Summary
Rheumatoid arthritis (RA) is associated with the presence of certain HLA class II genes. However, why some individuals carrying RA non‐associated alleles develop arthritis is still unexplained. The trans‐heterodimer between two RA non‐associated HLA genes can render susceptibility to develop arthritis in humanized mice, DQA1*0103/DQB1*0604, suggesting a role for DQ α chains in pathogenesis. In this study we determined the role of DQA1 in arthritis by using mice expressing DQA1*0103 and lacking endogenous class II molecules. Proximity ligation assay showed that DQA1*0103 is expressed on the cell surface as a dimer with CD74. Upon immunization with type II collagen, DQA1*0103 mice generated an antigen‐specific cellular and humoral response and developed severe arthritis. Structural modelling suggests that DQA1*0103/CD74 form a pocket with similarity to the antigen binding pocket. DQA1*0103 mice present type II collagen‐derived peptides that are not presented by an arthritis‐resistant DQA1*0103/DQB1*0601 allele, suggesting that the DQA1*0103/CD74 dimer may result in presentation of unique antigens and susceptibility to develop arthritis. The present data provide a possible explanation by which the DQA1 molecule contributes to susceptibility to develop arthritis.
Keywords: CD74, DQA1*0103, heterodimer, humanized mice, rheumatoid arthritis
Abbreviations
- CIA
collagen‐induced arthritis
- CII
type II collagen
- MIF
migration inhibitory factor
- RA
rheumatoid arthritis
Introduction
Rheumatoid arthritis (RA) is an autoimmune disease with a strong genetic predisposition. Among all the factors studied to date, HLA class II alleles provide the highest risk to develop arthritis. In various ethnic populations, DRB1*0401 has been associated with susceptibility to RA, though the role of DQ molecules in RA is limited to few studies.1, 2 Heterozygotes carrying two RA‐associated genotypes have higher relative risk of disease compared with those carrying homozygous susceptible genotypes, indicating an epistatic or synergistic effect.3 Structural studies have suggested that whereas a highly stable αβ dimer of the DQ6 molecule (DQA1*0103/DQB1*0601) results in protection from diabetes, an unstable αβ dimer of DQ8 (DQA1*0301/DQB1*0302) predisposes to diabetes.4 Similar to diabetes, DQ6 has been associated with resistance and DQ8 with susceptibility to RA.
CD74, invariant chain, binds MHC class II heterodimer and plays a crucial role in the expression and peptide loading.5 CD74 is also a receptor for macrophage migration inhibitory factor (MIF) and in complex with CD44 prolongs cell survival.6 An increase in the expression of CD74 has been observed in patients with α viral polyarthritis.7 HLA transgenic mice lacking the invariant chain exhibit low expression of HLA gene and do not generate antigen‐specific response and do not develop disease.8
Collagen‐induced arthritis (CIA) is an experimental model of autoimmune inflammatory polyarthritis sharing clinical and pathological features with RA.9 Transgenic mice expressing HLA genes, known to be associated with RA, mimic human disease in histopathology and sex‐bias.10, 11, 12 We have previously shown that DQ8 (DQA1*0301/DQB1*0302) mice are highly susceptible to CIA and mimic human RA,13, 14, 15 whereas DQA1*0103/DQB1*0601 (DQ6.1) mice do not develop CIA.16 The expression of trans‐heterodimer DQ molecules, DQA1*0103/DQB1*0604 (DQ6.4) makes mice susceptible to CIA.17 In this study we explored the role of DQA1 chain in arthritis by generating mice carrying only the DQA1*0103 chain. We demonstrate here that DQA1 can be expressed as a dimer with CD74 on the cell surface, generate antigen‐specific immune response and predispose transgenic mice to develop arthritis. Our study provides a possible mechanism by which patients carrying non‐susceptible HLA alleles may develop arthritis and highlights CD74 as a target for therapy.
Materials and methods
Mice
Transgenic mice expressing functional HLA‐DQ8.AEo (DQA1*0301/DQB1*0302) and DQ6.AEo (DQA1*0103/DQB1*0601) and DQA1*0103/DQB1*0604.AEo molecules were generated as described previously.14, 16, 17 Cosmids pPKQ5121 and pAKQ 4116 containing DQA1*0103 gene were a kind gift from Dr H. Inoko. The construct containing the DQA1*0103 gene was microinjected into (B6/J X B10.M) F2 embryos, resulting in B10.M‐DQ6α transgenic mice, which were bred on an FVB background and then on an AEo background to generate AEo.DQA1* 0103 mice. For convenience, mice expressing DQA1*0301/DQB1*0302 are referred to as DQ8, mice expressing DQA1*0103/DQB1*0604 as DQ6.4 and those expressing DQA1*0103/DQB1*0601.AEo as DQ6.1 mice in the manuscript.
Mice of both sexes (8–12 weeks of age) used in this study were bred and maintained in the pathogen‐free Immunogenetics Mouse Colony of Mayo Clinic. All experimental procedures were performed with the approval of the Institutional Animal Use and Care Committee.
