Summary
Decreased expression of CD3‐ζ chain, an adaptor protein associated with T‐cell signalling, is well documented in patients with oral cancer, but the mechanistic justifications are fragmentary. Previous studies in patients with oral cancer have shown that decreased expression of CD3‐ζ chain was associated with decreased responsiveness of T cells. Tumours are known to induce localized as well as systemic immune suppression. This study provides evidence that oral tumour‐derived factors promote immune suppression by down‐regulating CD3‐ζ chain expression. 2′5′‐Oligoadenylate synthetase 2 (OAS2) was identified by the proteomic approach and our results established a causative link between CD3‐ζ chain down‐regulation and OAS2 stimulation. The surrogate situation was established by over‐expressing OAS2 in a HEK293 cell line and cell‐free supernatant was collected. These supernatants when incubated with T cells resulted in down‐regulation of CD3‐ζ chain, which shows that the secreted OAS2 is capable of regulating CD3‐ζ chain expression. Incubation of T cells with cell‐free supernatants of oral tumours or recombinant human OAS2 (rh‐OAS2) induced caspase‐3 activation, which resulted in CD3‐ζ chain down‐regulation. Caspase‐3 inhibition/down‐regulation using pharmacological inhibitor or small interfering RNA restored down‐regulated CD3‐ζ chain expression in T cells induced by cell‐free tumour supernatant or rh‐OAS2. Collectively these results show that OAS2 leads to impairment in CD3‐ζ chain expression, so offering an explanation that might be applicable to the CD3‐ζ chain deficiency observed in cancer and diverse disease conditions.
Keywords: CD3‐ζ chain, humans, T cells, tumour immunology, tumour‐secreted factors
Abbreviations
- HIs
healthy individuals
- IFN
interferon
- MxA
myxovirus resistance gene A
- OAS2
2′5′‐oligoadenylate synthetase 2
- PBMCs
peripheral blood mononuclear cells
- rh‐OAS2
recombinant human OAS2
- TCR
T‐cell receptor
Introduction
The cancer immunoediting hypothesis stresses the dual role of the immune system: host protection and tumour shaping. The immune system, apart from eliminating the nascent malignant cells, also shapes the tumour through equilibrium and escape phases.1 The ability of tumour cells to escape obliteration by immune cells could be because of the plethora of strategies used to evade immune attack. One of these is represented by the production of soluble immunosuppressive factors that may prevent the pro‐inflammatory effects and promote T‐cell dysfunction in the tumour microenvironment. Immune dysfunction appears to be more frequent and profound in patients with cancer. Immune effector cells obtained from the peripheral blood of cancer patients, including oral cancer have been reported to have a variety of functional abnormalities, which may vary in magnitude from patient to patient and may be related to the extent of the disease.2, 3 These abnormalities include defects in T‐cell signalling via the T‐cell receptor (TCR), decreased tyrosine kinase activity following triggering with anti‐CD3 monoclonal antibodies, poor lymphocytic proliferative responses, defects in lytic capacity, and decreased ability for cytokine production.3, 4, 5, 6 The immune dysfunction is also associated with the down‐regulation of expression of the TCR‐ζ chain (CD3‐ζ) or of intracellular protein kinases in cancer‐bearing hosts.7
Decreased expression of the CD3‐ζ chain has been reported in several autoimmune, inflammatory and malignant diseases. It has been reported that cancer cells produce several ligands that function to prevent optimal T‐cell activation through CD3‐ζ chain down‐regulation and induces either T‐cell anergy or apoptosis.1, 8 Studies from our laboratory have shown that post‐translational down‐regulation is primarily responsible for decreased CD3‐ζ chain expression in the peripheral blood of patients with oral cancer whereas a dominant transcriptional defect is observed in the tumour compartment. The down‐regulation of CD3‐ζ chain culminates in impaired lymphocyte responses in these patients.9
The cytoplasmic domain of CD3‐ζ chain has several consensus target sequences for caspases, among which caspase‐3 and caspase‐7 have been shown to cleave in vitro translated CD3‐ζ chain.10 Caspase‐3, an effector caspase, is expressed during T‐cell anergy induction and recognizes proteins with a common DXXD motif and cleaves after the second aspartic residue.11, 12 Circumstantial evidence for a physiological involvement of active caspase‐3 in generating a CD3‐ζ‐chain‐deficient T‐cell phenotype has been described in patients with gastric and liver cancers.13, 14 T and B cells from Casp3−/− mice show hyperproliferative responses, which have been attributed to reduced activation‐induced cell death and to alterations of cell cycle regulation15, 16 in these cells, respectively. Caspase‐3 also regulates many non‐apoptotic cellular processes, such as cell proliferation, cell‐cycle regulation and cell differentiation.17, 18
The loss of CD3‐ζ chain is a common observation in cancer patients. However, the mechanism responsible for cancer‐associated decreased expression of CD3‐ζ chain remains controversial. This study reports the identification of a tumour‐secreted factor isolated from oral cancer patients that can mediate down‐regulation of CD3‐ζ chain expression. This study unravels the potential role of tumour‐secreted 2′5′‐oligoadenylate synthetase 2 (OAS2), identified by the proteomic approach, in down‐regulation of CD3‐ζ chain. Defining the mechanism, through which this factor modulates CD3‐ζ chain levels, might ultimately provide a therapeutic target leading to the generation of effective anti‐tumour cellular immune responses in patients with cancer.
Materials and methods
Study group
The study was approved by the institutional ethics committee. After written informed consent, surgically resected tumours (n = 31) were obtained from patients with newly diagnosed oral cancer (stage I–IV) before initiation of treatment. Blood specimens were obtained from healthy individuals (HIs). Peripheral blood mononuclear cells (PBMCs) were isolated by differential density gradient centrifugation (Ficoll–Hypaque, Sigma‐Aldrich, St Louis, MO) from HIs. The mononuclear cell fraction was washed twice with normal saline, counted and analysed.
Cell culture
The PBMCs isolated by Ficoll–Hypaque gradient were cultured with RPMI‐1640 medium supplemented with 10% fetal calf serum. The PBMCs from HIs were seeded in 24‐well plates at 1 × 106 cells/ml in each well. Oral tumour supernatants were added to HI PBMCs at a final dilution of 1 : 1 with RPMI‐1640 medium supplemented with 10% fetal calf serum. After incubation for different times, PBMCs were harvested and analysed for CD3‐ζ chain expression. The HI PBMCs were also stimulated with different concentrations of recombinant human OAS2 (rh‐OAS2) (Abnova, Taipei City, Taiwan) or rh‐OAS1 (Abnova; 3–96 ng/ml) for 24 hr. PBMCs were harvested and stained for CD3‐ζ chain and analysed by flow cytometry.
Tumours were collected in sterile plain RPMI medium (Invitrogen Life‐Technology, Carlsbad, CA) supplemented with antibiotics. The necrotic, haemorrhagic and fatty tissues were removed and tumour tissues were thoroughly washed with antibiotic‐containing plain RPMI. The tumour tissues were minced finely and incubated in plain RPMI containing an enzyme mixture (0·05% collagenase, 0·02% DNase, and 5 U/ml hyaluronidase) (Sigma‐Aldrich), at 37° for 2 hr with intermittent shaking. The tumour tissues were then passed through a 200‐gauge wire mesh. The cells were washed with plain RPMI medium and were tested for cell viability. Cells with > 90% viability were used to carry out further experiments.
Oral tumour‐derived cell lines (AW13516, AW8507 and AW9803) and HEK293 cells were cultured in 10% fetal calf serum supplemented Iscove's modified Dulbecco's medium and Dulbecco's modified Eagle's medium, respectively. These adherent cell lines were subcultured at 65–75% confluence.
Magnetic activated cell sorting
The PBMCs were isolated by differential density gradient centrifugation using Ficoll Hypaque (Sigma‐Aldrich). CD3+ T cells were purified from PBMCs using MicroBeads (Miltenyi Biotch, Bergisch Gladbach, Germany) by positive selection. The separation procedure was conducted according to the manufacturer's instructions. The purity of separated cells was > 95% as determined by flow cytometry (BD Biosciences, San Jose, CA).
Flow cytometry
Cells were either stained with FITC or phycoerythrin‐labelled mouse anti‐human CD3‐ε and CD3‐ζ antibodies (BD Biosciences, San Diego, CA) or with anti‐human p56Lck and ZAP70 antibodies (Santa Cruz Biotechnology, Santa Cruz, CA) for 45 min at 4°. Thereafter, cells were washed and incubated with FITC‐labelled goat anti‐mouse IgG (Sigma‐Aldrich) for another 45 min at 4°. Briefly, PBMCs (0·5 × 106 to 1 × 106) were rinsed in cold PBS and cold‐fixed in 1% paraformaldehyde in PBS for 10 min at 4°. The cells were washed and permeabilized for 15 min with 0·1% saponin in PBS. The permeabilized cells were stained with the cocktail of antibodies and 50 000 events/sample were acquired with a FACS Aria flow cytometer (Becton Dickinson, Mountain View, CA). For the above experiments, cells were gated on the basis of their forward and side scatter characteristics and the fluorescence intensity was measured. The analysis was performed using flowjo software (Tree Star, Ashland, OR).
