Abstract
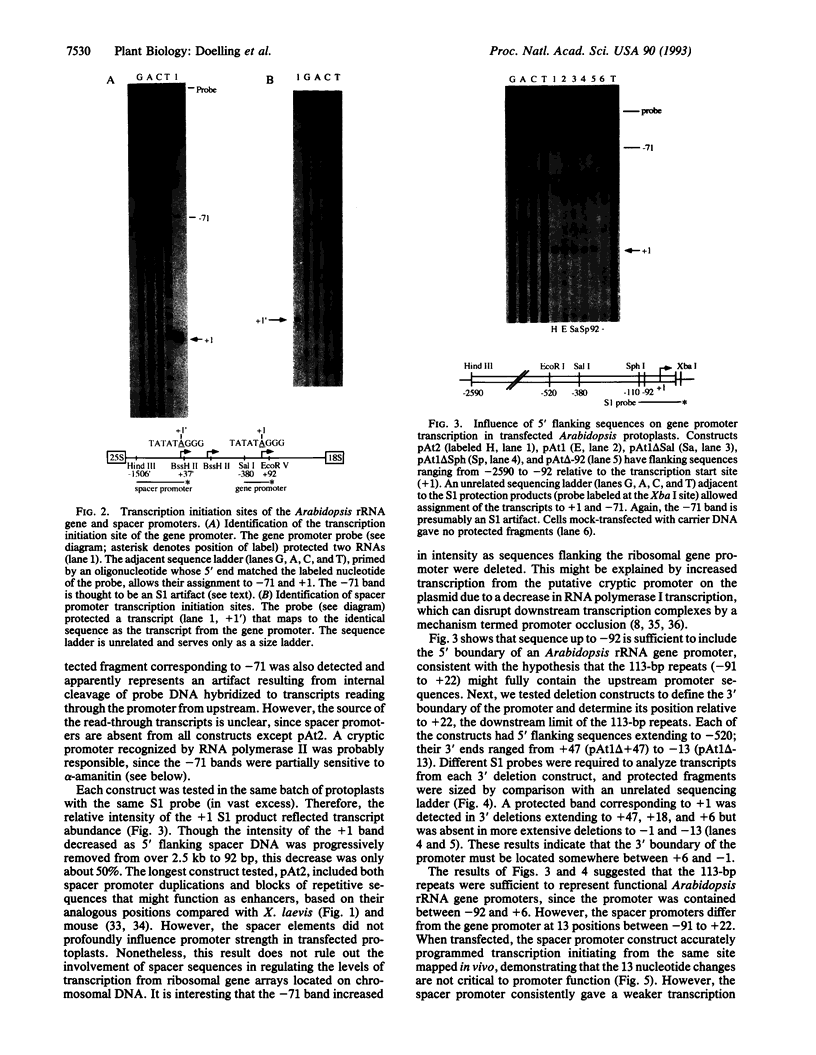
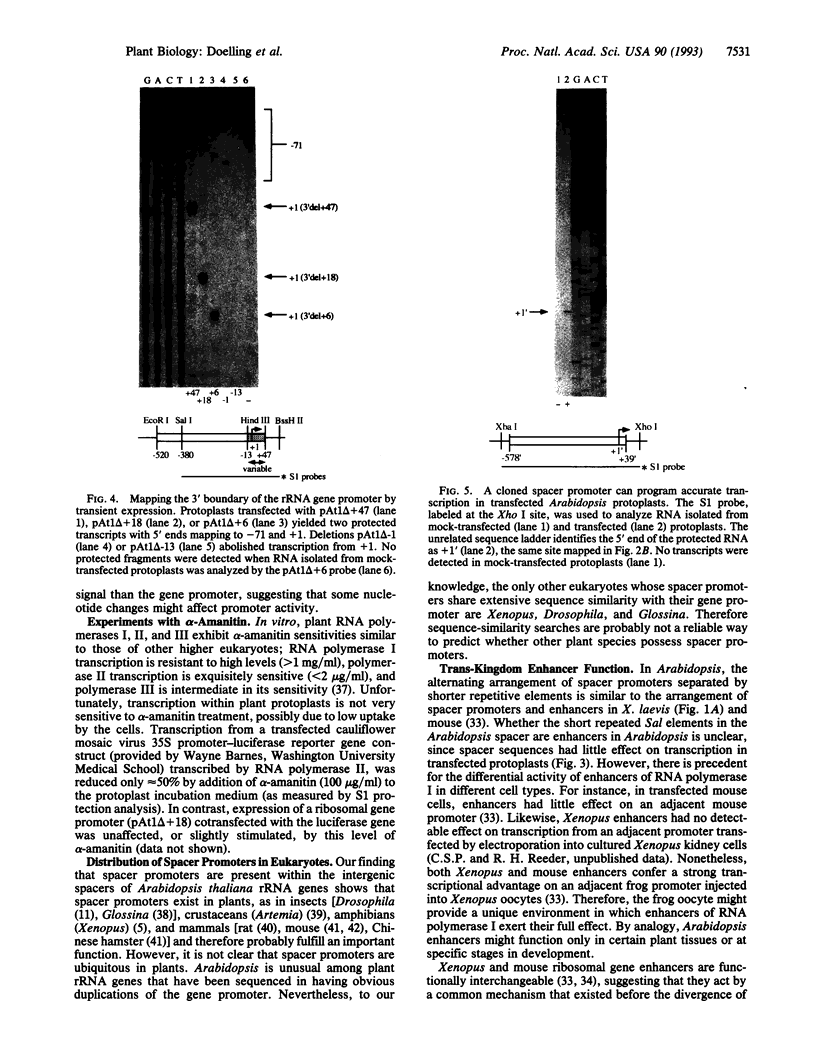
In eukaryotes, RNA polymerase I transcription is controlled by DNA elements located within the spacers that separate the tandemly arranged rRNA genes. Unlike rRNA coding sequences, the intergenic spacers evolve rapidly and have little sequence similarity even among closely related species. Nonetheless, the arrangement of functional elements, such as spacer promoters and enhancers, is thought to be highly conserved. Here, we identify spacer promoters in the plant Arabidopsis thaliana, thereby demonstrating their existence in both the plant and animal kingdoms. We also use an Arabidopsis transient expression system to perform transcriptional analysis of the ribosomal gene promoter. Spacer promoters share sequence similarity with the gene promoter from -91 to +22 relative to the transcription start site, +1. Deletion analysis shows that sequences required for RNA polymerase I transcription reside within these boundaries. Spacer sequences upstream of the gene promoter have only a small positive effect on transcription in transfected protoplasts but can increase transcription from a Xenopus ribosomal gene promoter in injected frog oocytes. This trans-kingdom enhancer effect further suggests that the functional elements within eukaryotic ribosomal genes are highly conserved.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker R. F., Harberd N. P., Jarvis M. G., Flavell R. B. Structure and evolution of the intergenic region in a ribosomal DNA repeat unit of wheat. J Mol Biol. 1988 May 5;201(1):1–17. doi: 10.1016/0022-2836(88)90434-2. [DOI] [PubMed] [Google Scholar]
- Bateman E., Paule M. R. Promoter occlusion during ribosomal RNA transcription. Cell. 1988 Sep 23;54(7):985–992. doi: 10.1016/0092-8674(88)90113-4. [DOI] [PubMed] [Google Scholar]
- Cassidy B. G., Yang-Yen H. F., Rothblum L. I. Additional RNA polymerase I initiation site within the nontranscribed spacer region of the rat rRNA gene. Mol Cell Biol. 1987 Jul;7(7):2388–2396. doi: 10.1128/mcb.7.7.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Cross N. C., Dover G. A. A novel arrangement of sequence elements surrounding the rDNA promoter and its spacer duplications in tsetse species. J Mol Biol. 1987 May 5;195(1):63–74. doi: 10.1016/0022-2836(87)90327-5. [DOI] [PubMed] [Google Scholar]