Induction and evaluation of collagen‐induced arthritis
Eight‐ to 12‐week‐old transgenic mice along with negative littermates were induced for CIA by immunizing with 100 μg chick type II collagen (CII; Chondrex Inc. Redmond, WA, USA) as described.10 The onset and progression of CIA was monitored and the arthritic severity of mice was evaluated as described.10 The mean arthritic score was determined using arthritic animals only. Mice were bled on day 35 post‐immunization and the level of anti‐chick CII in serum was determined using a standard ELISA technique as described previously.18
Proximity ligation assay
The proximity ligation assay was performed using splenic cells from naive transgenic mice by commercially available kit (Duolink assay kit; Olink Bioscience, Uppsala, Sweden) as per instructions. The primary antibodies used were SPV‐L3 (anti‐DQ) and anti‐CD74. The stained cells were then coated on to slides.19, 20 The slides were dried at room temperature, mounted with mounting medium (ProLong Gold with DAPI; Life Technologies, Grand Island, NY, USA), and stored at −20° until confocal images were taken. Cells were viewed with a confocal microscope (LAS‐AF; Leica, Wetzlar, Germany). Each fluorescent spot corresponds to a heterodimer. Images were taken at 40× and 60× under oil immersion. Slides were counted for total and dimer positive cells and percent positive for dimers from all cells was calculated. Mean florescence intensity (MFI) was measured using image j software (National Health Institutes, USA).
Molecular modelling and docking
The three‐dimensional structure of mouse CD74 extracellular domain consisting of residues 117–191 was generated by homology modelling using SWISSMODEL.21 The template structure used was from PDB ID 1IE.22 The three‐dimensional structure of the α chain of DQ was generated using template of HLA‐DQ2 (PDB ID 1S9V).23 CD74 was generated as a trimer. The three‐dimensional structures of monomer and trimer of CD74 was docked to DQA1 using ZDOCK24, 25 server and the first 10 docked structures were analysed. The docked structures were subjected to energy minimization and molecular dynamics simulations for 10 ps at 300 K using insightII (Accelrys Inc., San Diego, CA). Final structures were further energy minimized using a conjugate gradient method. The resulting structures were analysed for quality of the structure using molprobity.26 The three‐dimensional structure of the antigenic peptide was built using insightII (Accelrys Inc.). autodock software27 was used for docking antigenic peptide with the complex of CD74 and DQ. A grid box with dimensions 126 × 126 ×126 Å3 was built around the groove as the possible binding site of the peptide. Using a Lamarckian genetic algorithm, docking was performed and 10 million energy evaluations were performed using 150 starting conformations. Only 50 low‐energy structures were saved for final analysis. Structures with docking energy of 2 kcal/mol were considered for analysis from the lowest energy structure. Final docked structure was represented using pymol software (Schrodinger, Portland, OR).
Flow cytometry
Expression of HLA‐DQ, CD3, CD4, CD8, H2M (BD Biosciences, San Jose, CA) on splenic cells was analysed by flow cytometry using FACS IV as described earlier.12 Other antibodies used for staining were L227 (DQB1*0601), SPLV3, IVD12 (anti‐DQ), F23.2 (anti Vβ8.2) and CD74 (Santa Cruz Biotech, Dallas, TX). Flow cytometry data were analysed with flowjo software (FlowJo, Ashland, OR).
In vitro T‐cell proliferation
Mice were immunized with 200 μg of CII or human collagen II (HII)‐derived peptides28 emulsified 1 : 1 with complete Freund's adjuvant (Difco, Laboratories, Detroit, Michigan) and injected intradermally at the base of the tail. Ten days after immunization, spleen and draining popliteal, caudal and lumbar lymph nodes were removed and spleen/lymph nodes were cultured in vitro as described.10, 12
Reverse transcription polymerase chain reaction
Expression of MIF was analysed by RT‐PCR using RNAeasy columns (Qiagen, Maryland, MD, USA) and RNase H‐reverse transcriptase (Invitrogen, Carlsbad, CA). The cDNA was analyded by real‐time quantitative PCR in triplicates by using an SYBR® GreenER™ qPCR reagent system (Invitrogen) and primers (Applied Biosystems, Foster City, CA). The RNA transcript level was quantified using the threshold cycle (C t) method normalized for the housekeeping gene GAPDH.
Statistical analysis
Difference in incidence of arthritis between groups was analysed using chi‐squared test with Yates' correction. A P‐value of < 0·05 was considered significant. Cytokine levels, T‐cell proliferation, mRNA expression and arthritis severity were compared using two‐tailed Student's t‐test. Data are depicted as mean ± SEM.
Results
HLA‐DQA1*0103.AEo mice are susceptible to develop collagen‐induced arthritis
Our previous study using mice expressing a trans‐heterodimer between the two non‐associated HLA alleles, DQA1*0103/DQB1*0604 (DQ6.4), suggested a role of DQA1 gene in arthritis, though it did not prove its direct involvement.17 During the generation of DQ6.4 mice, we also generated mice expressing only DQA1*0103. We did not expect DQA1*0103.AEo mice to be functional without any beta chain. Since a functional HLA transgene leads to positive and negative selection of CD4 and CD8 T cells in mice, we tested if the presence of DQA1*0103 alone could rescue positive selection of CD4 T cells in transgenic mice. Introduction of DQA1*0103 transgene in AEo mice rescued CD4+ T cells similar to that in DQ6.1 and DQ6.4 mice (Table 1),17 suggesting that transgenic mice are functional. Since FVB/N mice are resistant to developing arthritis due to deletion of Vβ8.2,29 we tested for positive selection of Vβ8.2 T cells in DQA1*0103 mice. Higher percentages of Vβ8.2 cells were present in DQA1*0103 compared with FVB mice, suggesting that the transgene can positively select T‐cell receptor profile in thymus (8 ± 1·4 and 3 ± 2·8 in DQA1*0103 and FVB, respectively).
Table 1.