Cloning and cell transfection
The cDNA encoding OAS2 gene was amplified by PCR using a forward primer containing the PmeI restriction site and a reverse primer containing the SalI restriction site. The PCR with 35 cycles at 95° for 1 min, 58° for 2 min, and 72° for 2 min using the forward primer 5′‐AGTTTAAACCATGGGAAATGGGGAGTCCCAGCTGTC‐3′ and a reverse primer 5′‐ACCGTCGACTGATGACTTTTACCGGCACTTTC‐3′ was used. The PCR product was gel purified and by TA cloning inserted into pTZ57R/T vector. The pTZ57R/t vector was digested with EcoRI and SalI to release the OAS2 cDNA, which was purified and cloned into EcoRI‐ and SalI‐digested pEGFP‐N2 vector. The vector was sequenced and sequence confirmed the in‐frame addition of OAS2 gene with green fluorescent protein (GFP) present in the pEGFP‐N2 vector. The vector was transfected into HEK293 cells and supernatant was collected. The collected supernatant was added to the HI PBMCs to monitor CD3‐ζ chain down‐regulation.
Small interfering RNA
The PBMCs were transfected with small interfering RNA (siRNA) specific for caspase‐3 and fluorescent oligonucleotide control siRNA (Cell Signaling Technology, Danvers, MA) at a concentration of 100 nm using X‐tremeGENE HP transfection reagent (Roche Diagnostics, Indianapolis, IN). Briefly, cells were transfected with 100 nm control siRNA, or caspase‐3‐specific siRNA, or were left untransfected. After 40 hr, untransfected cells and caspase‐3 knockdown cells were stimulated with rh‐OAS2 (24 ng/ml) and at 60 hr post‐transfection cells were harvested; and samples were subjected to Western blot analysis for caspase‐3, CD3‐ζ chain or β‐actin. The inhibition of caspase 3 expression was assessed at the 60th hr post transfection.
Western blotting
Cells (PBMCs and oral tumour/cell lines) were washed and lysed using 1% Nonidet P‐40 lysis buffer containing 10 mm Tris–HCl, 50 mm NaCl, 5 mm EDTA, 1 mm PMSF, 10 μg aprotinin, 10 μg leupeptin and 1% Nonidet P‐40. The cells were sonicated at 4° then centrifuged for 10 min. Protein lysate was harvested and quantified using Bradford reagent (Sigma‐Aldrich). Samples were resolved on 12% SDS–PAGE and then transferred to nitrocellulose membrane (Amersham Pharmacia Biotech, Piscataway, NJ). The membrane was probed with the primary antibodies of CD3‐ζ chain (1 : 300) (6B10.2; Santa Cruz Biotechnology), OAS2 (1 : 500) (Santa Cruz Biotechnology), caspase‐3 (Cell Signaling Technology) and β‐actin (1 : 1000) (Sigma‐Aldrich) as loading control. Immunostaining was performed using appropriate secondary antibody at a dilution of 1 : 3000 and developed with ECL plus, a western blot detection system (Amersham Pharmacia).
Semi‐quantitative PCR
RNA was extracted from oral tumour and cell lines using Trizol reagent (Invitrogen Life Technologies, Grand Island, NY) in accordance with the company's instructions. RNA obtained from cells was reverse transcribed in the presence of 5 mm MgCl2, 1 × PCR Buffer II, 1 mm dNTPs, 25 u MuLV reverse transcriptase, 1 unit RNA guard Ribonuclease inhibitor (Amersham Pharmacia Biotech, Uppsala, Sweden). The OAS2 mRNA levels were measured by semi‐quantitative RT‐PCR. The forward and reverse primer pair for OAS2 5′‐TTAAATGATAATCCCAGCCCC‐3′ and 5′‐TCAGCGAGGCCAGTAATCTT‐3′, respectively.
Caspase‐3 detection by fluorometry
The PBMCs were washed in PBS, lysed in ice cold caspase lysis buffer for 30 min and the lysate was collected after centrifugation. The caspase enzymatic reaction was performed according to the manufacturer's instruction (Caspase‐3 assay kit, Fluorimetric CASP3F; Sigma‐Aldrich).
Mass spectroscopy
The cell‐free supernatant (1 ml) from surgically excised oral tumours (HPV‐negative), oral cancer cells lines AW8501, AW1351619 and medium alone (control) were lyophilized. The dried samples were dissolved in 0·5 ml of ammonium bicarbonate buffer (0·1 m, pH 8·5) and the samples were concentrated on a centricon (membrane with a molecular weight cut‐off of 3000). Samples were dried in a lyophilizer overnight and the dried samples were dissolved in dissolution buffer supplied with the i‐TRAQ kit (Applied Biosystems, Foster City, CA). Samples were digested with trypsin according to the manufacturer's protocol and the samples were labelled with the i‐TRAQ reagents 114, 115, 116 and 117. Tryptic peptides of medium control were labelled with 114, AW8501 peptides with 115, AW13516 peptides with 116 and finally tumour supernatants tryptic peptides with 117. Samples were pooled and dried in a speed‐vac. The dried samples were fractionated on strong cation exchange chromatography and fractions were collected using salt gradient. Each of these fractions was analysed using liquid chromatography tandem mass spectrometry (LC/MS/MS) on a QSTAR XL mass spectrometer. By differential labelling, peptides labelled with isotope 117 (threefold excess) were selected by subtracting the common peptides found in AW8501 and AW13516 cell lines from cell‐free supernatants of oral tumours. Mascot analysis was performed to identify the peptide and the protein from which the peptide was derived.
Annexin–propidium iodide staining
To analyse an effect of rh‐OAS2 on cell viability, propidium iodide (PI)/annexin V staining kit (BD PharMingen, San Diego, CA) was used. The PBMCs were stimulated with rh‐OAS2 and after 24 hr cells were harvested, suspended in binding buffer (10 mm HEPES pH 7·4, 150 mm NaCl, 0·25 mm CaCl2) and incubated with PI and FITC‐conjugated annexin V (BD Biosciences) in the dark for 15 min at room temperature. After incubation, 400 μl of binding buffer was added and cells were analysed immediately on FACSAria (10 000 events/sample).
Statistical analysis
Results are expressed as the mean ± standard error of mean (SEM). Statistical analysis was performed using prism software (Prism Software, Lake Forest, CA) and the P‐value was calculated using Student's t‐test. Two‐sided P‐values < 0·05 were considered statistically significant. Densitometry analysis was performed using imageJ software (National Institutes of Health, Bethesda, MD).
Results
Tumour‐derived factor from oral tumours down‐regulates CD3‐ζ chain in peripheral blood lymphocytes of healthy individuals
Our earlier published data demonstrated that compared with other T‐cell signalling molecules, (p56Lck, Zap70) CD3‐ζ chain is markedly decreased in peripheral blood T cells of patients with oral cancer.9 This study was proposed to investigate the role of an oral tumour‐derived factor in regulating CD3‐ζ chain expression. Single cell suspensions of oral tumours were prepared by enzyme disintegration. Oral tumour cells were cultured in serum‐free media and cell‐free supernatant (referred to as tumour supernatant) was collected after 72 hr in culture. The cell‐free supernatants of two cell lines derived from oral tumours (AW8501 and AW13516) were also used. The oral tumour supernatant (stage III patient) was added to the PBMCs of HI and incubated for 24–96 hr. As seen in Fig. 1(a) the expression of CD3‐ζ chain was monitored in HI PBMCs at defined time intervals after incubation with tumour supernatant. Time kinetics demonstrated that incubation of HI PMBCs with tumour supernatant led to the progressive decrease in CD3‐ζ chain expression compared with PBMCs incubated with medium alone (control). However, the supernatants derived from the AW8501 and AW13516 cell lines were not able to decrease the CD3‐ζ chain expression (Fig. 1b, c).
Figure 1.
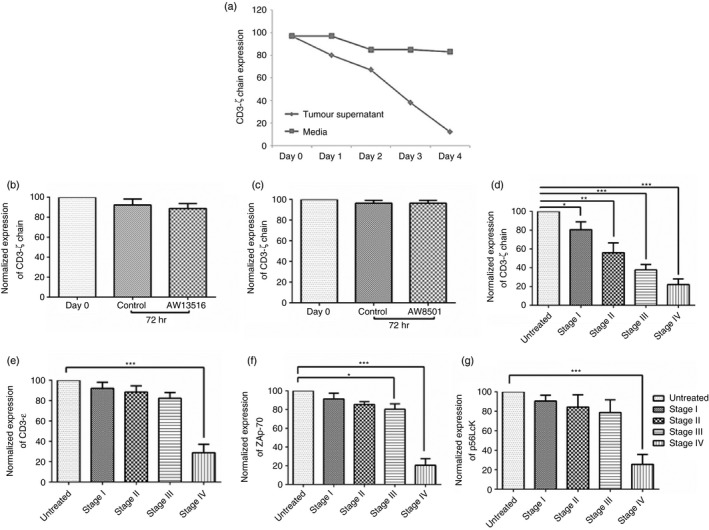
Healthy individual peripheral blood mononuclear cells (PBMCs) were cultured in the presence of tumour supernatant to monitor the expression of T‐cell signalling molecules. (a) Representative figure showing the expression of CD3‐ζ chain on gated CD3+ T cells in healthy individuals' (HI) PBMCs at different time‐points (24, 48, 72 and 96 hr) after incubation with cell‐free supernatant (diluted 1 : 1) obtained from patients with oral cancer (Stage III patient) and medium (control). (b, c) Cell‐free supernatants collected from oral tumour‐derived cell lines (AW13516 and AW8501) did not cause any change in CD3‐ζ chain expression after incubating with HI PBMCs for 72 hr. The graph shown is representative of four independent experiments. (d–g) Oral tumour supernatants were collected after 72 hr in plain RPMI from different tumour stages (Stage 1, n = 4; stage II, n = 3; stage III, n = 4; and stage IV, n = 6). Oral tumour supernatants were added to the HI PBMCs at a 1 : 1 ratio and after 72 hr the effects of tumour supernatant on CD3‐ζ chain and other T‐cell signalling molecules (CD3‐ε chain ZAP70 and p56Lck) were analysed. The graph indicates the expression (normalized median fluorescence intensity) of T‐cell signalling molecules on incubation with tumour supernatants obtained from different stage tumours. Fluorescence intensity was adjusted to 100 for control. Accordingly intensity for the test was calculated and represented as normalized fluorescence intensity. ***P < 0·0005, **P < 0·005 and *P < 0·05.
