Abstract
Objective
The assessment of potentially confounding factors affecting colon microbiota composition is essential to the identification of robust microbiome based disease markers. Here, we investigate the link between gut microbiota variation and stool consistency using Bristol Stool Scale classification, which reflects faecal water content and activity, and is considered a proxy for intestinal colon transit time.
Design
Through 16S rDNA Illumina profiling of faecal samples of 53 healthy women, we evaluated associations between microbiome richness, Bacteroidetes:Firmicutes ratio, enterotypes, and genus abundance with self-reported, Bristol Stool Scale-based stool consistency. Each sample’s microbiota growth potential was calculated to test whether transit time acts as a selective force on gut bacterial growth rates.
Results
Stool consistency strongly correlates with all known major microbiome markers. It is negatively correlated with species richness, positively associated to the Bacteroidetes:Firmicutes ratio, and linked to Akkermansia and Methanobrevibacter abundance. Enterotypes are distinctly distributed over the BSS-scores. Based on the correlations between microbiota growth potential and stool consistency scores within both enterotypes, we hypothesise that accelerated transit contributes to colon ecosystem differentiation. While shorter transit times can be linked to increased abundance of fast growing species in Ruminococcaceae-Bacteroides samples, hinting to a washout avoidance strategy of faster replication, this trend is absent in Prevotella-enterotyped individuals. Within this enterotype adherence to host tissue therefore appears to be a more likely bacterial strategy to cope with washout.
Conclusions
The strength of the associations between stool consistency and species richness, enterotypes and community composition emphasises the crucial importance of stool consistency assessment in gut metagenome-wide association studies.
Keywords: INTESTINAL BACTERIA, INTESTINAL MICROBIOLOGY
Significance of this study.
What is already known on this subject?
Stool consistency, measured by the Bristol Stool Scale (BSS), reflects differences in water content and activity in the colon ecosystem and is correlated with colon transit time. (low scores: firm stool and slow transit, high scores: loose stool and fast transit).
Species richness is proposed to be a major marker for gut health because of the idea that high bacterial richness and diversity often reflect ecosystem stability and resilience together with the consistent association between disease and a reduction in the number of species in a faecal sample.
Faecal metagenomes have been shown to stratify into enterotypes, splitting the human population around three possible constellations in terms of their gut microbial ecosystem; likewise Firmicutes:Bacteroidetes and Prevotellaceae:Bacteroidetes ratios have been put forward as important stratifiers for gut microbiomes.
What are the new findings?
Observed species richness declines with higher BSS scores reaching its minimum in individuals with loose stool.
Enterotypes are distinctly distributed over the BSS scores: the Prevotella (P) enterotype is more abundant in subjects with loose stool while the Ruminococcaceae-Bacteroides (RB) enterotype completely dominates the harder stool samples.
Within the RB enterotype, Methanobrevibacter and Akkermansia are positively correlated with colon transit time. A similar observation can be made for Oxalobacter and Butyricimonas, while Bacteroides is more abundant in loose stool.
Microbiota growth potential is positively correlated with BSS scores in the RB enterotype, hinting to transit time as a selective force on microbial life-strategies.
How might it impact on clinical practice in the foreseeable future?
Here we show that major alterations in species richness or community composition could partially be reflecting variation in stool consistency, hampering the identification, but especially robustness and reproducibility of disease markers. Proper confounder analysis is thus impossible without stool consistency measurements. This study therefore stresses the urgent necessity of these measurements in gut microbiota research and clinical studies.
Introduction
Despite recent efforts undertaken to characterise the healthy colon microbiota,1–4 many parameters potentially affecting microbial composition and metabolic capacity remain underexplored. The identification of such confounding factors, including diet,5 6 history of antibiotics intake,7 and inflammation status,8 9 is essential to define the boundaries of a healthy gut ecosystem and, hence, to identify robust disease markers in clinical microbiome association studies.
