Abstract
The contribution of specific molecular-genetic factors to muscle mass variation and sarcopenia remains largely unknown. To identify endogenous molecules and specific genetic factors associated with appendicular lean mass (APLM) in the general population, cross-sectional data from the TwinsUK Adult Twin Registry were used. Non-targeted mass spec-based metabolomic profiling was performed on plasma of 3953 females (mostly dizygotic and monozygotic twins). APLM was measured using dual-energy X-ray absorptiometry (DXA) and genotyping was genome-wide (GWAS). Specific metabolites were used as intermediate phenotypes in the identification of single-nucleotide polymorphisms associated with APLM using GWAS. In all, 162 metabolites were found significantly correlated with APLM, and explained 17.4% of its variation. However, the top three of them (unidentified substance X12063, urate, and mannose) explained 11.1% (P≤9.25 × 10−26) so each was subjected to GWAS. Each metabolite showed highly significant (P≤9.28 × 10−46) associations with genetic variants in the corresponding genomic regions. Mendelian randomization using these SNPs found no evidence for a direct causal effect of these metabolites on APLM. However, using a new software platform for bivariate analysis we showed that shared genetic factors contribute significantly (P≤4.31 × 10−43) to variance in both the metabolites and APLM – independent of the effect of the associated SNPs. There are several metabolites, having a clear pattern of genetic inheritance, which are highly significantly associated with APLM and may provide a cheap and readily accessible biomarker of muscle mass. However, the mechanism by which the genetic factor influences muscle mass remains to be discovered.
Introduction
Lean body mass (LBM) is one of the three major components of body composition, which also includes body fat and bone mass. All three components are essential for normal physiology and metabolism, and deviations from normal values may be associated with pathological conditions. One of them is sarcopenia, defined as age-related reduction in muscle mass and muscle strength is a particularly important clinical problem.1, 2 In the elderly, loss of LBM may be as much as 30% for women above 60 years old3 and it is often correlated with physical impairment and disability – with potentially severe clinical consequences including loss of mobility, osteoporosis, increased fracture risk, dyslipidemia, insulin resistance, and increased mortality.4 LBM is one of the major predictors of bone mineral density and is likely to be at least as important as fat body mass (FBM) in protection against post-menopausal bone loss.5, 6
Despite the undoubted clinical significance of LBM and the established strong familial component7, 8 there is a paucity of molecular genetic studies of this trait. This is in a stark contrast to the other two body composition components, the subject of many publications concerning candidate genes, whole genome and genome-wide association studies published during the last two decades. Currently, we are aware of only a few genome-wide linkage5 and association analyses.9, 10 Several potential candidate genes, including the FTO gene11 were reported to be associated with LBM, with small proportion of variance (1–3%) attributable to their effect.
Thus, the potential molecular genetic mechanisms regulating LBM remain almost unknown. The major aim of this study was therefore to investigate genetic factors affecting LBM. One way to approach this problem is through metabolomics. The recent published studies clearly suggest this may serve as a powerful proxy to discover tractable genetic polymorphisms involved in a variety of complex phenotypes.12, 13 If a small number of metabolites could be found to be associated with the primary phenotype (as indeed was observed in the present paper), then they could be used as a guide to a discovery of the functionally interpretable genetic polymorphisms associated with the primary phenotype.
Hence, in the present study we implemented a combination of ‘omics‘, including metabolomics and genomics in relation to appendicular lean mass (APLM). APLM was selected because it consists virtually entirely of skeletal muscular tissue, as compared with total lean mass, the measurement of which could be biased by other components of non-fat soft tissue, such as brain, lungs, liver, and other viscera. Because of this important advantage, APLM is currently widely used in assessment of a sarcopenic status of an individual14 as well as in variety of the genetic studies, for example.15, 16
We used a large sample of middle-aged UK female twins, who were assessed using whole body DXA, metabolomic (MTB) screening and genome-wide association scans imputed to 2.5 million variants. In addition, since we were interested in accurate estimation of common and variable-specific genetic and environmental effects on LBM and its associated metabolites, we also implemented a newly developed modification of bivariate analysis, reported here for the first time.
Materials and methods
Study sample
The data examined in the present study were from the TwinsUK Adult Twin Registry, described in detail elsewhere.17 The sample had been collected from the general population through national media campaigns in the Uniited Kingdom and without ascertainment for any of the individual characteristics, diseases, or traits. In the present study, 3953 individuals (101 singletons, 1156 dizygotic, and 770 monozygotic twin pairs) were included having all measurements of interest. All studied individuals were female with age range between 18 and 80 years, and average 46.9±12.3 years. The mean BMI was 25.8±4.6 kg/m2, and LBM ranged between 23.8 and 68.8 kg, with mean LBM=39.2±5.4 kg. All participants gave written informed consent before entering the study and the St Thomas' Hospital research ethics committee had approved the project.
