Abstract
Background
The eusocial Hymenoptera have radiated across a wide range of thermal environments, exposing them to significant physiological stressors. We reconstructed the evolutionary history of three families of Heat Shock Proteins (Hsp90, Hsp70, Hsp40), the primary molecular chaperones protecting against thermal damage, across 12 Hymenopteran species and four other insect orders. We also predicted and tested for thermal inducibility of eight Hsps from the presence of cis-regulatory heat shock elements (HSEs). We tested whether Hsp induction patterns in ants were associated with different thermal environments.
Results
We found evidence for duplications, losses, and cis-regulatory changes in two of the three gene families. One member of the Hsp90 gene family, hsp83, duplicated basally in the Hymenoptera, with shifts in HSE motifs in the novel copy. Both copies were retained in bees, but ants retained only the novel HSE copy. For Hsp70, Hymenoptera lack the primary heat-inducible orthologue from Drosophila melanogaster and instead induce the cognate form, hsc70-4, which also underwent an early duplication. Episodic diversifying selection was detected along the branch predating the duplication of hsc70-4 and continued along one of the paralogue branches after duplication. Four out of eight Hsp genes were heat-inducible and matched the predictions based on presence of conserved HSEs. For the inducible homologues, the more thermally tolerant species, Pogonomyrmex barbatus, had greater Hsp basal expression and induction in response to heat stress than did the less thermally tolerant species, Aphaenogaster picea. Furthermore, there was no trade-off between basal expression and induction.
Conclusions
Our results highlight the unique evolutionary history of Hsps in eusocial Hymenoptera, which has been shaped by gains, losses, and changes in cis-regulation. Ants, and most likely other Hymenoptera, utilize lineage-specific heat inducible Hsps, whose expression patterns are associated with adaptive variation in thermal tolerance between two ant species. Collectively, our analyses suggest that Hsp sequence and expression patterns may reflect the forces of selection acting on thermal tolerance in ants and other social Hymenoptera.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-015-0573-0) contains supplementary material, which is available to authorized users.
Keywords: Heat shock proteins, Heat shock response, Heat shock elements, Thermal tolerance, Gene expression, Cis-regulation, Comparative genomics
Background
Heat stress causes proteins to lose stability, misfold, and form aggregates, which can impair function and reduce organismal fitness [1–4]. To cope with macromolecular damage, the heat shock response (HSR) transcriptionally up-regulates thermally responsive genes such as heat shock proteins (Hsps), which maintain proteostasis by refolding or degrading denatured proteins and preventing aggregations [1, 2, 5]. Hsps are a set of highly conserved molecular chaperone proteins of diverse multigene families, named after their molecular weight (Hsp90, Hsp70, Hsp60, Hsp40, and small Hsps) [6, 7].
Although Hsps as a group are highly conserved, diversity within each Hsp gene family reflects evolutionary gains and losses of gene copies [8, 9]. Each Hsp protein family includes paralogues localized to different subcellular compartments (cytosol, endoplasmic reticulum, or mitochondria) that participate in housekeeping functions and/or respond to environmental stress [10–12]. For heat-inducible forms, the transcribed heat shock factors (HSF), bind to cis-regulatory elements known as heat shock elements (HSEs) and up-regulate Hsp transcription [13–15]. Patterns of variation in Hsp gene expression among taxa include expansion of additional Hsp genes [16] and shifts in the arrangement and position of HSE elements [14, 17, 18]. Among taxa, both the level of constitutive expression and the magnitude of Hsp induction are associated with adaptive variation in upper thermal limits [19–22]. Gene structure may also play a role in Hsp expression, but has not been well-studied. For example, genes with introns allow for more mRNA accumulation than do intronless genes [23–25].
