsutism, insulin resistance and increased risks of endometrial cancer, metabolic syndrome [4], type 2 diabetes (T2D), and cardiovascular disease [5, 6]. Thus, understanding the mechanism of this endocrine disease is imperative as a primary step to finding effective diagnostic and therapeutic targets.
Although the etiology of PCOS remains unclear, most researchers accept that PCOS is multifactorial and that genetic factors play pivotal roles in its development and maintenance [7, 8]. Many studies have reported gene expression profiles based on tissues from PCOS and control patients. These studies utilized cDNA microarrays in theca cells [9], whole ovaries [10, 11], oocytes [12], and cumulus cells [13]. The data from these studies revealed a number of the genes associated with PCOS are primarily involved in the insulin receptor signaling pathway, steroid biosynthesis, and the regulation of gonadotropin secretion [14].
However, the overwhelming majority of the transcriptional outputs of the mammalian genome were confirmed to be protein noncoding genes [15]. Over the past decade, noncoding RNAs have been proven to have important regulatory potential, both in transcription and post transcription, instead of just being “transcription noise” [16]. Noncoding RNAs are divided into short noncoding RNAs and long noncoding RNAs according to their length. Recent studies have revealed that microRNAs, the best-studied short noncoding RNAs, are differentially expressed in the follicular fluid of PCOS patients [17, 18]. Moreover, an increasing body of evidence indicates that lncRNAs longer than 200 nucleotides play key roles in both normal development and diseases [15]. Some lncRNAs have been identified in recent years that are involved in diseases closely related to PCOS. For example, HI-LNC25, a β cell-specific lncRNA, is dysregulated in type 2 diabetes, due to the down-regulation of the Kruppel-like zinc finger transcription factor (GLIS3) mRNA [19]. Braveheart (Bvht), a heart-associated lncRNA, is required for the progression of nascent mesoderm toward a cardiac fate [20]. One hundred and seventy-five lncRNAs are specifically regulated during adipogenesis [21]. PU.1 AS lncRNA binds to PU.1 mRNA to form an mRNA/AS lncRNA duplex in pre-adipocytes, thus promoting adipogenesis by preventing PU.1 mRNA translation [22]. Furthermore, estradiol (E2) can modulate lncRNA expression in E2 receptor (ER) α-positive epithelial ovarian cancer (EOC) cells, and certain lncRNAs have been correlated with advanced cancer progression [23]. However, the lncRNA expression profiles in PCOS have not yet been studied.
Although clinical and biochemical signs of PCOS are typically heterogeneous, abnormal folliculogenesis is regarded as a common characteristic of PCOS [24, 25]. Cumulus cells (CCs), a subset of granulosa cells, are intimately connected to oocytes and are responsible for providing several trophic and metabolic factors to the pre-ovulatory oocyte. Cumulus and granulosa cells are the major source of estradiol. For PCOS patients, the apoptosis level is increased in granulosa cells [26]. Moreover, PCOS granulosa cells, unlike normal granulosa cells, show a limited capacity to synthesize progesterone, either spontaneously or in response to follicle-stimulating hormone (FSH) stimulation [27]. However, the dysfunction of cumulus cells in PCOS and how lncRNAs regulate the pathogenesis of PCOS in cumulus cells remain unclear. In this study, to elucidate the abnormal development of oocytes or anovulation of PCOS, aberrantly expressed lncRNAs in the cumulus cells of PCOS patients were identified by using microarray to compare them with their expression levels in normal women.
Materials and methods
Patients’ characteristics and isolation of CCs
Seventy patients (35 PCOS patients and 35 normal patients) referred to our center for in vitro fertilization (IVF) were included in this study, after obtaining written informed consent. This study was approved by the institutional ethical review board of The Affiliated Hospital of Qingdao Medical University (Yuhuangding Hospital of Yantai). The PCOS patients were diagnosed by the presence of two or more of the following features: chronic oligo-ovulation or anovulation, androgen excess, and polycystic ovaries (ultrasound imaging). We excluded patients with Cushing’s syndrome, congenital adrenal hyperplasia, and androgen-secreting tumors. Normal patients had regular menstrual cycles and normal ovary sonographs, were non-diabetic, showed no clinical signs of hyperandrogenism, and only had tubal disease. The clinical characteristics of the PCOS and normal patients are indicated in Table 1. The age (<36 years), BMI (ranging between 20 and 28 kg/m2), and basal serum E2 (ranging between 35 and 46 pg/ml) did not significantly differ between the PCOS or normal patients. However, other clinical characteristics, including basal serum luteinizing hormone (LH), basal serum LH/FSH, serum androgen, and the number of obtained oocytes per cycle, significantly differed between the two groups. The average level of basal serum LH was 7.91 mIU/ml in PCOS patients and approximately 4.30 mIU/ml in the normal patients. In PCOS patients, the ratio of basal serum LH/FSH exceeded 2.0, and the level of serum androgen exceeded 0.5 ng/ml. For PCOS patients, the numbers of obtained oocytes ranged between 12 and 28 per cycle, while approximately 10 oocytes per cycle were obtained from normal patients.
Table 1.
Clinical characteristics in PCOS and normal patients
| Normal (n = 35) | PCOS (n = 35) | P value | |
|---|---|---|---|
| Age (years) | 34.6 ± 2.2 | 32.6 ± 3.1 | NS |
| BMI (kg/m2) | 21.4 ± 1.8 | 21.6 ± 1.5 | NS |
| E2 (pg/ml) | 36.3 ± 4.3 | 41.2 ± 9.8 | NS |
| LH (mIU/ml) | 4.30 ± 1.4 | 7.91 ± 1.6 | <0.05 |
| Basal LH/FSH | 0.65 ± 0.2 | 2.20 ± 0.7 | <0.001 |
| Androgen (ng/ml) | 0.13 ± 0.05 | 0.64 ± 0.05 | <0.001 |
| Number of obtained oocytes per cycle | 10.2 ± 1.1 | 24.1 ± 3.7 | <0.001 |
Data are the mean ± SEM
NS non-significant
Patients in both groups underwent controlled ovarian stimulation (COS) (combination of GnRH agonist protocols with recombinant FSH), as described previously [28]. Specifically, all patients received 0.05 mg/day of the GnRH agonist triptorelin acetate (diphereline, Ipsen Pharma Biotech, Paris, France), subcutaneously, starting at the mid-luteal phase. Once adequate pituitary down-regulation was confirmed (serum LH levels <3.0 ng/ml and serum E2 levels <30 pg/ml), the patients subcutaneously received 150–187.5 IU recombinant FSH (Gonal-f, follitropin alfa, Serono) for COS. When two or more follicles were at least 18 mm in diameter and the serum E2 levels were at least 300 pg/ml per dominant follicle, all patients received 250-μg human chorionic gonadotropin (hCG) (Profasi, Serono). Cumulus-oocyte complex (COC) retrieval was performed by vaginal puncture under ultrasound echo-guidance 36 h after hCG administration.