Phenotype
All three major body composition components, ie, bone mineral density (BMD), FBM and LBM were measured by using standard whole body DXA method,7 following the manufacturer's recommendations (QDR 4500 W system, Hologic Inc., Bedford, MA, USA) and performed by trained technicians using a standardized protocol of measurement, as described in our previous publications.18 Briefly, at installation, the manufacturer's engineer calibrated the instrument, and then daily quality control scans were performed using the spine phantom. Intra-scanner reproducibility, expressed as a coefficient of variation from duplicate measurements in healthy volunteers 1 week apart, was 0.8% at the lumbar spine. Twins of each pair were scanned on the same day. For the purposes of this study, we defined APLM as the sum of LBM measurements obtained at the four limbs. Since APLM is highly correlated (r=0.486, P«0.001) with body height (H), we used the relative APLM measure, RALM=APLM/H2, similar BMI as in previous studies.1
Metabolomics
Venous blood was taken in the seated/semi-recumbent position after >8 h fasting, between 0900 and 1400 h. Samples were taken from the co-twin pair within 10 min of each other. Non-targeted metabolomic profiling was performed on fasting plasma samples of TwinsUK participants by Metabolon Inc. using ultrahigh performance liquid chromatography and mass spectrometry (for detailed information on processing/QC, see Supplementary Material).19, 20 Raw data were median-normalized by dividing each MTB concentration by the day MTB median, then inverse normalized as MTB concentrations were not normally distributed. To avoid spurious false-positive associations due to small sample size, metabolic traits having >20% missing values were excluded.
Genomics
Twins UK subjects had been genotyped using a combination of Illumina arrays (Human Hap300 and the Human Hap610Q) as previously and repeatedly reported, for example.21 SNPs were excluded if call rate <97% (SNPs with minor allele frequency, MAF≥5%) or <99% (for 1%≤MAF<5%), Hardy–Weinberg P-values <10−6 and MAF<1%. Subjects were removed if genotyping failed in >2% SNP. The overall genotyping efficiency was 98.7%.
Design of the study and statistical analysis
The analysis of the data included the following stages and corresponding methods. First, we confirmed familial aggregation of the RALM in the study sample, computing zygosity-specific correlation between the twins. Next, we conducted linear regression analysis of all available MTBs, measured in at least 2500 individuals with RALM, using STATISTICA 7.1 software (www.statsoft.com). At this stage, we identified MTBs significantly correlated with RALM after correcting for multiple testing by FDR<0.05.22 Of these, using multiple regression analysis we selected MTBs that showed the highest and independent correlation with RALM. Highly associated MTBs were then subjected to GWAS conducted using the GenABEL software.23 This carries out association analysis, taking into account familial relationships. The most highly associated SNPs for each MTB were identified, and used in a Mendelian randomization procedure to test whether the selected metabolite(s) was causally associated with RALM.
The underlying idea of this method is that, where a genetic variant can be identified that alters the level of the intermediate phenotype (metabolite), it can be used to test whether a relationship between the metabolite and the outcome variable (RALM) is likely to be causal. As the genotype has been randomly assigned at conception (hence ‘Mendelian randomization') it can be used as a proxy for the intermediate variable (instrumental variable) to assess the effect of the exposure on the outcome, not the effect of a gene on an outcome.24, 25 To test the hypothesis of causal effect of the given metabolite, we conducted genetic variance component analysis26 of RALM, and each of the selected MTBs separately, as implemented in the statistical package MAN,27 which takes into account familial structure of the sample. Each analysis was conducted with simultaneous testing of the linear regression effect of the SNP, most significantly associated with the corresponding MTB variation. The causal effect of the MTB on RALM can be established by the ratio, ρ=βR/βM, where β2R and β2M are the least squares estimates of the SNP effect on the RALM and the MTB variations in the respective linear regression models.25 The test for the nullity of the causal effect ρ was based on the following approximation: ρ2/σρ2=χ2df=1, where σρ=(1/β2M)√(σ2Rβ2R+σ2M β2M).
To examine whether the metabolite/RALM correlation was caused by a shared genetic or environmental effect, we used a newly developed bivariate pedigree-based association analysis. This analysis simultaneously tests and adjusts variation of both dependent variables for any given SNP. This method, like variance component analysis, evaluates the contribution of the following factors to variation of the two correlated variables: VSNP, VADs, VADc, VCFs, and VCFc. Here, V represents the proportion of the phenotypic variance attributable to linear additive effect of the SNP; the trait specific (s) and common (c) additive genetic effect (AD); and common familial (twins') environment (CF) specific for the given trait (s) or shared by both phenotypes (c), respectively. Unexplained residual variation is defined as VRS. The estimates are based on discrete approximation of additive genetic and familial factors. The use of the discrete model allows the direct computation of likelihood from the pedigree data and does not require N-dimensional normal distribution of the likelihood (LH) for evaluation of a pedigree of N individuals.28, 29 The method is applicable for genetic analysis of both quantitative and qualitative phenotypes, and is fully described in the Supplementary Materials B.
Public data access
The data from this study can be accessed through the following website: http://www.ncbi.nlm.nih.gov/clinvar/?term=SCV000212129.