The eusocial Hymenoptera (wasps, ants, and bees) occupy diverse thermal environments from low to high latitudes [26] and elevations [27, 28], suggesting that temperature may have played an important selective role in their evolution [29]. Species employ a variety of behavioral [30, 31] and physiological strategies [32] to reduce individual and colony-level exposure to thermal stress. However, individual foragers that leave the nest each day and immobile brood that develop in the nest are likely to encounter sufficiently high temperatures to trigger the HSR [33, 34]. Although key members of Hsp90 and Hsp70 have been identified in a few species of Hymenoptera [33–36], the diversity, functional properties, and regulation of molecular chaperones underlying adaptive variation in Hymenopteran thermal tolerance are poorly understood.
In this study, we evaluated the diversity and evolutionary history of Hsps across 12 species of Hymenoptera and five outgroup species (Culex quinquefasciatus, Drosophila melanogaster, Bombyx mori, Tribolium castaneum, Acyrthosiphon pisum) spanning four insect orders. We analyzed recently published genomes of multiple species of ants [37–42], bees (Apis [43] and Bombus [44]), and the jewel wasp (Nasonia vitripennis [45]) to identify orthologues within each major Hsp gene family and to characterize the upstream regulatory motifs governing their transcription (HSEs). We reconstructed molecular evolutionary relationships within each Hsp multigene family to identify evolutionary gains and losses and tested for positive or purifying selection for each homologous Hsp among lineages and across sites. To characterize the evolution of cis-regulation and identify Hsps involved in the HSR, we identified cis-regulatory HSEs within the promoter region for each homologous Hsp. We then tested whether HSE presence and configuration successfully predicted inducibility in two species of ants that experience different thermal environments: the hot-climate Pogonomyrmex barbatus, which inhabits deserts of the southwestern United States [46], and the cool-climate Aphaenogaster picea, which inhabits temperate deciduous forests of the eastern United States [47]. We found that ants, and probably other Hymenoptera, harbor unique, lineage-specific sets of heat inducible Hsps that were shaped by evolutionary gains, losses, and shifts in cis-regulation. Expression patterns of these heat-inducible Hsps reflect adaptive variation in thermal tolerance between P. barbatus and A. picea.
Results
Identification of conserved Hsp and cis-regulatory HSEs
We recovered conserved Hsps from all of the major gene families (Hsp90, Hsp70, Hsp60, Hsp40, small Hsps; Table 1). Three paralogues within the Hsp90 gene family (trap1, gp93, and hsp83) were found across all surveyed insects. We recovered five of the six Drosophila melanogaster Hsp70 homologues (CG2918, hsc70-3 (BIP), hsc70-4, hsc70-5, and hsp70CB; Table 1) for Hymenoptera. With the exception of Nasonia vitripennis, the Hymenopteran taxa all lacked the heat-inducible orthologue hsp70 (Table 1). For all species, we recovered two paralogues of Hsp60 (Table 1). Hsp40 gene families are one of the most diverse Hsps, but we narrowed our search to DnaJ-1, which is the known heat-inducible paralogue of D. melanogaster (Table 1). We did not recover a DnaJ-1 paralogue from any of the insects surveyed and found the best BLAST match to be D. melanogaster CG5001 (Table 1). Forward BLAST searching for D. melanogaster sHsps (hsp22, hsp23, hsp26, hsp27) yielded no reciprocal BLAST hits; instead, the closest match was lethal 2 essential for life (l(2)efl), for which there were 3–9 copies in the Hymenoptera, and 1–17 copies in other members of the outgroup (Table 1).
Table 1.