Transit time is a key determinant of the gut microbial habitat, affecting nutrient and water absorption along the intestine as well as setting the pace of luminal microbial population clearance during egestion.10 As direct measurement of transit time is often impractical or mildly invasive, it has generally been neglected in microbiome studies so far. However, stool consistency as categorised by the Bristol Stool Scale (BSS) scores has been put forward as a proxy for colonic transit rate,10–12 allowing assessment of its impact even when applying home-based sampling protocols. The use of the BSS is widespread in clinical studies and has been advised for the assessment of constipation and diarrhoea in functional bowel disorders.13–15 The BSS classifies human faeces into seven consistency categories, with highest scores corresponding to loose stools and fast transit, while lower scores stand for hard stools and longer colon transit times.10–12 16 Each consistency category reflects differences in moisture content of faecal material, with decreased water activity—associated with prolonged intestinal transit—limiting microbial growth through reduced nutrient mobility and hampered enzymatic activity.17 Hence, BSS categorisation summarises the impact of two major—and obviously related—selective forces shaping the gut ecosystem: rate of intestinal transit and water activity.
Here, we used self-reported BSS scores combined with 16S rDNA Illumina amplicon profiles of faecal samples of 53 healthy volunteers to assess potential associations between microbiota composition and stool consistency. In a healthy Western population, more than half of the stools passed are predicted to belong to the central BSS categories 3 and 4.11 However, increased occurrence of harder stools has been reported to be more common among women.11 12 18 Hence, in order to increase chances of sampling a more uniform distribution of stools over all BSS categories, we limited inclusion to female volunteers.
Methods
Sample data
Fifty-three healthy women, aged 20–55 years (median 42.5), were recruited as part of the Flemish Gut Flora Project. None were diagnosed with cancer or IBD, or had taken diarrhoea inhibitors, laxatives or prebiotics in the week before sampling, nor antibiotics within 3 months before sampling (see online supplementary table S1). The aims of the project and the commitments required were explained by means of an information brochure and all participants signed a statement of informed consent. The participants’ general practitioner recorded their medical history, together with height, weight, and waist and hip circumferences. Volunteers recorded time of defaecation and BSS and reported this information together with information about general diet and health status in a questionnaire.
Characterisation of the bacterial component of the gut microbiota by variable region 4 rDNA sequencing
Faecal samples were frozen at −20°C immediately after collection by the participants. After frozen transport to a collection point in their neighbourhood and subsequent transport on dry ice, samples were stored at −80°C within 72 h after delivery at the collection point until DNA extraction. DNA extraction was performed according to Godon et al.19 To amplify the variable region 4 of the 16S rRNA gene, we used the 515F and 806R primers (GTGCCAGCMGCCGCGGTAA and GGACTACHVGGGTWTCTAAT, respectively) modified to contain Illumina adapters and barcode sequences to allow for directional sequencing. Amplifications were performed in triplicate as 25 μL reactions containing 2 μL of diluted template (1:10), 2.5 μL of 10X AccuPrime PCR Buffer I, 0.1 μL of AccuPrime Taq High Fidelity (5 U/μL), and 0.5 μL of 515F and 1.0 μL of 806R primer (10 μM of each primer). Thermal cycling consisted of an initial denaturation step (3 min at 94°C), followed by 30 cycles of denaturation (45 s at 94°C), annealing (60 s at 50°C) and 90 s extension at 72°C. Final extension consisted of 5 min at 72°C. Amplicons were quantified on the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, California, USA) and pooled in equimolar concentrations. Fragment size was selected (∼400 bp) by cutting the main band from the agarose gel to reduce non-specific products of amplification. A final library size and quantification was also conducted with the Agilent Bioanalyzer. Sequencing was performed on the Illumina MiSeq platform (MiSeq Reagent Kit V.2, 500 cycles) according to the manufacturer's specifications to generate paired-end reads of 250 bases in length in each direction. The overlapping paired-end reads were merged using fastq-join20 and processed with MacQIIME V.1.8. Only Illumina reads with a length >250 bp and an average quality score above 30, were retained for further analysis. Reads were assigned to operational taxonomic units (OTUs) by de novo OTU picking through the QIIME pipeline. This way 99.6%, 87.6%, 53.4% and 12.5% of the reads were assigned to order, family, genus and species levels, respectively. Closed OTU picking with QIIME against the Greengenes 2013 database was performed in addition for a more stringent taxonomic assignment. From OTU abundance and their respective taxonomic classifications, feature abundance matrices were calculated at different taxonomic levels, representing OTU and taxa abundance per sample. To compare the different samples, sample counts were rarefied to 26 260 reads for the de novo OTU picking data set and 26 024 reads for closed OTU picking and trimmed for the consequently absent OTUs with the phyloseq package based on the minimum of the sum of taxa abundances in RV.3.0.1.
