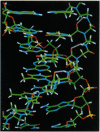
Abstract
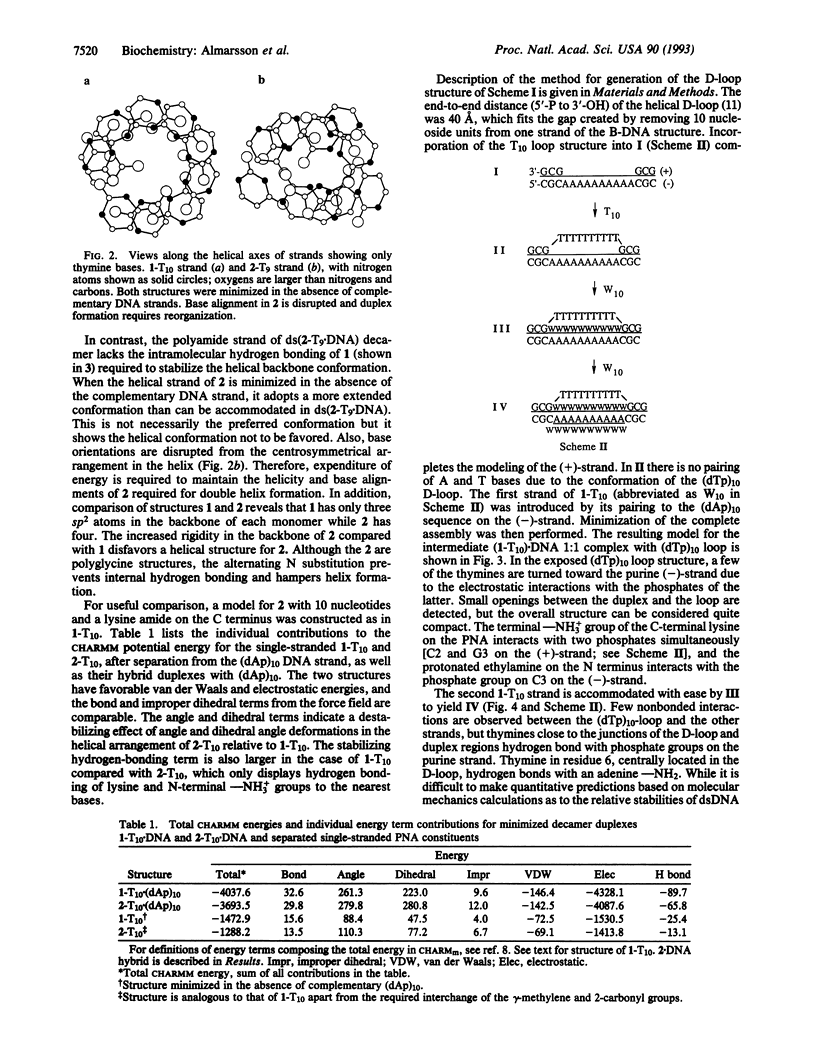
Polyamide nucleic acids (PNAs) have emerged as useful agents for recognition of single- and double-stranded nucleic acids. Interresidue hydrogen bonds between the amide carbonyl nearest the nucleobase and chain NH moieties provide inherent stability to the helical conformation of PNA 1. Moving the amide carbonyl away from the nucleobase to the backbone, and replacing it with a methylene group, results in 2 lacking the stabilizing hydrogen bond. Oligomers of 2 do not interact with DNA. Modeling suggests that 2 displays a more extended conformation than 1, and nucleobase orientation is disrupted in 2 in the absence of a complementary DNA strand. This is in contrast to 1, which retains a centrosymmetric arrangement of nucleobases. Structures for 1-T10.DNA and (1-T10)2.DNA species spanned by a pyrimidine strand (D-loop) were constructed. In the triple helical (1-T10)2.DNA structure, the two PNA strands form the complementary Watson-Crick paired strand and the Hoogsteen base-paired strand in the major groove of the 1.DNA duplex. The PNA strands are proposed to bind antiparallel to one another in (1-T10)2.DNA structure. The factors suggested to account for the stability of this 2:1 complex are (i) a hydrophobic attraction between two PNA backbones and (ii) a favorable electrostatic effect resulting from replacement of a phosphodiester backbone by a neutral peptide backbone.