The effect of tumour supernatant on other T‐cell signalling molecules (p56Lck, Zap70 and CD3ε) was also studied after incubating for 72 hr. The supernatants collected from the patients (Stage I–III) led to a marked decrease in CD3‐ζ chain expression whereas its effect on CD3‐ε chain, ZAP‐70 and p56Lck expression was marginal (Fig. 1d–g). However, there was a decrease in expression of all these signalling molecules when incubated with the tumour supernatant collected from stage IV patients (Fig. 1d–g). Interestingly, the effect was best observed on the CD3‐ζ chain levels and it inversely correlated with stage of oral cancer in patients, as shown in Fig. 1(d). The broad range effect of supernatant collected from the patients with advanced stage cancer (stage IV) on all the T‐cell signalling molecules (CD3‐ζ chain, p56Lck, ZAP‐70) could be attributed to a general state of immunosuppression in these patients contributed by the accumulation of immunosuppressive cells and the secreted immune suppressive factors.
To identify the oral tumour cell‐derived factor (s) that degrade CD3‐ζ chain, a pragmatic approach was adopted. The cell‐free supernatants derived from oral tumours and cell lines (AW8501 and AW13516) were subjected to LC‐MS/MS. The identities of proteins observed exclusively in cell‐free tumour supernatants were shortlisted (Table 1). The proteins – Nuclear migration protein nudC, SET‐binding protein, Pleckstrin homology‐like domain family B member 2, Kinesin‐like protein KIF14, OAS2, Protocadherin β16 precursors – had a higher score (> 23). OAS2 belongs to the 2′5′‐oligoadenylate synthetase family of antiviral proteins consisting of OAS1, OAS2, OAS3 and OASL proteins, and was taken for further analysis.20 It is known to control cellular apoptosis;21 which may alter the expression of caspase‐3 and thereby regulate CD3‐ζ chain expression. OAS2 is also characterized by different subcellular locations and enzymatic parameters, suggesting that this protein might have distinct roles.22 Hence, based on LC‐MS/MS data and previously documented literature OAS2 was shortlisted as the key molecule present in the cell‐free supernatants of the oral tumours that may play a role in CD3‐ζ chain down‐regulation.
Table 1.
Screening of proteins present exclusively in tumour supernatant by liquid chromatography tandem mass spectrometry
| Description of protein | Peptide sequence | Mass/Score |
|---|---|---|
| Nuclear migration protein nudC | K.SMGLPTSDEQKK.Q | 885·1/23 |
| Dedicator of cytokinesis protein 7 | R.SYTEDWAIVIRK.Y | 885/19 |
| SET‐binding protein (SEB) | R.SYEGFGTYREK.D | 884·9/23 |
| Dual specificity tyrosine‐phosphorylation‐regulated kinase 4 | R.NLKPQPRPQTLRK.S | 860·8/18 |
| Toll‐like receptor 6 precursor (CD286 antigen) | K.SIVENIINCIEK.S | 860·4/15 |
| Microtubule‐associated proteins 1A/1B light chain 3C precursor (Microtubule‐associated protein 1 light chain 3γ) (MAP1A/MAP1B LC3 C) (MAP1A/1B light chain 3 C) (MAP1 light chain 3‐like protein 3) (Autophagy‐related protein LC3 C). | K.SLVSMSATMAEIYR.D | 860·4/17 |
| U6 snRNA‐associated Sm‐like protein LSm2 | K.SLVGKDVVVELK.N | 860·1/19 |
| SWI/SNF‐related matrix‐associated actin‐dependent regulator of chromatin subfamily A containing DEAD/H box 1 (EC 3·6·1.‐) | K.LNYAIFDEGHMLK.N | 855·4/15 |
| Protein GPR89A (Putative MAPK‐activating protein PM01) | R.RLLQTMDMIISK.K | 590·6/20 |
| Ephexin‐1 (Eph‐interacting exchange protein) | K.SVNEPLTLNIPWSR.M | 590·3/15 |
| Semenogelin‐1 precursor | K.EQTSVSGAQK.G | 590·3/17 |
| Aryl hydrocarbon receptor nuclear translocator 2 (ARNT protein 2) | R.SGMDFDDEDGEGPSK.F | 582·3/22 |
| Pleckstrin homology‐like domain family B member 2 | R.SGAASMPSSPK.Q | 582/23 |
| Kinesin‐like protein KIF14 | R.SGHLTTKPTQSK.L | 534·0/16 |
| Chromatin‐modifying protein 4c | K.KHGTQNK.R | 478·3/16 |
| PHD finger protein 21B | R.HQNGDLK.K | 478·3/16 |
| CD109 antigen precursor (p180) | R.TYTWLK.G | 478·3/16 |
| Ankyrin repeat and SOCS box protein 1 (ASB‐1) | K.WESLGPESRGR.R | 473·3/14 |
| Interleukin‐1 family member 8 (IL‐1F8) | R.TNIGMPGR.M | 502·8/11 |
| Mastermind‐like protein 3 (Mam‐3) | R.SLQGMPGR.T | 502·8/11 |
| Kinesin‐like protein KIF14 | R.SGHLTTKPTQSK.L | 524·9/27 |
| Microtubule‐associated proteins 1A/1B light chain 3C precursor | K.SLVSMSATMAEIYR.D | 860·4/15 |
| Integrin α‐L precursor | R.RGLFPGGR.H | 503·8/14 |
| Ras‐related protein Rab‐6B | M.SAGGDFGNPLRK.F | 503·3/14 |
| Kelch domain‐containing protein 5 | R.SNFKLVAVNSK.L | 450·8/11 |
| 2′‐5′‐oligoadenylate synthetase 2 (EC 2·7·7.‐) | R.ILNNNSK.R + | 473·3/23 |
| Epididymal secretory protein E1 precursor | K.TYSYLNK.L | 517·3/19 |
| Pericentrin (Pericentrin B) (Kendrin) | R.SLTEQQGR.L | 531·2/22 |
| Ubiquitin‐conjugating enzyme E2 J1 (EC 6·3·2·19) | R.GPPDSDFDGGVYHGR.I | 573·8/16 |
| Protocadherin β16 precursor (PCDH‐β16) | R.VIDINDHSPMFTEKEMILK. | 855·1/23 |
Expression of OAS2 in oral tumours and tumour‐derived cell lines
To show the presence of OAS2 in the tumour microenvironment, the expression of OAS2 at both protein and mRNA level was analysed in oral tumours. The expression analysis of OAS2 by western blotting showed that oral tumours (Stage II) express OAS2 (69 000 MW) at the protein level as shown in Fig. 2(a). For OAS2 mRNA expression, total RNA was extracted from surgically excised oral tumours and adjacent normal tissue. Complementary DNA (cDNA) was prepared from the total RNA using MuLV RT enzyme. Gene‐specific PCR was performed for GAPDH and OAS2. The heterogeneous expression of OAS2 mRNA was observed in oral tumours (Stage II and III) (Fig. 2b). The overall expression of mRNA for OAS2 remained high in oral tumours with two out of three tumours showing higher mRNA of OAS2 compared with the histologically adjacent normal tissues (Fig. 2c). In one of the oral tumour samples, mRNA expression of OAS2 was not observed but in its adjacent normal tissue OAS2 expression was noted. However, in oral tumour‐derived cell lines (AW13516, AW8507 and AW9803) OAS2 expression was observed only at mRNA level (Fig. 2d) but not at the protein level. This may suggest that the tumour microenvironment contributes to OAS2 expression.
Figure 2.