Statistical analysis
Statistical analysis was conducted on the rarefied and trimmed feature abundance matrices using RV.3.0.1. Clustering of the samples into enterotypes was done using the cluster package21 according to instructions available on http://enterotyping.embl.de. Optimal number of clusters was determined by the Calinski-Harabasz index. In addition, enterotyping was performed with Dirichlet multinomial mixtures (DMM) using the DirichletMultinomial 1.6.0 package in R.22 Laplace was used to determine the optimal number of clustering by penalising model complexity. Clusters were assigned the enterotype Ruminoccoccaceae-Bacteroides or Prevotella based on the taxa dominating the enterotype as in the original article of Arumugam et al23 (see online supplementary table S2).
Species richness (Observed, Chao1) and α diversity measurements (Shannon) were calculated using the phyloseq package.24
Each sample’s microbiota growth potential was estimated as the average of the maximum growth rates (maxGRs) of the genera, weighted by their abundance in the sample. The maxGRs of gut-reference species (IMG v4Ref) were estimated from genomic traits25 and the median was assigned to the genus.
Correlations between species richness, microbiota growth potential, and relative bacterial taxa abundance with BSS were assessed by Spearman's rank order correlation, as implemented in R. p Values were corrected for multiple testing with Benjamini-Hochberg false discovery rate correction (q value). Spearman's rank order correlation was used to check the correlation between enterotype and transit time (BSS), based on the percentage of each enterotype in each BSS category. Significance of differences in microbiota growth potential between enterotypes was assessed by Wilcoxon signed rank test.
Results
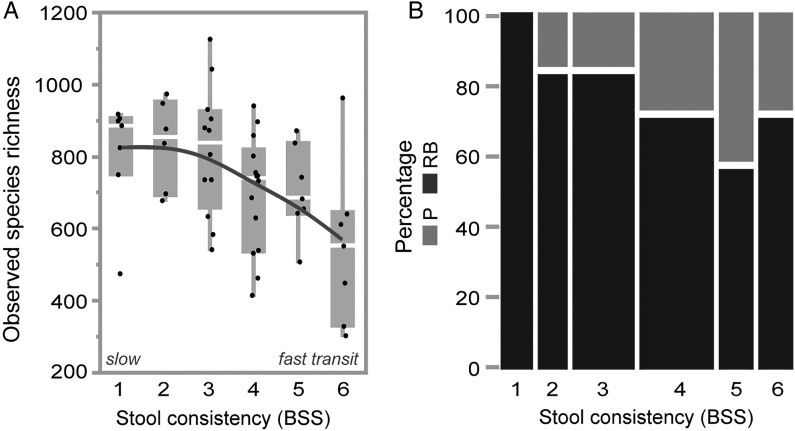
A healthy gut microbial ecosystem is generally thought to be characterised by high bacterial richness and diversity, presumed to reflect ecosystem stability and resilience.26 27 Faecal microbiome analyses have revealed a seemingly consistent association between disease and a reduction in the observed or estimated number of species in a sample, suggesting bacterial richness to be a major marker for gut health.26 27 Here, we identify stool consistency to be strongly associated to faecal microbial richness. Indeed, observed species’ richness significantly declines with stool firmness (Spearman's r=−0.45, p=0.0007), reaching its minimum in diarrhoea-afflicted individuals (figure 1A). Estimation of total species richness (Chao1) confirms this negative correlation (r=−0.41, p=0.003, data not shown) and the signal remains significant with the application of a more stringent taxonomical assignment (closed reference OTU picking, see Methods; trend also confirmed in a data set excluding patients with IBS, online supplementary figure S7, and in a data set of 24 men, online supplementary figure S8). These results are in line with anecdotal reports of decreased microbial richness associated with osmotic diarrhoea.28
Figure 1.
Stool consistency variation drives species richness and human enterotypes. Correlation between (A) observed species richness and stool consistency, defined by Bristol Stool Scale (BSS) (Spearman's correlation, r=−0.45, p=0.0007) and (B) enterotype distribution and stool consistency (BSS); Blue: Ruminococcaceae-Bacteroides (RB) enterotype (r=−0.88, p=0.019), green: P enterotype (r=0.88, p=0.019).