Results
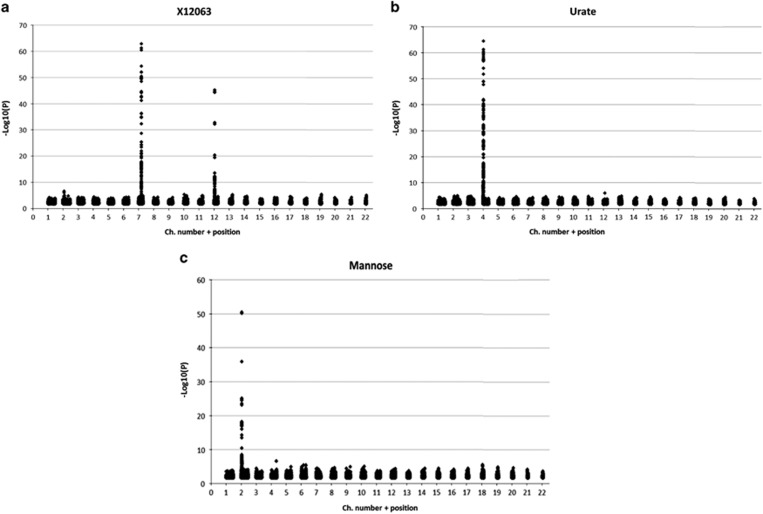
Descriptive statistics of the study sample are provided in Table 1, and include the distribution characteristics of RALM. The intra-class correlations of MZ and DZ twins for RALM were significant: 0.795±0.022 and 0.460±0.026, respectively, suggestive of genetic influence. We conducted a series of regression analyses of all available MTB levels with RALM: 162 MTBs were significantly correlated with RALM (P<0.05, by FDR), and explained 17.4% of its variation. However, this set of MTBs was found to be largely redundant. Implementing stepwise multiple regression analysis we found that one MTB explains 6.7%, three – 11.1% and ten – 13.2% of the RALM variation. The 10 MTBs most highly associated with lean mass are listed in Table 2. Their correlations were also consistently observed with other measurements of lean mass. These 10 MTBs were subjected to GWAS, with simultaneous adjustment of each variable for age. Three MTBs, unknown substance X12063, urate, and mannose showed clear and strong association with specific genomic regions (Figure 1). X12063 was associated with two genomic regions, 7q22.1:98736000-99335000 (the top SNPs were mapped to pseudogene CYP3AP1 (Cytochrome P450, family 3, subfamily A)) and 12p12:20955000-21343000 (protein coding SLCO1B1& SLCO1A2 (Solute carrier organic anion transporter family) genes). As seen in Figure 1, each genomic region association is marked by numerous SNPs retaining statistical significance at FDR<0.005. Urate and mannose were associated each with, 4p16.1:9529000-10026000 and 2p23:27372000-27967000, respectively. The genotyping results of the most polymorphic SNP among the top 10 in each region were selected for the next stage of the study and given in Table 3.
Table 1. Basic descriptive statistics of the study sample.
| Variable | Mean | Minimum | Maximum | SD |
|---|---|---|---|---|
| Age (years) | 46.91 | 17.70 | 80.00 | 12.30 |
| Height (cm) | 162.19 | 141.00 | 191.00 | 6.19 |
| Weight (kg) | 66.10 | 35.10 | 140.90 | 12.29 |
| Fat body mass (kg) | 23.10 | 5.24 | 67.51 | 8.65 |
| Lean body mass (kg) | 39.18 | 23.57 | 68.76 | 5.37 |
| APLM (kg) | 17.72 | 10.47 | 32.92 | 2.88 |
| BMI (kg/m2) | 25.14 | 13.92 | 52.71 | 4.55 |
| RALM (kg/m2) | 0.67 | 0.42 | 1.17 | 0.10 |
Abbreviations: APLM, appendicular lean mass; BMI, body mass index; RALM, relative appendicular lean mass.
Table 2. Linear correlations between the ten top MTBs and lean mass variables.
| Metabolite | LBM | RLBM | RALM | P-valuea |
|---|---|---|---|---|
| X12063 | 0.200 | 0.214 | 0.259 | 2.85E−42 |
| Urate | 0.177 | 0.223 | 0.235 | 6.31E−48 |
| Glutamate | 0.159 | 0.204 | 0.213 | 2.11E−27 |
| Mannose | 0.181 | 0.197 | 0.207 | 9.25E−26 |
| Gamma-glutamylleucine | 0.163 | 0.179 | 0.204 | 5.40E−34 |
| Gamma-glutamylvaline | 0.150 | 0.179 | 0.203 | 2.27E−30 |
| Valine | 0.158 | 0.174 | 0.190 | 1.78E−30 |
| Isoleucine | 0.165 | 0.164 | 0.184 | 1.67E−27 |
| Leucine | 0.158 | 0.153 | 0.177 | 4.37E−29 |
| Kynurenine | 0.161 | 0.158 | 0.176 | 1.55E−24 |
Abbreviations: LBM, total lean mass; RLBM (=LBM/H2), relative lean body mass.
Significance of the correlations between MTB and RALM.
Figure 1.
GWAS results obtained for X12063 (a), urate (b), and mannose (c) variations.
Table 3. Basic characteristics of the SNPs selected in GWAS of the three MTBs.
| SNP | A1 | A2 | F_A1 | Chr. | Position | MTB | P-value* |
|---|---|---|---|---|---|---|---|
| rs4646450 | A | G | 0.1614 | 7q22.1 | 99104254 | X12063 | 4.987E−50 |
| rs4363657 | T | C | 0.8283 | 12p12 | 21259989 | 9.287E−46 | |
| rs737267 | T | G | 0.2550 | 4p16.1 | 9543842 | Urate | 2.13E−59 |
| rs1260326 | T | C | 0.4088 | 2p23 | 27584444 | Mannose | 5.97E−56 |
Basic characteristics of the SNPs corresponds to NCBI Build 36 (hg18 in HGVC nomenclature). F_A1, sample frequency of allele A1.
*P-values – statistical significance of the family-based association test for the given SNP with the respective metabolite variation. The A2 allele in each SNP was associated with greater levels of the MTB, and was considered as a reference '+' allele in this analysis.