- De Winter R. F., Moss T. Spacer promoters are essential for efficient enhancement of X. laevis ribosomal transcription. Cell. 1986 Jan 31;44(2):313–318. doi: 10.1016/0092-8674(86)90765-8. [DOI] [PubMed] [Google Scholar]
- Delcasso-Tremousaygue D., Grellet F., Panabieres F., Ananiev E. D., Delseny M. Structural and transcriptional characterization of the external spacer of a ribosomal RNA nuclear gene from a higher plant. Eur J Biochem. 1988 Mar 15;172(3):767–776. doi: 10.1111/j.1432-1033.1988.tb13956.x. [DOI] [PubMed] [Google Scholar]
- Gerstner J., Schiebel K., von Waldburg G., Hemleben V. Complex organization of the length heterogeneous 5' external spacer of mung bean (Vigna radiata) ribosomal DNA. Genome. 1988 Oct;30(5):723–733. doi: 10.1139/g88-120. [DOI] [PubMed] [Google Scholar]
- Grimaldi G., Di Nocera P. P. Multiple repeated units in Drosophila melanogaster ribosomal DNA spacer stimulate rRNA precursor transcription. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5502–5506. doi: 10.1073/pnas.85.15.5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimaldi G., Fiorentini P., Di Nocera P. P. Spacer promoters are orientation-dependent activators of pre-rRNA transcription in Drosophila melanogaster. Mol Cell Biol. 1990 Sep;10(9):4667–4677. doi: 10.1128/mcb.10.9.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruendler P., Unfried I., Pascher K., Schweizer D. rDNA intergenic region from Arabidopsis thaliana. Structural analysis, intraspecific variation and functional implications. J Mol Biol. 1991 Oct 20;221(4):1209–1222. doi: 10.1016/0022-2836(91)90929-z. [DOI] [PubMed] [Google Scholar]
- Gruendler P., Unfried I., Pointner R., Schweizer D. Nucleotide sequence of the 25S-18S ribosomal gene spacer from Arabidopsis thaliana. Nucleic Acids Res. 1989 Aug 11;17(15):6395–6396. doi: 10.1093/nar/17.15.6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilfoyle T. J. Purification and Characterization of DNA-dependent RNA Polymerases from Cauliflower Nuclei. Plant Physiol. 1976 Oct;58(4):453–458. doi: 10.1104/pp.58.4.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S., Sollner-Webb B. A transcriptional terminator is a novel element of the promoter of the mouse ribosomal RNA gene. Cell. 1986 Dec 26;47(6):891–900. doi: 10.1016/0092-8674(86)90804-4. [DOI] [PubMed] [Google Scholar]