Previously, faecal metagenomes have been shown to stratify into enterotypes,23 splitting the human population around three possible constellations in terms of their gut microbial community structure. As enterotypes have also been linked to richness gradients,8 9 we analysed the distribution of enterotypes over stool consistency scores. Applying multiple clustering approaches, we find that our current data set optimally separates into two distinct clusters (see online supplementary table S2). Named after the dominating taxa as in the reference publication,23 we refer to the clusters observed as the Ruminococcaceae-Bacteroides (RB) and the Prevotella (P) enterotype (77% and 23% of total samples, respectively; partitioning around medoids (PAM) clustering with Jensen-Shannon distance). The enterotypes observed are distinctly distributed over BSS scores: while the P enterotype is more abundant in individuals with loose stools (r: 0.88, p=0.019), the RB enterotype completely dominates firmer samples (r: −0.88, p=0.019; figure 1B; trend also confirmed in men, online supplementary figure S8). These observations are substantiated by the analysis of the Firmicutes:Bacteroidetes and Prevotellaceae:Bacteroidetes abundance ratios (both put forward as alternatives to enterotyping29) over stool score (respectively r=0.42, p=0.001 and r=−0.04, p=0.77; see online supplementary figure S1A, B). Of note, within the RB enterotype, Ruminococcaceae abundance positively correlates with BSS scores (r=−0.37, p=0.016), while Bacteroides populations increase in looser stools (r=0.43, p=0.004) (see online supplementary figure S2). Hence, the enhanced prevalence of the RB enterotype in the harder stool categories is driven by the samples with a high Ruminococcaceae:Bacteroides ratio, which would classify them as Ruminococcaceae enterotype according to the terminology originally suggested by Arumugam et al.23 As even within a single enterotype low richness samples are more abundant in looser stool (see online supplementary figure S3), the richness signal is thus not a mere consequence of enterotype distribution across consistency scores. Although the identification of environmental factors shaping or contributing to enterotype stratification is still ongoing, the potentially diverging impact of diet has been emphasised previously.5 30 More specifically, the occurrence of the P enterotype has been linked to a rural, fibre-rich diet.5 31 As non-fermentable fibre consumption is thought to increase water content and plasticity of stool,32 this observation fits with the increased prevalence of the P enterotype in the loose stool categories. However, the question whether dietary fibre consumption or the resulting changes in transit rate or water activity are driving Prevotella blooms, remains currently unanswered.
Stool consistency variation is associated with shifts in faecal microbiota composition at the level of community structure and diversity, and it correlates with abundance gradients of individual genera (see online supplementary table S3). Within the RB enterotype, Methanobrevibacter and Akkermansia populations increase with stool firmness (and are thus more prevalent in slow transit individuals). A similar observation can be made for Oxalobacter and Butyricimonas, while Bacteroides is more abundant in loose stools (see online supplementary figure S4). Most of these associations remain significant in the total data set (see table 1 and online supplementary figure S5) or when applying a more stringent taxonomic assignment (see online supplementary table S4).
Table 1.
Genera abundances significantly correlated with stool consistency
| Genus | Total data set | RB enterotype | P enterotype | |||
|---|---|---|---|---|---|---|
| r | q Value | r | q Value | r | q Value | |
| Akkermansia | −0.504 | 0.0722 | −0.528 | 0.0342 | −0.078 | 0.6696 |
| Bacteroides | 0.177 | 0.6134 | 0.460 | 0.0718 | −0.048 | 0.7803 |
| Butyricimonas | −0.348 | 0.0722 | −0.406 | 0.0718 | −0.264 | 0.8620 |
| Methanobrevibacter | −0.126 | 0.0722 | −0.095 | 0.0718 | −0.134 | 0.7978 |
| Methanosphaera | −0.305 | 0.0966 | −0.307 | 0.1318 | NA | NA |
| Odoribacter | −0.094 | 0.0966 | −0.048 | 0.2232 | −0.284 | 0.8551 |
| Oxalobacter | −0.424 | 0.0722 | −0.460 | 0.0350 | −0.051 | 0.6127 |
Genera abundances significantly correlated with stool consistency (BSS) (q<0.1) in the total data set, the RB enterotype, or the P enterotype. Spearman's rank order correlation with Benjamini-Hochberg false discovery rate correction.
BSS, Bristol Stool Scale; NA, not assigned; RB, Ruminococcaceae-Bacteroides.