Summary of orthologous HSPs from the combination of reciprocal BLAST and HMMER searches using D. melanogaster as the reference
| Outgroup | Hymenoptera | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene family | Gene | C. quinque. | T. castaneum | B. mori | A. pisum | Ants | Bees | N. vitripennis |
| Hsp90 | trap1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| gp93 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | |
| hsp83 | 3 | 3 | 1 | 2 | 1–2* | 2 | 2 | |
| Hsp70 | CG2918 | 1 | 1 | 1 | 3 | 1 | 1 | 1 |
| hsc70-3 (BIP) | 1 | 1 | 1 | 1 | 1* | 1 | 1 | |
| hsc70-4 | 2 | 1 | 1 | 2 | 2* | 2 | 2 | |
| hsc70-5 | 1 | 1 | 1 | 1 | 1* | 1 | 1 | |
| hsp70 | 6 | 1 | 2 | 3 | 0 | 0 | 1 | |
| hsp70CB | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |
| Hsp60 | tcp-1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| hsp60 | 1 | 1 | 1 | 1 | 1* | 1 | 1 | |
| Hsp40 | CG5001 | 1 | 1 | 1 | 1 | 1* | 1 | 1 |
| small Hsps | hsp23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| hsp24 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| hsp26 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| hsp27 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| l(2)efl | 8 | 10 | 17 | 1 | 3–6* | 4–9 | 7 | |
Each entry is the number of orthologous HSPs detected. The astericks (*) indicate orthologues that were detectable by qPCR. For l(2)efl, only one paralogue was detectable by qPCR. C. quinque = Culex quinquefasciatus, T. castaneum = Tribolium castaneum. B. mori = Bombyx mori, A. pisum = Acyrthosiphon pisum, N. vitripennis = Nasonia vitripennis. See text for further details of ants and bees used for analysis
Of the Hsp homologues, eight were quantifiable by qPCR and were subsequently searched for cis-regulatory HSEs (Table 1, indicated with asterisks). Local alignment of the promoter regions of hsp83, hsc70-4 (h1 and h2), and hsp40 across species indicated conserved location, conformation, and arrangement of cis-regulatory HSEs (Figs. 1, 2 and 3), whereas hsc70-3 (BIP), hsc70-5, hsp60, and l(2)efl had less conserved HSEs (Additional file 1: Figures S1, Additional file 2: Figures S2, Additional file 3: Figures S3; data not shown for l(2)efl). For Hsps with conserved HSEs, 193 HSE motifs were annotated, including 114 head types (‘nGAAn’) and 79 tail types (‘nTTCn’; Figs. 1, 2 and 3). Across all sampled insects, we found no consistent preference for head or tail motifs in hsp83 (exact binomial test, p = 0.055), significant preference for the head motif in hsc70-4 (p < 0.001), and significant preference for the tail motif in hsp40 (p < 0.05).
Fig. 1.

Evolutionary gains and losses in hsp83 within Hymenoptera, followed by diversification in cis-regulatory HSEs. Relationships of homologous hsp83 were reconstructed with PhyML for 17 insect species (rooted with A. pisum) using a JTT substitution model with 1000 bootstrap replicates (>90 bootstrap support indicated; left). Branches of the outgroup taxa are colored in blue and black, while well-supported paralogues of Hymenopteran branches are colored in orange (h1) and red (h2). Statistically significant episodic diversifying selection using Branch-Rel is indicated along the branch (+ corresponds to p < 0.05; * = p < 0.01; ** = p < 0.001). Cis-regulatory HSEs in the promoter region spanning 400 bps from the transcription start site (TSS; right) are mapped onto the phylogeny and are annotated by their length and motif type
Fig. 2.