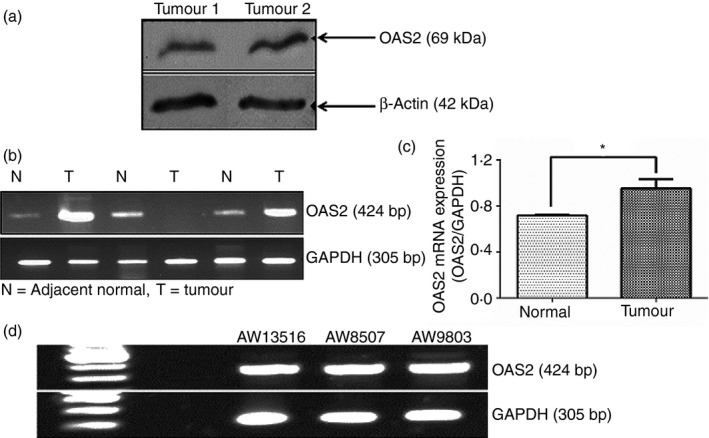
Expression of 2′5′‐oligoadenylate synthetase 2 (OAS2) in oral tumour cells. (a) Total protein extract from oral tumours was used to detect the levels of OAS2 and β‐actin by Western blotting. The protein extracts (40–60 μg) prepared from oral tumours (n = 2) was loaded into the SDS–PAGE and then transferred to the nitrocellulose membrane. The results showed a presence of the OAS2 (p69) in these tumours, β‐actin was used as internal control. (b) Total RNA (150–300 ng/μl) was isolated from the oral tumours and cell lines of oral tumour origin. The isolated RNA was converted into cDNA and cDNA was subjected to PCR, as described in the Materials and methods, to detect OAS2 and GAPDH transcripts. The expression of OAS2 transcript in oral tumours (T) and its adjacent normal tissues (N) is shown. (c) Band densitometry illustrating total OAS2 transcript expression is higher in oral tumours compared with the adjacent normal tissue. (d) Expression of OAS2 mRNA in oral tumour‐derived cell lines (AW13516, AW8507 and AW9803). *P < 0.05
Secreted OAS2 mediates CD3‐ζ chain down‐regulation in PBMCs of healthy individuals
To analyse the role of extracellular OAS2 in degrading CD3‐ζ chain, the key T‐cell signalling molecule, a surrogate situation, was created by transfecting vector control (pEGFP‐N2) or an OAS2‐encoding pEGFP‐N2 vector (pEGFP‐N2 + OAS2) into HEK293 cells. Total RNA was extracted from transfected cells, and mRNA expression levels were analysed by RT‐PCR. As shown in Fig. 3(a; upper panel) OAS2 expression was higher in cells transfected with (pEGFP‐N2 + OAS2) compared with the untransfected or vector‐control‐transfected HEK293 cells. Transfected cells were also analysed by flow cytometry for GFP positivity, which indirectly also confirms over‐expression of OAS2 protein as depicted in Fig. 3(a; lower panel).
Figure 3.
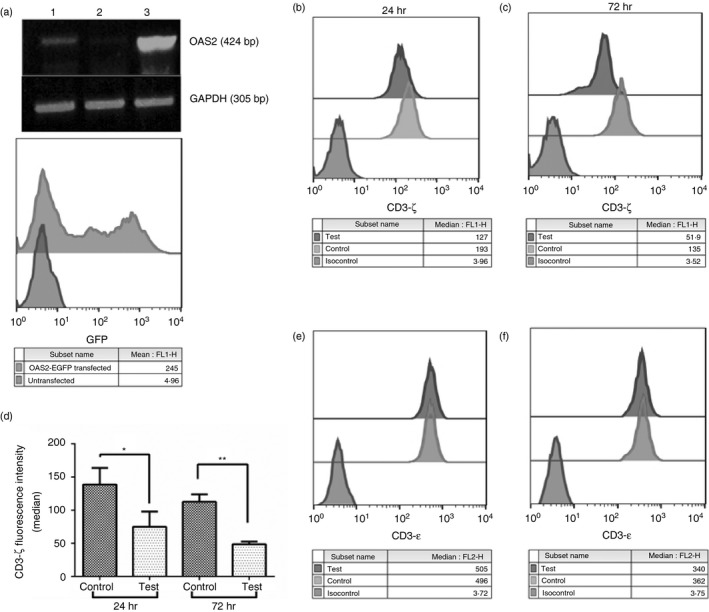
Effect of supernatant collected from 2′5′‐oligoadenylate synthetase 2 (OAS2) over‐expressing HEK293 cells on T‐cell signalling molecules. (a) HEK293 cells were transfected with either vector control (pEGFP‐N2) or vector encoding the OAS2 cDNA (pEGFP‐N2 + OAS2). Total RNA was extracted from transfected cells, and mRNA expression levels were analysed by RT‐PCR (upper panel) using gene‐specific primers for OAS2 and GAPDH. Lanes 1, 2 and 3 represent untransfected HEK293, vector control transfected HEK293 and pEGFP‐N2 + OAS2 transfected HEK293 cells, respectively. The pEGFP‐N2 + OAS2 transfected cells were also monitored for GFP expression by flow cytometry (lower panel). (b, c) Cell‐free supernatant collected from pEGFP‐OAS2 transfected HEK293 cells led to the decrease in expression of CD3‐ζ chain on gated CD3+ T cells in healthy individuals' (HI) peripheral blood mononuclear cells (PBMCs) at different time‐points (24 and 72 hr) compared with the supernatant collected from vector control transfected cells. (d) The median fluorescence intensity of CD3‐ζ chain staining in HI PBMCs treated with cell‐free supernatant of vector control and pEGFP‐OAS2 transfected HEK293 cells at 24 and 72 hr. (e, f) Unaffected fluorescence intensity (median) of CD3‐ε chain after incubating HI PBMCs with cell‐free supernatant collected from vector control and pEGFP‐OAS2 transfected HEK293 cells at different time intervals 24 and 72 hr, respectively. The data represented here are the mean of three experiments carried out using supernatants collected from three independent transfections. **P < 0·005 and *P < 0·05.
Supernatants were collected from OAS2 (pEGFP‐N2 + OAS2) and vector control (pEGFP‐N2) ‐transfected HEK293 (referred to as control) cells after 48 hr. The collected supernatants from control‐ and OAS2‐transfected HEK293 cells were added to the HI PBMCs at 1 : 1 [supernatant: medium (RPMI)] dilution for different time intervals. As shown in Fig. 3(b, c), the CD3‐ζ chain expression was observed in gated CD3+ T cells after 24 and 72 hr of incubation, respectively. The results confirmed that at both time‐points there was a decreased expression of CD3‐ζ chain when incubated with supernatant collected from OAS2 over‐expressing HEK293 cells compared with control HEK293 cells (vector‐control‐transfected) as shown in Fig. 3(d). On the other hand, CD3‐ε expression at both time intervals 24 and 72 hr remained unaltered on incubation with supernatant collected from OAS2‐transfected HEK293 cells as shown in Fig. 3(e, f). This confirms that the effect of secreted OAS2 was specific and altered only CD3‐ζ chain expression. The presence of OAS2 in supernatants was confirmed by the immuno‐dot blot assay (data not shown).
Recombinant human OAS2 protein decreases CD3‐ζ chain expression in PBMCs of healthy individuals
To elucidate the role of OAS family proteins in down‐regulating the CD3‐ζ chain expression, HI PBMCs were stimulated with different concentrations of rh‐OAS2 and rh‐OAS1 (3–96 ng/ml) for 24 hr. The PBMCs were harvested and expression of CD3‐ζ and CD3‐ε chain expression was examined by flow cytometry.
The HI PBMCs stimulated with rh‐OAS2 showed decreased expression of CD3‐ζ chain on a gated population of CD3+ T cells (Fig. 4a). The treatment of PBMCs with rh‐OAS2 at 6, 12 and 24 ng/ml concentrations for 24 hr led to a significant decrease in CD3‐ζ chain expression as shown in Fig. 4(a, c), although a dose–response was not evident. However, rh‐OAS2 treatment was not able to induce the changes in CD3‐ε expression as studied in Fig. 4(b), which is consistent with the earlier findings. The data clearly indicated that as observed with the supernatant collected from OAS2 transfected HEK293 cells, rh‐OAS2 was unable to affect the CD3‐ε chain expression on CD3+ T cells, again confirming that the effect of rh‐OAS2 was specific in modulating only CD3‐ζ chain expression.
Figure 4.
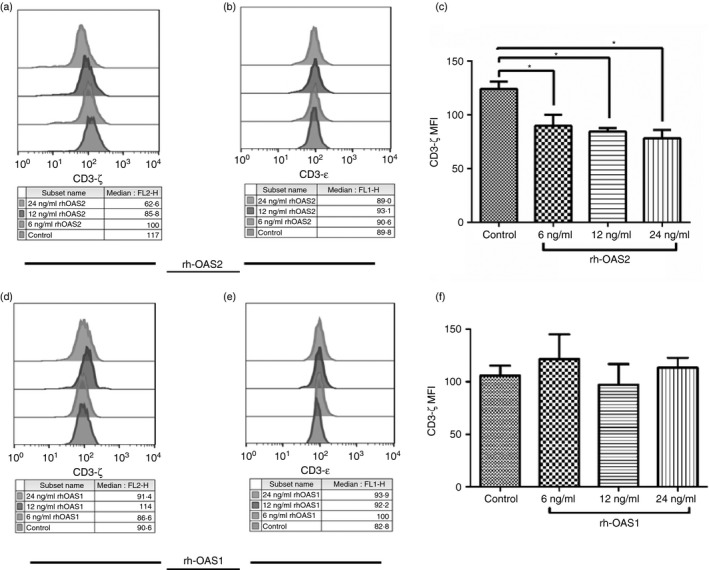
Effect of different concentrations of recombinant human 2′5′‐oligoadenylate synthetase 2 (rh‐OAS2) and rh‐OAS1 on the expression of CD3‐ζ and CD3‐ε chain on CD3+ T cells of healthy individuals' (HI) peripheral blood mononuclear cells (PBMCs). (a, c) The fluorescence intensity (median) of CD3‐ζ chain decreased after incubation of HI PBMCs with rh‐OAS2. Significant reduction in the expression of CD3‐ζ chain was observed on gated CD3+ T cells when treated with 6, 12 and 24 ng/ml of rh‐OAS2. (b) The fluorescence intensity (median) of CD3‐ε after incubation of HI PBMCs with rh‐OAS2 did not show any change. (d, f) the fluorescence intensity (median) of CD3‐ζ chain upon rh‐OAS1 stimulation of HI PBMCs remained unaltered on CD3+ T cells compared with the medium control used. (e) CD3‐ε expression was not affected on CD3+ T cells after treatment with rh‐OAS1 at different concentrations (6, 12 and 24 ng/ml). The data represented here are the mean of four independent experiments (*P < 0·05).