The increased abundance of methanogens such as Methanobrevibacter in harder stools confirms previous reports of elevated methane production in constipated individuals.33 It has been suggested that methane plays an active role in the delay of transit by slowing down intestinal motility.34 Moreover, hydrogen removal through methane production alters the whole of gut fermentation processes, which could potentially affect colon peristalsis.33 Alternatively, increased abundance could reflect the fitness of a genus to grow in conditions of slowed-down intestinal transit. As firmer stool consistencies correspond with reduced ecosystem water activity, associated fluctuations in microbial abundances could also result from species-specific resistance to water stress.
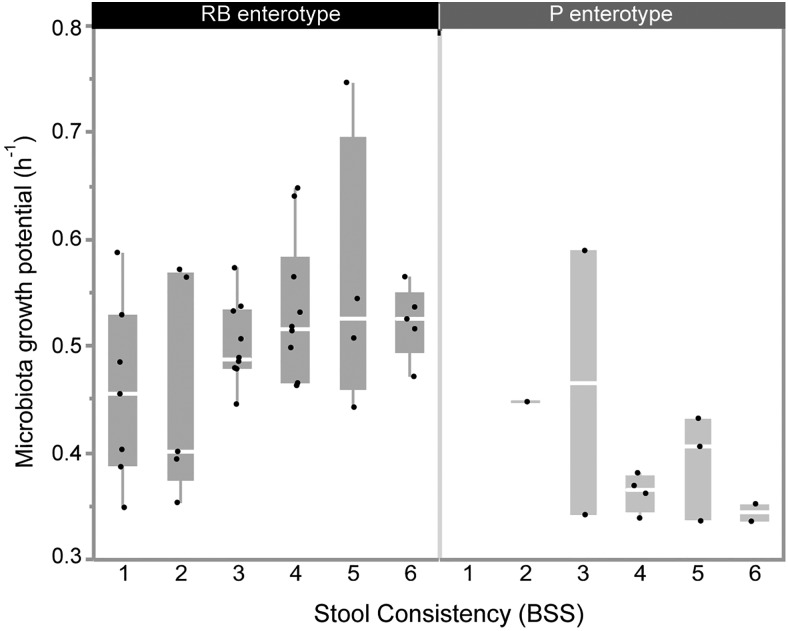
The human colon ecosystem is an open, nutrient-rich and continuous-flow environment. In order to avoid washout, the residing bacteria can either reproduce at a sufficiently high growth rate or attach to or colonise host tissues.35 Hence, transit time may act as a selective force on gut bacterial growth rates. Indeed, higher fluctuations in nutrient availability and microbiota population size induced by decreased colon transit time would be the text-book selection pressure for fast growing bacteria, outgrowing their slow growing counterparts whenever resources are available (r-selection).36 Also enhanced water activity and the associated increase of nutrient mobility in loose stools will contribute to the selective force imposed by accelerated transit. Indications for the suggested correlation between bacterial growth rates and rates of passage come from in vitro work with continuous flow fermenters37–39—where dilution rates determine pace of growth of the bacteria cultured—and in vivo observations of higher faecal bacterial biomass associated with shorter transit times.40 41 To assess this hypothesis, we estimated each sample’s microbiota growth potential as the average of the maxGR25 (see methods) of the genera, weighted by their abundance in the sample (based on de novo OTU as well as closed reference OTU assignment; online supplementary table S5). We find that microbiota growth potential is indeed positively correlated with stool score, and hence colon transit time, in the RB enterotype (r=0.34, p=0,028; figure 2). However, this correlation is absent in the P enterotype (figure 2). Strikingly, the average growth rate of the P enterotype samples is lower than the one of the RB samples (median microbiota growth potential 0.38/h vs 0.52/h, respectively, Wilcoxon p<10−4; online supplementary figure S6). This is largely driven by the higher abundance of relatively slow-growing Prevotella spp in these samples (median relative abundance of Prevotella of 0.48671 vs 0.00026 in the P-enterotype and RB-enterotype, respectively). On one hand, the absence of correlation between growth rate and colon transit time in the P enterotype could suggest that species in this enterotype resort to an alternative strategy to avoid washout, namely a higher degree of adherence to host tissues. The fact that Prevotella is indeed able to bind collagen and degrade mucin oligosaccharides support this hypothesis.42 43 Alternatively, our results could indicate that stool consistency and hence water activity in Prevotella individuals are independent of accelerated transit, and mainly reflect increased faecal water-binding capacity, for example, related to fibre consumption. Overall, as faeces and library dilution series have been shown not to affect compositional readouts in 16S amplicon analyses44—and the results obtained are thus not due to technical issues arising from the variation in water content—our results hint to transit time as one of the determining selective forces on microbial life-strategies.
