Abstract
Background and objective
The composition of the salivary microbiota, as determined using various molecular methods, has been reported to differentiate oral health from diseases. Thus, the purpose of this study was to utilize the newly developed molecular technique HOMINGS (Human Oral Microbe Identification using Next Generation Sequencing) for comparison of the salivary microbiota in patients with periodontitis, patients with dental caries, and orally healthy individuals. The hypothesis was that this method could add on to the existing knowledge on salivary bacterial profiles in oral health and disease.
Design
Stimulated saliva samples (n=30) were collected from 10 patients with untreated periodontitis, 10 patients with untreated dental caries, and 10 orally healthy individuals. Salivary microbiota was analyzed using HOMINGS and statistical analysis was performed using Kruskal–Wallis test with Benjamini–Hochberg's correction.
Results
From a total of 30 saliva samples, a mean number of probe targets of 205 (range 120–353) were identified, and a statistically significant higher mean number of targets was registered in samples from patients with periodontitis (mean 220, range 143–306) and dental caries (mean 221, range 165–353) as compared to orally healthy individuals (mean 174, range 120–260) (p=0.04 and p=0.04). Nine probe targets were identified with a different relative abundance between groups (p<0.05).
Conclusions
Cross-sectional comparison of salivary bacterial profiles by means of HOMINGS analysis showed that different salivary bacterial profiles were associated with oral health and disease. Future large-scale prospective studies are needed to evaluate if saliva-based screening for disease-associated oral bacterial profiles may be used for identification of patients at risk of acquiring periodontitis and dental caries.
Keywords: saliva, HOMINGS, periodontitis, dental caries
A specific oral microbiota is associated with oral health, and a symbiotic relationship between the resident oral microbiota and the host is believed to be essential for maintenance of oral health. Furthermore, local compositional changes of the oral microbiota in relation to ecological perturbations, or dysbiosis, are suggested as a crucial component of the pathogenesis of periodontitis and dental caries (1–4).
The advent of DNA-based identification of oral bacteria has demonstrated that the oral microbiota is far more complex and diverse than originally anticipated (5, 6). Therefore, today more than 700 different predominant bacterial species have been identified in the oral cavity, out of which 35% are still unculturable (7). In concert, several studies using microarray-based techniques (8–10) and next generation sequencing (11–15) have shown that alterations of local bacterial communities in relation to periodontitis and dental caries are far more complex than was previously believed.
The salivary microbiota is an integral part of the oral microbiota, which has been characterized in orally healthy conditions (16). Furthermore, the salivary microbiota in oral disease has been analyzed by means of contemporary cloning-independent methods, including PCR-based detection (17) and next generation sequencing methods (18), which have collectively reported that the salivary microbiota of patients with periodontitis and dental caries can be differentiated from the salivary microbiota associated with oral health. In addition, we have previously compared the salivary microbiota from patients with periodontitis and dental caries with orally healthy individuals using the Human Oral Microbe Identification Microarray (HOMIM) and reported that periodontitis (19) and dental caries (20) were associated with a salivary bacterial profile different from that of oral health. Furthermore, different salivary bacterial profiles were observed in patients with periodontitis and dental caries, respectively (21). However, as microbial data may be heavily influenced both by the DNA-extraction methods and the molecular technique employed for bacterial identification (22, 23), it is pivotal to continually refine molecular methods for comprehensive analysis of the salivary microbiota.
Consequently, Human Oral Microbe Identification using Next Generation Sequencing (HOMINGS), which is the successor to HOMIM, is a recently developed Illumina-based molecular technique, which enables simultaneous species-level identification of nearly 600 oral bacterial taxa (24). Thus, in this study, we used HOMINGS for comparison of salivary bacterial profiles in periodontitis, dental caries, and oral health, as we hypothesized that this technology would provide more comprehensive information on the salivary microbiota, and thus add on to existing knowledge on salivary bacterial profiles in oral health and disease.