Percent positive of CD3, CD4 and CD8 T cells and Vβ8.2‐positive T cells in mice
| Mice | CD3 | CD4 | CD8 | Vβ8.2 |
|---|---|---|---|---|
| DQB1a0604/DQA1*0103 | 26·5 ± 4·9 | 15·5 ± 2·7 | 19·5 ± 2·1 | 8 ± 1·4 |
| DQA1*0103 | 24·5 ± 7·7 | 13·8 ± 1·5 | 20·8 ± 1·6 | 11 ± 6 |
| FVB | 25·0 ± 4·2 | 20·0 ± 2·8 | 11·5 ± 2·1 | 3 ± 2·8a |
The cells were enumerated in splenocytes of naive mice, n = 4 per strain. The numbers show % ± SD.
P < 0·05, FVB versus DQA1*0103 and DQB1*0604/DQA1*0103.
As this was a surprising observation, we tested DQA1*0103 mice for antigen‐specific response and CIA (Fig. 1a). All DQA1*0103 mice (n = 12) and 75% of DQ6.4 mice (n = 12) developed arthritis whereas DQ6.1 mice (n = 10) and FVB mice (n = 12) were resistant to developing CIA (10% and 8%, respectively). DQA1*0103 mice also developed much more severe disease compared with DQ6.4 mice (P < 0·05) and DQ6.1 mice (P < 0·001) (Fig. 1b). DQ8 and DQA1*0103 mice produced anti‐CII antibodies, which were significantly higher compared with CIA‐resistant DQ6.1 and FVB mice (Fig. 1c). In vitro DQA1*0103 mice generated a robust CII‐specific T‐cell immune response compared with DQ6.1 mice, P < 0·03 (Fig. 1d). The response was mediated by CD4+ T cells as observed by inhibition studies (not shown). Cytokines measured from the supernatants of in vitro culture showed that DQA1*0103 mice produced levels of interferon‐γ and interleukin‐12(p40) similar to those in DQ8 mice, but that were significantly higher compared with DQ6.1 mice (Fig. 1e). The relative expression of mRNA of MIF showed significantly lower amounts in DQA1*0103 mice compared with DQ8 and DQ6.1 mice (P < 0·0001) (Fig. 1f). Next we compared response to CII peptides known to be presented by arthritis‐resistant DQ6.1 and arthritis‐susceptible DQ8 mice.28 There are few CII‐derived peptides that are not presented by CIA‐resistant DQ6.1 mice but that are presented by CIA‐susceptible DQ8 mice, suggesting that they might be important in pathogenesis. DQA1*0103 mice generated response to the CII‐derived peptides known to be DQ8‐restricted but not presented by DQ6.1 mice (Fig. 1g), and the response was dose dependent (not shown).
Figure 1.
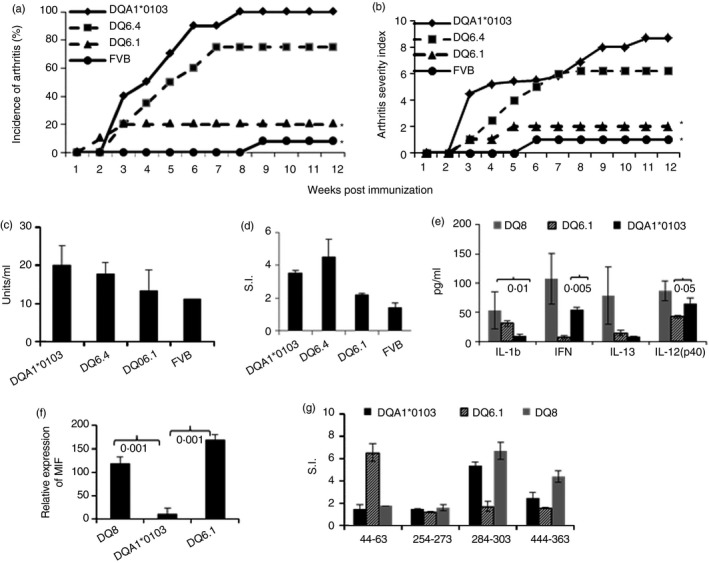
DQA1*0103 mice are susceptible to collagen‐induced arthritis. (a) Per cent incidence of arthritis (DQA1*0103, n = 12; DQ6.1, n = 10; DQ6.4, n = 12; FVB, n = 12). (b) Disease severity, *P < 0·001 DQA1*0103 versus DQ6.1 and FVB. DQA1*0103 mice generate CII‐specific cellular and humoral response. (c) Anti‐CII antibodies, DQA1*0103 versus DQ6.4, P < 0·05; DQA1*0103 versus DQ6.1 and FVB, P < 0·02; DQ6.4 versus DQ6.1, P < 0·05; DQ6.4 versus FVB, P < 0·02. (d) In vitro response to recall challenge with CII, DQA1*0103 versus DQ6.1 and FVB, P < 0·04. (e) Cytokines measured from the supernatants of culture in (c). (f) Relative expression of mRNA transcripts of macrophage inhibitory factor (MIF) in splenic cells of transgenic mice. (g) In vitro peptide specific recall response using lymph node cells from mice primed with individual CII‐peptide, and n = 3 to n = 5 mice per strain for all in vitro experiments.
DQA1/CD74 dimers are expressed on cell surface
In vivo data suggested that DQA1 molecules are expressed on the cell surface. Splenic cells from DQA1*0103 mice tested positive with anti‐DQ antibody, SPLV3 and IVD12, but not with L227 while DQB1*0601/A1*0103 mice showed positivity with all the antibodies (Fig. 2a). Next we determined if DQA1*0103 is being expressed on cells as a heterodimer paired with H2M or CD74. All transgenic mice demonstrated positivity with H2M (Fig. 2b). However, 30–40% of splenic cells from DQA1*0103 mice exhibited positive expression for both DQ and CD74 compared with 20–25% of cells in DQ8 and 15–20% in DQ6.1 mice.