Figure 2.
Microbiota growth potential correlates to faster intestinal transit in the Ruminococcaceae-Bacteroides (RB) enterotype. Microbiota growth potential over stool consistency (Bristol Stool Scale (BSS)), which is proposed as a proxy for transit time, in the RB-enterotype (r=0.34, p=0.028).
The results of the present study indicate that major alterations in species richness or community composition could partially be reflecting variation in stool consistency, hampering the identification, and affecting robustness and reproducibility of disease markers. While constipation and or diarrhoea are often seen as indicators of a dysbiotic colon microbiota potentially contributing to disease symptoms, onset or evolvement, stool consistency or transit time are not always taken into account as confounders. We illustrate this concern using a recent study by Scheperjans et al45 evaluating the role of the microbiota in Parkinson's disease. The authors detected a lower abundance of Prevotellaceae combined with an increased Ruminococcaceae population in patients compared with healthy controls. However, as constipation has been reported as an early symptom of Parkinson’s disease46 and given the overlap between the microbiome signal observed and our findings of BSS-associated fluctuations in microbial abundances, there might be a risk that the microbiota signature proposed is a mere consequence of stool consistency differences between healthy and diseased individuals. To their credit, the authors did include the Wexner constipation score in the study—yet this score is primarily focused on long-term assessment of severe constipation (ie, extreme BSS 1) and would not compensate for day-to-day variation in BSS in patients as well as controls.47 Inclusion of stool consistency records or colon transit time measurements as a confounder would have allowed better disentangling of signals exclusively associated to the disease and those associated to stool consistency.
Conclusion
Together, our results demonstrate a profound association between stool consistency and all major readouts of gut microbiota composition. As increased colon transit time has been linked to enhanced proteolytic fermentation48–50 and associated production of potentially deleterious metabolites, the observed negative correlation between BSS-assessed passage rate and microbiome richness as reported here challenges the currently dominating view of high richness being directly associated to host health. Furthermore, we here identify gut microbial compositional differences with stool consistency on community scale and genus level and evaluate a hypothesised biological mechanism of how transit time might shape the gut microbiota through selective pressure on microbial life-strategies. Although longitudinal studies combining stool score records and direct transit time measurements are necessary to consolidate the observed correlations and proposed hypotheses, the strength of the associations between BSS and species richness, enterotypes and community composition emphasise the crucial importance of stool consistency assessment and confounder analysis in gut microbiota research and clinical studies.
Supplementary Material
Acknowledgments
The authors thank two anonymous reviewers for their constructive comments on an earlier version of this manuscript as well as members of the Raes lab for lively discussions and feedback.
Footnotes
Twitter: Follow Marie Joossens at @JoossensM
Contributors: DV participated in data collection, performed the analyses and wrote the paper. GF designed the study, collected the data and participated in data analysis and writing of the paper. SVS participated in data analysis and writing of the paper. RT performed data preprocessing. MJ collected the data, participated in study design and writing of the paper. JR participated in study design and writing of the paper and was responsible for overall study coordination.
Funding: Fund for Scientific Research Flanders (FWO), KU Leuven, Marie Curie Actions (FP7 People COFUND—Proposal 267139), Flemish government (IWT130359), Agency for Innovation by Science and Technology (IWT), Metacardis (FP7 METACARDIS HEALTH-F4-2012-305312), Rega institute for Medical Research.
Competing interests: None declared.