The cumulus cells were retrieved, and the oocytes were cultured and inseminated as described previously [29]. In brief, after COC retrieval, a proportion of the CCs surrounding a single oocyte was removed using a sharp needle, lysed in 80 μl of RLT buffer (RNeasy Mini Kit, Qiagen) supplemented with 1 % (v/v) 2-β- mercaptoethanol (M-3148; Sigma, Lyon, France), and stored at −80 °C until RNA extraction. The oocytes were then inseminated, and embryos were individually cultured in 20-μl droplets of sequential SAGE medium (CooperSurgical, Leisegang Medical, Berlin) covered by mineral oil. The embryos were transferred or vitrified on day 3, and the other embryos were cultured to the blastula stage (until day 5–6). The cumulus cells isolated from ten patients (five PCOS and five normal patients) were used for the microarray analysis of lncRNAs, and those isolated from the other 60 patients (30 PCOS and 30 normal patients) were used for an extra evaluation by quantitative RT-PCR (qRT-PCR).
RNA extraction
The cumulus cells isolated from a single patient were pooled together to extract the total RNA. The total RNA was isolated using the Qiagen RNeasy Mini Kit (Qiagen, Hilder, Germany) according to the manufacturer’s instructions. This RNA isolation kit significantly reduces contamination from both genomic DNA and proteins. The purity and concentration of RNA were determined from OD260/280 readings using a spectrophotometer (NanoDrop ND-1000). The RNA integrity was determined with 1 % formaldehyde denaturing gel electrophoresis.
RNA labeling and array hybridization
The purified RNA, total RNA with the ribosomal RNA (rRNA) removed, was amplified and transcribed into fluorescent cDNA for hybridization to the Agilent human lncRNA + mRNA Array v2.0 (4 × 180 K format). Each array contains probes that interrogate approximately 39,000 human lncRNAs and approximately 32,000 mRNAs. These lncRNA and mRNA target sequences were merged from multiple authoritative databases: 4765 from RefSeq, 12,754 from ENSEMBL, 8195 lincRNA from John Rinn’s lab [30], 1289 from NRED (ncRNA Expression Database), 17,203 from H-InvDB, 2975 from ENCODE, 529 from CombinedLit, 1053 from Antisense ncRNA pipeline, 407 Hox ncRNAs, 481 UCRs, and 848 from Chen Ruisheng’s laboratory (Institute of Biophysics, Chinese Academy of Science). The array also contains 4974 Agilent control probes. These probes were used to detect each RNA twice as follows: cDNA was labeled with a fluorescent dye (Cy3-dCTP) using Eberwine’s linear RNA amplification method and a subsequent enzymatic reaction. This procedure was improved using the CapitalBio cRNA Amplification and Labeling Kit (CapitalBio) to increase the yields of labeled cDNA. In detail, double-stranded cDNAs (containing the T7 RNA polymerase promoter sequence) were synthesized from 1 μg of total RNA using the CbcScript reverse transcriptase with a cDNA synthesis system and the T7 Oligo (dT) and T7 Oligo (dN) primers according to the manufacturer’s protocol (CapitalBio). After completing the double-stranded cDNA (dsDNA) synthesis using DNA polymerase and RNase H, the dsDNA products were purified using a PCR NucleoSpin Extract II Kit (MN) and eluted with 30 μL of elution buffer. The eluted double-stranded cDNA products were vacuum-evaporated to 16 μL and transcribed in vitro in a total volume of 40 μL at 37 °C for 14 h using a T7 Enzyme Mix. The amplified cRNA was purified using the RNA Clean-up Kit (MN). The Klenow enzyme labeling strategy was adopted after reverse transcription using CbcScript II reverse transcriptase. Briefly, 2 μg of amplified RNA was mixed with 4 μg of random nanomers. The RNA was then denatured at 65 °C for 5 min and cooled on ice. Subsequently, 5 μL of 4× first-strand buffer, 2 μL of 0.1 M DTT, and 1.5 μL of CbcScript II reverse transcriptase were added. The mixtures were incubated at 25 °C for 10 min, then at 37 °C for 90 min. The cDNA products were purified using a PCR NucleoSpin Extract II Kit (MN) and vacuum-evaporated to 14 μL. The cDNA was mixed with 4 μg of random nanomers, heated to 95 °C for 3 min, and snap-cooled on ice for 5 min. Subsequently, 5 μL of Klenow buffer, dNTPs, and Cy3-dCTP (GE Healthcare) were added to final concentrations of 240-μM dATP, 240-μM dGTP, 240-μM dTTP, 120-μM dCTP, and 40-μM Cy-dCTP. Klenow enzyme (1.2 μL) was then added, and the reaction was performed at 37 °C for 90 min. The labeled cDNA was purified with a PCR NucleoSpin Extract II Kit (MN) and re-suspended in elution buffer. The labeled controls and test samples labeled with Cy3-dCTP were dissolved in 80 μL of hybridization solution containing 3× SSC, 0.2 % SDS, 5× Denhardt’s solution, and 25 % formamide. The DNA in the hybridization solution was denatured at 95 °C for 3 min prior to being loaded onto a microarray. The arrays were hybridized in an Agilent hybridization oven overnight at a rotation speed of 20 rpm and a temperature of 42 °C. The arrays were then washed in a solution containing 0.2 % SDS and 2× SSC at 42 °C for 5 min, then in 0.2× SSC for 5 min at room temperature. The microarray was analyzed by CapitalBio Corporation, Beijing, P. R. China.