Upon stimulation with another member of OAS family, i.e. rh‐OAS1, PBMCs were unable to decrease the CD3‐ζ chain expression on gated population of CD3+ T cells as shown in Fig. 4(d). Data in Fig. 4(f) also show that the treatment of PBMCs with different concentrations of rh‐OAS1 did not alter the CD3‐ζ chain expression. Similar to rh‐OAS2, rh‐OAS1 was also incapable of decreasing the CD3‐ε chain expression on T cells as shown in Fig. 4(e). These data confirmed that OAS2 but not OAS1 was capable of modulating the CD3‐ζ chain expression.
Tumour supernatant and rh‐OAS2 decreases CD3‐ζ chain expression in sorted T cells
To understand whether oral tumour supernatants or rhOAS2 directly acted on T cells to down‐regulate CD3‐ζ chain expression, experiments were performed on sorted T cells (CD3+ T‐cell purity 99·5%, Fig. 5a). Incubation of tumour supernatant (diluted 1 : 1) with purified T cells decreased the CD3‐ζ chain expression in T cells. The decrease in CD3‐ζ chain expression observed in purified T cells after incubation with tumour supernatant for 24 hr was comparable to that observed in gated CD3+ T cells in PBMCs (Fig. 5b, d). Incubation of T cells with the various concentration of rh‐OAS2 (6, 12, 24 ng/ml) also led to the decrease in CD3‐ζ chain expression in accordance with the earlier data obtained with PBMCs incubated with rh‐OAS2 (Fig. 5c, e). These results demonstrate that tumour supernatant and rh‐OAS2 specifically acted on CD3‐ζ chain expressed in T cells.
Figure 5.
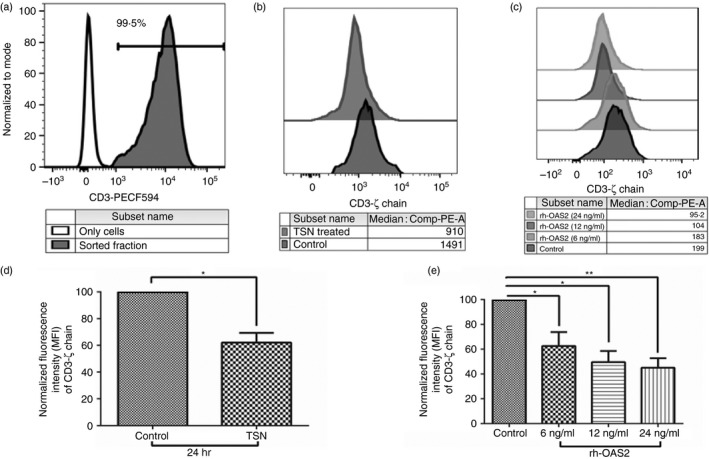
Effect of tumour supernatant and recombinant human 2′5′‐oligoadenylate synthetase 2 (rh‐OAS2) on the sorted CD3+ T cells. (a) The purity of CD3+ T cells isolated from the peripheral blood mononuclear cells (PBMCs). The shaded histogram is the sorted fraction (99·5% purity) whereas the unfilled histogram represents only cells. (b) Representative figure showing the expression of CD3‐ζ chain in CD3+ T cells after incubation with cell‐free supernatant (TSN) (diluted 1 : 1) obtained after culturing oral tumours or with medium (control). (c) Representative figure showing that fluorescence intensity (median) of CD3‐ζ chain decreased after incubation of CD3+ T cells with different concentrations of rh‐OAS2. Significant reduction in the expression of CD3‐ζ chain (MFI) was observed on CD3+ T cells when treated with 6, 12 and 24 ng/ml of rh‐OAS2. (d, e) Expression (normalized median fluorescence intensity) of CD3‐ζ chain on incubation with tumour supernatants obtained from oral tumours (d) or rh‐OAS2 (e). Fluorescence intensity was adjusted to 100 for control. Accordingly intensity for the test was calculated and represented as normalized fluorescence intensity. The graph shown is representative of three independent experiments (**P < 0·005 and *P < 0·05).
Tumour‐derived supernatants and rh‐OAS2 increased caspase‐3 levels in T cells of healthy individuals
Single‐cell suspension of oral tumours prepared by enzyme digestion was cultured in serum‐free medium and cell‐free tumour supernatants were collected after 72 hr of culture. The tumour supernatants were added to the HI PBMCs and incubated for 24 hr. Treatment of HI PBMCs with H2O2 served as positive control for caspase‐3 induction. As shown in Fig. 6(a) a marked increase in caspase‐3 levels was observed in HI PBMCs after H2O2 treatment (positive control). The addition of the tumour supernatant was also effectively able to induce caspase‐3 levels in HI PBMCs, indicating that tumour supernatant may alter CD3‐ζ expression by increasing caspase‐3 levels.
Figure 6.
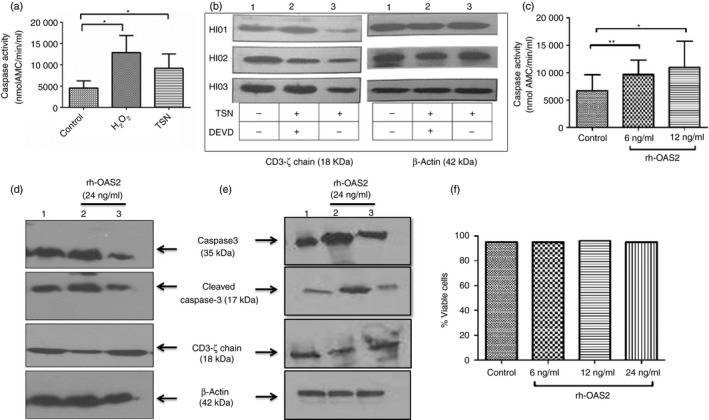
Caspase‐3 levels after treatment with tumour supernatant and recombinant human 2′5′‐oligoadenylate synthetase 2 (rh‐OAS2). (a) The healthy individuals' (HI) peripheral blood mononuclear cells (PBMCs) were treated with H2O2 (positive control for caspase‐3 induction) or tumour supernatant (TSN of stage III patients) or left untreated. H2O2 used as positive control to induce caspase‐3 levels showed higher levels of caspase‐3 activation. The addition of TSN resulted in the increase of caspase‐3 levels in the PBMCs. The data represented here are the mean of three independent experiments (*P < 0·05). (b) Healthy individuals' (HI) PBMCs on incubation with tumour supernatant showed a drastic reduction in CD3‐ζ chain expression (lane 3) compared with untreated (lane 1). On addition of caspase‐3 inhibitor (DEVD‐CHO), TSN was ineffective in inducing the CD3‐ζ chain down‐regulation (lane 2), i.e. caspase‐3 inhibitor prevented TSN mediated down‐regulation of CD3‐ζ chain levels. The data shown are PBMCs of three HIs treated with TSN in the presence or absence of caspase‐3 inhibitor (c) Addition of rh‐OAS2 (6 and 12 ng/ml) on HI PBMCs resulted in the increase in caspase‐3 levels. The data represented here are the mean of four independent experiments (**P < 0·005 and *P < 0·05). (d) Western blotting was performed on cell lysates prepared from PBMCs transfected with 100 nm control small interfering RNA (siRNA) (Lane 1) or PBMCs transfected with caspase‐3‐specific siRNA and incubated with rh‐OAS2 (lane 3) or untransfected PBMCs stimulated with rh‐OAS2 (lane 2) using antibodies for caspase‐3 (detects total caspase along with cleaved caspase‐3), CD3‐ζ chain or β‐actin (β‐actin was used as loading control). Stimulation of PBMCs with rh‐OAS2 induced caspase‐3 activation causing CD3‐ζ chain down‐regulation in them. This down‐regulation of CD3‐ζ chain in PBMCs was rescued by the caspase‐3 knockdown. (e) Western blotting was performed on cell lysates prepared from purified T cells transfected with 100 nM control siRNA (lane 1) or T cells transfected with caspase‐3‐specific siRNA and incubated with rh‐OAS2 (lane 3) or untransfected T cells stimulated with rh‐OAS2 (lane 2) using antibodies for caspase‐3, CD3‐ζ chain or β‐actin. In purified T cells, a pronounced effect was observed on caspase‐3 activation leading to CD3‐ζ down‐regulation on rh‐OAS2 stimulation, which was recovered back to normal levels on caspase‐3 knockdown. (f) PBMCs were treated with rh‐OAS2 to examine the percentage of viable cells (annexin V/PI negative cells). The rh‐OAS2 (6, 12 and 24 ng/ml) did not alter the viability of PBMCs in culture, indicating that increased caspase‐3 levels were not associated with apoptosis.
To validate the role of caspase‐3 in decreasing CD3‐ζ chain expression, PBMCs of HI were co‐incubated with the tumour supernatant and caspase‐3 inhibitor (DEVD‐CHO). As shown in Fig. 6(b), CD3‐ζ chain expression was decreased after incubation with cell‐free oral tumour supernatant (lane 3), which was almost restored back to its normal levels after addition of the caspase‐3 inhibitor, DEVD‐CHO (Lane 2). These data clearly suggest that caspase‐3 is involved in the regulation of CD3‐ζ chain expression.