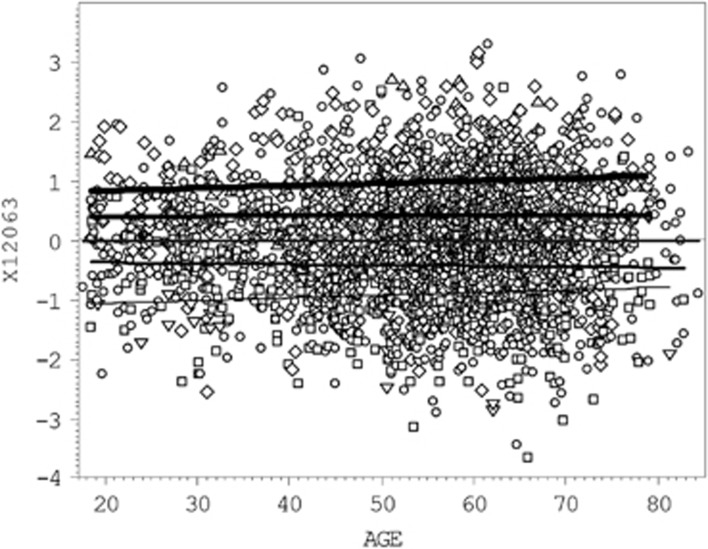
The three MTBs, X12063, urate, and mannose, collectively explained 11.07% of the variation of lean mass variable RALM. By likelihood ratio test (LRT) their combined effect in the multiple regression model, adjusting for relatedness, was extremely highly significant at P=1.03E−71. Therefore, the Mendelian randomization approach was then used to test for evidence of any causal association between metabolites and RALM using each corresponding SNP in turn. The genotype-dependent variation of X12063 levels, in relation to age is demonstrated in Figure 2, showing the combined effect of the two independent SNPs for this MTB (Figure 1, Table 3). The genotypes of each SNP were numbered from 0 to 2 according to the dose of ‘+' allele. In case of the X12063 levels, the individuals were scored from 0 to 4 (a sum of the ‘+' alleles by two SNPs) and displayed clear dose-dependent discrimination between the regression lines. The urate and mannose demonstrated similar dose dependence on ‘+' allele at rs737267:G>T and rs1260326:T>C, respectively (Supplementary Material A: Supplementary Figures S1 and S2). Results of the Mendelian randomization tests are detailed in Supplementary material A: Supplementary Table S1. The test ratio ρ=βR/βM did not deviate significantly from the nullity of the effect, and we therefore rejected the working hypothesis of causal association between RALM and selected MTBs. We looked for evidence that the correlation observed between them (see Table 2) is caused by other common genetic and/or environmental effects.
Figure 2.
Multiple scatterplot of X12063 values vs age. Five black regression lines correspond to the X12063 average values in individuals carrying from 0 (the lower thin line) to 4 (upper heavy line) ‘+' alleles in rs4646450-rs4363657 genotype. The regression line corresponded to carries of two ‘+' alleles (double heterozygotes) overlay the population average (the middle line). Comparison of the genotype-specific regression lines gives statistically highly significant difference by LRT (P=2.3E−62) and the ‘+' allele explains 10.52% of the X12063 variation. The ‘+' allele for each of two SNPs is defined in Table 3.
To test this hypothesis, we undertook the bivariate variance pedigree-based component analysis of RALM and each of the three MTBs separately. The best fitting and most parsimonious model (column ‘MP-model') confirms a highly significant association of rs4646450:G>A with X12063 levels, but not with RALM (Table 4). However, it shows that RALM variation is governed by additive genetic factors which are both phenotype specific and shared with X12063, explaining 39.6% and 34.5% of its variation respectively. The effects were highly significant by LRT. The significance of the effect of the common genetic factors shared by RALM and X12063 reached P=4.3E−43; however, the phenotype-specific effect of the common genetic factors on each of the variables variation was not equal in size: 0.348 (±0.063) vs 0.139 (±0.011). We found no detectable contribution of the environmental factors simultaneously affecting both phenotypes. The residual correlation between RALM and X12063, after adjustment for common genetic effects remained significant, 0.210 (±0.057) with P=1.54E−08. That is, about 12.5% of the phenotypic correlation (Rph=0.240) between these two traits is attributable to their common genetic factors. Similar estimates of the additive genetic and family effects were obtained when another marker rs4363657:T>C was included in the analysis (not shown). The SNP genotype explained about 4.1% (P=1.44E−26) of the X12063 variance, while its effect on RALM was not significant. The common genetic (but not the environmental) factors were significant and their estimates were virtually the same as above.