Microarray imaging and data analysis
The lncRNA + mRNA array data were analyzed to summarize and normalize the data and control their quality using the GeneSpring software V11.5 (Agilent). To select the differentially expressed genes, we used threshold values of ≥2.0- and ≤ −2.0-fold changes and a Benjamini-Hochberg-corrected P value of 0.05. The data were log2 transformed and median centred by genes using the Adjust Data function of the CLUSTER 3.0 software. The data were then further analyzed using hierarchical clustering with average linkage [31]. Finally, we visualized the tree using Java Treeview (Stanford University School of Medicine, Stanford, CA, USA). The analysis was performed by CapitalBio Corporation, Beijing, P. R. China.
lncRNA classification and subgroup analysis
Recently, some classes or clusters of lncRNAs, such as human homeobox transcription factors (HOX) lncRNAs and lncRNAs with enhancer-like functions, have been found to play specific roles in human cells. To further understand the features of differentially expressed lncRNAs isolated in this study, we classified the lncRNAs as follows: lncRNAs with enhancer-like functions, Rinn’s lincRNAs, and HOX cluster lncRNAs. Of these classes, lncRNAs with enhancer-like functions were identified using the GENCODE annotation of human genes [30, 32]; transcripts mapped to the exons and introns of annotated protein-coding genes, natural antisense transcripts that overlap the protein-coding genes and all known transcripts were excluded. Rinn’s lincRNA and HOX cluster lncRNAs were identified according to Rinn’s reports [33–35].
Construction of the coding-non-coding gene co-expression network
The differentially expressed mRNAs and lncRNAs were selected to construct the co-expression network as described in previous reports [36, 37]. The network construction procedures included the following: (1) pre-processing data: for one mRNA with different transcripts, the median value was used to represent the gene expression, without special treatment for lncRNA expression values; (2) screening data: subsets of data were removed according to the lists of the differential expression of lncRNA and mRNA; (3) Pearson’s correlation coefficient was calculated, and the P value was used to calculate the correlation coefficient between lncRNAs and mRNAs; and (4) screening based on Pearson’s correlation coefficient: Pearson’s correlation coefficients greater than 0.99 were considered meaningful and used to draw the lncRNA/mRNA co-expression network with the Cytoscape software (V2.8.3, http://www.cytoscape.org/) [38]. The blue node represents differentially expressed lncRNAs, and the green node represents differentially expressed mRNAs.
Confirmation of differentially expressed lncRNAs by real-time quantitative RT-PCR
qRT-PCR was performed to confirm the differential expression of lncRNAs identified with the microarray analysis. Briefly, the first-strand complementary synthesis reaction was performed using a M-MLV Reverse Transcriptase kit (Invitrogen). Amplification reactions were conducted using Power SYBR Green PCR Master Mix (Applied Biosystems) and an Applied Biosystems 7900HT system. The lncRNA-specific qRT-PCR primers used in this study are listed in Table S1 at the end of this article. GAPDH served as an internal control to normalize the loading of the template cDNA. The PCR thermal cycling conditions were 95 °C for 10 min to activate the polymerase and denature the template, followed by 40 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 60 s, and extension at 72 °C for 30 s. A melting curve analysis was recorded at the end of the amplification to evaluate the presence of contaminants or primer dimers. Each set of qRT-PCR reactions was repeated three times, and the fold change in the expression of each lncRNA of interest was analyzed using the DDCt method [39]. Student’s t test of independent data was used for the statistical analyses. The differences between groups were considered significant when the P-value was <0.05.
Results
Overview of lncRNA and mRNA profiles
In our experiment, we tested thousands of differentially expressed human lncRNAs and mRNAs from RefSeq, ENSEMBL, John Rinn’s lab, NRED, H-InvDB, ENCODE, CombinedLit, Antisense ncRNA pipeline, Hox ncRNAs, UCRs, and Chen Ruisheng’s laboratory. The differentially expressed lncRNAs and mRNAs were selected as follows: the fold change (the log2 value of the absolute fold change of PCOS/normal (P/N)) cut-off was 2.0, and a positive value indicated up-regulation, while a negative value indicated down-regulation. The fold change and P value were calculated from the normalized expression. We compared the lncRNAs and mRNA expression levels between cumulus cells isolated from five PCOS and five normal patients based on the microarray data. The raw microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and can be accessed using the GEO series accession number GSE65746.
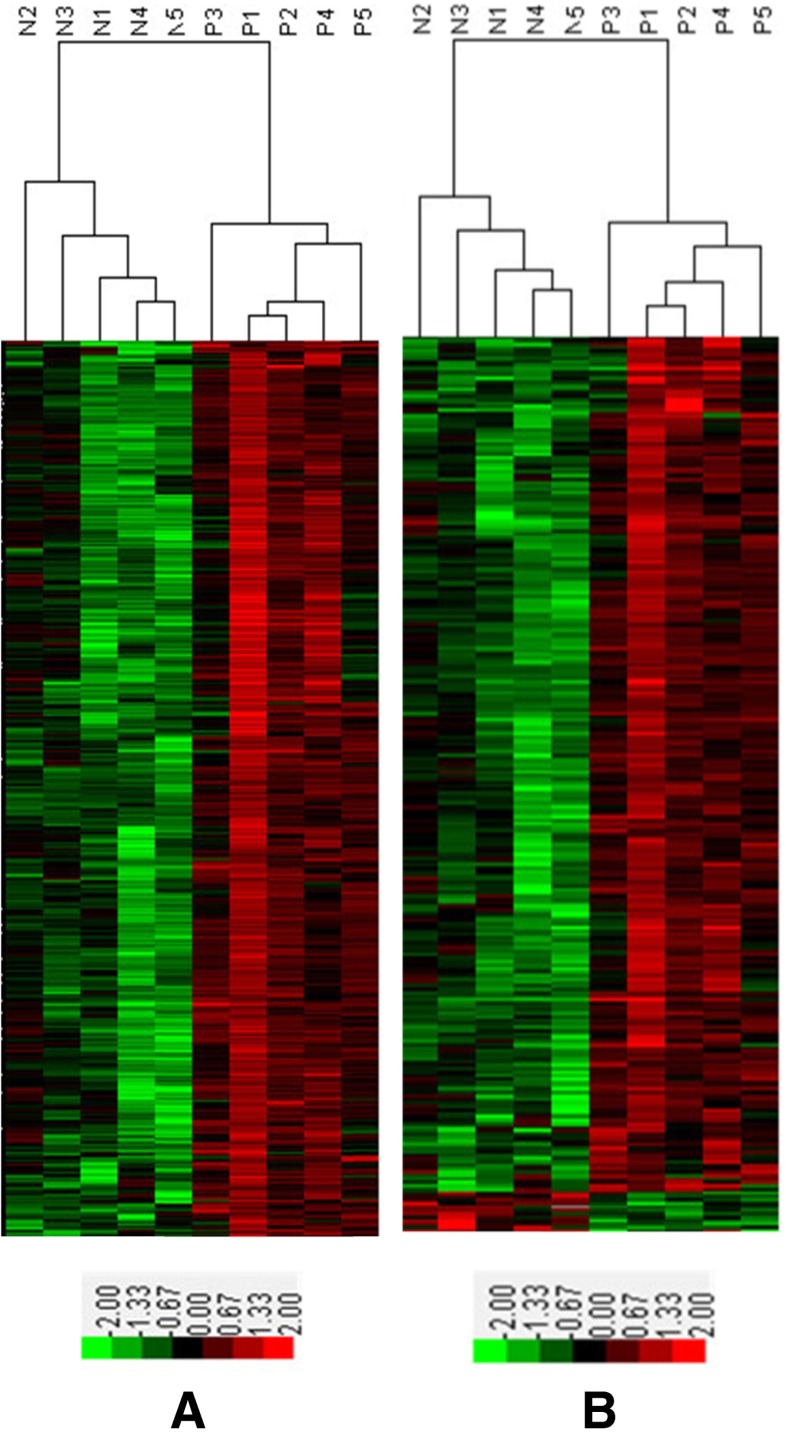
In this study, 24,292 lncRNAs and 30,184 coding transcripts were examined in the cumulus cells of PCOS and normal patients. Of these transcripts, 623 lncRNAs and 260 mRNAs were significantly up- or down-regulated, which could be used to discriminate the cumulus cells of PCOS patients from those of normal patients (≥2-fold) (Tables S2 and S4). A clustering analysis of the arrays based on the differentially expressed lncRNAs or mRNAs perfectly clustered the PCOS and normal groups (Fig. 1).
