Abstract
Background:
Escherichia coli (E. coli) is the most extensively used host for the production of recombinant proteins. However, most of the eukaryotic proteins are typically obtained as insoluble, misfolded inclusion bodies that need solubilization and refolding. Reteplase as a highly disulfide-bonded recombinant protein is an example of difficult to express protein in E. coli.
Methods:
In this study, a codon optimized reteplase gene was synthetically prepared and cloned under the control of an IPTG inducible T7 promoter. The vector was simultaneously transformed and expressed in three different E. coli strains. The ability of strains for expression of this recombinant pharmaceutical was compared. Also, an attempt was made to increase the soluble production of reteplase in SHuffle T7 E. coli with alterations of expression condition like temperature, inducer concentration and oxygen supply.
Results:
High amounts of reteplase were expressed as inclusion bodies in all three strains. BL21 (DE3) showed the highest level of expression in inclusion bodies followed by Rosetta-gami (DE3) and Shuffle T7. Changes of expression conditions were insufficient for soluble expression of reteplase in SHuffle T7 as a genetically engineered host for production of disulfide bonded proteins.
Conclusion:
The oxidizing cytoplasm of Rosetta-gami and Shuffle T7 in addition to alterations of cultivation parameters could not result in soluble production of reteplase, although the inclusion bodies produced in these two strains might increase the rate of refolding procedure likely due to formation of folding intermediates.
Keywords: Escherichia coli, Recombinant proteins, Reteplase, Tissue plasminogen activator
Introduction
Thrombolysis is the first choice of therapy for Acute Myocardial Infarction (AMI) 1. Tissue Plasminogen Activator (tPA) which derives from fibrinolytic system of blood vessel endothelial cells is a thrombolytic agent that activates plasminogen to form plasmin 2,3. Native tPA is a 70 kDa serine protease composed of 527 amino acid residues with five structural domains and 17 disulfide bonds 2,4.
Reteplase is a recombinant non-glycosylated form of human tPA produced in Escherichia coli (E. coli). It has been modified to contain 355 (1–3 and 176–527) of the 527 amino acids of the original protein with a total molecular mass of 39 kDa. This deleted variant of tPA has lost three structural domains of Kringle I, Finger, and EGF, while retaining the thrombolytic Kringle II and protease domains and contains 9 disulfide bonds. However, because of the deletion of three structural domains as well as the carbohydrate side chains, the plasma half-life of this mutated variant is increased to 13–16 min when compared to full length tPA 5–7.
E. coli is by far the most widely used host organism for biopharmaceutical production of heterologous recombinant proteins. E. coli expression platform is favored due to its ability to quickly reach high cell densities, simplicity, low cost and Food and Drug Administration (FDA) approved status for human applications 8,9. However, production of reteplase in E. coli remains challenging. Presence of several disulfide bonds, rare codon usage and cytotoxicity are the major drawbacks of reteplase expression in E. coli 9,10. Here, a synthetic reteplase gene optimized for expression in E. coli was prepared and its expression was induced under the control of strong T7 promoter in BL21 (DE3), Rosetta-gami (DE3) and SHuffle T7 strains. Also, an attempt was made to compare the ability of the three different strains in the T7-based production of this recombinant pharmaceutical.
Several research groups have also reported the expression of reteplase in E. coli strains, resulting in the formation of inactive inclusion bodies which need to be renatured to gain biological activity. Since there are 9 disulfide bonds, renaturation of reteplase is very difficult with low efficiency 11–14. Furthermore, the refolding procedures may not fully restore the native protein fold and may reduce recombinant protein function 15,16. At the same time, there are few reports which claim soluble production of reteplase in E. coli. For instance, display of reteplase on the surface of polyhydroxybutyrate granule using phasin as the affinity tag prevented the aggregation of this protein but the final protein needed a thrombin cleavage step for isolation of reteplase 2. Also, translocation of reteplase to priplasmic region of E. coli Top10 under the control of an arabinose promoter has produced negligible amounts of soluble protein 13. Since these methods are inefficient for high level production of soluble reteplase, still refolding of inclusion bodies is the method of choice.
Therefore, in this study, common approaches were used to increase the yield of soluble and active reteplase production in SHuffle T7 as a genetically engineered E. coli which is suitable for production of disulfide-bonded proteins 17. Moreover, the effect of isopropyl-beta-D-thiogalactopyranoside (IPTG) concentrations, temperature of expression and oxygen supply were investigated in order to find a way to produce remarkable amounts of biologically active and soluble form of reteplase in E. coli.