Figure 2.
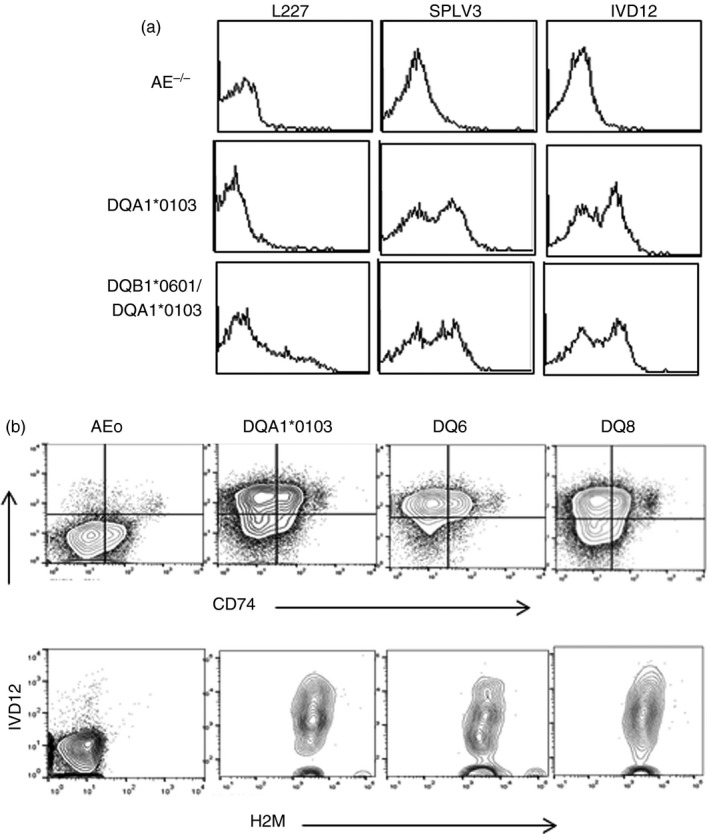
DQA1*0103 and CD74 are co‐expressed on the cell surface. (a) Expression of DQ molecule on splenic cells of transgenic mice. A representative histogram showing expression of the DQ molecule using DQ‐specific antibodies (SPLV3 and IVD12) and DQ6.1‐specific antibody (L227). (b) CD19+ cells were gated from splenic cells of transgenic mice and analysed for expression of DQA1*0103 and CD74 (Top) and DQA1*0103 and H2M (bottom) by FACS and analysed by flowjo software (n = 5 per strain).
DQA1*0103 and CD74 pairing was confirmed with a proximity ligation assay test on the splenic cells of naive transgenic mice as well as confocal microscopy. Very few cells in DQ6.1 mice (< 1%) demonstrated staining for DQA1*0103/CD74, which was mainly intracellular and not on the cell surface (Fig. 3a). On the other hand, DQA1*0103/CD74 dimers were observed in the cytoplasm as well as on the surface of splenic cells in DQA1*0103 mice (3·7% positive cells, DQA1*0103 compared with DQ6.1, P < 0·01). Interestingly, DQ8 mice also showed the presence of DQA1/CD74 dimers, although they were observed mainly in the cytoplasm (2·5% positive cells), with only a few cells expressing it on the cell surface. However, the MFI was significantly higher for DQA1*0103 mice compared with DQ8 and DQ6.1 cells (P = 6·2E‐03 and P = 2·6E‐05, respectively) (Fig. 3b). DQ8 mice exhibited significantly higher MFI for dimers compared with DQ6.1, P < 0·001. These observations suggest that the HLA‐DQA1*0103 chain can be expressed as a dimer with CD74. This is the first time a functional DQA1*0103/CD74 dimer has been described in the pathogenesis of arthritis.
Figure 3.
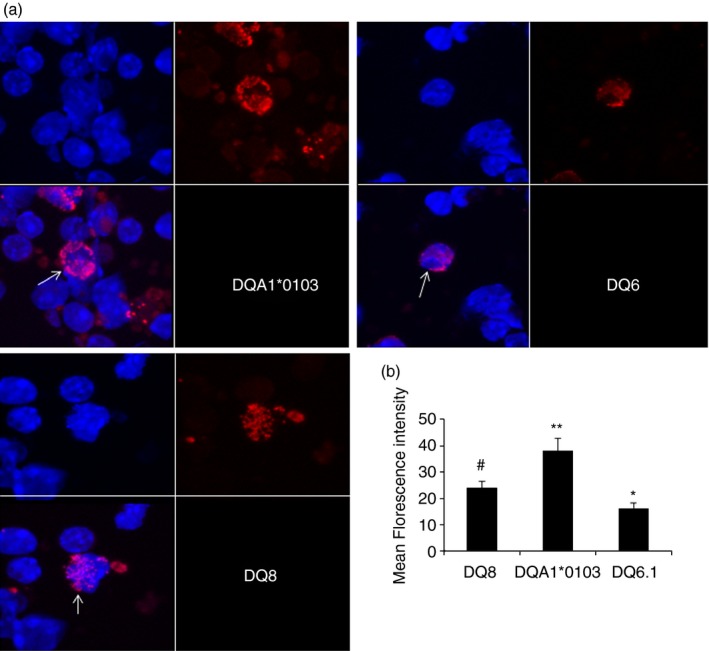
Proximity ligation assay demonstrated DQ and CD74 as a dimer on the cell surface of mice. (a) The slides were visualized by confocal microscopy, blue depicts nuclei staining using DAPI and red is the probe for the proteins; both pictures were merged in the lower left side for each strain. The picture is representative of three experiments, magnification 60×. Arrow points to the cells positive for the CD74/DQA1 heterodimer in each strain. (b) Quantitative analysis of mean fluorescence intensity of the dimers shown in (a). *P < 0·001 DQ8 versus DQ6.1, #DQA1*0103 versus DQ8, P < 0·0006 and **DQA1*0103 versus DQ6.1, P < 2·6E‐05.