Fig. 1.
Cluster of lncRNAs (a) and mRNAs (b) overexpressed in cumulus cells (CCs) obtained from PCOS or normal patients. a Of the 623 differentially expressed lncRNAs, 620 lncRNAs were up-regulated in PCOS cumulus cells, while only three lncRNAs were down-regulated. b Of the 260 differentially expressed mRNAs, 248 mRNAs were up-regulated, while 12 mRNAs were down-regulated. The supervised hierarchical clustering of genes overexpressed in CCs obtained from PCOS or normal patients are shown. Distinct signatures were observed in the PCOS and normal groups. The value of each gene was adjusted with a median-centering algorithm on a log scale; the colors indicate the relative gene expression in the red-green heat map. 0.00 is indicated by pure black and represents no change from the median gene expression level in all samples. −2.00 is indicated by pure green and represents lower expression. +2.00 is indicated by pure red and represents relatively higher expression. N1-N5 represents the CC samples from five normal patients. P1-P5 represents the CC samples from five PCOS patients
Of the 623 differentially expressed lncRNAs, 620 lncRNAs were up-regulated in PCOS cumulus cells while only three lncRNAs were down-regulated (Table S3). The data indicate that up-regulated lncRNAs were more common than down-regulated ones. Among these lncRNAs, ENST00000433673 (log2 fold change P/N = 8.350521) was the most up-regulated lncRNA, while NR_027436 (log2 fold change P/N = −2.47488) was the most down-regulated.
Among the 260 differentially expressed mRNAs, 248 mRNAs were up-regulated, while 12 mRNAs were down-regulated (Table S5). Of these mRNAs, SGCZ (NM_139167) (log2 fold change P/N = 4.719) was the most up-regulated mRNA, and HOXA3 (NM_153631) (log2 fold change P/N = −3.943) was the most down-regulated.
Distribution of differentially expressed lncRNAs in human chromosomes
The lengths of differentially expressed lncRNAs ranged from 201 to 11,869 bp, and they were transcribed from the + or – strand of the corresponding chromosomes. Furthermore, these differentially expressed lncRNAs were distributed in all human chromosomes. Except for the regions of three lncRNAs (p0249_imsncRNA392, p0829_imsncRNA629, and UCSC_1350_3661) that were uncertain, 74 lncRNAs were transcribed from chromosome 2, while only seven lncRNAs were from chromosome 22. The numbers of lncRNAs transcribed from different chromosomes are displayed in Table S6 and Fig. 2.
Fig. 2.

The numbers of lncRNAs transcribed from different chromosomes. The distribution of lncRNAs was not uniform in human chromosomes. The x-axis indicates different human chromosomes, and the y-axis indicates the number of differentially expressed lncRNAs transcribed from different chromosomes
lncRNA classification and subgroup analysis
In the current study, the differentially expressed lncRNAs were classified into three subgroups: HOX loci lncRNA, enhancer-like lncRNA, and lincRNA. (1) HOX loci lncRNA subgroup: 174 lncRNAs were detected in the human HOX loci in this microarray, and only two (int-HOXA3-8 and nc-HOXA9-77) of these lncRNAs were differentially expressed in PCOS samples. All related data are shown in Table S7. (2) Enhancer-like lncRNAs subgroup:2352 enhancer-like lncRNAs were detected, 87 of which were differentially expressed (Table S8). (3) lincRNA subgroup: 798 lincRNAs were detected, six of which were differentially expressed in the cumulus cells of PCOS patients (Table S9).
Construction of the coding-non-coding gene co-expression network
A coding-non-coding gene co-expression network was constructed according the correlation of the differentially expressed mRNAs with the lncRNAs. Within the co-expression network, 43 lncRNAs and 29 mRNAs comprised the CNC network nodes. Most pairs positively correlated (Table S10). The co-expression network indicated that one mRNA correlated with one or more lncRNAs. For example, NPY1R (neuropeptide Y receptor Y1) was related to approximately 20 different lncRNAs and four mRNAs (Fig. 3a). Furthermore, a previously reported lncRNA, XLOC_011402 (Prader-Willi region non-protein-coding RNA 2, PWRN2), was co-expressed with ATP6V1G3 (ATPase, H+ transporting, lysosomal 13 kDa, V1 subunit G3) (Fig. 3b). The co-expression network suggests that lncRNAs and mRNAs are inter-regulated in the cumulus cells of PCOS patients.
Fig. 3.