Table 4. Bivariate variance component analysis of the genetic and familial effects on RALM and X12063 variation.
| G-model | MP-model | Parameter significance | ||||
|---|---|---|---|---|---|---|
| Variance components | RALM | X12063 | RALM | X12063 | RALM | X12063 |
| VSNP(rs4646450) | 0.050 | 6.857 | 0 | 6.924 (0.561) | 5.00E−38 | |
| VADs | 37.730 | 42.608 | 37.999 (4.544) | 47.134 (4.199) | 9.57E−18 | 8.91E−32 |
| VADc | 35.242 | 13.397 | 34.785 (6.322) | 13.891 (1.055) | 3.11E−04* | 4.30E−43** |
| VCFs | 7.281 | 4.686 | 7.518 (3.403) | 0 | 1.80E−02 | |
| VCFc | 0 | 0 | 0 | 0 | ||
| VRS | 19.698 | 32.451 | 19.698 (5.991) | 32.051 (3.708) | 4.29E−04 | 1.67E−19 |
| Rrs | 0.21 | 0.210 (0.057) | 1.54E−08 | |||
VSNP, VADs, VADc, VCFs, VCFc, and VRS are variance components attributable to the SNP (rs4646450) effect, trait specific (s), and common for both phenotypes (c) additive genetic effect (AD); common familial (twins') environment (CF) specific for the given trait (s) or shared by both phenotypes (c), and unexplained residual variation (RS), respectively. RRS is correlation between residuals after accounting for the contribution of the significant variance components. G-model is general model with all parameters estimated; MP-model is most parsimonious model with standard error estimates, in parenthesis. Parameter significance is estimated by likelihood ratio test, in step-by-step uni-parametric restriction. *The P-value for the hypothesis assuming equal VADc estimate for both variables, and **VADc=0 for both traits.
Results of bivariate analysis of RALM with urate and mannose and their corresponding SNPs, rs737267:G>T and rs1260326:T>C, were similar to the above (Supplementary material A: Supplementary Tables S2 and S3). They confirm the significance of the association of the SNPs with their respective MTBs, but not with RALM. The best fitting models unequivocally suggest a significant common effect for the additive genetic factors on MTB/RALM variation. It comprised 34.47±5.10% (P=2.5E−23) and 39.724±2.983 (P=1.76E−42) of the RALM total variation in the analyses of urate and mannose, respectively.
Since RALM is one of the two major components of soft tissue mass, we tested whether its correlation with the selected metabolites was unique to lean tissue, by computing their correlation with relative appendicular fat mass (RAFM), and also with BMI. This was particularly important because of the recognized correlation between lean and fat mass adjusted for height2 (ref. 30) (also apparent in this study); and the fact that the associated metabolites and genes are strongly linked to obesity and the metabolic syndrome. All three metabolites were significantly correlated with BMI (between 0.244 (P=1.46E−46) and 0.353 (P=1.28E−89), and RAFM (between 0.207 (P=1.67E−25) and 0.291 (P=7.19E−45). As both body composition components are highly correlated with BMI (r=0.719 for RALM and r=0.857 for RAFM) and inter-correlated (r=0.4166, P=5.25E−128), to test which of RALM or RAFM was associated with study metabolites, we adjusted BMI to each of them separately and for both simultaneously, and then computed the correlations between the metabolites and adjusted BMI again. Adjustment for each RALM and RAFM led to substantial decrease in magnitude and statistical significance of all the correlations, however, they remained statistically significant in all tests (P≤4.24E−13). Only when BMI was adjusted for both RALM and RAFM the correlations were eliminated, suggesting that both fat and lean mass are independently associated with the three metabolites. Confirming this, canonical correlation analysis (www.statsoft.com) showed comparable contributions of each of the metabolites (‘left set') to variation of their canonical variable, and similarly of RALM and RAFM (‘right set') to their canonical variable (Supplementary material A: Supplementary Table S4). The corresponding canonical correlation was 0.41; χ2(6)=411.9, P<0.000001 (Supplementary material A: Supplementary Figure S3).
Discussion
The contribution of genetic and metabolic factors to human muscular mass is not currently well established. However, significant heritability estimates have been reported5, 8 as well as the identification of some candidate genes.10, 11 These, however, explain only a minor portion of the muscular mass variation. In this study, we implemented a combined metabolomics and genomics approach, aiming to identify metabolically interpretable genetic determinants of muscle mass variation. With muscular mass, this approach is probably undertaken for the first time, although attempts to detect circulating molecules that discriminate healthy lean from healthy obese individuals have previously been undertaken.31
Our analysis identified several metabolic factors, in particular urate, mannose, and a currently unidentified metabolite, X12063, that were highly significantly associated with skeletal muscle mass, as assessed by APLM relative to body height (RALM) (P=2.11E−27–6.31E−48). Over 160 MTBs displayed pairwise significant correlations with RALM. Variations of many of these MTBs were not independent, and their combined association with RALM explained 17.4% of its phenotypic variation. The three aforementioned MTBs were virtually independent, and explained >11%. Of these, the major contributor was X12063, alone explaining 6.7% of the variation. GWAS demonstrated that X12063 was associated with two independent genomic regions with top P-values 4.987E−50 and 9.287E−46, on chromosomes 7q22.1 and 12p12 correspondingly. Significant association signals of the two other MTBs were located in at 4p16.1 for urate (best P=2.13E−59) and 2p23 for mannose (best P=5.97E−56). In all instances, the association signals were mapped to genomic regions with known function. Specifically, X12063 variation was associated with gene CYP3A5 on chromosome 7. This gene encodes a member of the cytochrome P450 superfamily of enzymes involved in drug metabolism and synthesis of cholesterol, steroids, and other lipids related to the lipid-lowering efficacy of simvastatin32 and the metabolism of diverse drugs ibuprofen.33 Strong association between CYP3A5 and X12063 was recently reported in a GWA study on the KORA sample.34 X12063 was also associated with genetic variation of protein coding SLCO1B1 gene involved in hepatic uptake of statins and in statin-induced myopathy.35 Urate is a product of purine metabolism and produced by muscle and fat breakdown, with strong links to metabolic syndrome and cardiovascular disease.36 It was strongly associated with SLC2A9 (Solute carrier family 2, facilitated glucose transporter).37 This locus is associated with various phenotypes of obesity, in particular waist-to-hip ratio, visceral adiposity, and abdominal obesity, and others.38 Strong association of serum urate concentrations with SLC2A9 was reported in previous studies and is in good agreement with our present findings.19, 34, 39 It should be mentioned that although the last two studies used also twin's sample, we compared our results with the estimates obtained in the independent KORA sample also reported in these two papers. Finally, we found that mannose was significantly associated with protein coding GCKR (glucokinase (hexokinase 4) regulator) gene, which is also in a good agreement with previously published work.19 Expression of this gene controls activity and intracellular location of glucokinase, a key enzyme of glucose metabolism,40 with polymorphisms associated with lipid levels41 and chronic kidney disease.42 The last 3 loci were listed among the top 37 genetic loci associated with blood MTB concentrations.19