Materials and Methods
Bacterial strains and plasmids
E. coli DH5α was used as the cloning host for propagation of expression vector. Three different E. coli expression strains BL21(DE3), Rosetta-gami and SHuffle T7 were prepared and transformed using the standard protocols.
pET-21a was used as the expression vector. The gene coding for reteplase was synthesized by Bio-basic (Canada). Restriction enzymes and T4 DNA ligase were from Fermentas. Taq DNA polymerase master mix was obtained from Ampliqon. Pfu DNA polymerase was purchased from iNtron. Complete protease inhibitor cocktail tablets were purchased from Roche. All chemicals were purchased from Sigma and Merck.
Construction of the expression vector
The gene coding for reteplase was synthetically prepared with codon preference of E. coli in pUC57 plasmid. The gene was amplified using the primers ret-F 5′ CATATGTCTTACCAGGGTAACAGC 3′ and ret-R 5′ AAGCTTCGGGCGCATGTTATCGC 3′. The forward and reverse primers contained NdeI and HindIII recognition sites, respectively. PCR was performed with the following conditions: denaturation at 94°C for 5 min, 30 cycles of 95°C for 45 s, 66°C for 45 s and 72°C for 1 min followed by a final extension of 10 min at 72°C. The PCR product was double digested with NdeI and HindIII restriction enzymes and subsequently cloned in pET-21a previously digested with the same enzymes. The ligation mixture was transformed into E. coli DH5α and selected on LB agar medium containing 100 μg/ml of ampicillin. The authenticity of the cloning procedure was confirmed by sequencing. The resulting plasmid was named pET-ret.
Expression of reteplase in different E. coli hosts
The pET-ret plasmid was transformed into chemically competent expression hosts. Single colonies from transformed cells were used for inoculation of 5 ml pre-culture medium containing ampicillin. The cultures were incubated for 12 hr at 37°C with the shake of 180 rpm. 200 μl of the pre-cultures were used for inoculation of 20 ml of LB medium at 37°C until reaching the OD600 of 0.8. For protein expression, IPTG was added to the final concentration of 1 mM. Expression was continued at 37°C for 20 hr and cells were harvested within desired intervals. Quantification of protein expression was carried out by image analysis of the SDS-PAGE scans using ImageJ software.
Determination of cultivation parameters
For temperature optimization, cells were grown at 37°C until reaching the desired turbidity of 0.8, then the temperature was reduced to 25°C and 18°C and cells were incubated for 30 min. After reaching the desired temperature, IPTG was added and expression was continued for 4 hr.
For optimization of IPTG concentration, two different concentrations of inducer were used. IPTG was added to the final concentration of 1 mM and 0.1 mM after reaching the optimal density of bacterial growth. The expression was continued for four hours and samples were withdrawn with one hour intervals.
In order to find the effect of oxygen supply, two different shaking speeds were checked. After induction of expression with IPTG, the shaking speed of incubator was fixed to 180 or 120 rpm. All experiments were carried out in duplicates.
SDS-PAGE analysis
For analysis of reteplase production, cells were harvested by centrifugation at 5000 rpm for 5 min. The bacterial pellets were resuspended in lysis buffer containing 1X protease inhibitor cocktail and disrupted by 5 cycles of sonication (30 s) with 10 s of rest interval. The whole steps were carried out on ice. The supernatant was separated from cell debris by centrifugation at 13000 rpm for 30 min at 4°C. Both fractions were analyzed on 5% stacking, 10% resolving SDS-PAGE. The gels were stained with Coomassie Blue R-250 dye for 2 hr and destained overnight.
Results
Construction of the expression vector
The reteplase gene was cloned in NdeI and HindIII recognition sites of pET-21a, therefore located under the control of IPTG inducible T7 promoter. The constructed pET-ret plasmid was extracted from E. coli DH5α cells and cloning was confirmed by PCR. The 1065 bp fragment of reteplase gene was successfully amplified using pET-ret plasmid (Figure 1). Sequencing additionally proved the authenticity of cloning procedure.
Figure 1.
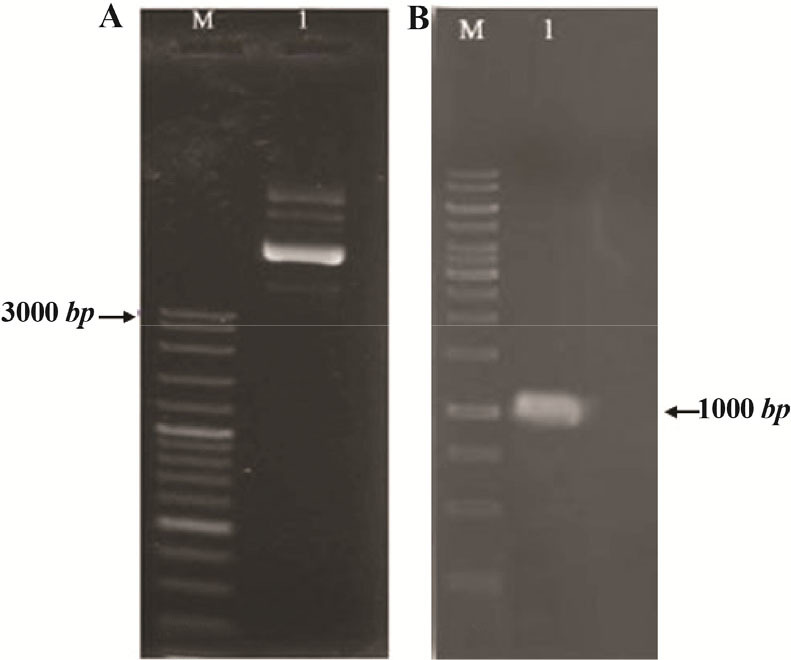
Constructed pET-ret plasmid after cloning of reteplase gene in pET-21; A) Purification from E. coli DH5α. M: molecular weight marker, 1:3 μl of plasmid on 1% agarose gel. B) PCR amplification of reteplase gene (1065 bp) using pET-ret as the template, M: molecular weight marker, 1:5 μl of the PCR product on 1% agarose gel.