Material and methods
Study population
In April 2015, a total of 30 adults (10 periodontitis patients, 10 dental caries patients, and 10 orally healthy individuals) fulfilling the inclusion criteria participated in the study. Inclusion criteria were as follows: individuals aged ≥18 years seeking dental treatment at the Department of Odontology, University of Copenhagen. Exclusion criteria were as follows: current daily smoking, presence of any systemic disease, use of any kind of medication, and treatment with local or systemic antibiotics within the last 3 months prior to participation. In addition, all participants had at least 20 natural teeth, and they had neither removable prosthesis nor were undergoing concurrent orthodontic treatment.
Smoking habit, systemic health, and use of medication and antibiotics were addressed by questionnaire. Clinical screening of untreated periodontitis and untreated dental caries was performed by the same examiner (DB) in combination with analysis of bite-wing x-rays. DMFT (decayed–missed–filled-teeth) and DT (decayed teeth) were recorded based on clinical and radiographic examination. Based on our previous analysis, periodontitis was defined as bleeding on probing ≥25% of total sites+minimum two teeth with clinical attachment level ≥4 mm+minimum two teeth with probing depth ≥6 mm, and dental caries was defined as manifest untreated ≥3 surfaces caries (19–21).
The study population comprised 30 adult individuals (13 males and 17 females) with a mean age of 37.6 years (22–70 years). The three groups were as follows: periodontitis group (mean age: 53.5 years, range: 42–70 years), dental caries group (mean age: 32.2 years, range: 22–50 years), and control group (mean age: 27.1 years, range: 24–39 years). The gender distribution across these groups was as follows: periodontitis group (six males and four females), dental caries group (seven males and three females), and control group (10 females).
All participants signed an informed consent prior to participation, and the study was approved by the regional ethical committee (H-15000856) and reported to the Danish Data Authorization (2015-54-0970).
Collection of saliva samples
Stimulated saliva samples were collected between 8:00 AM and 11:00 AM and before any dental treatment was performed to avoid bleeding and contamination of saliva. Collection of stimulated saliva samples was performed according to a previously described protocol (25). In brief, participants started by flushing thoroughly with water followed by chewing on paraffin gum for 1 min. The subjects expectorated for 1 min, and the saliva was discarded. For an additional 3 min, subjects continued to expectorate, and the saliva was collected in a plastic cup and stored at −80°C.
Human Oral Microbe Identification using Next Generation Sequencing
DNA isolation was performed following specifications of the protocol: Pathogen_Universal_200 (Roche, Mannheim, Germany) (26), and the HOMINGS technique was used for microbial analysis (24). In brief, HOMINGS is a recently developed molecular technology, using Illumina-based next generation sequencing in conjunction with a customized BLAST program (ProbeSeq for HOMINGS, Forsyth Institute, Boston, MA) for bacterial analysis. ProbeSeq contains sequences of species-specific custom made 16S rDNA probes (17 to 40 bases), based on the HOMD database (27), most of which were originally developed for the HOMIM (8). Thus, to obtain nearly complete coverage, bacterial identification was based on 598 oligonucleotide probes targeting individual oral bacterial species or, in some cases, a few closely related species and 94 genus-specific probes (a probe with a sequence that identifies a varied number of closely related species within the same genus). A complete list of probes present in the probeseq database is presented in Supplementary file.
The laboratory procedures of HOMINGS were carried out by specifications of a modified protocol previously described (28). In brief, V3-V4 forward (341F) AATGATACGGCGACCACCGAGATCTACACTATGGTAATTGTCCTACGGGAGGCAGCAG and reverse (806R) CAAGCAGAAGACGGCATACGAGATNNNNNNNNNNNNAGTCAGTCAGCCGGACTACHVGGGTWTCTAAT primers were employed for PCR-amplification of 10–50 ng of DNA, followed by purification using AMPure beads. Next, a library of 100 ng was pooled, gel-purified, and subsequently quantified by qPCR. Finally, 12 pM of the library was added 20% Phix and sequenced by use of MiSeq (Illumina, San Diego, CA). In this study, an average of 54,055 sequences of about 441 bp per sequence were obtained after bad reads and chimeric sequences had been removed from the analyses.