Ethics approval: The Research Ethics Committee of the Vrije Universiteit Brussel (BUN 143201215505).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1.Qin J, Li R, Raes J, et al. . A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010;464:59–65. 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gevers D, Knight R, Petrosino JF, et al. . The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol 2012;10:e1001377 10.1371/journal.pbio.1001377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costello EK, Lauber CL, Hamady M, et al. . Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–7. 10.1126/science.1177486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J, de Vos WM, DiStefano PS, et al. . Translating the human microbiome. Nat Biotechnol 2013;31:304–8. 10.1038/nbt.2543 [DOI] [PubMed] [Google Scholar]
- 5.Wu GD, Chen J, Hoffmann C, et al. . Linking long-term dietary patterns with gut microbial enterotypes. Science 2011;334:105–8. 10.1126/science.1208344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David LA, Maurice CF, Carmody RN, et al. . Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014;505:559–63. 10.1038/nature12820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho I, Yamanishi S, Cox L, et al. . Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012;488:621–6. 10.1038/nature11400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Chatelier E, Nielsen T, Qin J, et al. . Richness of human gut microbiome correlates with metabolic markers. Nature 2013;500:541–6. 10.1038/nature12506 [DOI] [PubMed] [Google Scholar]
- 9.Hildebrand F, Nguyen TLA, Brinkman B, et al. . Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 2013;14:R4 10.1186/gb-2013-14-1-r4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis SJ, Heaton KW. Stool form scale as a useful guide to intestinal transit time. Scand J Gastroenterol 1997;32:920–4. 10.3109/00365529709011203 [DOI] [PubMed] [Google Scholar]
- 11.Heaton KW, Radvan J, Cripps H, et al. . Defecation frequency and timing, and stool form in the general population: a prospective study. Gut 1992;33:818–24. 10.1136/gut.33.6.818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Degen LP, Phillips SF. How well does stool form reflect colonic transit? Gut 1996;39:109–13. 10.1136/gut.39.1.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Longstreth GF, Thompson WG, Chey WD, et al. . Functional Bowel Disorders. Gastroenterology 2006;130:1480–91. 10.1053/j.gastro.2005.11.061 [DOI] [PubMed] [Google Scholar]
- 14.Videlock EJ, Cheng V, Cremonini F. Effects of linaclotide in patients with irritable bowel syndrome with constipation or chronic constipation: a meta-analysis. Clin Gastroenterol Hepatol 2013;11:1084–92. 10.1016/j.cgh.2013.04.032 [DOI] [PubMed] [Google Scholar]
- 15.Collado Yurrita L, San Mauro Martín I, Ciudad-Cabañas MJ, et al. . Effectiveness of inulin intake on indicators of chronic constipation; a meta-analysis of controlled randomized clinical trials. Nutr Hosp 2014;30:244–52. 10.3305/nh.2014.30.2.7565 [DOI] [PubMed] [Google Scholar]
- 16.Saad RJ, Rao SSC, Koch KL, et al. . Do stool form and frequency correlate with whole-gut and colonic transit? Results from a multicenter study in constipated individuals and healthy controls. Am J Gastroenterol 2010;105:403–11. 10.1038/ajg.2009.612 [DOI] [PubMed] [Google Scholar]
- 17.Schiraldi A, Fessas D, Signorelli M. Water activity in biological systems—a review. Polish J Food Nutr Sci 2012;62:5–13. 10.2478/v10222-011-0033-5 [DOI] [Google Scholar]
- 18.Degen LP, Phillips SF. Variability of gastrointestinal transit in healthy women and men. Gut 1996;39:299–305. 10.1136/gut.39.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godon JJ, Zumstein E, Dabert P, et al. . Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol 1997;63:2802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aronesty E. Comparison of Sequencing Utility Programs. Open Bioinforma J 2013;7:1–8. 10.2174/1875036201307010001 [DOI] [Google Scholar]
- 21.Maechler AM, Analysis DC, Struyf A. cluster: Cluster Analysis Basics and Extensions. R package version 1.14.4 2013.