Expression of reteplase in E. coli BL21 (DE3) strain
E. coli BL21 (DE3) is the most widely used prokaryotic expression host which is deficient for two main proteases OmpT and Lon. This strain has remained as the gold standard among expression hosts since commercialization of them started 18,19. Therefore, the pET-ret plasmid was first transformed and expressed in this strain. When the cell density reached to OD600 of 0.8, IPTG was added to the final concentration of 1 mM and expression was continued for 20 hr at 37°C. The shaking speed of incubator was 180 rpm during the growth and induction phases. As shown in figure 2, a clear band of overexpression was observed with the molecular mass of 39 kDa. Obviously, all of the expressed protein was deposited in the insoluble fraction of cells and no expression was found in the soluble fraction (Figure 2). Expression was also carried out at decreased temperatures (25°C and 18°C) but no accumulation of reteplase was observed in soluble fraction of cell lysates. Induction of expression at lower temperatures reduced the total amount of reteplase expression in inclusion bodies (Data not shown).
Figure 2.
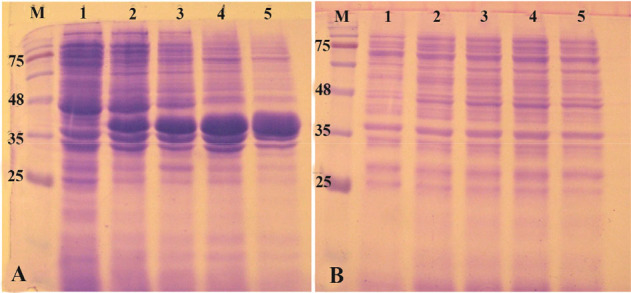
Analysis of recombinant reteplase expressed in E. coli BL21 (DE3) at 37°C and 1 mM IPTG. A) The insoluble (pellet) and B) soluble fractions of cell lysates were separated and loaded on 10% SDS-PAGE. M: prestained molecular weight marker, fractions were withdrawn before induction of expression (lane 1), at 1 hr (lane 2), 2 hr (lane 3), 4 hr (lane 4) and 20 hr (lane 5) after induction.
Expression of reteplase in E. coli Rosetta-gami (DE3) strain
Rosetta-gami B (DE3) is designed to enhance the expression of eukaryotic proteins in E. coli by overcoming the problem of codon bias. The strain supplies six tRNAs for rare codons. In addition, due to the mutation in glutaredoxin reductase and thioredoxin reductase (Δgor ΔtrxB), correct folding of disulfide-bonded recombinant protein in cytosolic fraction of E. coli is enhanced 18,19.
As described above, reteplase contains 9 disulfide bonds, therefore, Rosetta-gami as the expression host was used and the effect of oxidizing cytoplasm on expression of reteplase was found. Expression was induced with 1 mM IPTG at 37°C and 180 rpm shaking. As shown in figure 3, recombinant protein was expressed and accumulated in the insoluble fraction. No overexpression bands were observed in soluble fraction of cell lysates. Reducing the temperature of expression to 25 and 18°C did not also increase the expression of reteplase as a soluble protein (Data not shown).
Figure 3.
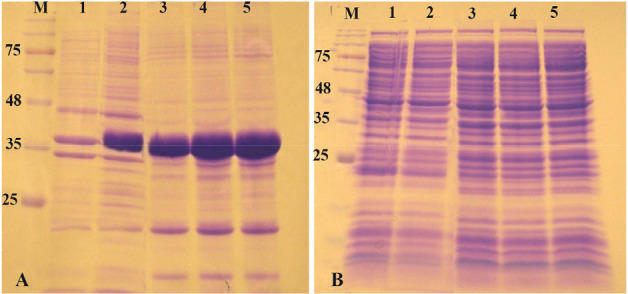
Analysis of recombinant reteplase expressed in E. coli Rosetta-gami (DE3) at 37°C and 1 mM IPTG. A) The insoluble and B) soluble fractions of cell lysates were separated and loaded on 10% SDS-PAGE. M: prestained molecular weight marker, fractions were withdrawn before induction of expression (lane 1), at 1 hr (lane 2), 2 hr (lane 3), 4 hr (lane 4) and 20 hr (lane 5) after induction.