To validate further, HI PBMCs were also incubated with rh‐OAS2 to analyse caspase‐3 activation. As observed with tumour supernatant, rh‐OAS2 also induced caspase‐3 activation in the PBMCs of HI compared with the untreated controls (Fig. 6c). The synthetic siRNA sequences targeting the caspase‐3 were transfected into the PBMCs (Fig. 6d) and purified CD3+ T cells (Fig. 6e). PBMCs were transfected with 100 nm of control siRNA or caspase‐3‐specific siRNA duplexes or left untransfected. Forty hours post‐transfection, cells were stimulated with rh‐OAS2 (24 ng/ml) and at 60 hr cells were harvested and lysates were prepared. Caspase‐3 expression was compared in unstimulated control siRNA transfected PBMCs, PBMCs stimulated with rh‐OAS2, and in PBMCs transfected with caspase‐3 siRNA and stimulated with rh‐OAS2 (Fig. 6d). The result showed that compared with control siRNA transfected PBMCs (unstimulated, lane 1) caspase‐3 expression after rh‐OAS2 stimulation appeared to be marginally increased in these cells (lane 2). Transfection with caspase‐3 siRNA led to the reduction in the expression of active caspase‐3 (lane 3) compared with untransfected PBMCs (lane 2) (Fig. 6d). Silencing of caspase‐3 prevented rh‐OAS2 induced down‐regulation of CD3‐ζ chain expression in PBMCs (lane 3 versus lane 2; Fig. 6d). This clearly demonstrates that expression of CD3‐ζ chain was restored and remained comparable to that observed with control siRNA. In purified CD3+ T cells, rh‐OAS2 stimulation had a more pronounced effect on caspase‐3 activation leading to CD3‐ζ chain down‐regulation. This down‐regulation of CD3‐ζ chain in purified T cells was rescued by knockdown of caspase‐3 and was comparable to the expression observed in control siRNA transfected cells (Fig. 6e). The results confirm that caspase‐3 is the key downstream molecule induced by OAS2 present in the tumour supernatant that is responsible for CD3‐ζ chain down‐regulation in T cells.
Additionally, the effect of rh‐OAS2 on apoptotic/necrotic cell death of PBMCs was analysed to rule out the possibility that the CD3‐ζ chain down‐regulation is not a result of apoptosis. To verify, the effect of rh‐OAS2 stimulation on the frequency of apoptotic cells was compared with unstimulated cells. Results showed no significant differences in the frequency of early apoptotic (annexin V‐positive), late apoptotic (annexin V‐positive PI‐positive) and necrotic (PI‐positive) cells in untreated HI PBMCs or rh‐OAS2‐treated HI PBMCs (Fig. 6f). This shows that caspase‐3 activity was not associated with the induction of cell death or apoptosis. It can be concluded that activation of caspase‐3 and the associated CD3‐ζ cleavage is a consequence of OAS2 signalling and not due to apoptosis of lymphocytes.
Discussion
Immunosuppression is a hallmark of oral cancer patients, a state in which established tumour escapes immune attack. A number of phenotypic and functional alterations, including down‐regulation of the CD3‐ζ chain, contributes to immunosuppression.23 It is well established that down‐regulation of the CD3‐ζ chain limits the favourable T helper type 1 response needed for controlling tumour growth.9, 24 Decreased or impaired CD3‐ζ chain expression was also observed in other malignancies, such as lymphoma, ovarian cancer and gastric carcinomas, leading to ineffectiveness of the anti‐tumour immune response against the autologous tumour.25, 26, 27 Several distinct mechanisms that contribute to a rapid turnover of CD3‐ζ chain have been proposed that may be responsible for differences observed in CD3‐ζ chain expression in T cells of patients with cancer.28, 29, 30 In the present study, it was demonstrated that treatment of HI PBMCs with cell‐free supernatants derived from oral tumours led to the down‐regulation of CD3‐ζ chain expression in T cells without affecting the other T‐cell signalling molecules (ZAP70, CD3‐ε and p56Lck). LC‐MS/MS proteomic approach was therefore adopted and OAS2 (member of OAS gene family) was identified as one of the factors secreted by the oral tumour cells. The transcription of OAS genes is induced by both virus infection and interferon (IFN) stimulation conferring protective and anti‐proliferative properties.31, 32 Among the OAS genes, OAS2 has the highest level of induction by interferons (IFN‐α and IFN‐β). Several studies have reported the presence of OAS2 in the sera of patients with viral infections and even after IFN treatment.33, 34, 35 The success of IFN therapy was also correlated with levels of OAS gene activity in the sera of patients undergoing IFN therapy.36, 37 This correlation is interesting and suggests a biological relevance of extracellular OAS.
A variable number of factors, including tumour‐secreted 14 000 MW protein, semaphorin A, NKG2D, are reported to be responsible for CD3‐ζ chain down‐regulation in different malignancies.26, 29, 30 The present study reported how extrinsic soluble mediator OAS2 secreted by oral tumour cells modulate CD3‐ζ chain expression in T cells.38 The 2′5′‐OAS, apart from its antiviral action is involved in other cellular processes such as cell growth and differentiation, gene regulation and apoptosis.39 It has been demonstrated that the IFN‐β‐mediated signalling pathways as well as its upstream regulators are up‐regulated in oral squamous cell carcinoma lesions.40 It therefore appears that IFN‐β signalling may act as a key factor responsible for OAS2 expression in oral tumours. Interferon‐β signalling is also known for its immune regulatory properties, suggesting its importance in tumour progression either directly or through modulation of the immune system through its downstream mediators like OAS2.
The observation of OAS in the sera of patients with infection or undergoing IFN treatment37 prompted us to look at the role of OAS in modulating the immune system. The differential expression of OAS2 has been reported in prostate and breast cancers.41, 42 Our study also confirmed the presence of OAS2 in oral tumours at both mRNA and protein levels. The significance of OAS2 in the tumour microenvironment can be appreciated by the observation that certain tumours exist in an antiviral state and hence are resistant to oncolytic‐virus‐based virotherapy used to obliterate tumour cells. These tumours are reported to have higher expression of the Janus kinase/signal transducer and activator of transcription pathway and IFN‐stimulated genes (ISG).43 It was also reported that over‐expression of myxovirus resistance gene A (MxA) and OAS mRNAs were involved in the suppression of hepatitis B virus replication mediated by IL‐17A in a non‐cytopathic manner.44 Hence, these tumours with a functional IFN‐responsive pathway and expression of a downstream gene (OAS2) may be sensitive to IFN stimulation, thereby conferring the virus‐resistant phenotype that shields tumours from virotherapy. The source of IFN in the tumour microenvironment is likely to be either the epithelial cells or the tumour‐infiltrating immune cells, such as type 2 dendritic cells and macrophages.40, 45 The IFN secreted by these cells will activate the ISG genes and will execute their pro‐tumour function either intracellularly or in secreted form by modulating the immune system. However, the source of OAS2 in oral tumours needs to be further investigated.
In our study, a surrogate model where OAS2 was over‐expressed showed that secreted OAS2 could modulate the expression of CD3‐ζ chain in T cells. This was further validated by stimulating HI PBMCs with rh‐OAS2 which led to the specific down‐regulation of CD3‐ζ chain expression in T cells. Up‐regulation of type I IFN response genes has also been reported in peripheral blood cells of patients with autoimmune diseases, like systemic lupus erythematosus, multiple sclerosis and rheumatoid arthritis and these diseases are associated with decreased CD3‐ζ chain expression.46, 47, 48 Type 1 IFNs have been shown to have anti‐proliferative activity on activated CD4+ and CD8+ T cells.49 Protein and mRNA levels of OAS genes are low in circulating mononuclear cells during clinically active, untreated multiple sclerosis. The IFN‐β therapy treatment of multiple sclerosis and experimental autoimmune encephalomyelitis to decrease the levels of T helper type 17 cells is associated with increased expression of OAS2 protein.37 Hence, OAS2 might be acting in a paracrine manner in the tumour microenvironment to decrease the expression of CD3‐ζ chain in T lymphocytes.
The present data showed that on incubation of PBMCs with tumour supernatant or rh‐OAS2 leads to increase in caspase‐3 levels. Increased caspase‐3 levels are observed in T cells under suboptimal T‐cell stimulation leading to the induction of T‐cell unresponsiveness without inducing cell death.50 Caspase‐3 activity is high in CD3ζ low/− T cells from a patient with systemic lupus erythematosus and treatment of these T cells with caspase inhibitors restores CD3‐ζ chain expression.51 2′5′‐OASs lead to cytochrome c release into the cytoplasm and then to caspase‐3 activation.52 Interferon‐α also increases caspase‐3 mRNA levels in activated T cells without modulating activation‐induced cell death.53 In our study, induction of caspase‐3 in non‐apoptotic cells was observed on stimulation with rh‐OAS2. This observation supports that caspase‐3‐dependent proteolytic inactivation of CD3‐ζ chain is essential to maintain T‐cell tolerance in cancer patients.53 The amino acid sequence of translated CD3‐ζ chain contains putative caspase‐3 cleavage sites DVLD and DTYD (five in number).10 Hence, the selective loss of CD3‐ζ chain and not of CD3‐ε could be because of the caspase‐3 activation, as observed upon stimulation with rh‐OAS2 or tumour supernatant. However, the downstream consequence of CD3‐ζ down‐regulation on the rh‐OAS2 challenge was not found to be dose‐dependent but might depend on induction of caspase‐3. To alter the expression of CD3‐ζ chain on HI PBMCs in a dose‐dependent manner, caspases are expected to vary in logarithmic values. In our study, such change in caspase‐3 value was not observed upon rh‐OAS2 stimulation. It has been shown that OAS that is exogenously added to cell cultures can be internalized to exert its effect. This exogenous OAS can exert its effect either by binding to the nucleotide‐binding and oligomerization domain‐2 to activate RNaseL or independent of this to induce caspase cascade.36, 54 Hence, the internalization of OAS2 inside the cells might be the limiting factor in inducing the caspase‐3 levels in a dose‐dependent manner.