Coding-non-coding gene co-expression networks were constructed according the correlation analysis between the differentially expressed mRNAs and lncRNAs using the Cytoscape software (V2.8.3, http://www.cytoscape.org/). a lncRNAs–NPY1R subnetwork: NPY1R (neuropeptide Y receptor Y1) was related to approximately 20 different lncRNAs and four mRNAs. b lncRNAs–ATP6V1G3 subnetwork: XLOC_011402 (Prader-Willi region non-protein-coding RNA 2, PWRN2) was co-expressed with ATP6V1G3 (ATPase, H+ transporting, lysosomal 13 kDa, V1 subunit G3). Genes colored in green are protein-coding RNAs, and genes colored in blue are lncRNAs
Quantitative RT-PCR validation
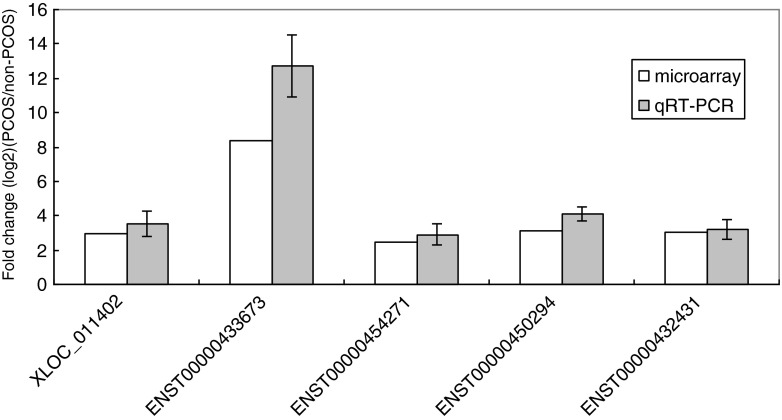
According to the fold change (FC) values and the subgroups to which the differentially expressed lncRNAs belong, five lncRNAs (XLOC_011402, ENST00000433673, ENST00000454271, ENST00000450294, and ENST00000432431) were selected to validate the microarray results by quantitative RT-PCR. Except for XLOC_011402, which is a previously reported lncRNA [40], ENST00000433673 (with the highest FC value), ENST00000454271, ENST00000450294, and ENST00000432431 were enhancer-like lncRNAs. LncRNAs (ENST00000454271, ENST00000450294, and ENST00000432431) with a high signal value in the microarray were selected randomly. The expression levels of the five lncRNAs indicated above, determined by qRT-PCR, agreed with the microarray data for the samples used for the microarray analysis. Specifically, a 3.55-fold change (2.98-fold in the microarray analysis) was observed for XLOC_011402, a 12.72-fold change (8.35-fold in the microarray analysis) was observed for ENST00000433673, a 2.91-fold change (2.47-fold in the microarray analysis) was observed for ENST00000454271, a 4.12-fold change (3.11-fold in the microarray analysis) was observed for ENST00000450294, and a 3.17-fold change (3.00-fold in the microarray analysis) was observed for ENST00000432431 (Fig. 4).
Fig. 4.
Relative expression of five lncRNAs (XLOC_011402, ENST00000454271, ENST00000433673, ENST00000450294, and ENST00000432431) in cumulus cells from PCOS and normal patients. The qRT-PCR results agreed with the microarray data set. The gray bars show the relative lncRNA expression measured using qRT-PCR. The white bars show the relative lncRNA expression measured using microarrays. The same samples were used for the qRT-PCR and microarray analyses
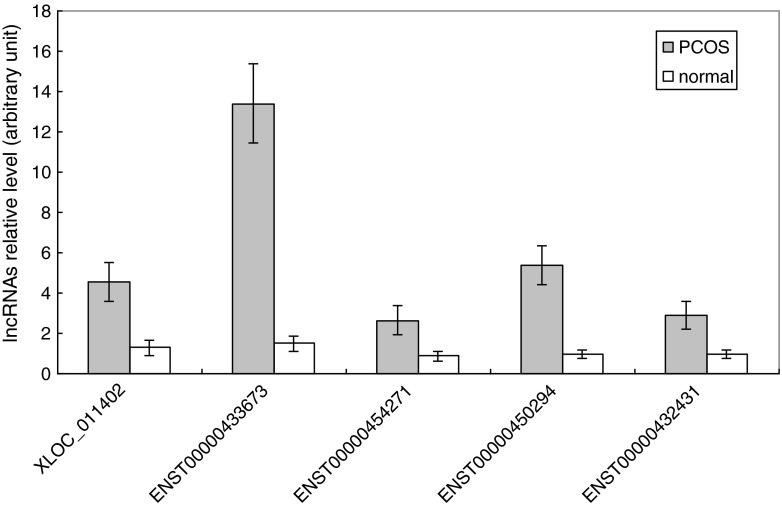
In addition, the expression levels of the five selected lncRNAs were also examined by qRT-PCR in a cohort of CC samples isolated from an additional 30 PCOS and 30 normal patients. A comparison between the CCPCOS and CCnon-PCOS groups showed that the mean transcript levels of the five lncRNAs were significantly higher in CCs isolated from PCOS patients, with 3.54-fold, 9.06-fold, 3.01-fold, 5.56-fold, and 3.07-fold increases for XLOC_011402 (4.57 ± 0.97 vs. 1.29 ± 0.4), ENST00000433673 (13.41 ± 1.95 vs. 1.48 ± 0.38), ENST00000454271 (2.65 ± 0.74 vs. 0.88 ± 0.24), ENST00000450294 (5.39 ± 0.99 vs. 0.97 ± 0.18), and ENST00000432431 (2.92 ± 0.69 vs. 0.95 ± 0.21), respectively. The qRT-PCR results for different samples further suggest that the fold changes observed in the microarray analysis were reasonable (Fig. 5).
Fig. 5.
The expression levels of the five selected lncRNAs (XLOC_011402, ENST00000454271, ENST00000433673, ENST00000450294, and ENST00000432431) were examined by qRT-PCR in a cohort of CC samples isolated from an additional 30 PCOS and 30 normal patients. The expression levels of the five lncRNAs were up-regulated in the cumulus cells of PCOS patients compared to those of normal patients (p < 0.01). The five different lncRNAs are shown on the x-axis. The signal intensity for each lncRNA is shown on the y-axis in arbitrary units determined by qRT-PCR analysis with GAPDH as an endogenous reference. The gray bars show the relative lncRNA expression measured in cumulus cells of PCOS patients. The white bars show the relative lncRNA expression measured in the cumulus cells of normal patients. The results are presented as the means ± SEM
Discussion
Recent advances in sequencing technologies have revealed that the genome is extensively transcribed, yielding a large repertoire of noncoding RNAs, which are emerging as a new class of RNAs with important roles in a variety of biological processes. The lncRNAs that have been characterized have been proven or assumed to play roles in the endocrine systems and endocrine-related diseases [41]. PCOS is the most common endocrine abnormality in women of reproductive age. The lncRNA expression profiles have not yet been described in PCOS, or in cumulus cells, which play pivotal roles in oocyte maturation and metabolism. Although sufficient oocytes are usually retrieved from PCOS patients who are under COS, the number of high-quality mature oocytes is limited. Therefore, in-depth genetic characterizations will help to understand the occurrence and development of PCOS and identify new personalized therapeutic strategies to improve the oocyte quality of PCOS patients who are receiving IVF.