Structural binding of CII‐peptide with CD74/DQA1 dimer
The sequences from the extracellular domain of murine CD74 were used to model the CD74/DQA1 pair. The resulting structure was a trimer with two α helices connected by a short β‐strand. The C‐terminal end of CD74 was an extended/flexible structure with kinks because of several proline residues. The DQA1 structure denotes sequences from the α‐chain of human sequence and was similar to DRA1 chain with one helix and a β‐sheet that creates a peptide‐binding groove. The complex of CD74 and DQ molecules was modelled using ZDOCK24 for the monomer as well as trimer of CD74. ZDOCK produced several possible complexes. All the monomer complexes produced were similar in terms of the conformation of the complex; 92% of all residues were in the favoured region and 98% of all residues were in the allowed region of the Ramachandran map. The monomer of CD74 complexes with DQA1 in such a way that one of the helices of CD74 (residues Gly41 to Asn62) occurred nearly parallel with the DQ helix and aligned with β‐sheets of the α‐chain of DQ creating a groove similar to MHC complex. However, because only the DQA1 chain is present for the DQ molecule, all the pockets in the groove are not present in the structure. In the case of the trimer of CD74–DQ complex, the interaction of CD74 and DQ molecules was similar; however, instead of a helical structure aligning the groove, the C‐terminal extended structure aligned with the β‐sheet and helical structure of DQA1 (not shown).
The antigenic CII‐peptide consisting of 12 amino acids, AQGPRGEPGTPG was docked to the CD74 monomer–DQ complex using autodock.27 The peptide was docked with the N‐terminal near the interaction site of CD74‐DQ and the C‐terminal extended into the groove formed by the CD74–DQ molecules (Fig. 4). The peptide formed three hydrogen‐bonding interactions with the complex, namely, the NH of Gly 6 of peptide with Try83 of DQ, side chain of Glu7 with side chain of Lys61 of CD74, and NH of Gly12 of the peptide with Ser63 of CD74. In terms of interaction with pockets in the groove, Gly3 was in the P3 pocket surrounded by Phe128, Trp118 and Phe126 of the DQ chain as well as Trp50 of CD74. This pocket formed a hydrophobic interaction. Major hydrophobic interaction was from pocket P4, where Pro4 of the peptide inserted into the pocket formed by Phe128, Phe107, Tyr97, Leu123 and His99 from DQ β‐sheets and Phe123 from the helical region of DQ. Gly6 of the peptide was in the pocket formed by Try83 of DQ and Phe57 of CD74. The C‐terminal part of the peptide was extended out of the groove and formed interaction with helix of CD74. Overall, the major contribution for the binding was observed by Pro4 at the P4 pocket.
Figure 4.
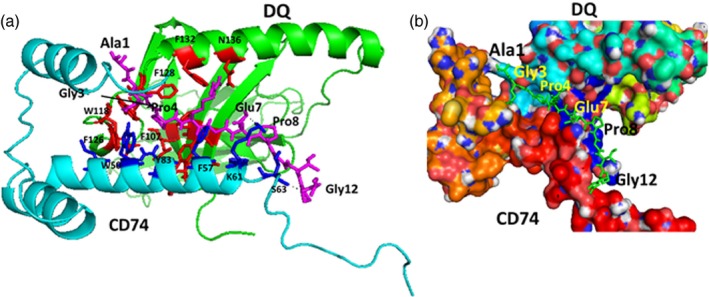
Proposed model of CD74‐DQ interaction with antigen‐binding groove. (a) CD74 is shown in cyan and DQ in green. A 12mer antigenic peptide is shown in the groove as sticks in magenta. Notice the Gly3 and Pro4 in the pocket formed by DQ. The C‐terminal part of the peptide is extended out from the groove and interacts with CD74 helix. Amino acids from DQ that form the pocket are shown as red sticks and those from CD74 are shown as blue sticks. The C‐terminal proline‐rich part of CD74 extends out of the structure of the complex. (b) Surface representation of the proposed model for CD74‐DQ with a 12mer antigenic peptide in the groove shown as sticks (green). The C‐terminal part of the peptide near Gly12 is extended out from the groove and interacts with CD74 helix. Amino acid residues from the antigenic peptide are labelled with a three‐letter code and those from DQ and CD74 are shown with a single‐letter code.
Discussion
Despite the known association between HLA and RA,30 no known mechanism explaining this association exists. Moreover, not all patients carry RA‐associated genes, suggesting a role of other genetic and environmental factors. A role of trans‐heterodimer between two non‐associated DQ alleles suggested a significant but indirect role of the DQA1 gene in CIA in transgenic mice.17, 31 In this study, we show for the first time that the DQA1*0103 gene can cause transgenic mice to develop severe CIA. The possibilities by which DQA1*0103 predisposes to CIA include (i) formation of homodimers capable of presenting antigens, (ii) expression as a heterodimer with H2M that can present antigen, or (iii) DQA1*0103 chains bind CD74 in the cytoplasm but in the absence of the β‐chain, CD74 is not cleaved off and is expressed as a dimer on the cell surface. It is unknown if structurally it is possible for DQA1 molecules to generate a homodimer. Crystallographic analysis of HLA‐DR1 molecules has shown that two α‐chains can form a dimer via a salt bridge between amino acids at positions 88 and 111 and that amino acid 88 in the DRA chain is important for antigen‐specific T‐cell proliferation.32 A similar situation can be speculated with DQA1*0103 molecules, although a crystallographic analysis for such a probability needs to be proven. Our data support the presence and expression of DQA1*0103/CD74 dimer as observed by proximity ligation assay and FACS analysis. One explanation for the HLA association with diseases is the positive and negative selection of T cells in the thymus. An isoform of CD74 is believed to play a key role in T‐cell selection in the thymus.33 Our data showing positive selection of Vb8.2 T cells compared with controls suggests that DQA1/CD74 may be able to select T cells in the thymus.