Statistical analyses
Differences between clinical and epidemiological parameters were assessed by analyses of variance ANOVA and for these analyses a p<0.05 was considered statistically significant. Comparisons between groups at probe level were performed by Kruskal–Wallis test and false discovery rate (FDR) with Benjamini–Hochberg correction being used to control for multiple hypothesis (29). For this analysis, a p<0.05 in combination with a FDR value of 0 was considered statistically significant. Graphpad prism 5 (San Diego, CA) and MeV 4_8_1 (30) were used as statistical software.
Results
Oral disease status
A comparable mean number of natural teeth were recorded in the periodontitis group (mean: 28, range: 21–32), the dental caries group (mean: 29, range: 27–32), and the control group (mean; 30, range: 28–32) (p>0.05).
A significantly higher DMFT value was observed in the dental caries group (mean: 11.4, range: 8–15) and the periodontitis group (mean: 10.7, range: 0–27) when compared with the control group (mean: 5.2, range: 0–12) (p<0.05). Furthermore, a significantly higher DT level was observed in the dental caries group (mean: 10.3, range: 6–15) compared with the periodontitis group (mean: 0.2, range 0–1) and the control group (mean: 0.1, range: 0–1), respectively (p<0.05).
The mean percentage of periodontal pockets ≥5 mm was 64.3% (range: 27–100%) in the periodontitis group, whereas no pockets ≥5 mm were identified in the dental caries group and the control group, respectively (p<0.05). In addition, bleeding on probing (BOP) ranged from 75 to 100% in the periodontitis group compared with a mean BOP<25% in the dental caries and the control group, respectively (p<0.05).
General findings
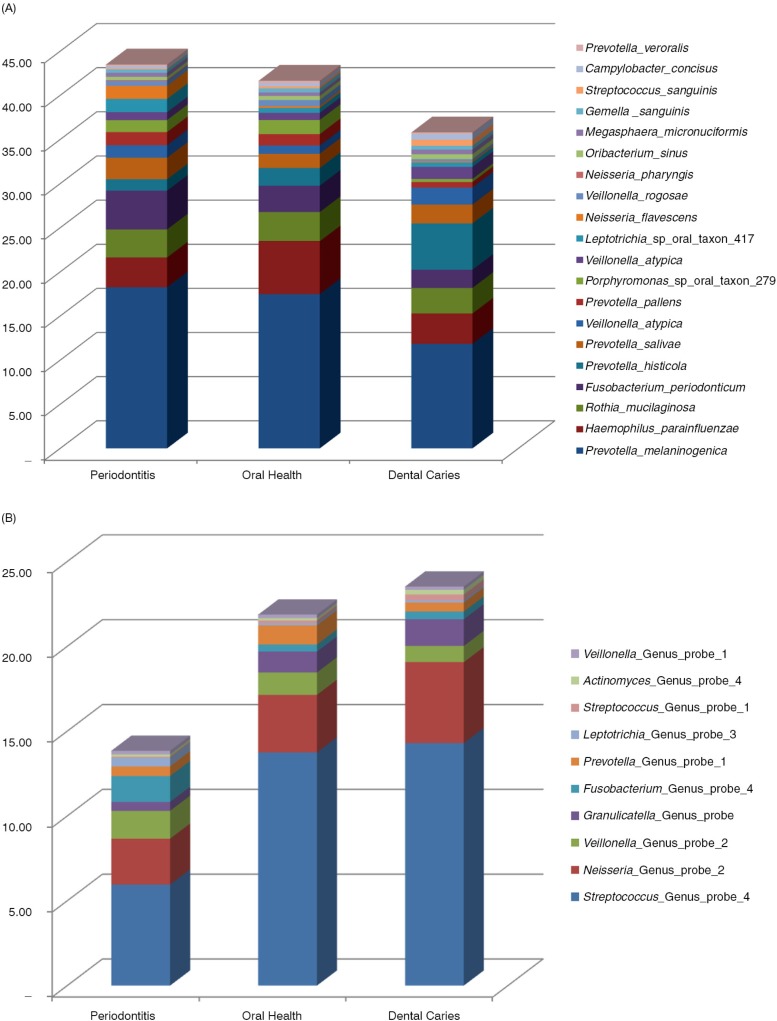
In the 30 stimulated saliva samples from 10 patients with untreated periodontitis, 10 patients with untreated dental caries, and 10 orally healthy individuals, positive identification for targets of 532 probe-sequences were observed (447 identifying a species-level taxon and 85 identifying a bacterial genus) corresponding to a coverage of 69% of the 768 probe sequences present in the probeseq database. The mean number of positive identifications was 205 (range 