- Kohorn B. D., Rae P. M. Nontranscribed spacer sequences promote in vitro transcription of Drosophila ribosomal DNA. Nucleic Acids Res. 1982 Nov 11;10(21):6879–6886. doi: 10.1093/nar/10.21.6879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller H. T., Frondorf K. A., Maschner P. D., Vaughn J. C. In vivo transcription from multiple spacer rRNA gene promoters during early development and evolution of the intergenic spacer in the brine shrimp Artemia. Nucleic Acids Res. 1987 Jul 10;15(13):5391–5411. doi: 10.1093/nar/15.13.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Deppert U., Grummt I. A 140-base-pair repetitive sequence element in the mouse rRNA gene spacer enhances transcription by RNA polymerase I in a cell-free system. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7527–7531. doi: 10.1073/pnas.87.19.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Grummt I. A novel promoter in the mouse rDNA spacer is active in vivo and in vitro. EMBO J. 1987 Nov;6(11):3487–3492. doi: 10.1002/j.1460-2075.1987.tb02673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn A., Normann A., Bartsch I., Grummt I. The mouse ribosomal gene terminator consists of three functionally separable sequence elements. EMBO J. 1988 May;7(5):1497–1502. doi: 10.1002/j.1460-2075.1988.tb02968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. A 12-base-pair sequence is an essential element of the ribosomal gene terminator in Xenopus laevis. Mol Cell Biol. 1987 May;7(5):1900–1905. doi: 10.1128/mcb.7.5.1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labhart P., Reeder R. H. Characterization of three sites of RNA 3' end formation in the Xenopus ribosomal gene spacer. Cell. 1986 May 9;45(3):431–443. doi: 10.1016/0092-8674(86)90329-6. [DOI] [PubMed] [Google Scholar]
- McStay B., Reeder R. H. An RNA polymerase I termination site can stimulate the adjacent ribosomal gene promoter by two distinct mechanisms in Xenopus laevis. Genes Dev. 1990 Jul;4(7):1240–1251. doi: 10.1101/gad.4.7.1240. [DOI] [PubMed] [Google Scholar]
- Moss T., Birnstiel M. L. The putative promoter of a Xenopus laevis ribosomal gene is reduplicated. Nucleic Acids Res. 1979 Aug 24;6(12):3733–3743. doi: 10.1093/nar/6.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry K. L., Palukaitis P. Transcription of tomato ribosomal DNA and the organization of the intergenic spacer. Mol Gen Genet. 1990 Mar;221(1):103–112. doi: 10.1007/BF00280374. [DOI] [PubMed] [Google Scholar]