Together with the genotyping, the data presented here suggest that HLA‐DQA1*0103 chain, in the absence of other MHC class II proteins, can be expressed as a dimer with CD74, adopting a conformation that shares some similarity to α‐chains in αβ heterodimers. Although this study does not directly prove that the CD74/DQA1 dimer molecules predispose mice to arthritis, the lack of endogenous class II molecules and the expression of DQA1*0103/CD74 dimers on the cell surface support the notion. H2M was expressed with DQA1 in all transgenic mice excluding the possibility of its role in pathogenesis. It is possible that DQA1*0103/CD74 heterodimer may be unstable. A highly stable αβ dimer of DQ6 has been suggested to be protective whereas an unstable αβ dimer DQ8 predisposes to RA and diabetes.1, 34, 35, 36 Based on the present and previous17 data, we suggest that DQA1*0103 has a very strong influence on arthritis predisposition. MHC class II constructs have been shown to inhibit MIF/CD74 binding and downstream effects.37 Our data confirm those of the previous study as we observed very low levels of MIF in DQA1*0103 mice.
DQA1*0102, which generally occurs in linkage disequilibrium with arthritis‐resistant DQB1*0604, differs from DQA1*0103 at four positions including a critical position at α22Y. This position has a water‐mediated hydrogen bond to the peptide backbone that is important for peptide stability.38, 39 Allelic variation in DQ6 alleles has been shown to result in change in T‐cell receptor recognition and disease association.4, 40, 41 Hence, DQA1*0102 may not contribute to disease susceptibility as DQA1*0102/DQB1*0604 has not been associated with RA in humans whereas mice expressing DQA1*0103/DQB1*0604 are susceptible to developing arthritis.17 Indeed, an increased occurrence of DQA1*0103 was observed in juvenile idiopathic seropositive arthritis.42 One can speculate that individuals carrying haplotypes with DQA1*0103 may be able to form trans‐heterodimers17 as well as dimers with CD74 and present arthritogenic peptides and generate a pro‐inflammatory response. Anti‐CD74 antibody is in phase II clinical trials for chronic lymphocytic leukaemia43 and is a candidate for B‐cell neoplasms44 and other B‐cell‐dependent diseases. It has been shown to alter migration and proliferation of B cells. B cells are important antigen‐presenting cells and antibody producers in the causation of arthritis, which is underscored by the efficacy of B‐cell depletion therapy in patients.13, 45 DQA1*0103 is also associated with DQB1*05 and DQB1*0603, alleles not associated with RA. It is possible that during an infection or certain conditions, DQA1*0103 molecules can form trans‐heterodimers or are expressed on the cell surface with CD74 molecules, although in normal conditions it is expected that DQA1 will pair with DQB1. This situation is reminiscent of HLA‐B27, which is expressed on the cell surface in a shorter time span compared with other class I molecules.46 Although this allows for clearance of infections, it also probably causes some molecules to misfold or form homodimers that can cause a stress immune response and pathogenesis.47, 48 One can speculate that similar to the observations in transgenic mice, individuals carrying these haplotypes may be able to form trans‐heterodimers with DQB1 as well as dimers with CD74 and present arthritogenic peptides. However, pairing of the DQA1 chain with other molecules, like CD74, may occur occasionally and so the relative risk for these HLA alleles to be considered as susceptible alleles may be low in patients with RA. These speculations need to be proven in future studies. Our data provide an explanation by which non‐associated HLA alleles render susceptibility. We highlight a role of CD74 in arthritis with a speculation of it as a target candidate for treating some patients with RA.
Author contribution
VT was responsible for the concept and design of the animal studies and writing the manuscript; LD, AG and MB performed the experiments; SJ designed and wrote the molecular modelling and docking studies; AJ helped with confocal microscopy and HL was responsible for defining arthritis in mice.
Disclosures
None of the authors declare any financial conflicts.
Acknowledgements
The authors thank Dr Chella David, Mayo Clinic, for HLA transgenic mice and the Louisiana Optical Network Initiative (LONI) for providing the computational facility. This study was supported by NIH grants AR30752 and AI075262 to VT. SJ was supported by an Institutional Development Award (IDeA) from NIGMS grant 8P20GM103424.