120–353) with significantly more targets identified in samples from patients with periodontitis (mean 220, range 143–306) and dental caries (mean 221, range 165–353) compared with orally healthy individuals (mean 174, range 120–260) (p=0.04 and p=0.04). The 20 most predominant species-level probe targets based on relative abundance are presented in Fig. 1a, and the 10 most predominant genus-level probe targets are presented in Fig. 1b, and a complete list of bacterial probes identified in samples from this study is presented in the Supplementary file. On average, 54,055 sequences (range 24,699–119,707) per sample were generated, out of which 49.4% (range 31.5–65.8%) and 21.6% (range 10.6–41.7%) were identified at the species-level and genus-level, respectively. In addition, an average of 29.0% (range 13.7–52.0%) of the sequences generated could not be assigned to either a species-specific or a genus-specific probe sequence–based on blast against the probeseq database. Comparable numbers of sequences identified at species- and genus-level, as well as proportions of unmatched sequences, were observed in all three groups (p>0.05).
Fig. 1.
Relative abundance of predominant species-level and genus-level probe targets. (a) Relative abundance of the 20 most predominant species-level probe targets in each group. (b) Relative abundance of the 10 most predominant genus-level probe targets in each group.
Specific salivary probe targets associates with untreated periodontitis and untreated dental caries
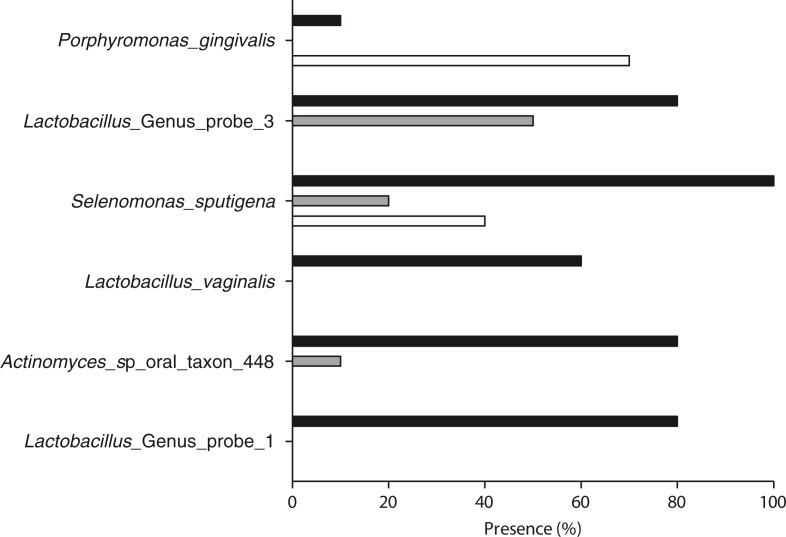
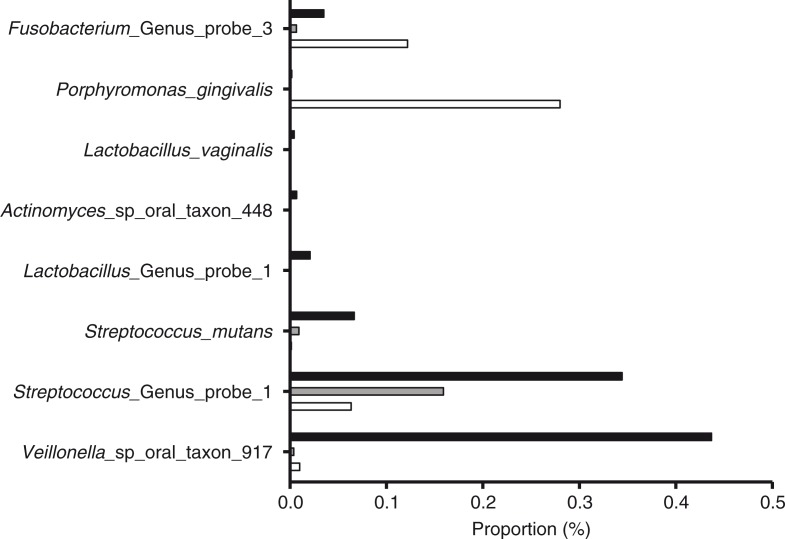
Based on comparison at the probe level, six probes were present with a significantly different frequency in the three groups. Samples from periodontitis patients were significantly associated with Porphyromonas gingivalis and samples from patients with dental caries were significantly associated with Lactobacillus Genus probe 1, Lactobacillus Genus probe 3, Lactobacillus vaginalis, Actinomyces sp oral taxon 448, and Selenomonas sputigena (p<0.05, Fig. 2). In addition, nine probes were identified with a different relative abundance between groups as samples from periodontitis patients were significantly associated with P. gingivalis and Fusobacterium Genus probe 3, and samples from patients with dental caries were significantly associated with Streptococcus mutans, Streptococcus Genus probe 4, Streptococcus Genus probe 1, Lactobacillus Genus probe 1, L. vaginalis, Actinomyces sp oral taxon 448, and Veillonella sp oral taxon 917 (p<0.05, Fig. 3).