In this study, 623 lncRNAs and 260 mRNAs were differentially expressed between CC samples from PCOS and normal patients. The differentially expressed mRNAs (n = 260) isolated in this study were not identical to those published in previous reports [2, 13]. This difference may be due to differences in the samples (oocyte, ovarian tissue, or cumulus cells) and the gene chips between ours and the previous studies. Notably, because previous reports have shown that the lean PCOS group is the source of the unique characteristics of “authentic” PCOS and obesity may be regarded as modifier of the syndrome or as a separate pathological mechanism that results in similar consequences [13], we selected lean PCOS patients with lower BMI (21.6 ± 1.5) in this study to reduce the differences in gene expression caused by obesity. In fact, PCOS in lean and obese individuals should be regarded as separate subentities. Subgrouping PCOS patients into more homogeneous groups to do further large-scale molecular research will provide a better understanding of the molecular pathophysiology of this syndrome. Additionally, in this study, we selected patients with tubal factor infertility as a control group. Tubal disease could be mediated by endometriosis or pelvic inflammatory disease. Previous reports have shown that endometriosis could affect the lncRNA expression patterns in the ovaries [42]. Thus, whether the differentially expressed lncRNAs between the PCOS and control groups were induced by a chronic inflammatory state needs further research.
Among the 623 differentially expressed lncRNAs, 620 lncRNAs were up-regulated in PCOS samples, while three lncRNAs were down-regulated. The fact that most differentially expressed lncRNAs (n = 74) were transcribed from chromosome 2 is remarkable. Notably, a recent genome-wide association study (GWAS) identified three of 11 susceptibility loci for PCOS on chromosome 2, including the 2p16.3 (rs13405728), 2p21 (rs13429458), and FSHR genes [1, 43]. Our data and previously published studies suggest that the genes located on chromosome 2 play important roles in the development and progression of PCOS. Moreover, because lncRNAs are almost evolutionarily conserved in mammalian genomes and thus function in diverse biological processes, we classified the lncRNAs into different subgroups, lncRNAs with enhancer-like function, Rinn’s lincRNAs, and HOX (human homeobox transcription factors) cluster lncRNAs, by comparing them to previous reports [30, 33–35]. This classification identified most lncRNAs as enhancer-like lncRNAs (n = 87), while the numbers of HOX lncRNAs (n = 2) and lincRNAs (n = 6) were limited. As for the classical enhancers, enhancer-like lncRNAs are orientation-independent and require a minimal promoter in their target genes to enhance their transcription. The function of enhancer-like lncRNAs may be investigated by depleting them and detecting the expression changes of their neighboring protein-coding genes.
Several lncRNAs, such as SRA (steroid receptor RNA activator), GAS5 (growth arrest-specific 5), and CTBP1-AS (C-terminal binding protein 1-anstisense), play important roles in the endocrine systems and endocrine-related diseases. Of these lncRNAs, SRA and GAS5 are two well-characterized lncRNAs that play roles in the endocrine system by mediating their functions as regulators of nuclear receptor (NR) signaling. SRA modulates the effects of steroid hormones on physiology and development [44], while GAS5 lncRNA regulates glucocorticoid signaling in metabolic and inflammatory pathways [45]. Unlike SRA and GAS5, which have been linked to many NRs, another lncRNA, CTBP1-AS, is an androgen-responsive (AR) lncRNA and associated with the AR signaling pathway [46]. We detected all three lncRNAs, but their expression levels did not significantly differ between the PCOS or normal groups. This lack of difference may be attributed to the different pathogenic molecular mechanism between PCOS and other endocrine-related diseases.
Notably, the coding-non-coding gene co-expression network provided us with valuable insight. First, approximately 20 lncRNAs were co-expressed with the neuropeptide Y1 receptor (NPY1R). Neuropeptide Y (NPY) interacts with the Y1 receptor (NPY1R) to control adrenergic activity and blood pressure (BP) [47]. Furthermore, NPY1R was identified as a positional candidate gene for both obesity and T2D [48]. A recent study demonstrated that the Y1 receptor plays an essential role in glucose transporter (GLUT4) translocation and may be a potential therapeutic target for type 2 diabetes [49]. PCOS is also associated with endocrine-metabolic derangements that lead to a broad range of adverse sequelae, which include dyslipidemia, atherosclerosis, insulin resistance, and type 2 diabetes [4–6]. Therefore, we inferred that lncRNAs that were co-expressed with NPY1R play vital roles in the occurrence of PCOS. An improved understanding of these lncRNAs will broaden our perspectives of molecular endocrinology and may facilitate the discovery of new therapeutic agents for PCOS.
Second, we identified XLOC_011402 for the first time, which represents the Prader-Willi region non-protein-coding RNA 2 (PWRN2) that is up-regulated in the CCs of PCOS patients. A previous study [40] reported that PWRN2 is only expressed in the testes and is up-regulated after meiosis during spermatogenesis. Consequently, the expression of PWRN2 may be more accurately described to occur in reproduction-related tissues. Furthermore, the coding-non-coding gene co-expression network identified XLOC_011402 and a protein-coding gene (ATP6V1G3) coding for one of the isoforms of a subunit of vacuolar-H+ ATPase (V-ATPase), which were co-expressed in cis. The V-ATPase is a multi-subunit enzyme that couples ATP hydrolysis to proton pumping across membranes and regulates the pH of extracellular compartments [50]. In our research, the ATP6V1G3 mRNA level was up-regulated in the CCs of PCOS patients compared to normal patients. We postulated that the high level of ATP6V1G3 mRNA enhances active proton transport and induces the low pH of the follicular microenvironment. A previous report demonstrated that a reduced intracellular pH is associated with changes in the organization or stability of the meiotic metaphase spindle and influences the development of oocytes, even causing aneuploidy [51]. Thus, the higher expression levels of XLOC_011402 and ATP6V1G3 are more likely to cause oocyte dysplasia in PCOS patients by down-regulating the pH level of the follicular microenvironment.