We undertook Mendelian randomization procedure24 to test whether any of the associations seen between metabolites and RALM are likely to be causal. This study found no evidence for causal association between the variants and muscle mass. In the case of urate at least, this was expected – urate levels are strongly associated with increased body mass, especially visceral fat mass, due to increased production of urate and reduced excretion in insulin resistant and insulinemic states.43 Weight reduction interventions lead to reductions in urate levels.44 However, it remains possible that circulating levels of urate, as well as of the two other aforementioned metabolites may also be influenced to some extent by medications, metabolic disease and/or smoking, eating behavior and other factors, which were not examined in the present study. This problem, however, would require a separate and detail analysis, which is out of scope this study, but represents one of its limitations.
Since our sample included both, MZ and DZ twins, we were able to examine whether MTBs share genetic and/or common environmental factors with RALM. For this purpose, we developed a new method of bivariate association analysis (Supplementary material B). This analysis supported the assertion that the association of the metabolites and RALM was in each case due to shared genetic factors but not, in fact, due to the associated genetic variants we had identified. No evidence was found for shared environmental factors contributing to the correlations between metabolites and RALM. For example, after adjustment of X12063 variation for rs4646450:G>A, the null hypothesis of no common genetic factor was rejected with P=4.30E−43. These results are unequivocal, and demonstrate that other genetic or epigenetic factors are involved in the covariation of these MTBs and RALM.
Our analysis also showed significant and independent association of the three metabolite levels with RAFM. The corresponding correlations were of the similar magnitude and statistical significance when RAFM and RALM were compared, thus indicating that these metabolites are probably markers of soft tissue in general and not muscle and fat specifically. This was illustrated in canonical correlation analysis (Supplementary material A: Supplementary Figure S3). The question is then what these metabolites mark in soft tissue physiology? In healthy subjects, fat mass and lean mass are significantly correlated,30 and mechanisms leading to increases in both fat and lean mass may act through dynamically changing trophic hormones such as growth hormone, as well as cortisol and leptin.45 With increasing age, the correlation between fat and lean mass appears to lessen – greater fat mass at baseline has been reported to be significantly related to greater subsequent loss of lean mass, and loss of muscle quality (defined as strength per unit mass),30 potentially leading to the deleterious state of sarcopenic obesity. The participants of this study were community dwelling healthy volunteers with an average age of 46.9, and results here largely represent the midlife period. Future studies are planned to investigate the metabolomic and genomic correlates of lean and fat mass longitudinally.
As with any study, the present one has limitations. A major limitation is that despite the large sample size (>3000 individuals), it was conducted on single sample of the data. Nevertheless, as mentioned, the available published data support our findings.19, 34, 39
This project has also a number of advantages, besides sample size. All the individuals in the study were assessed for all the variables of interest, which included DXA evaluation of the body composition, the genome-wide association genotyping and non-targeted metabolomics of >360 endogenous molecules. We developed a special statistical tool that allows accurate estimating of the contribution of the common and specific genetic and environmental factors. The model of bivariate pedigree analysis implemented in this study has several important features in comparison with traditional models.26, 46 It does not require commonly used suggestion of N-dimensional normal distribution of the likelihood (LH) for evaluation a pedigree of N individuals.28, 29 The method allows direct computation of the likelihood on pedigree data, and for the pair of the tested phenotypes; it estimates the common and the trait-specific genetic and family effects. It also estimates the phenotype-specific SNP effects. The model allows the inference of the correlations between the genetic, family, and residual variance components of the pair of the phenotypes. The approach is also applicable for genetic analysis of qualitative traits, and does not require assumption of the underlying liability with threshold to the binary condition.26 It should be mentioned that despite the quite different mathematical algorithms the implementation of the two models gives comparable results when we compare adequate variance/covariance components (eg, heritability, common twin environment, and genetic correlations). More detail comparison of the two models is given in Supplementary materials, Supplementary Table S5.
Additional limitation of this study is that it focused on females only. However, sex differences in human body composition including muscle mass and its age-related changes are profound and well known. The analysis of one-sex sample is therefore not prone to potential genotype × sex interactions, which are likely the rule rather than exception.47, 48
In summary, the present study found highly significant correlations of skeletal muscular mass variation with a few endogenous metabolic factors, each of which showed extremely significant association with just one or two genomic regions. The association of these metabolites and lean mass was shown not to be causal, but instead driven by shared genetic etiology. Using the twin structure of the cohort, we found substantial common causation by additive genetic factors shared by APLM and metabolites after adjusting for the effect of key associated SNPs. The nature of these common genetic effects remains to be discovered.