Fig. 2.
Periodontitis- and dental caries–associated probe targets: Periodontitis- and dental caries–associated probe targets based on the frequency of detection in samples from periodontitis patients (white bars), dental caries patients (black bars), and orally healthy individuals (gray bars) (p<0.05).
Fig. 3.
Periodontitis- and dental caries–associated probe targets: Periodontitis- and dental caries associated–probe targets based on relative abundance in samples from periodontitis patients (white bars), dental caries patients (black bars). and orally healthy individuals (gray bars) (p<0.05).
Discussion
The purpose of the present investigation was to compare salivary bacterial profiles of patients with periodontitis, patients with dental caries, and orally healthy individuals based on microbial analysis performed by the newly developed next generation sequence-based molecular technique, known as HOMINGS. The main finding was that periodontitis and dental caries each were associated with characteristic salivary bacterial profiles different from that of oral health.
In this study, significantly higher mean numbers of bacterial taxa were identified in stimulated saliva samples from patients with periodontitis (n=220) and dental caries (n=221) than in orally healthy individuals (n=174), which indicates that salivary bacterial profiles are more diverse with established oral disease in the oral cavity. A higher bacterial diversity in saliva samples from periodontitis patients are in accordance with a previous HOMIM-based analysis (19), whereas both a higher (18) and a lower (20, 31) bacterial diversity in saliva samples from patients with dental caries have been reported, relative to samples from orally healthy individuals. However, different molecular methods used for bacterial identification, as well as variations across study populations may account for these discrepancies.