Although the number of identified lncRNAs is growing quickly, further study will be needed to explore their molecular and biological functions. To understand the roles of lncRNAs in the pathogenesis of PCOS, subsequent loss-of-function experiments using RNAi-mediated knockdown in cell lines or animal models will validate associations between lncRNAs and corresponding functional pathways and physiological outcomes. Moreover, the subcellular localization of lncRNAs which could be detected by RNA fluorescence in situ hybridization will provide clues to their biological functions [52]. Ultimately, the differential expression of lncRNAs maybe facilitate the classification of PCOS disease into distinct subtypes and the development of lncRNA-based therapies.
Electronic supplementary material
(DOC 31 kb)
(XLS 14066 kb)
(XLS 514 kb)
(XLS 30253 kb)
(XLS 311 kb)
(XLS 760 kb)
(XLS 124 kb)
(XLS 1401 kb)
(XLS 467 kb)
(XLSX 89 kb)
Acknowledgments
We gratefully acknowledge the CapitalBio Corporation for conducting the RNA extractions and microarrays.
Compliance with ethical standards
This study was approved by the institutional ethical review board of The Affiliated Hospital of Qingdao Medical University (Yuhuangding Hospital of Yantai).
Financial disclosure
This study was supported by the National Natural Science Foundation of China (Grant 81401172 and 81170622) and the Natural Science Foundation of Shandong Province (Grant ZR2013HQ004).
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Capsule
Our study is the first to determine genome-wide lncRNA expression patterns in cumulus cells isolated from PCOS patients by microarray.
Contributor Information
Xin Huang, Phone: 0086-535-6691999-83911, Email: huangxin92129@163.com.
Cuifang Hao, Phone: 0086-535-6691999-83905, Email: cuifanghao@aliyun.com.
Hongchu Bao, Email: bhch1005@126.com.
Meimei Wang, Email: wangmeimei76@163.com.
Huangguan Dai, Email: 459643750@qq.com.
References
- 1.Shi Y, Zhao H, Cao Y, Yang D, Li Z, Zhang B, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44(9):1020–5. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 2.Diao FY, Xu M, Hu Y, Li J, Xu Z, Lin M, et al. The molecular characteristics of polycystic ovary syndrome (PCOS) ovary defined by human ovary cDNA microarray. J Mol Endocrinol. 2004;33(1):59–72. doi: 10.1677/jme.0.0330059. [DOI] [PubMed] [Google Scholar]
- 3.Group REA-SPCW Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81(1):19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Kandaraki E, Christakou C, Diamanti-Kandarakis E. Metabolic syndrome and polycystic ovary syndrome… vice versa. Arq Bras Endocrinol Metabol. 2009;53(2):227–37. doi: 10.1590/S0004-27302009000200014. [DOI] [PubMed] [Google Scholar]
- 5.Carmina E. Cardiovascular risk and events in polycystic ovary syndrome. Climacteric. 2009;12(Suppl 1):22–5. doi: 10.1080/13697130903003842. [DOI] [PubMed] [Google Scholar]
- 6.Wild S, Pierpoint T, Jacobs H, McKeigue P. Long-term consequences of polycystic ovary syndrome: results of a 31 year follow-up study. Hum Fertil (Camb) 2000;3(2):101–5. doi: 10.1080/1464727002000198781. [DOI] [PubMed] [Google Scholar]
- 7.Diamanti-Kandarakis E, Piperi C. Genetics of polycystic ovary syndrome: searching for the way out of the labyrinth. Hum Reprod Update. 2005;11(6):631–43. doi: 10.1093/humupd/dmi025. [DOI] [PubMed] [Google Scholar]
- 8.Vink JM, Sadrzadeh S, Lambalk CB, Boomsma DI. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91(6):2100–4. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 9.Wood JR, Ho CK, Nelson-Degrave VL, McAllister JM, Strauss JF., 3rd The molecular signature of polycystic ovary syndrome (PCOS) theca cells defined by gene expression profiling. J Reprod Immunol. 2004;63(1):51–60. doi: 10.1016/j.jri.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Jansen E, Laven JS, Dommerholt HB, Polman J, van Rijt C, van den Hurk C, et al. Abnormal gene expression profiles in human ovaries from polycystic ovary syndrome patients. Mol Endocrinol. 2004;18(12):3050–63. doi: 10.1210/me.2004-0074. [DOI] [PubMed] [Google Scholar]
- 11.Oksjoki S, Soderstrom M, Inki P, Vuorio E, Anttila L. Molecular profiling of polycystic ovaries for markers of cell invasion and matrix turnover. Fertil Steril. 2005;83(4):937–44. doi: 10.1016/j.fertnstert.2004.10.037. [DOI] [PubMed] [Google Scholar]
- 12.Wood JR, Dumesic DA, Abbott DH, Strauss JF., 3rd Molecular abnormalities in oocytes from women with polycystic ovary syndrome revealed by microarray analysis. J Clin Endocrinol Metab. 2007;92(2):705–13. doi: 10.1210/jc.2006-2123. [DOI] [PubMed] [Google Scholar]
- 13.Kenigsberg S, Bentov Y, Chalifa-Caspi V, Potashnik G, Ofir R, Birk OS. Gene expression microarray profiles of cumulus cells in lean and overweight-obese polycystic ovary syndrome patients. Mol Hum Reprod. 2009;15(2):89–103. doi: 10.1093/molehr/gan082. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed-Hussein ZA, Harun S. Construction of a polycystic ovarian syndrome (PCOS) pathway based on the interactions of PCOS-related proteins retrieved from bibliomic data. Theor Biol Med Model. 2009;6:18. doi: 10.1186/1742-4682-6-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 16.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–9. doi: 10.1126/science.1231776. [DOI] [PubMed] [Google Scholar]
- 17.Roth LW, McCallie B, Alvero R, Schoolcraft WB, Minjarez D, Katz-Jaffe MG. Altered microRNA and gene expression in the follicular fluid of women with polycystic ovary syndrome. J Assist Reprod Genet. 2014;31(3):355–62. doi: 10.1007/s10815-013-0161-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sang Q, Yao Z, Wang H, Feng R, Zhao X, Xing Q, et al. Identification of microRNAs in human follicular fluid: characterization of microRNAs that govern steroidogenesis in vitro and are associated with polycystic ovary syndrome in vivo. J Clin Endocrinol Metab. 2013;98(7):3068–79. doi: 10.1210/jc.2013-1715. [DOI] [PubMed] [Google Scholar]