Type I IFNs (IFN‐α and IFN‐β) could exert broad dual effects on the immune system, reflecting both immune stimulatory and immune‐suppressive activities. Immune‐stimulatory activities relate to the activation of myeloid dendritic cells, chemokines, chemokine receptors, co‐stimulatory molecules (CD40, CD80 and CD86), and humoral responses. Immune‐suppressive effects are reflected by Th2 cell skewing and anti‐proliferative and pro‐apoptotic effects. The role of IFNs in disease may range from beneficial to detrimental. However, it may be postulated that OAS2 plays a major immunoregulatory role based on its expression in oral tumours and autoimmune diseases where CD3‐ζ chain expression is down‐regulated.9, 55
Type I IFNs are promising but incompletely understood anticancer agents. Clinical trials have demonstrated benefit in both haematological and solid tumours, although the effectiveness is somewhat modest in some cancers.56, 57 Therapy with IFN‐α has been suggested to be effective in a subgroup of patients with oral cancer.58, 59 Further, it has also been demonstrated that the up‐regulation of ISGs (Mx1, OAS3, IFI44, IFI44L, OAS2, USP18 and RSAD2) in the chronic hepatitis C virus‐infected liver is related to a poor treatment response to pegylated IFN‐α therapy.60 Interferon‐α may be important in the prevention of carcinogenesis and high expression of OAS2 might be impairing cellular response to IFN‐α and promotes oral tumour progression by modulating anti‐tumour immune response. However, the role of others ISGs in modulating the IFN‐α response cannot be ruled out. Identification of molecular biomarkers like OAS2, which can also identify oral cancer patients sensitive to Type I IFN therapy would be very helpful in the clinic.
Hence, the data presented here indicate, for the first time, a potentially important function of tumour‐derived OAS2 as a paracrine negative regulator of T‐cell functions. Our data highlight OAS2 as a novel molecular target for the manipulation of T‐cell‐dependent immunity with important implications for cancer immunotherapy.
Disclosures
The authors declare no conflict of interest.
Acknowledgements
We are grateful to the Council for Scientific and Industrial research (CSIR), India for providing AAD with a fellowship. Help extended by Dr Robin Mukhopadhyaya and Dr Ajit Chande in cloning experiments is gratefully acknowledged.
References
- 1. Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: integrating immunity's roles in cancer suppression and promotion. Science 2011; 331:1565–70. [DOI] [PubMed] [Google Scholar]
- 2. Reichert TE, Strauss L, Wagner EM, Gooding W, Whiteside TL. Signaling abnormalities, apoptosis, and reduced proliferation of circulating and tumor‐infiltrating lymphocytes in patients with oral carcinoma. Clin Cancer Res 2002; 8:3137–45. [PubMed] [Google Scholar]
- 3. Zea AH, Curti BD, Longo DL, Alvord WG, Strobl SL, Mizoguchi H et al Alterations in T cell receptor and signal transduction molecules in melanoma patients. Clin Cancer Res 1995; 1:1327–35. [PubMed] [Google Scholar]
- 4. Whiteside TL. Immune cells in the tumor microenvironment. Mechanisms responsible for functional and signaling defects. Adv Exp Med Biol 1998; 451:167–71. [PubMed] [Google Scholar]
- 5. Lockhart DC, Chan AK, Mak S, Joo HG, Daust HA, Carritte A et al Loss of T‐cell receptor‐CD3ζ and T‐cell function in tumor‐infiltrating lymphocytes but not in tumor‐associated lymphocytes in ovarian carcinoma. Surgery 2001; 129:749–56. [DOI] [PubMed] [Google Scholar]
- 6. Choi SH, Chung EJ, Whang DY, Lee SS, Jang YS, Kim CW. Alteration of signal‐transducing molecules in tumor‐infiltrating lymphocytes and peripheral blood T lymphocytes from human colorectal carcinoma patients. Cancer Immunol Immunother 1998; 45:299–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mizoguchi H, O'Shea JJ, Longo DL, Loeffler CM, McVicar DW, Ochoa AC. Alterations in signal transduction molecules in T lymphocytes from tumor‐bearing mice. Science 1992; 258:1795–8. [DOI] [PubMed] [Google Scholar]
- 8. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140:883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kulkarni DP, Wadia PP, Pradhan TN, Pathak AK, Chiplunkar SV. Mechanisms involved in the down‐regulation of TCR ζ chain in tumor versus peripheral blood of oral cancer patients. Int J Cancer 2009; 124:1605–13. [DOI] [PubMed] [Google Scholar]
- 10. Gastman BR, Johnson DE, Whiteside TL, Rabinowich H. Caspase‐mediated degradation of T‐cell receptor ζ‐chain. Cancer Res 1999; 59:1422–7. [PubMed] [Google Scholar]
- 11. Thornberry NA, Lazebnik Y. Caspases: enemies within. Science 1998; 281:1312–6. [DOI] [PubMed] [Google Scholar]
- 12. Rathmell JC, Thompson CB. The central effectors of cell death in the immune system. Annu Rev Immunol 1999; 17:781–828. [DOI] [PubMed] [Google Scholar]
- 13. Takahashi A, Kono K, Amemiya H, Iizuka H, Fujii H, Matsumoto Y. Elevated caspase‐3 activity in peripheral blood T cells coexists with increased degree of T‐cell apoptosis and down‐regulation of TCR ζ molecules in patients with gastric cancer. Clin Cancer Res 2001; 7:74–80. [PubMed] [Google Scholar]
- 14. Maki A, Matsuda M, Asakawa M, Kono H, Fujii H, Matsumoto Y. Decreased expression of CD28 coincides with the down‐modulation of CD3ζ and augmentation of caspase‐3 activity in T cells from hepatocellular carcinoma‐bearing patients and hepatitis C virus‐infected patients. J Gastroenterol Hepatol 2004; 19:1348–56. [DOI] [PubMed] [Google Scholar]
- 15. Woo M, Hakem R, Furlonger C, Hakem A, Duncan GS, Sasaki T et al Caspase‐3 regulates cell cycle in B cells: a consequence of substrate specificity. Nat Immunol 2003; 4:1016–22. [DOI] [PubMed] [Google Scholar]
- 16. Alam A, Cohen LY, Aouad S, Sekaly RP. Early activation of caspases during T lymphocyte stimulation results in selective substrate cleavage in nonapoptotic cells. J Exp Med 1999; 190:1879–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Algeciras‐Schimnich A, Barnhart BC, Peter ME. Apoptosis‐independent functions of killer caspases. Curr Opin Cell Biol 2002; 14:721–6. [DOI] [PubMed] [Google Scholar]
- 18. Denis F, Rheaume E, Aouad SM, Alam A, Sekaly RP, Cohen LY. The role of caspases in T cell development and the control of immune responses. Cell Mol Life Sci 1998; 54:1005–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tatake RJ, Rajaram N, Damle RN, Balsara B, Bhisey AN, Gangal SG. Establishment and characterization of four new squamous cell carcinoma cell lines derived from oral tumors. J Cancer Res Clin Oncol 1990; 116:179–86. [DOI] [PubMed] [Google Scholar]
- 20. Rebouillat D, Hovnanian A, Marie I, Hovanessian AG. The 100‐kDa 2′,5′‐oligoadenylate synthetase catalyzing preferentially the synthesis of dimeric pppA2′p5′A molecules is composed of three homologous domains. J Biol Chem 1999; 274:1557–65. [DOI] [PubMed] [Google Scholar]
- 21. Ghosh A, Sarkar SN, Rowe TM, Sen GC. A specific isozyme of 2′‐5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl‐2 family. J Biol Chem 2001; 276:25447–55. [DOI] [PubMed] [Google Scholar]
- 22. Hovanessian AG. On the discovery of interferon‐inducible, double‐stranded RNA activated enzymes: the 2′‐5′oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev 2007; 18:351–61. [DOI] [PubMed] [Google Scholar]
- 23. Rapidis AD, Wolf GT. Immunotherapy of head and neck cancer: current and future considerations. J Oncol 2009; doi:10.1155/2009/346345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kiessling R, Wasserman K, Horiguchi S, Kono K, Sjöberg J, Pisa P et al Tumor‐induced immune dysfunction. Cancer Immunol Immunother 1999; 48:353–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frydecka I, Kaczmarek P, Bocko D, Kosmaczewska A, Morilla R, Catovsky D. Expression of signal‐transducing ζ chain in peripheral blood T cells and natural killer cells in patients with Hodgkin's disease in different phases of the disease. Leuk Lymphoma 1999; 35:545–54. [DOI] [PubMed] [Google Scholar]
- 26. Taylor DD, Bender DP, Gercel‐Taylor C, Stanson J, Whiteside TL. Modulation of TcR/CD3‐ζ chain expression by a circulating factor derived from ovarian cancer patients. Br J Cancer 2001; 84:1624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ishigami S, Natsugoe S, Tokuda K, Nakajo A, Higashi H, Iwashige H et al CD3‐ζ chain expression of intratumoral lymphocytes is closely related to survival in gastric carcinoma patients. Cancer 2002; 94:1437–42. [DOI] [PubMed] [Google Scholar]