Interestingly, mean numbers of 20–25 bacterial taxa identified per sample have been reported based on HOMIM analysis of stimulated saliva samples from patients with periodontitis, patients with dental caries, and orally healthy controls (19–21), which corresponds to only 10% of the number of bacterial taxa identified in this study by HOMINGS. Thus, the main advantages of HOMINGS when compared with HOMIM are that HOMINGS is more comprehensive than HOMIM, and furthermore that in HOMINGS probe targets are expressed as actual proportions of total sequences recorded which provide valuable information about interspecies relations in the samples analyzed. In this study, however, a mean of 29% of the generated sequences could not be identified by a reference sequence in the probeseq database, which means that some part of the salivary microbiota still remains unknown by this molecular analysis. Other pipelines, such as QIIME still result in unidentified taxa. However, identifications can often be determined at higher taxonomic levels. Nevertheless, the main advantage of the Probeseq database is the ability of a rapid, species-level identification of nearly 600 oral bacterial taxa, which makes HOMINGS proficient for comparisons of bacterial profiles between groups.
It is noteworthy that in the present investigation periodontitis and dental caries were associated with increased salivary abundance of nine specific probe targets (Fig. 3), which are in accordance with previous HOMIM-based analysis of the salivary microbiota (19–21). In this study, periodontitis was associated with increased salivary proportions of P. gingivalis and dental caries was associated with increased salivary proportions of S. mutans, which is in line with previous investigations (17, 32–34). However, collectively these investigations employed microbial methods that specifically addressed presence or proportions of a very limited number of oral bacterial taxa, which hamper the possibility to proportionally compare these findings to the total salivary bacterial profiles in the individuals investigated. In contrast, we have previously analyzed salivary bacterial community profiles in samples from patients with periodontitis and dental caries by means of HOMIM, and in these studies P. gingivalis and S. mutans were only sporadically identified (19, 21). Thus, these findings suggest an increased depth and sensitivity of HOMINGS analysis when compared with HOMIM. Traditionally, P. gingivalis and S. mutans have been suggested as key pathogens associated with periodontitis and dental caries, respectively (35, 36). Thus, it is interesting that in the present study, salivary presence of these specific oral bacterial species associates with established oral disease (Figs. 2 and 3). Recent literature suggests that periodontitis and dental caries may evolve as a consequence of ecological perturbations, which stress the composition of local microbial communities, rather than the presence of specific bacterial species (1–4). However, we identified nine probe targets that differed significantly between samples from patients with periodontitis and dental caries, including genus-level probes recognizing the genera of Lactobacillus and Veillonella, which are well-known to be associated with local caries lesions (10). Furthermore, genus-level identification of Fusobacterium, which has also been reported to be associated with local periodontitis lesions (8), was prominent in saliva samples from periodontitis patients. Collectively, these findings suggest that bacterial accumulation in local periodontal and caries lesions entails salivary identification of detached bacteria from local oral diseased sites, which may mirror local compositional changes associated with oral disease progression.
Even though the focus of this study was to demonstrate the utility of HOMINGS, we were able to illustrate major compositional changes of the salivary microbiota in association with oral disease status. For example, there was a three-fold decrease in the relative abundance of oral streptococci in saliva samples from patients with periodontitis, in comparison with samples from patients with dental caries and orally healthy controls (Fig. 1b). These findings were accompanied by a two-fold increase in the relative abundance of Fusobacterium periodonticum in samples from periodontitis patients, a two-fold increase in relative proportion of the genera of Neisseria in samples from patients with dental caries and a two-fold increase of Haemophilus parainfluenzae in samples from orally healthy individuals (Fig. 1a and b). These associations failed to reach statistical significance after adjustment of multiple dependent assumptions, which was most likely influenced by the modest number of samples included in the analysis. Based on these results, we are confident that with an adequate sample size, next generation sequence-based methods, such as HOMINGS, will fully appreciate the magnitude of compositional changes of the salivary microbiota associated with oral health and disease.