Expression of reteplase in E. coli SHuffle T7 strain
SHuffel T7 is an E. coli engineered by New England Biolabs specifically for expression of disulfide-bonded recombinant proteins controlled by T7 promoter. In SHuffle strain, not only glutaredoxin reductase and thioredoxin reductase are deleted (Δgor ΔtrxB), but also a version of the periplasmic disulfide bond isomerase DsbC which lacks its signal sequence is expressed in cytoplasm. It has been claimed that the strain is able to express recombinant proteins with multiple disulfide bonds and also correct the mis-oxidized bonds and promote proper folding 17.
The pET-ret plasmid was introduced into E. coli SHuffle T7 and expression was induced with 1 mM IPTG at 37°C and 180 rpm. The expression was continued for 20 hr. As can be seen in figures 4A and B in all fractions that were analyzed on SDS-PAGE, the overexpression band of the recombinant protein was deposited in inclusion bodies and no expression band was observed in soluble fraction.
Figure 4.
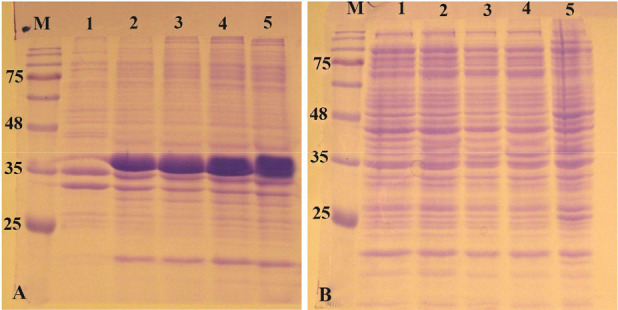
Analysis of recombinant reteplase expressed in E. coli SHuffle T7 at 37°C and 1 mM IPTG. A) The insoluble and B) soluble fractions of cell lysates were separated and loaded on 10% SDS-PAGE. M: prestained molecular weight marker, fractions were withdrawn before induction of expression (lane 1), at 1 hr (lane 2), 2 hr (lane 3), 4 hr (lane 4) and 20 hr (lane 5) after induction.
Comparison of E. coli strains on the expression of reteplase in aggregated form
In this experiment, E. coli strains BL21, Rosetta-gami and SHuffle were simultaneously induced with 1 mM IPTG at 37°C and 180 rpm and expression was continued for 2 and 4 hr. After sonication, pellets were isolated and equal amounts of total proteins were loaded on SDS-PAGE. Figure 5 shows the comparison of reteplase expression in insoluble fraction of the three strains. E. coli BL21 produced the highest amount of reteplase in inclusion bodies in comparison to Rosetta-gami and SHuffle T7. In addition, total amount of reteplase expression was higher in Rosetta-gami in comparison to SHuffle T7. Also, the amount of recombinant expression was quantified using ImageJ software version 1.49 (National Institutes of Health, Bethesda, MD). The percent of recombinant reteplase band to the total insoluble fraction of cells is shown in table 1. SHuffle T7 expressed reteplase in almost 50% of the insoluble fraction after 4 hr. This amount of expression is the lowest concentration of reteplase among strains that were used. BL21 (DE3) was able to express reteplase as 70% of the insoluble fraction after 4 hr which shows the high ability of this strain in expression of codon optimized reteplase gene. Rosetta-gami showed an average ability in expression of reteplase. Although the strain had the advantage of expressing rare tRNAs, it was capable of expressing reteplase in lower concentrations in comparison to BL21 (DE3).
Figure 5.
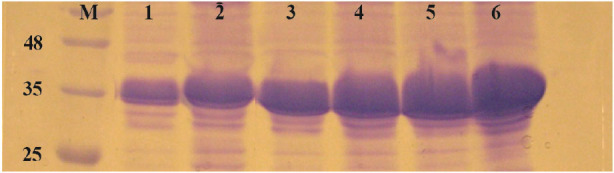
Comparison of E. coli BL21 (DE3), Rosatta-gami (DE3) and SHuffle T7 for production of reteplase in insoluble fraction of cell lysate. M: prestained molecular weight marker, SHuffle T7 2 hr (lane 1) and 4 hr (lane 2) after induction. Rosetta-gami (DE3) 2 hr (lane 3) and 4 hr (lane 4) after induction. BL21 (DE3) 2 hr (lane 5) and 4 hr (lane 6) after induction.
Table 1.