References
- 1. Taneja V, Mehra NK, Chandershekaran AN, Ahuja RK, Singh YN, Malaviya AN. HLA‐DR4‐DQw8, but not DR4‐DQw7 haplotypes occur in Indian patients with rheumatoid arthritis. Rheumatol Int 1992; 11:251–5. [DOI] [PubMed] [Google Scholar]
- 2. Newton JL, Harney SM, Wordsworth BP, Brown MA. A review of the MHC genetics of rheumatoid arthritis. Genes Immun 2004; 5:151–7. [DOI] [PubMed] [Google Scholar]
- 3. Taneja V, Mehra NK, Kailash S, Anand C, Malaviya AN. Protective and risk DR phenotypes in Asian Indian patients with rheumatoid arthritis. Indian J Med Res 1992; 96:16–23. [PubMed] [Google Scholar]
- 4. Ettinger RA, Kwok WW. A peptide binding motif for HLA‐DQA1*0102/DQB1*0602, the class II MHC molecule associated with dominant protection in insulin‐dependent diabetes mellitus. J Immunol 1998; 160:2365–73. [PubMed] [Google Scholar]
- 5. Lotteau V, Teyton L, Peleraux A, Nilsson T, Karlsson L, Schmid SL et al Intracellular transport of class II MHC molecules directed by invariant chain. Nature 1990; 348:600–5. [DOI] [PubMed] [Google Scholar]
- 6. Shi X, Leng L, Wang T, Wang W, Du X, Li J et al CD44 is the signaling component of the macrophage migration inhibitory factor–CD74 receptor complex. Immunity 2006; 25:595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Herrero LJ, Sheng KC, Jian P et al Macrophage migration inhibitory factor receptor CD74 mediates alphavirus‐induced arthritis and myositis in murine models of alphavirus infection. Arthritis Rheum 2013; 65:2724–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Behrens M, Smart M, Luckey D, Luthra H, Taneja V. To B or not to B: role of B cells in pathogenesis of arthritis in HLA transgenic mice. J Autoimmun 2011; 37:95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wooley PH, Luthra HS, Stuart JM, David CS. Type II collagen‐induced arthritis in mice. I. Major histocompatibility complex (I region) linkage and antibody correlates. J Exp Med 1981; 154:688–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Taneja V, Behrens M, Mangalam A, Griffiths MM, Luthra HS, David CS. New humanized HLA‐DR4‐transgenic mice that mimic the sex bias of rheumatoid arthritis. Arthritis Rheum 2007; 56:69–78. [DOI] [PubMed] [Google Scholar]
- 11. Taneja V, Taneja N, Paisansinsup T, Behrens M, Griffiths M, Luthra H et al CD4 and CD8 T cells in susceptibility/protection to collagen‐induced arthritis in HLA‐DQ8‐transgenic mice: implications for rheumatoid arthritis. J Immunol 2002; 168:5867–75. [DOI] [PubMed] [Google Scholar]
- 12. Behrens M, Trejo T, Luthra H, Griffiths M, David CS, Taneja V. Mechanism by which HLA‐DR4 regulates sex‐bias of arthritis in humanized mice. J Autoimmun 2010; 35:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Taneja V, Krco CJ, Behrens MD, Luthra HS, Griffiths MM, David CS. B cells are important as antigen presenting cells for induction of MHC‐restricted arthritis in transgenic mice. Mol Immunol 2007; 44:2988–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nabozny GH, Baisch JM, Cheng S, Cosgrove D, Griffiths MM, Luthra HS et al HLA‐DQ8 transgenic mice are highly susceptible to collagen‐induced arthritis: a novel model for human polyarthritis. J Exp Med 1996; 183:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taneja V, Griffiths M, Behrens M, Luthra HS, David CS. Auricular chondritis in NOD.DQ8.Abetao (Ag7–/–) transgenic mice resembles human relapsing polychondritis. J Clin Invest 2003; 112:1843–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bradley DS, Nabozny GH, Cheng S, Zhou P, Griffiths MM, Luthra HS et al HLA‐DQB1 polymorphism determines incidence, onset, and severity of collagen‐induced arthritis in transgenic mice. Implications in human rheumatoid arthritis. J Clin Invest 1997; 100:2227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Behrens M, Papadopoulos GK, Moustakas A, Smart M, Luthra H, David CS et al Trans heterodimer between two non‐arthritis‐associated HLA alleles can predispose to arthritis in humanized mice. Arthritis Rheum 2011; 63:1552–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Griffiths MM, Nabozny GH, Hanson J et al Collagen‐induced arthritis and TCRs in SWR and B10.Q mice expressing an Ek α transgene. J Immunol 1994; 153:2758–68. [PubMed] [Google Scholar]
- 19. Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM et al Protein detection using proximity‐dependent DNA ligation assays. Nat Biotechnol 2002; 20:473–7. [DOI] [PubMed] [Google Scholar]
- 20. Trifilieff P, Rives ML, Urizar E et al Detection of antigen interactions ex vivo by proximity ligation assay: endogenous dopamine D2‐adenosine A2A receptor complexes in the striatum. Biotechniques 2011; 51:111–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS‐MODEL workspace: a web‐based environment for protein structure homology modelling. Bioinformatics 2006; 22:195–201. [DOI] [PubMed] [Google Scholar]
- 22. Jasanoff A, Wagner G, Wiley DC. Structure of a trimeric domain of the MHC class II‐associated chaperonin and targeting protein Ii. EMBO J 1998; 17:6812–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kim CY, Quarsten H, Bergseng E, Khosla C, Sollid LM. Structural basis for HLA‐DQ2‐mediated presentation of gluten epitopes in celiac disease. Proc Natl Acad Sci U S A 2004; 101:4175–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pierce BG, Hourai Y, Weng Z. Accelerating protein docking in ZDOCK using an advanced 3D convolution library. PLoS One 2011; 6:e24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein–protein docking benchmark version 3.0. Proteins 2008; 73:705–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen VB, Arendall WB 3rd, Headd JJ et al MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 2010; 66(Pt 1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS et al AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 2009; 30:2785–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Krco CJ, Pawelski J, Harders J, McCormick D, Griffiths M, Luthra HS et al Characterization of the antigenic structure of human type II collagen. J Immunol 1996; 156:2761–8. [PubMed] [Google Scholar]