In this study, significant age and gender differences were observed between the three study groups, which may potentially influence the differences in salivary bacterial profiles reported. However, based on previous studies using HOMIM analysis of stimulated saliva samples, bacterial profiles of saliva have been reported to be comparable between individuals with different age and gender (26). However, as mentioned above, the focus of this study was to demonstrate the utility of the HOMINGS methodology rather than to conduct an actual clinical study with sufficient sample size, in which analyses can be adjusted for age.
In conclusion, within the limitations of the cross-sectional study design employed, this study demonstrates the utility of HOMINGS resulting in a more comprehensive comparative analysis of the salivary bacterial profiles that differentiate oral health and disease. Future large-scale prospective analysis is required to evaluate whether the composition of the salivary microbiota legitimately portrays structural changes associated with periodontal and caries lesions.
Supplementary Material
Conflict of interest and funding
This study was supported by external financial support by: the Danish Dental Association, the Danish Foundation of Mutual Efforts in Dental Care, Trygfonden, and the Simon Spies Foundation. Partly funded by NIH (DE021565).
References
- 1.Jenkinson HF, Lamont RJ. Oral microbial communities in sickness and in health. Trends Microbiol. 2005;13:589–95. doi: 10.1016/j.tim.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 2.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–43. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 3.Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809–20. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 5.Teles R, Teles F, Frias-Lopez J, Paster B, Haffajee A. Lessons learned and unlearned in periodontal microbiology. Periodontol 2000. 2013;62:95–162. doi: 10.1111/prd.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paster BJ, Dewhirst FE. Molecular microbial diagnosis. Periodontol 2000. 2009;51:38–44. doi: 10.1111/j.1600-0757.2009.00316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–32. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preza D, Olsen I, Willumsen T, Boches SK, Cotton SL, Grinde B, et al. Microarray analysis of the microflora of root caries in elderly. Eur J Clin Microbiol Infect Dis. 2009;28:509–17. doi: 10.1007/s10096-008-0662-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Torlakovic L, Klepac-Ceraj V, Ogaard B, Cotton SL, Paster BJ, Olsen I. Microbial community succession on developing lesions on human enamel. J Oral Microbiol. 2012;4:16125. doi: 10.3402/jom.v4i0.16125. doi: http://dx.doi.org/10.3402/jom.v4i0.16125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B, Faller LL, Klitgord N, Mazumdar V, Ghodsi M, Sommer DD, et al. Deep sequencing of the oral microbiome reveals signatures of periodontal disease. PLoS One. 2012;7:e37919. doi: 10.1371/journal.pone.0037919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffen AL, Beall CJ, Campbell JH, Firestone ND, Kumar PS, Yang ZK, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–85. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crielaard W, Zaura E, Schuller AA, Huse SM, Montijn RC, Keijser BJ. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulze-Schweifing K, Banerjee A, Wade WG. Comparison of bacterial culture and 16S rRNA community profiling by clonal analysis and pyrosequencing for the characterization of the dentine caries-associated microbiome. Front Cell Infect Microbiol. 2014;4:164. doi: 10.3389/fcimb.2014.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obata J, Takeshita T, Shibata Y, Yamanaka W, Unemori M, Akamine A, et al. Identification of the microbiota in carious dentin lesions using 16S rRNA gene sequencing. PLoS One. 2014;9:e103712. doi: 10.1371/journal.pone.0103712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, et al. Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol. 2012;13:R42. doi: 10.1186/gb-2012-13-6-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paju S, Pussinen PJ, Suominen-Taipale L, Hyvonen M, Knuuttila M, Kononen E. Detection of multiple pathogenic species in saliva is associated with periodontal infection in adults. J Clin Microbiol. 