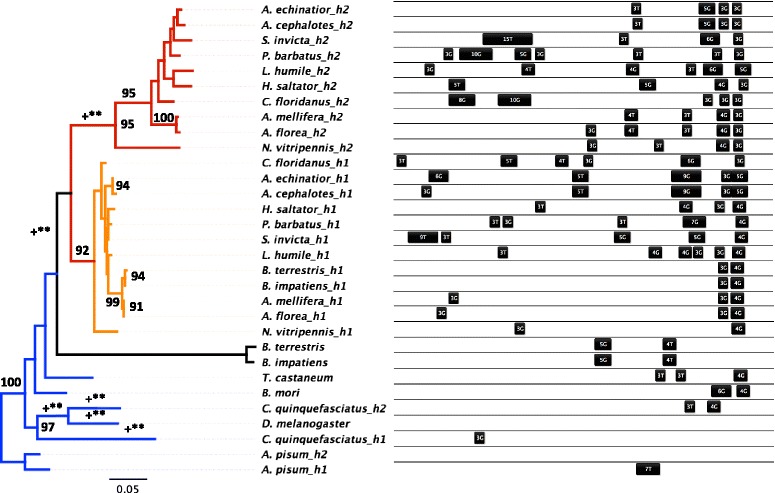
Evolutionary conservation of two copies of hsc70-4 within Hymenoptera, but both copies harbor an extraordinary amount of diversity in cis-regulatory HSEs. Relationships of homologous hsc70-4 were reconstructed with PhyML for 17 insect species (rooted on A. pisum) using a JTT substitution model with 1000 bootstrap replicates (>90 bootstrap support indicated; left). Branches of the outgroup taxa are colored in blue, while well-supported paralogues of Hymenopteran branches are colored in orange (h1) and red (h2). Statistically significant episodes of positive selection identified with Branch-Rel are indicated along the branch(+ corresponds to p < 0.05; * = p < 0.01; ** = p < 0.001). Cis-regulatory HSE elements in the promoter region spanning 570 bps from the transcription start site (TSS; right side) are mapped onto the phylogeny and are annotated by their length and motif type
Fig. 3.

Evolutionary conservation of hsp40 copy number and cis-regulatory HSEs. Relationships of homologous hsp40 were reconstructed with PhyML for 17 insect species (rooted on A. pisum) using a JTT substitution model with 1000 bootstrap replicates (>90 support indicated). The outgroup and Hymenopteran branches are indicated in blue and red, respectively. Statistically significant episodes of positive selection using Branch-Rel are indicated along the branch (+ corresponds to p < 0.05; * = p < 0.01; ** = p < 0.001). Cis-regulatory HSE elements in the promoter region spanning 370 bps from the transcription start site (TSS; right side) are mapped onto the phylogeny and are annotated by their length and motif type. S. invicta did not provide enough sequence information for the identification of cis-regulatory HSEs
Heat shock protein (Hsp) and cis-regulatory heat shock element (HSE) evolution
Hsp83
Phylogenetic reconstruction of hsp83 revealed multiple duplications and losses in both the outgroup and Hymenoptera (Fig. 1). An early duplication event in a common ancestor of the Hymenoptera generated two paralogues of hsp83 (h1 and h2 in Fig. 1). Although both paralogues are present in bees and wasps, only one paralogue (h2) exists in ants, indicating a secondary loss. A second duplication of the h2 orthologue occurred in Linepithema humile. Selection analysis along the length of the gene sequence indicated that most sites (608/714 and 625/714, Single likelihood ancestor counting (SLAC) and Relative effects likelihood (REL) analyses, respectively, Table 2) identified purifying selection; there was no evidence for episodic diversifying selection in branches leading to Hymenopteran paralogues (Branch-REL, p > 0.5; Fig. 1).
Table 2.
Summary of selection analyses for three HSP genes
| Global ω | ω-/ω+ | ||||||
|---|---|---|---|---|---|---|---|
| Gene | N | Codons | SLAC | REL | SLAC | FEL | MEME |
| hsp83 | 25 | 714 | 0.0603 | 0.071 | 608/0 | 625/0 | NA/1 |
| hsc70-4 | 31 | 710 | 0.0549 | 0.051 | 608/0 | 610/0 | NA/7 |
| hsp40 | 17 | 384 | 0.1147 | 0.100 | 253/0 | 284/0 | NA/1 |
For each gene, the number of sequences and number of codons were used for detecting positive selection. The mean global ω is shown for SLAC and REL methods. The number of sites that are negatively or positively selected are shown under ω -/ ω + for SLAC, FEL. P-values were set to default (p = 0.1) for SLAC, FEL, REL. MEME provides evidence for episodic positive selection at individual branches and sites (p < 0.01) and the number of negatively selected sites are non applicable (NA)
In spite of overall sequence conservation, Hymenopteran hsp83 h2 differs in genomic structure and cis-regulation from Hymenopteran hsp83 h1 and from outgroup species in three ways. First, Hymenopteran hsp83 h1 and most outgroup species completely lack introns, whereas hsp83 h2 has two introns; Apis mellifera hsp83 h1 is the exception, with one intron in hsp83 h1 (Additional file 4: Figure S4). Second, Hymenopteran hsp83 h2 has a split HSE arrangement (4–6 and 3 HSE motifs), whereas both hsp83 Hymenopteran h1 and the outgroup have a contiguous HSE arrangement (6–9 HSE motif length) at the proximal end of the molecule (30–100 bps upstream TSS; Fig. 1). Third, there is a preference in head-type motifs only in Hymenopteran hsp83 h2 (Fisher’s Exact Test, p <0.001; Fig. 1).