- Pikaard C. S., Pape L. K., Henderson S. L., Ryan K., Paalman M. H., Lopata M. A., Reeder R. H., Sollner-Webb B. Enhancers for RNA polymerase I in mouse ribosomal DNA. Mol Cell Biol. 1990 Sep;10(9):4816–4825. doi: 10.1128/mcb.10.9.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikaard C. S., Reeder R. H. Sequence elements essential for function of the Xenopus laevis ribosomal DNA enhancers. Mol Cell Biol. 1988 Oct;8(10):4282–4288. doi: 10.1128/mcb.8.10.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt R. E., Meyerowitz E. M. Characterization of the genome of Arabidopsis thaliana. J Mol Biol. 1986 Jan 20;187(2):169–183. doi: 10.1016/0022-2836(86)90226-3. [DOI] [PubMed] [Google Scholar]
- Reeder R. H. Regulatory elements of the generic ribosomal gene. Curr Opin Cell Biol. 1989 Jun;1(3):466–474. doi: 10.1016/0955-0674(89)90007-0. [DOI] [PubMed] [Google Scholar]
- Toloczyki C., Feix G. Occurrence of 9 homologous repeat units in the external spacer region of a nuclear maize rRNA gene unit. Nucleic Acids Res. 1986 Jun 25;14(12):4969–4986. doi: 10.1093/nar/14.12.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tower J., Henderson S. L., Dougherty K. M., Wejksnora P. J., Sollner-Webb B. An RNA polymerase I promoter located in the CHO and mouse ribosomal DNA spacers: functional analysis and factor and sequence requirements. Mol Cell Biol. 1989 Apr;9(4):1513–1525. doi: 10.1128/mcb.9.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremousaygue D., Laudie M., Grellet F., Delseny M. The Brassica oleracea rDNA spacer revisited. Plant Mol Biol. 1992 Mar;18(5):1013–1018. doi: 10.1007/BF00019222. [DOI] [PubMed] [Google Scholar]
- Valvekens D., Van Montagu M., Van Lijsebettens M. Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci U S A. 1988 Aug;85(15):5536–5540. doi: 10.1073/pnas.85.15.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincentz M., Flavell R. B. Mapping of ribosomal RNA transcripts in wheat. Plant Cell. 1989 Jun;1(6):579–589. doi: 10.1105/tpc.1.6.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentgraf U., Ganal M., Hemleben V. Length heterogeneity of the rRNA precursor in cucumber (Cucumis sativus). Plant Mol Biol. 1990 Sep;15(3):465–474. doi: 10.1007/BF00019163. [DOI] [PubMed] [Google Scholar]