Quantification of the overexpressed reteplase bands to the total insoluble fraction for three strains
| Host cell | Time of induction | (Overexpressed band/pellet) |
|---|---|---|
| SHuffle T7 | 2 hr | 43.7% |
| 4 hr | 55.67% | |
| Rosetta-gami | 2 hr | 58.46% |
| 4 hr | 64.34% | |
| BL21 | 2 hr | 61.75% |
| 4 hr | 70.03% |
Effect of expression parameters on solubilization of reteplase
Among E. coli strains used here, SHuffle T7 is the most promising host for soluble expression of reteplase in cytoplasm. Since expression of reteplase at 37°C and 180 rpm resulted in complete accumulation of reteplase in inclusion bodies, investigating the role of expression factors on solubilization of this protein in SHuffle T7 host was the main purpose. In order to find the effect of temperature, induction was carried out with 1 mM IPTG at 25°C and 18°C and 180 rpm. In spite of low temperature, most of the gene product still aggregated in inclusion bodies and no improvement was reached in soluble expression of reteplase (Figures 6A and 6B). Lowering the temperature also reduced the total amount of reteplase accumulated in insoluble fraction.
Figure 6.
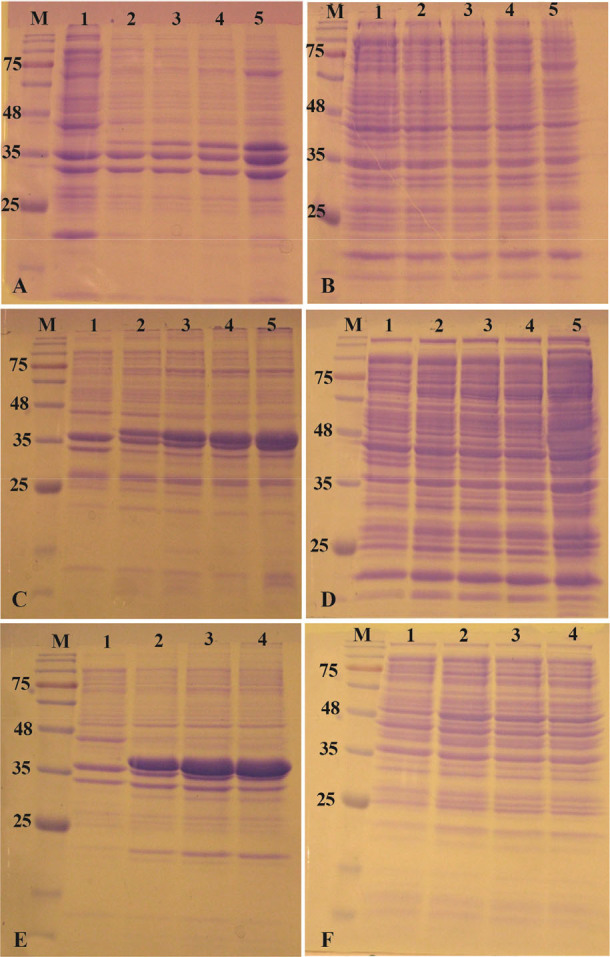
Alteration of expression condition in SHuffle T7 E. coli. A) The insoluble and B) soluble fractions of cell lysates after induction of expression at 18°C with 1 mM IPTG. C) The insoluble and D) soluble fractions of cell lysates after induction of expression at 37°C with 120 rpm shaking. E) The insoluble and F) soluble fractions of cell lysates after induction of expression at 18°C and 120 rpm with 0.1 mM IPTG. M: prestained molecular weight marker, fractions were withdrawn before induction of expression (lane 1), at 1 hr (lane 2), 2 hr (lane 3), 4 hr (lane 4) and 20 hr (lane 5) after induction.
The concentration of IPTG was also reduced to 0.1 mM in order to reduce the amount of reteplase production which might be beneficial for correct folding of the protein. The temperature of expression was adjusted to 37°C with 180 rpm shake. In this strategy, the amount of reteplase accumulation in insoluble fraction was decreased but still no significant amount of reteplase was observed in soluble fraction of cell lysate (Data not shown).
The effect of oxygen supply was also investigated on reteplase production in SHuffle T7 E. coli. The speed of shaking was reduced to 120 rpm when 1 mM IPTG was added to the expression flask. The expression was carried out at 37°C. Figures 6C and 6D show that reduction of aeration cannot increase the expression of reteplase in soluble fraction. Interestingly, the accumulation of reteplase was increased in insoluble fraction.
Finally, an expression was carried out with all three modifications in induction protocol. 0.1 mM IPTG was added to culture medium at 18°C and 120 rpm. Simultaneous modification of all three parameters was also unable to suppress the aggregation of reteplase in SHuffle T7 E. coli and no protein band was observed in supernatant fraction of cell lysate (Figures 6E and 6F).