Hsc70-4
Phylogenetic reconstruction of hsc70-4 indicates multiple duplication events both within species (C. quinquefasciatus and A. pisum) and in a common ancestor of the Hymenoptera, leading to two paralogues (h1 and h2; Fig. 2). Each paralogue forms a strongly supported clade, with the exception of the two Bombus species, in which the h1 paralogue is nested within the h1 clade but the second copy does not group with either Hymenopteran paralogue (Fig. 2). There is evidence of episodic diversifying selection along the branch preceding the hsc70-4 duplication in the Hymenoptera and also in the Hymenopteran hsc70-4 h2 lineage (Branch-REL, p <0.001 in both cases; Fig. 2), even though most individual sites (608/710 and 610/710, SLAC and REL analyses, respectively) were under purifying selection (Table 2).
Hymenopteran hsc70-4 differs in genomic structure and cis-regulatory HSEs from that of D. melanogaster. The orthologue of hsc70-4 in D. melanogaster lacks introns and cis-regulatory HSEs (Additional file 5: Figure S5; Fig. 2). In contrast, Hymenopteran hsc70-4 h1 has one intron, with the exception of N. vitripennis, which has two introns. Hymenopteran hsc70-4 h2 also has two introns, with the exception of Bombus (Additional file 5: Figure S5). Compared to the hsc70-4 in members of the outgroup (Fig. 2, right), both Hymenopteran hsc70-4 paralogues showed high diversification in cis-regulatory HSEs, particularly at the more distal positions ( >120 bps upstream TSS). At the proximal position (30–115 bps upstream TSS), however, HSEs of Hymenopteran hsc70-4 aligned locally with the inducible D. melanogaster hsp70 gene (data not shown).
Hsp40
Both sequence and copy number of hsp40 were phylogenetically conserved across all insect species (Fig. 3). Most sites were under purifying selection (Table 2), and there was no evidence of episodic diversifying selection along branches leading to the Hymenoptera (Fig. 3). Cis-regulatory HSEs of hsp40 were concentrated in one conserved proximal block of 3–7 HSE subunits that were located 35–100 bps upstream of the TSS, although in D. melanogaster HSEs were located 255–285 bps upstream (Fig. 3). However, the genetic structure appears less conserved, ranging from zero to three introns (Additional file 6: Figure S6).
Inducible Hsp expression
We tested whether the presence or absence of conserved cis-regulatory HSEs successfully predicted Hsp gene induction in response to experimental heat shock. The four Hsp genes with conserved HSEs were all significantly up-regulated in response to increasing temperature treatments (hsp83 (F5,12 = 8.48; p < 0.01), hsc70-4 h1 (F5,12 = 3.74; p < 0.05), hsc70-4 h2 (F5,12 = 10.6; p < 0.001), and hsp40 (F5,12 = 6.97, p < 0.01); Fig. 4a–d). The other four Hsps, which lacked conserved HSEs, were not significantly up-regulated after heat shock (hsc70-5 (F5,12 = 2.17; p = 0.13), hsc70-3 (F5,12 = 1.91; p = 0.17), hsp60 (F5,12 = 2.86; p = 0.063), and l(2)efl (F5,12 = 0.223; p = 0.946); Fig. 5a–d).
Fig. 4.