Acknowledgments
This study was supported by Israel Science Foundation (grant number #1018/13) and the Wellcome Trust, the EU and the UK Department of Health via the National Institute for Health Research (NIHR) comprehensive Biomedical Research Centre award to Guy's & St Thomas' NHS Foundation Trust in partnership with King's College London. We would like to thank all the twins who participated in the study, staff in the Department of Twin Research and Genetic Epidemiology, King's College London, UK.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
Supplementary Material
References
- 1Janssen I, Baumgartner RN, Ross R, Rosenberg IH, Roubenoff R: Skeletal muscle cutpoints associated with elevated physical disability risk in older men and women. Am J Epidemiol 2004; 159: 413–421. [DOI] [PubMed] [Google Scholar]
- 2Cruz-Jentoft AJ, Baeyens JP, Bauer JM et al: Sarcopenia: European consensus on definition and diagnosis. Age and Ageing 2010; 39: 412–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3Doherty TJ: Aging and sarcopenia. J Appl Physiol 2003; 95: 1717–1727. [DOI] [PubMed] [Google Scholar]
- 4Karakelides H, Nair KS: Sarcopenia of aging and its metabolic impact. Curr Top Dev Biol 2005; 68: 123–148. [DOI] [PubMed] [Google Scholar]
- 5Livshits G, Kato BS, Wilson SG, Spector TD: Linkage of genes to total lean body mass in normal women. J Clin Endocrinol Metab 2007; 92: 3171–3176. [DOI] [PubMed] [Google Scholar]
- 6Gjesdal CG, Halse JI, Eide GE, Brun JG, Tell GS: Impact of lean mass and fat mass on bone mineral density: The Hordaland Health Study. Maturitas 2008; 59: 191–200. [DOI] [PubMed] [Google Scholar]
- 7Arden NK, Spector TD: Genetic influences on muscle strength, lean body mass, and bone mineral density: a twin study. J Bone Miner Res 1997; 12: 2076–2081. [DOI] [PubMed] [Google Scholar]
- 8Hsu FC, Lenchik L, Nicklas BJ et al: Heritability of body composition measured by DXA in the diabetes heart study. Obes Res 2005; 13: 312–319. [DOI] [PubMed] [Google Scholar]
- 9Hai R, Pei YF, Shen H et al: Genome-wide association study of copy number variation identified gremlin1 as a candidate gene for lean body mass. J Hum Genet 2012; 57: 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Guo YF, Zhang LS, Liu YJ et al: Suggestion of GLYAT gene underlying variation of bone size and body lean mass as revealed by a bivariate genome-wide association study. Hum Genet 2013; 132: 189–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Livshits G, Malkin I, Moayyeri A, Spector TD, Hammond CJ: Association of FTO gene variants with body composition in UK twins. Ann Hum Genet 2012; 76: 333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12Madsen RK, Lundstedt T, Gabrielsson J et al: Diagnostic properties of metabolic perturbations in rheumatoid arthritis. Arthritis Res Ther 2011; 8: 13, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13Liu ML, Zheng P, Liu Z et al: GC-MS based metabolomics identification of possible novel biomarkers for schizophrenia in peripheral blood mononuclear cells. Mol Biosyst 2014; 10: 2398–2406. [DOI] [PubMed] [Google Scholar]
- 14Merlini L, Vagheggini A, Cocchi D: Sarcopenia and sarcopenic obesity in patients with muscular dystrophy. Front Aging Neurosci 2014; 6: 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Sun L, Tan LJ, Lei SF et al: Bivariate genome-wide association analyses of femoral neck bone geometry and appendicular lean mass. PLoS ONE 2011; 6: e27325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16Ran S, Liu YJ, Zhang L et al: Genome-wide association study identified copy number variants important for appendicular lean mass. PLoS One 2014; 9: e89776, PLoS One 2011; 6: e27325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Moayyeri A, Hammond CJ, Hart DJ, Spector TD: The UK Adult Twin Registry (TwinsUK Resource). Twin Res Hum Genet 2013; 16: 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Moayyeri A, Hammond CJ, Hart DJ, Spector TD: Effects of age on genetic influence on bone loss over 17 years in women: the Healthy Ageing Twin Study (HATS). J Bone Miner Res 2012; 27: 2170–2178. [DOI] [PubMed] [Google Scholar]
- 19Suhre K, Shin SY, Petersen AK et al: Human metabolic individuality in biomedical and pharmaceutical research. Nature 2011; 477: 54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20Reitman ZJ, Jin G, Karoly ED et al: Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome. Proc Nat Acad Sci USA 2011; 108: 3270–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Burri A, Hysi P, Clop A, Rahman Q, Spector TD: A genome-wide association study of female sexual dysfunction. PLoS ONE 2012; 7: e35041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22Benjamini Y, Yekutieli D: Quantitative trait loci analysis using the false discovery rate. Genetics 2005; 171: 783–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Aulchenko YS, Ripke S, Isaacs A, van Duijn CM: GenABEL: an R library for genome-wide association analysis. Bioinformatics 2007; 23: 1294–1296. [DOI] [PubMed] [Google Scholar]