Discussion
As a second-generation recombinant tissue type plasminogen activator, reteplase is developed to treat thrombotic diseases like myocardial infarction, ischemic stroke, peripheral arterial occlusion and venous thromboembolism 20–22. Reteplase shows decreased affinity to fibrin which allows free diffusion and better penetrance in blood clots instead of binding only on the surface 23.
Reteplase is famous as an aggregation-prone protein in E. coli since contains multiple disulfide bonds and reductive environment of cytoplasm does not let correct disulfide bond formation resulting in generally misfolded and biologically-inactive inclusion bodies 2,24. Refolding of inclusion bodies elongates the time of drug production. The efficiency of refolding may be low and refolded enzymes might be unstable due to incomplete refolding or incomplete removal of detergent and denaturants used in this process. Therefore, it is often desirable to obtain high degrees of soluble protein. Till now there is no universal approach for efficient soluble production of aggregation-prone recombinant proteins 25,26 but several strategies like introduction of molecular chaperons to the host bacteria, modulation of expression level, optimization of culture medium, reduction of temperature, addition of chemical chaperons have helped in preventing protein aggregation 27.
In the current study, an attempt was made to express reteplase in three different strains of E. coli and compare their ability for soluble expression and production yield of this recombinant protein. E. coli BL21 (DE3) is the most common prokaryotic strain used for expression of recombinant proteins till now 19,28. Previous reports on tPA expression in BL21 (DE3) strain show that common strategies failed to produce soluble protein in this strain but aggregated inclusion bodies are extracted for refolding procedure 29.
Rosetta-gami B (DE3) is a hybrid strain engineered to enhance the expression of genes containing rare co-dons. At the same time, it can enhance disulfide bond formation in the cytosolic fraction due to mutations of trxB and gor genes. This strain can yield 10-fold more active protein than other hosts even when overall expression levels are similar 18,19,28. The protease domain of tPA has been expressed as a soluble protein in this strain but there is no report on expression of reteplase which contains kringle II and protease domains in this strain yet 10.
SHuffle T7 is a recently prepared E. coli capable of expressing disulfide bonded proteins in its cytoplasm. In addition to mutations in trxB and gor genes, SHuffle expresses a signal sequenceless DsbC gene in the cytoplasm. Using this strain, production of several disulfide-bond containing recombinant proteins have been dramatically increased in soluble fraction of cell lysates. Expression of vtPA as a truncated mutant of tPA which contains 6 disulfide bonds -formed between 12 cysteins- was also investigated in this strain and presence of vtPA in the soluble fraction of cell lysate was confirmed 17. There are no reports on expression of reteplase in this novel strain and there remains the answer whether SHuffle T7 may favor soluble expression of reteplase.
In the present study, the induction of reteplase expression with 1 mM IPTG at 37°C and 180 rpm in all three strains resulted in high level expression of reteplase in aggregated fraction of cell lysates. Obviously, BL21 (DE3) showed the highest expression in comparison to Rosetta-gami and SHuffle T7 possibly due to its protease deficient genotype (lon−, OmpT−). Supplying rare tRNAs in cytoplasm of Rosetta-gami was not effective for increasing the expression of reteplase since a codon-optimized reteplase gene was prepared here synthetically 18,19. In none of the strains, protein band with the molecular weight of reteplase was observed in soluble fraction. As expected, the whole recombinant protein expressed in normal reducing cytoplasm of BL21 (DE3) was accumulated in inclusion bodies but also the engineered cytoplasms of Rosetta-gami and SHuffle T7 were inefficient for correct folding of this recombinant protein.
The effect of environmental factors such as temperature of expression, concentration of the inducer and oxygen supply was also investigated on reteplase production in SHuffle T7 strain since it provides a more favorable cytoplasmic condition for soluble expression of disulfide-bonded proteins. Decreasing the temperature of expression is a well known technique that limits the aggregation of recombinant proteins. Since hydrophobic interactions are highly temperature dependant, lower temperature will favor correct folding and solubilization of the target protein 8,26. In the present work, even in the lowest temperature tested (18°C, no soluble expression band was observed and the whole recombinant protein was accumulated in inclusion bodies. Obviously, with decreasing the temperature the amount of reteplase was reduced in inclusion bodies which shows higher yield of protein production at 37°C.
Another known strategy for reduction of aggregation during recombinant protein production is optimization of inducer concentration. In high concentrations of inducer, the expression of the protein is fully induced and with respect to the limited capacity of cellular environment for correct folding, the precipitation of aggregation-prone recombinant proteins will be increased 25. Here, two different concentrations of inducer were used but reduction of IPTG concentration to 0.1 mM could not result in solubilization of reteplase although the total amount of reteplase was reduced.