- 29. Osman GE, Hannibal MC, Anderson JP, Lasky SR, Ladiges WC, Hood L. FVB/N (H2q) mouse is resistant to arthritis induction and exhibits a genomic deletion of T‐cell receptor Vβ gene segments. Immunogenetics 1999; 49:851–9. [DOI] [PubMed] [Google Scholar]
- 30. Stastny P. Association of the B‐cell alloantigen DRw4 with rheumatoid arthritis. N Engl J Med 1978; 298:869–71. [DOI] [PubMed] [Google Scholar]
- 31. Taneja V, David CS. Role of HLA class II genes in susceptibility/resistance to inflammatory arthritis: studies with humanized mice. Immunol Rev 2010; 233:62–78. [DOI] [PubMed] [Google Scholar]
- 32. Goodman S, Sawada T, Barbosa JA, Cole B, Pergolizzi R, Silver J et al Mutational analysis of two DR α residues involved in dimers of HLA‐DR molecules. J Immunol 1995; 155:1210–7. [PubMed] [Google Scholar]
- 33. Wright RJ, Bikoff EK, Stockinger B. The Ii41 isoform of invariant chain mediates both positive and negative selection events in T‐cell receptor transgenic mice. Immunology 1998; 95:309–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Taneja V, Giphart MJ, Verduijn W, Naipal A, Malaviya AN, Mehra NK. Polymorphism of HLA‐DRB, ‐DQA1, and ‐DQB1 in rheumatoid arthritis in Asian Indians: association with DRB1*0405 and DRB1*1001. Hum Immunol 1996; 46:35–41. [DOI] [PubMed] [Google Scholar]
- 35. Laivoranta‐Nyman S, Mottonen T, Hermann R et al HLA‐DR‐DQ haplotypes and genotypes in Finnish patients with rheumatoid arthritis. Ann Rheum Dis 2004; 63:1406–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sanjeevi CB, Landin‐Olsson M, Kockum I, Dahlquist G, Lernmark A. The combination of several polymorphic amino acid residues in the DQα and DQβ chains forms a domain structure pattern and is associated with insulin‐dependent diabetes mellitus. Ann N Y Acad Sci 2002; 958:362–75. [DOI] [PubMed] [Google Scholar]
- 37. Benedek G, Meza‐Romero R, Andrew S et al Partial MHC class II constructs inhibit MIF/CD74 binding and downstream effects. Eur J Immunol 2013; 43:1309–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sidney J, del Guercio MF, Southwood S, Sette A. The HLA molecules DQA1*0501/B1*0201 and DQA1*0301/B1*0302 share an extensive overlap in peptide binding specificity. J Immunol 2002; 169:5098–108. [DOI] [PubMed] [Google Scholar]
- 39. Godkin A, Friede T, Davenport M, Stevanovic S, Willis A, Jewell D et al Use of eluted peptide sequence data to identify the binding characteristics of peptides to the insulin‐dependent diabetes susceptibility allele HLA‐DQ8 (DQ 3.2). Int Immunol 1997; 9:905–11. [DOI] [PubMed] [Google Scholar]
- 40. Ettinger RA, Papadopoulos GK, Moustakas AK, Nepom GT, Kwok WW. Allelic variation in key peptide‐binding pockets discriminates between closely related diabetes‐protective and diabetes‐susceptible HLA‐DQB1*06 alleles. J Immunol 2006; 176:1988–98. [DOI] [PubMed] [Google Scholar]
- 41. Routsias J, Papadopoulos GK. Polymorphic structural features of modelled HLA‐DQ molecules segregate according to susceptibility or resistance to IDDM. Diabetologia 1995; 38:1251–61. [DOI] [PubMed] [Google Scholar]
- 42. Thomson W, Barrett JH, Donn R et al Juvenile idiopathic arthritis classified by the ILAR criteria: HLA associations in UK patients. Rheumatology (Oxford) 2002; 41:1183–9. [DOI] [PubMed] [Google Scholar]
- 43. Frolich D, Blassfeld D, Reiter K et al The anti‐CD74 humanized monoclonal antibody, milatuzumab, which targets the invariant chain of MHC II complexes, alters B‐cell proliferation, migration, and adhesion molecule expression. Arthritis Res Ther 2012; 14:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Stein R, Mattes MJ, Cardillo TM, Hansen HJ, Chang CH, Burton J et al CD74: a new candidate target for the immunotherapy of B‐cell neoplasms. Clin Cancer Res 2007; 13(Pt 2):5556s–63s. [DOI] [PubMed] [Google Scholar]
- 45. Cohen SB, Emery P, Greenwald MW et al Rituximab for rheumatoid arthritis refractory to anti‐tumor necrosis factor therapy: results of a multicenter, randomized, double‐blind, placebo‐controlled, phase III trial evaluating primary efficacy and safety at twenty‐four weeks. Arthritis Rheum 2006; 54:2793–806. [DOI] [PubMed] [Google Scholar]
- 46. Zernich D, Purcell AW, Macdonald WA et al Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J Exp Med 2004; 200:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Turner MJ, Delay ML, Bai S, Klenk E, Colbert RA. HLA‐B27 up‐regulation causes accumulation of misfolded heavy chains and correlates with the magnitude of the unfolded protein response in transgenic rats: implications for the pathogenesis of spondylarthritis‐like disease. Arthritis Rheum 2007; 56:215–23. [DOI] [PubMed] [Google Scholar]
- 48. Kollnberger S, Bird L, Sun MY, Retiere C, Braud VM, McMichael A et al Cell‐surface expression and immune receptor recognition of HLA‐B27 homodimers. Arthritis Rheum 2002; 46:2972–82. [DOI] [PubMed] [Google Scholar]