- 19.Moran I, Akerman I, van de Bunt M, Xie R, Benazra M, Nammo T, et al. Human beta cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 2012;16(4):435–48. doi: 10.1016/j.cmet.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klattenhoff CA, Scheuermann JC, Surface LE, Bradley RK, Fields PA, Steinhauser ML, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun L, Goff LA, Trapnell C, Alexander R, Lo KA, Hacisuleyman E, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A. 2013;110(9):3387–92. doi: 10.1073/pnas.1222643110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pang WJ, Lin LG, Xiong Y, Wei N, Wang Y, Shen QW, et al. Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation. J Cell Biochem. 2013;114(11):2500–12. doi: 10.1002/jcb.24595. [DOI] [PubMed] [Google Scholar]
- 23.Qiu JJ, Ye LC, Ding JX, Feng WW, Jin HY, Zhang Y, et al. Expression and clinical significance of estrogen-regulated long non-coding RNAs in estrogen receptor alpha-positive ovarian cancer progression. Oncol Rep. 2014;31(4):1613–22. doi: 10.3892/or.2014.3000. [DOI] [PubMed] [Google Scholar]
- 24.Christakou C, Diamanti-Kandarakis E. Polycystic ovary syndrome—phenotypes and diagnosis. Scand J Clin Lab Invest Suppl. 2014;244:18–22. doi: 10.3109/00365513.2014.936675. [DOI] [PubMed] [Google Scholar]
- 25.Fux Otta C, Fiol de Cuneo M, Szafryk de Mereshian P. [Polycystic ovary syndrome: physiopathology review] Rev Fac Cien Med Univ Nac Cordoba. 2013;70(1):27–30. [PubMed] [Google Scholar]
- 26.Onalan G, Selam B, Baran Y, Cincik M, Onalan R, Gunduz U, et al. Serum and follicular fluid levels of soluble Fas, soluble Fas ligand and apoptosis of luteinized granulosa cells in PCOS patients undergoing IVF. Hum Reprod. 2005;20(9):2391–5. doi: 10.1093/humrep/dei068. [DOI] [PubMed] [Google Scholar]
- 27.Erickson GF, Magoffin DA, Garzo VG, Cheung AP, Chang RJ. Granulosa cells of polycystic ovaries: are they normal or abnormal? Hum Reprod. 1992;7(3):293–9. doi: 10.1093/oxfordjournals.humrep.a137638. [DOI] [PubMed] [Google Scholar]
- 28.Huang X, Hao C, Shen X, Liu X, Shan Y, Zhang Y, et al. Differences in the transcriptional profiles of human cumulus cells isolated from MI and MII oocytes of patients with polycystic ovary syndrome. Reproduction. 2013;145(6):597–608. doi: 10.1530/REP-13-0005. [DOI] [PubMed] [Google Scholar]
- 29.Huang X, Hao C, Shen X, Zhang Y, Liu X. RUNX2, GPX3 and PTX3 gene expression profiling in cumulus cells are reflective oocyte/embryo competence and potentially reliable predictors of embryo developmental competence in PCOS patients. Reprod Biol Endocrinol. 2013;11:109. doi: 10.1186/1477-7827-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143(1):46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl 1):1–9. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458(7235):223–7. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–72. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, et al. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS One. 2012;7(8):e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39(9):3864–78. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 40.Wawrzik M, Spiess AN, Herrmann R, Buiting K, Horsthemke B. Expression of SNURF-SNRPN upstream transcripts and epigenetic regulatory genes during human spermatogenesis. Eur J Hum Genet. 2009;17(11):1463–70. doi: 10.1038/ejhg.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun M, Kraus WL. Minireview: long noncoding RNAs: new “links” between gene expression and cellular outcomes in endocrinology. Mol Endocrinol. 2013;27(9):1390–402. doi: 10.1210/me.2013-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun PR, Jia SZ, Lin H, Leng JH, Lang JH. Genome-wide profiling of long noncoding ribonucleic acid expression patterns in ovarian endometriosis by microarray. Fertil Steril. 2014;101(4):1038–46. doi: 10.1016/j.fertnstert.2013.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Chen ZJ, Zhao H, He L, Shi Y, Qin Y, Li Z, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43(1):55–9. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 44.Xu B, Gerin I, Miao H, Vu-Phan D, Johnson CN, Xu R, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One. 2010;5(12):e14199. doi: 10.1371/journal.pone.0014199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci Signal. 2010;3(107):ra8. doi: 10.1126/scisignal.2000568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takayama K, Horie-Inoue K, Katayama S, Suzuki T, Tsutsumi S, Ikeda K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32(12):1665–80. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang L, Rao F, Zhang K, Mahata M, Rodriguez-Flores JL, Fung MM, et al. Neuropeptide Y(1) Receptor NPY1R discovery of naturally occurring human genetic variants governing gene expression in cella as well as pleiotropic effects on autonomic activity and blood pressure in vivo. J Am Coll Cardiol. 2009;54(10):944–54. doi: 10.1016/j.jacc.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elbers CC, Onland-Moret NC, Franke L, Niehoff AG, van der Schouw YT, Wijmenga C. A strategy to search for common obesity and type 2 diabetes genes. Trends Endocrinol Metab. 2007;18(1):19–26. doi: 10.1016/j.tem.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 49.Gericke MT, Schroder T, Kosacka J, Nowicki M, Kloting N, Spanel-Borowski K. Neuropeptide Y impairs insulin-stimulated translocation of glucose transporter 4 in 3T3-L1 adipocytes through the Y1 receptor. Mol Cell Endocrinol. 2012;348(1):27–32. doi: 10.1016/j.mce.2011.07.028. [DOI] [PubMed] [Google Scholar]
- 50.Pietrement C, Sun-Wada GH, Silva ND, McKee M, Marshansky V, Brown D, et al. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod. 2006;74(1):185–94. doi: 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- 51.Van Blerkom J. The influence of intrinsic and extrinsic factors on the developmental potential and chromosomal normality of the human oocyte. J Soc Gynecol Investig. 1996;3(1):3–11. doi: 10.1016/1071-5576(95)00041-0. [DOI] [PubMed] [Google Scholar]
- 52.Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, et al. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322(5908):1717–20. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 31 kb)
(XLS 14066 kb)
(XLS 514 kb)
(XLS 30253 kb)
(XLS 311 kb)
(XLS 760 kb)
(XLS 124 kb)
(XLS 1401 kb)
(XLS 467 kb)
(XLSX 89 kb)