Oxygen supply is known for variable effects on protein expression since can increase or decrease the production of proteins. Therefore, the effect of oxygen supply is protein specific and should be individually investigated for each specific protein. It was previously shown that with increased aeration, the amount of expression for a tPA derivative (K2P) will be decreased in BL21 (DE3) cells. Limited oxygen supply results in lower acetate and higher ammonia accumulation and prepares more K2P expression 30. Here, in agreement to Wang et al, a higher amount of reteplase was expressed in SHuffle T7 strain with decreasing oxygen availability but in both 180 and 120 rpm shaking, the whole expression was accumulated in inclusion bodies. Good or poor oxygen availability did not affect solubilization of reteplase but obviously changed the total amount of recombinant expression.
Long et al reported soluble expression of full-length tPA in three different E. coli strains, BL21 (DE3), Rosetta and Origami 2 using an auto-induction procedure. In this article, they have claimed that auto-induction is a reliable method to reach at least 50% of tPA expression in soluble fraction of E. coli strains 9. With respect to 17 disulfide bonds present in full length tPA, it is possible that the same strategy results in soluble production of reteplase which has a less complicated structure with nine disulfide bonds. Therefore, it should be experimentally investigated whether the auto-induction might be an effective strategy for production of reteplase or not.
Till now, isolation and renaturation of inclusion bodies remains the solution for active reteplase production. Finding new refolding techniques or improving existing methods can increase the efficiency and lower the cost of bioactive reteplase production. Therefore, production and accumulation of high concentrations of reteplase in inclusion bodies will be beneficial. Here, it was shown that among the three strains, BL21 (DE3) seems to be the preferred host with respect to the highest amount of reteplase accumulation but it is also well known that inclusion bodies in the cytoplasm grow not only from completely unfolded proteins, but also several folding intermediates can be trapped in these moie-ties. In folding intermediates, some of the disulfide bonds are likely to happen between correct cysteine pairs. The engineered cytoplasms of Rosetta-gami and SHuffle T7 might increase the folding intermediates of reteplase in inclusion bodies. In renaturation procedure, these folding intermediates will rapidly transit to native and active state of proteins, therefore the yield of re-folding will be increased 25,31,32. Therefore, the lower concentration of reteplase obtained from Rosetta-gami and SHuffle T7 might have a suitable yield of refolding in comparison to higher concentrations of BL21.
Conclusion
It is concluded that for remarkable production of soluble reteplase in E. coli, strategies other than engineered hosts or environmental parameters should be tested. Manipulating the strength of promoter and reducing the expression level might be helpful. Also, it is shown that BL21 (DE3) is the most efficient expression system with respect to its high level of expression, but maximum recovery of the soluble and active protein from inclusion bodies produced in each strain during the refolding procedure needs to be investigated.
Acknowledgement
The authors express their gratitude to the Iran National Science Foundation for the financial support during the course of this project.
References
- 1. Mandi N, Sundaram KR, Tandra SK, Bandyopadhyay S, Padmanabhan S. Asn and asn: critical residues for in vitro biological activity of reteplase. Adv Hematol 2010; 2010: 172484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Geng Y, Wang S, Qi Q. Expression of active recombinant human tissue-type plasminogen activator by using in vivo polyhydroxybutyrate granule display. Appl Environ Microbiol 2010; 76 (21): 7226– 7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Topol EJ. A comparison of reteplase with alteplase for acute myocardial infarction. The Global Use of Strategies to Open Occluded Coronary Arteries (GUSTO III) Investigators. N Engl J Med 1997; 337 (16): 1118– 1123. [DOI] [PubMed] [Google Scholar]
- 4. Qiu J, Swartz JR, Georgiou G. Expression of active human tissue-type plasminogen activator in Escherichia coli. Appl Environ Microbiol 1998; 64 (12): 4891– 4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baruah DB, Dash RN, Chaudhari MR, Kadam SS. Plasminogen activators: a comparison. Vascul Pharmacol 2006; 44 (1): 1– 9. [DOI] [PubMed] [Google Scholar]
- 6. Kunadian V, Gibson CM. Recombinant tissue-type plasminogen activators: “time matters”. Drugs Today (Barc) 2011; 47 (7): 559– 570. [DOI] [PubMed] [Google Scholar]