- 24Bochud M, Rousson V: Usefulness of Mendelian randomization in observational epidemiology. Int J Environ Res Public Health 2010; 7: 711–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25Brion MJA, Shakhbazov K, Visscher PM: Calculating statistical power in Mendelian randomization studies. Int J Epidemiol 2013; 42: 1497–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26Falconer DS, Mackay TFC: Introduction to Quantitative Genetics 4th edn. Harlow, UK: Addison Wesley Longman, 1996. [Google Scholar]
- 27Malkin I, Ginsburg E: Program Package for Pedigree Analysis (MAN-2013): Manual. Tel Aviv, Israel: Department of Anatomy and Anthropology, Tel Aviv University, 2013. [Google Scholar]
- 28Boehnke M, Moll PP, Lange K, Weidman WH, Kottke BA: Univariate and bivariate analyses of cholesterol and triglyceride levels in pedigrees. Am J Med Genet 1986; 23: 775–792. [DOI] [PubMed] [Google Scholar]
- 29Ginsburg E, Malkin I, Elston RC: Theoretical Aspects of Pedigree Analysis. Tel Aviv, Israel: Ramot Publishing, Tel Aviv University, 2006. [Google Scholar]
- 30Koster A, Ding J, Stenholm S et al: Does the amount of fat mass predict age-related loss of lean mass, muscle strength, and muscle quality in older adults? J Gerontol A Biol Sci Med Sci 2011; 66: 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31Oberbach A, Blüher M, Wirth H et al: Combined proteomic and metabolomic profiling of serum reveals association of the complement system with obesity and identifies novel markers of body fat mass changes. J Proteome Res 2011; 10: 4769–4788. [DOI] [PubMed] [Google Scholar]
- 32Li YP, Zhang LR, Jia M, Hu XJ: CYP3AP1*3 allele is associated with lipid-lowering efficacy of simvastatin and atorvastatin in Chinese women. J Clin Pharmacol 2011; 51: 181–188. [DOI] [PubMed] [Google Scholar]
- 33Baris D, Karagas MR, Koutros S et al: Nonsteroidal anti-inflammatory drugs and other analgesic use and bladder cancer in northern New England. Int J Cancer 2013; 132: 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34Shin SY, Fauman EB, Peterson AK et al: An atlas of genetic influences on human blood metabolites. Nat Genet 2014; 46: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35Carr DF, O'Meara H, Jorgensen AL et al: SLCO1B1 genetic variant associated with statin-induced myopathy: a proof-of-concept study using the clinical practice research datalink. Clin Pharmacol Ther 2013; 94: 695–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36Gagliardi AC, Miname MH, Santos RD: Uric acid: a marker of increased cardiovascular risk. Atherosclerosis 2009; 202: 11–17. [DOI] [PubMed] [Google Scholar]
- 37Richette P, Bardin T: Gout. Lancet 2010; 375: 318–328. [DOI] [PubMed] [Google Scholar]
- 38Brandstätter A, Kiechl S, Kollerits B et al: Sex-specific association of the putative fructose transporter SLC2A9 variants with uric acid levels is modified by BMI. Diabetes Care 2008; 31: 1662–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Köttgen A, Albrecht E, Teumer A et al: Genome-wide association analyses identify 18 new loci associated with serum urate concentrations. Nat Genet 2013; 45: 145–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Lynedjian PB: Molecular physiology of mammalian glucokinase. Cell Mol Life Sci 2009; 66: 27–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Alfred TY, Ben-Shlomo Y, Cooper R et al: Associations between a polymorphism in the pleiotropic GCKR and Age-related phenotypes: the HALCyon programme. PLoS ONE 2013; 8: e70045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42Hishida A, Takashima N, Turin TC et al: GCK, GCKR polymorphisms and risk of chronic kidney disease in Japanese individuals: data from the J-MICC Study. J Nephrol 2014; 27: 143–149. [DOI] [PubMed] [Google Scholar]
- 43Takahashi S, Yamamoto T, Moriwaki Y, Yamakita J, Higashino K: Close correlation between visceral fat accumulation and uric acid metabolism in healthy men. Metabolism 1997; 46: 1162–1165. [DOI] [PubMed] [Google Scholar]
- 44Serpa Neto A, Rossi FM, Valle LG, Teixeira GK, Rossi M: Relation of uric acid with components of metabolic syndrome before and after Roux-en-Y gastric bypass in morbidly obese subjects. Arq Bras Endocrinol Metabol 2011; 55: 38–45. [DOI] [PubMed] [Google Scholar]
- 45Waters DL, Qualls CR, Dorin RI, Veldhuis JD, Baumgartner RN: Altered growth hormone, cortisol, and leptin secretion in healthy elderly persons with sarcopenia and mixed body composition phenotypes. J Gerontol A Biol Sci Med Sci 2008; 63: 536–541. [DOI] [PubMed] [Google Scholar]
- 46Almasy L, Blangero J: Variance component methods for analysis of complex phenotypes. Cold Spring Harb Protoc 2010; 5: pdb.top77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Weiss LA, Pan L, Abney M, Ober C: The sex specific genetic architecture of quantitative traits in humans. Nat Genet 2006; 38: 218–222. [DOI] [PubMed] [Google Scholar]
- 48Vink JM, Bartels M, van Beijsterveldt TC, van Dongen J, van Beek JH, Distel MA: Sex differences in genetic architecture of complex phenotypes? PLoS ONE 2012; 7: e47371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.