Full text
PDF
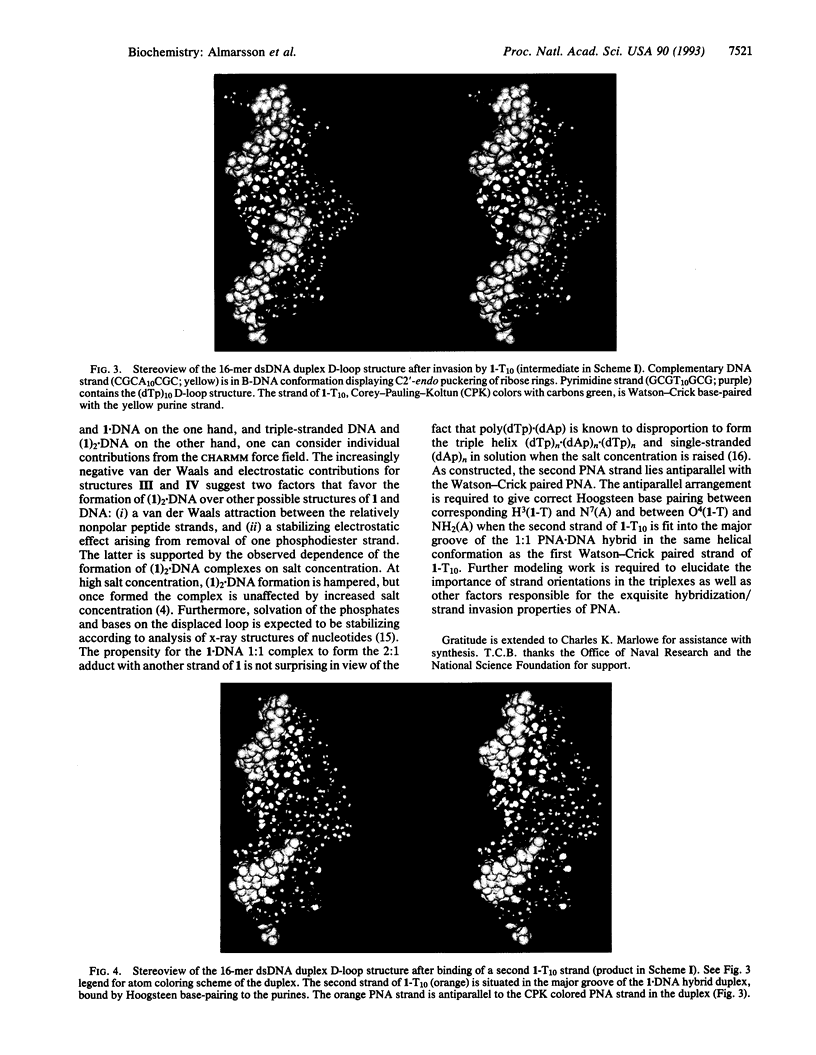

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnott S., Selsing E. The conformation of C-DNA. J Mol Biol. 1975 Oct 15;98(1):265–269. doi: 10.1016/s0022-2836(75)80115-x. [DOI] [PubMed] [Google Scholar]
- Camerman N., Fawcett J. K., Cameran A. Molecular structure of a deoxyribose-dinucleotide, sodium thymidylyl-(5' yields to 3')-thymidylate-(5') hydrate (pTpT), and a possible structural model for polythymidylate. J Mol Biol. 1976 Nov 15;107(4):601–621. doi: 10.1016/s0022-2836(76)80086-1. [DOI] [PubMed] [Google Scholar]
- Hanvey J. C., Peffer N. J., Bisi J. E., Thomson S. A., Cadilla R., Josey J. A., Ricca D. J., Hassman C. F., Bonham M. A., Au K. G. Antisense and antigene properties of peptide nucleic acids. Science. 1992 Nov 27;258(5087):1481–1485. doi: 10.1126/science.1279811. [DOI] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Sequence specific inhibition of DNA restriction enzyme cleavage by PNA. Nucleic Acids Res. 1993 Jan 25;21(2):197–200. doi: 10.1093/nar/21.2.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen P. E., Egholm M., Berg R. H., Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991 Dec 6;254(5037):1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- Raghunathan G., Miles H. T., Sasisekharan V. Symmetry and molecular structure of a DNA triple helix: d(T)n.d(A)n.d(T)n. Biochemistry. 1993 Jan 19;32(2):455–462. doi: 10.1021/bi00053a009. [DOI] [PubMed] [Google Scholar]
- Schneider B., Cohen D., Berman H. M. Hydration of DNA bases: analysis of crystallographic data. Biopolymers. 1992 Jul;32(7):725–750. doi: 10.1002/bip.360320703. [DOI] [PubMed] [Google Scholar]
- Siegel S. A., Lin T. S. Biological activity of two novel inhibitors of uridine phosphorylase. Biochem Pharmacol. 1985 Apr 1;34(7):1121–1124. doi: 10.1016/0006-2952(85)90621-5. [DOI] [PubMed] [Google Scholar]