- 22.Holmes I, Harris K, Quince C. Dirichlet multinomial mixtures: generative models for microbial metagenomics. PLoS ONE 2012;7:e30126 10.1371/journal.pone.0030126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arumugam M, Raes J, Pelletier E, et al. . Enterotypes of the human gut microbiome. Nature 2011;473:174–80. 10.1038/nature09944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013;8:e61217 10.1371/journal.pone.0061217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vieira-Silva S, Rocha EPC. The systemic imprint of growth and its uses in ecological (meta)genomics. PLoS Genet 2010;6:e1000808 10.1371/journal.pgen.1000808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone CA, Stombaugh JI, Gordon JI, et al. . Diversity, stability and resilience of the human gut microbiota. Nature 2012;489:220–30. 10.1038/nature11550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet 2012;13:260–70. 10.1038/nrg3182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gorkiewicz G, Thallinger G, Trajanoski S. Alterations in the colonic microbiota in response to osmotic diarrhea. PLoS ONE 2013;8:e55817 10.1371/journal.pone.0055817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yatsunenko T, Rey FE, Manary MJ, et al. . Human gut microbiome viewed across age and geography. Nature 2012;486:222–7. 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salonen A, de Vos WM. Impact of diet on human intestinal microbiota and health. Annu Rev Food Sci Technol 2014;5:239–62. 10.1146/annurev-food-030212-182554 [DOI] [PubMed] [Google Scholar]
- 31.De Filippo C, Cavalieri D, Di Paola M, et al. . Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–6. 10.1073/pnas.1005963107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mudgil D, Barak S. Composition, properties and health benefits of indigestible carbohydrate polymers as dietary fiber: a review. Int J Biol Macromol 2013;61:1–6. 10.1016/j.ijbiomac.2013.06.044 [DOI] [PubMed] [Google Scholar]
- 33.Sahakian AB, Jee S-R, Pimentel M. Methane and the gastrointestinal tract. Dig Dis Sci 2010;55:2135–43. 10.1007/s10620-009-1012-0 [DOI] [PubMed] [Google Scholar]
- 34.Pimentel M, Lin HC, Enayati P, et al. . Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol 2006;290:1089–95. 10.1152/ajpgi.00574.2004 [DOI] [PubMed] [Google Scholar]
- 35.Flint H, Duncan S, Scott KP, et al. . Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol 2007;9:1101–11. 10.1111/j.1462-2920.2007.01281.x [DOI] [PubMed] [Google Scholar]
- 36.Koch a L. Oligotrophs versus copiotrophs. Bioessays 2001;23:657–61. 10.1002/bies.1091 [DOI] [PubMed] [Google Scholar]
- 37.James TW. Continous culture of microorganisms. Annu Rev Microbiol 1960:27–46.
- 38.Hobson PN. Continuous culture of some anerobic and facultatively anaerobic rumen bacteria. J Gen Microbiol 1965;38:167–80. 10.1099/00221287-38-2-167 [DOI] [PubMed] [Google Scholar]
- 39.Isaacson HR, Hinds FC, Bryant MP, et al. . Efficiency of energy utilization by mixed rumen bacteria in continuous culture. J Dairy Sci 1975;58:1645–59. 10.3168/jds.S0022-0302(75)84763-1 [DOI] [PubMed] [Google Scholar]
- 40.Stephen a M, Wiggins HS, Cummings JH. Effect of changing transit time on colonic microbial metabolism in man. Gut 1987;28:601–9. 10.1136/gut.28.5.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stephen M, Cummings JH. The microbial contribution to human fecal mass. J Med Microbiol 1980;13:45–56. 10.1099/00222615-13-1-45 [DOI] [PubMed] [Google Scholar]
- 42.Grenier D. Collagen-binding activity of Prevotella intermedia measured by a microtitre plate adherence assay. Microbiology 1996;142:1537–41. 10.1099/13500872-142-6-1537 [DOI] [PubMed] [Google Scholar]
- 43.Wright D, Rosendale D, Robertson A. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol Lett 2000;190:73–9. 10.1111/j.1574-6968.2000.tb09265.x [DOI] [PubMed] [Google Scholar]
- 44.Santiago A, Panda S, Mengels G, et al. . Processing faecal samples: a step forward for standards in microbial community analysis. BMC Microbiol 2014;14:112 10.1186/1471-2180-14-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheperjans F, Aho V, Pereira P a. B, et al. . Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord 2014;30:350–8. 10.1002/mds.26069 [DOI] [PubMed] [Google Scholar]
- 46.Kaye J, Gage H, Kimber A, et al. . Excess burden of constipation in Parkinson's disease: a pilot study. Mov Disord 2006;21:1270–3. 10.1002/mds.20942 [DOI] [PubMed] [Google Scholar]
- 47.Agachan F, Chen T, Pfeifer J, et al. . A constipation scoring system to simplify evaluation and management of constipated patients. Dis Colon Rectum 1996;39:681–5. 10.1007/BF02056950 [DOI] [PubMed] [Google Scholar]
- 48.Lewis SJ, Heaton KW. Increasing butyrate concentration in the distal colon by accelerating intestinal transit. Gut 1997;41:245–51. 10.1136/gut.41.2.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lewis SJ, Heaton KW. The metabolic consequences of slow colonic transit. Am J Gastroenterol 1999;94:2010–16. 10.1111/j.1572-0241.1999.01271.x [DOI] [PubMed] [Google Scholar]
- 50.Burkitt D, Walker A, Painter N. Effect of dietary fibre on stools and transit times and its role in the causation of disease. Lancet 1972;300:1408–11. 10.1016/S0140-6736(72)92974-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.