- 7. Nordt TK, Bode C. Thrombolysis: newer thrombolytic agents and their role in clinical medicine. Heart 2003; 89 (11): 1358– 1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marisch K, Bayer K, Cserjan-Puschmann M, Luchner M, Striedner G. Evaluation of three industrial Escherichia coli strains in fed-batch cultivations during high-level SOD protein production. Microb Cell Fact 2013; 12: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Long X, Gou Y, Luo M, Zhang S, Zhang, Bai L, et al. Soluble expression, purification, and characterization of active recombinant human tissue plasminogen activator by auto-induction in E. coli. BMC Biotechnol 2015; 15: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee HJ, Im H. Soluble expression and purification of human tissue-type plasminogen activator protease domain. Bull Korean Chem Soc 2010; 31 (9): 2607– 2612. [Google Scholar]
- 11. Zhao Y, GE W, Kong Y, Zhang Ch. Cloning, expression, and renaturation studies of reteplase. J Microbiol Biotechnol 2003; 13 (6): 989– 992. [Google Scholar]
- 12. Aghaabdollahian S, Rabbani M, Ghaedi K, Sadeghi HM. Molecular cloning of Reteplase and its expression in E. coli using tac promoter. Adv Biomed Res 2014; 3: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khodabakhsh F, Dehghani Z, Zia MF, Rabbani M, Sadeghi HM. Cloning and Expression of Functional Reteplase in Escherichia coli TOP10. Avicenna J Med Biotechnol 2013; 5 (3): 168– 175. [PMC free article] [PubMed] [Google Scholar]
- 14. Gao L, Zhang C, Li L, Liang L, Deng X, Wu W, et al. Construction, expression and refolding of a bifunctional fusion protein consisting of C-terminal 12-residue of hirudin-PA and reteplase. Protein J 2012; 31 (4): 328– 336. [DOI] [PubMed] [Google Scholar]
- 15. Nausch H, Huckauf J, Koslowski R, Meyer U, Broer I, Mikschofsky H. Recombinant production of human inter-leukin 6 in Escherichia coli. PLoS One 2013; 8 (1): e54933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rabhi-Essafi I, Sadok A, Khalaf N, Fathallah DM. A strategy for high-level expression of soluble and functional human interferon alpha as a GST-fusion protein in E. coli. Protein Eng Des Sel 2007; 20 (5): 201– 209. [DOI] [PubMed] [Google Scholar]
- 17. Lobstein J, Emrich CA, Jeans C, Faulkner M, Riggs P, Berkmen M. SHuffle, a novel Escherichia coli protein expression strain capable of correctly folding disulfide bonded proteins in its cytoplasm. Microb Cell Fact 2012; 11: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Novagen, pET system manual, 10th edition, May, 2003. Available at: http://download.bioon.com.cn/upload/201011/25/141105fw9i8zjm1jfm19m8.attach.pdf.
- 19. Rosano GL, Ceccaelli EA. Recombinant protein expression in Escherichia coli: advances and challenges. Front Microbiol 2014; 5 (172): 1– 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kunadian V, Gibson CM. Thrombolytics and myocardial infarction. Cardiovasc Ther 2012; 30 (2): e81– 88. [DOI] [PubMed] [Google Scholar]
- 21. Collen D, Lijnen HR. The tissue-type plasminogen activator story. Arterioscler Thromb Vasc Biol 2009; 29 (8): 1151– 1155. [DOI] [PubMed] [Google Scholar]
- 22. Collen D, Lijnen HR. Thrombolytic agents. Thromb Haemost 2005; 93 (4): 627– 630. [DOI] [PubMed] [Google Scholar]
- 23. Wooster MB, Luzier AB. Reteplase: a new thrombolytic for the treatment of acute myocardial infarction. Ann Pharmacother 1999; 33 (3): 318– 324. [DOI] [PubMed] [Google Scholar]
- 24. Ershadi S, Rashedi H, Fazeli A. A study on the mechanism of aggregation of therapeutic reteplase protein by using the monomer-loss model. J Appl Biotechnol Rep 2015; 2 (1): 191– 197. [Google Scholar]
- 25. Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat Biotechnol 2004; 22 (11): 1399– 1408. [DOI] [PubMed] [Google Scholar]
- 26. Sørensen HP, Mortensen KK. Soluble expression of recombinant proteins in the cytoplasm of Escherichia coli. Microb Cell Fact 2005; 4 (1): 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. de Marco A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb Cell Fact 2009; 8: 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Makino T, Skretas G, Georgiou G. Strain engineering for improved expression of recombinant proteins in bacteria. Microb Cell Fact 2011; 10: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mirnajd-Gerami S, Farajnia S, Mahboudi F, Babaei H. Optimizing refolding condition for recombinant tissue plasminogen activator. Iran J Biotech 2011; 9 (4): 253– 259. [Google Scholar]
- 30. Wang H, Wang F, Wei D. Impact of oxygen supply on rtPA expression in Escherichia coli BL21 (DE3): ammonia effects. Appl Microbiol Biotechnol 2009; 82 (2): 249– 259. [DOI] [PubMed] [Google Scholar]
- 31. Rudolph R, Lilie H. In vitro folding of inclusion body proteins. FASEB J 1996; 10 (1): 49– 56. [PubMed] [Google Scholar]
- 32. Liu YD, Zhang GF, Li JJ, Chen J, Wang YJ, Ding H, et al. Identification of an oxidative refolding intermediate of recombinant consensus interferon from inclusion bodies and design of a two-stage strategy to promote correct disulfide-bond formation. Biotechnol Appl Biochem 2007; 48 (Pt 4): 189– 198. [DOI] [PubMed] [Google Scholar]