- 28. Ersek B, Molnar V, Balogh A, Matko J, Cope AP, Buzas EI et al CD3ζ‐chain expression of human T lymphocytes is regulated by TNF via Src‐like adaptor protein‐dependent proteasomal degradation. J Immunol 2012; 189:1602–10. [DOI] [PubMed] [Google Scholar]
- 29. Catalano A, Caprari P, Moretti S, Faronato M, Tamagnone L, Procopio A. Semaphorin‐3A is expressed by tumor cells and alters T‐cell signal transduction and function. Blood 2006; 107:3321–9. [DOI] [PubMed] [Google Scholar]
- 30. Hanaoka N, Jabri B, Dai Z, Ciszewski C, Stevens AM, Yee C et al NKG2D initiates caspase‐mediated CD3ζ degradation and lymphocyte receptor impairments associated with human cancer and autoimmune disease. J Immunol 2010; 185:5732–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Benech P, Vigneron M, Peretz D, Revel M, Chebath J. Interferon‐responsive regulatory elements in the promoter of the human 2′,5′‐oligo(A) synthetase gene. Mol Cell Biol 1987; 7:4498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Melchjorsen J, Kristiansen H, Christiansen R, Rintahaka J, Matikainen S, Paludan SR et al Differential regulation of the OASL and OAS1 genes in response to viral infections. J Interferon Cytokine Res 2009; 29:199–207. [DOI] [PubMed] [Google Scholar]
- 33. Mihm U, Ackermann O, Welsch C, Herrmann E, Hofmann WP, Grigorian N et al Clinical relevance of the 2′‐5′‐oligoadenylate synthetase/RNase L system for treatment response in chronic hepatitis C. J Hepatol 2009; 50:49–58. [DOI] [PubMed] [Google Scholar]
- 34. Kim KI, Kim SR, Sasase N, Taniguchi M, Harada S, Kinoshita K et al 2′‐,5′‐Oligoadenylate synthetase response ratio predicting virological response to PEG‐interferon‐α2b plus ribavirin therapy in patients with chronic hepatitis C. J Clin Pharm Ther 2006; 31:441–6. [DOI] [PubMed] [Google Scholar]
- 35. Shindo M, Hamada K, Morikawa T, Harano Y, Nakajima T, Okuno T. In vivo interferon system assessed by 2′‐5′ oligoadenylate synthetase activity in chronic hepatitis C virus patients treated with pegylated interferon and ribavirin. Hepatol Res 2008; 38:1213–20. [DOI] [PubMed] [Google Scholar]
- 36. Kristiansen H, Scherer CA, McVean M, Iadonato SP, Vends S, Thavachelvam K et al Extracellular 2′‐5′ oligoadenylate synthetase stimulates RNase L‐independent antiviral activity: a novel mechanism of virus‐induced innate immunity. J Virol 2010; 84:11898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feng X, Petraglia AL, Chen M, Byskosh PV, Boos MD, Reder AT. Low expression of interferon‐stimulated genes in active multiple sclerosis is linked to subnormal phosphorylation of STAT1. J Neuroimmunol 2002; 129:205–15. [DOI] [PubMed] [Google Scholar]
- 38. Sanda C, Weitzel P, Tsukahara T, Schaley J, Edenberg HJ, Stephens MA et al Differential gene induction by type I and type II interferons and their combination. J Interferon Cytokine Res 2006; 26:462–72. [DOI] [PubMed] [Google Scholar]
- 39. Hovanessian AG, Justesen J. The human 2′‐5′oligoadenylate synthetase family: unique interferon‐inducible enzymes catalyzing 2′‐5′ instead of 3′‐5′ phosphodiester bond formation. Biochimie 2007; 89:779–88. [DOI] [PubMed] [Google Scholar]
- 40. Chi LM, Lee CW, Chang KP, Hao SP, Lee HM, Liang Y et al Enhanced interferon signaling pathway in oral cancer revealed by quantitative proteome analysis of microdissected specimens using 16O/18O labeling and integrated two‐dimensional LC‐ESI‐MALDI tandem MS. Mol Cell Proteomics 2009; 8:1453–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bower JE, Greendale G, Crosswell AD, Garet D, Sternlieb B, Ganz PA et al Yoga reduces inflammatory signaling in fatigued breast cancer survivors: a randomized controlled trial. Psychoneuroendocrinology 2014; 43:20–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vaarala MH, Porvari K, Kyllonen A, Vihko P. Differentially expressed genes in two LNCaP prostate cancer cell lines reflecting changes during prostate cancer progression. Lab Invest 2000; 80:1259–68. [DOI] [PubMed] [Google Scholar]
- 43. Liu YP, Suksanpaisan L, Steele MB, Russell SJ, Peng KW. Induction of antiviral genes by the tumor microenvironment confers resistance to virotherapy. Sci Rep 2013; 3:2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang B, Zhao XP, Fan YC, Zhang JJ, Zhao J, Wang K. IL‐17A but not IL‐22 suppresses the replication of hepatitis B virus mediated by over‐expression of MxA and OAS mRNA in the HepG2.2.15 cell line. Antiviral Res 2013; 97:285–92. [DOI] [PubMed] [Google Scholar]
- 45. Lallemand C, Lebon P, Rizza P, Blanchard B, Tovey MG. Constitutive expression of specific interferon isotypes in peripheral blood leukocytes from normal individuals and in promonocytic U937 cells. J Leukoc Biol 1996; 60:137–46. [DOI] [PubMed] [Google Scholar]
- 46. Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ et al Interferon‐inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 2003; 100:2610–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. van Baarsen LG, van der Pouw Kraan TC, Kragt JJ, Baggen JM, Rustenburg F, Hooper T et al A subtype of multiple sclerosis defined by an activated immune defense program. Genes Immun 2006; 7:522–31. [DOI] [PubMed] [Google Scholar]
- 48. van der Pouw Kraan TC, Wijbrandts CA, van Baarsen LG, Voskuyl AE, Rustenburg F, Baggen JM et al Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis 2007; 66:1008–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tough DF, Sun S, Zhang X, Sprent J. Stimulation of naive and memory T cells by cytokines. Immunol Rev 1999; 170:39–47. [DOI] [PubMed] [Google Scholar]
- 50. Puga I, Rao A, Macian F. Targeted cleavage of signaling proteins by caspase 3 inhibits T cell receptor signaling in anergic T cells. Immunity 2008; 29:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Krishnan S, Kiang JG, Fisher CU, Nambiar MP, Nguyen HT, Kyttaris VC et al Increased caspase‐3 expression and activity contribute to reduced CD3ζ expression in systemic lupus erythematosus T cells. J Immunol 2005; 175:3417–23. [DOI] [PubMed] [Google Scholar]
- 52. Rusch L, Zhou A, Silverman RH. Caspase‐dependent apoptosis by 2′,5′‐oligoadenylate activation of RNase L is enhanced by IFN‐β . J Interferon Cytokine Res 2000; 20:1091–100. [DOI] [PubMed] [Google Scholar]
- 53. Ludwiczek O, Kaser A, Koch RO, Vogel W, Cruikshank WW, Tilg H. Activation of caspase‐3 by interferon α causes interleukin‐16 secretion but fails to modulate activation induced cell death. Eur Cytokine Netw 2001; 12:478–86. [PubMed] [Google Scholar]
- 54. Dugan JW, Albor A, David L, Fowlkes J, Blackledge MT, Martin TM et al Nucleotide oligomerization domain‐2 interacts with 2′‐5′‐oligoadenylate synthetase type 2 and enhances RNase‐L function in THP‐1 cells. Mol Immunol 2009; 47:560–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Takeuchi T, Suzuki K, Kondo T, Yoshimoto K, Tsuzaka K. CD3 ζ defects in systemic lupus erythematosus. Ann Rheum Dis 2012; 71(Suppl. 2):i78–81. [DOI] [PubMed] [Google Scholar]
- 56. Clavien PA. Interferon: the magic bullet to prevent hepatocellular carcinoma recurrence after resection? Ann Surg 2007; 245:843–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Sun HC, Tang ZY, Wang L, Qin LX, Ma ZC, Ye QH et al Postoperative interferon α treatment postponed recurrence and improved overall survival in patients after curative resection of HBV‐related hepatocellular carcinoma: a randomized clinical trial. J Cancer Res Clin Oncol 2006; 132:458–65. [DOI] [PubMed] [Google Scholar]
- 58. Urba SG, Forastiere AA, Wolf GT, Amrein PC. Intensive recombinant interleukin‐2 and α‐interferon therapy in patients with advanced head and neck squamous carcinoma. Cancer 1993; 71:2326–31. [DOI] [PubMed] [Google Scholar]
- 59. Mukherjee KK, Bose A, Ghosh D, Sarkar K, Goswami S, Pal S et al IFNα2b augments immune responses of cisplatin+5‐fluorouracil treated tongue squamous cell carcinoma patients – a preliminary study. Indian J Med Res 2012; 136:54–9. [PMC free article] [PubMed] [Google Scholar]
- 60. Honda M, Sakai A, Yamashita T, Nakamoto Y, Mizukoshi E, Sakai Y et al Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology 2010; 139:499–509. [DOI] [PubMed] [Google Scholar]