2009;47:235–8. doi: 10.1128/JCM.01824-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang F, Zeng X, Ning K, Liu KL, Lo CC, Wang W, et al. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 2012;6:1–10. doi: 10.1038/ismej.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belstrom D, Fiehn NE, Nielsen CH, Kirkby N, Twetman S, Klepac-Ceraj V, et al. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J Clin Periodontol. 2014;41:104–12. doi: 10.1111/jcpe.12190. [DOI] [PubMed] [Google Scholar]
- 20.Belstrom D, Fiehn NE, Nielsen CH, Holmstrup P, Kirkby N, Klepac-Ceraj V, et al. Altered bacterial profiles in saliva from adults with caries lesions: a case-cohort study. Caries Res. 2014;48:368–75. doi: 10.1159/000357502. [DOI] [PubMed] [Google Scholar]
- 21.Belstrom D, Fiehn NE, Nielsen CH, Klepac-Ceraj V, Paster BJ, Twetman S, et al. Differentiation of salivary bacterial profiles of subjects with periodontitis and dental caries. J Oral Microbiol. 2015;7:27429. doi: 10.3402/jom.v7.27429. doi: http://dx.doi.org/10.3402/jom.v7.27429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarevic V, Whiteson K, Gaia N, Gizard Y, Hernandez D, Farinelli L, et al. Analysis of the salivary microbiome using culture-independent techniques. J Clin Bioinforma. 2012;2:4. doi: 10.1186/2043-9113-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lazarevic V, Gaia N, Girard M, Francois P, Schrenzel J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. PLoS One. 2013;8:e67699. doi: 10.1371/journal.pone.0067699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gomes BP, Berber VB, Kokaras AS, Chen T, Paster BJ. Microbiomes of endodontic–periodontal lesions before and after chemomechanical preparation. J Endod. 2015;41:1975–84. doi: 10.1016/j.joen.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kongstad J, Ekstrand K, Qvist V, Christensen LB, Cortsen B, Gronbaek M, et al. Findings from the oral health study of the Danish Health Examination Survey 2007–2008. Acta Odontol Scand. 2013;71:1560–9. doi: 10.3109/00016357.2013.776701. [DOI] [PubMed] [Google Scholar]
- 26.Belstrom D, Holmstrup P, Nielsen CH, Kirkby N, Twetman S, Heitmann BL, et al. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J Oral Microbiol. 2014;6:23609. doi: 10.3402/jom.v6.23609. doi: http://dx.doi.org/10.3402/jom.v6.23609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen T, Yu WH, Izard J, Baranova OV, Lakshmanan A, Dewhirst FE. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database (Oxford) 2010;2010 doi: 10.1093/database/baq013. baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4516–22. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Stat Med. 1990;9:811–18. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 30.Saeed AI, Bhagabati NK, Braisted JC, Liang W, Sharov V, Howe EA, et al. TM4 microarray software suite. Methods Enzymol. 2006;411:134–93. doi: 10.1016/S0076-6879(06)11009-5. [DOI] [PubMed] [Google Scholar]
- 31.Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbiol. 2007;45:81–7. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kononen E, Paju S, Pussinen PJ, Hyvonen M, Di TP, Suominen-Taipale L, et al. Population-based study of salivary carriage of periodontal pathogens in adults. J Clin Microbiol. 2007;45:2446–51. doi: 10.1128/JCM.02560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Relvas M, Coelho C, Velazco HC, Ramos E. Cariogenic bacteria and dental health status in adolescents: the role of oral health behaviours. Eur J Paediatr Dent. 2014;15:281–7. [PubMed] [Google Scholar]
- 34.Wennerholm K, Emilson CG. Comparison of Saliva-Check Mutans and Saliva-Check IgA Mutans with the Cariogram for caries risk assessment. Eur J Oral Sci. 2013;121:389–93. doi: 10.1111/eos.12069. [DOI] [PubMed] [Google Scholar]
- 35.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 36.Parisotto TM, Steiner-Oliveira C, Silva CM, Rodrigues LK, Nobre-dos-Santos M. Early childhood caries and mutans streptococci: a systematic review. Oral Health Prev Dent. 2010;8:59–70. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.