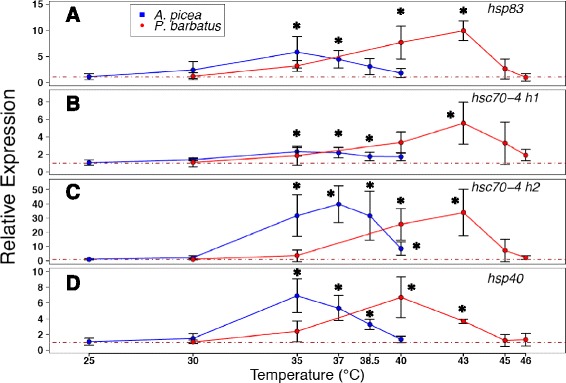
Relative fold increase in gene expression (+/− SD) for four inducible HSPs in A. picea and P. barbatus across different temperature treatment. Relative expression of hsp83 (a), hsc70-4 h1 (b), hsc70-4 h2 (c), and hsp40 (d) were normalized to the 18 s rRNA and β-actin, 18 s rRNA and GAPDH in A. picea (N = 4 per treatment) and P. barbatus (N = 3 per treatment), respectively. Significant up-regulation from 25 °C (A. picea) and 30 °C (P. barbatus) is denoted by ‘*’ from post hoc Tukey tests (p < 0.05)
Fig. 5.

Relative fold change in gene expression (+/− SD) for four non-inducible Hsps in A. picea and P. barbatus across different temperature treatment. Relative expression of hsc70-5 (a), hsc70-3 (b), hsp60 (c), and l(2)efl (d) were normalized to the 18 s rRNA and β-actin and 18 s rRNA and GAPDH for A. picea (N = 4 per treatment) and P. barbatus (N = 3 per treatment), respectively
Species comparisons
We then tested whether variation in thermal tolerances between two ant species was accompanied by changes in Hsp inducibility. The median lethal temperature 50 (LT50) of the warm-climate P. barbatus (median LT50 = 46.9 °C) was significantly higher than the LT50 of the cool-climate A. picea (median LT50 = 38.78 °C; generalized linear model (GLM) with a binomial response variable: influence of species, p < 0.001; Additional file 7: Figure S7). These survivorship differences were matched by patterns of Hsp gene expression: P. barbatus shifted its expression profile toward higher temperatures than did A. picea for all inducible Hsps (Fig. 4a–d). For hsp83, hsc70-4 h1, and hsc70-4 h2, P. barbatus showed peak expression at 43 °C, whereas A. picea showed peak expression at 35–38.5 °C (Fig. 4a–c). For hsp40, peak expression was 40 and 35 °C for P. barbatus and A. picea, respectively (Fig. 4d). P. barbatus exhibited significantly higher constitutive expression of hsc70-4 h1 (ANOVA, F1,5 = 87.8, p < 0.01) and l(2)efl (F1,5 = 6.92, p < 0.05), and significantly lower constitutive expression of hsc70-3 (F1,5 = 596, p < 0.01), hsc70-5 (F1,5 = 24.3, p < 0.001), and hsp60 (F1,5 = 31.2, p < 0.01) than did A. picea (Fig. 6). Among the inducible Hsps, there was a positive relationship between relative basal expression levels and relative inducibility (linear regression, r2 = 0.918, p < 0.05; Fig. 7).
Fig. 6.
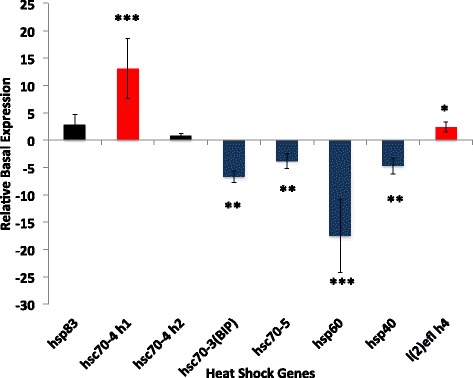
Relative basal heat shock gene (target) expression (+/− SD) between P. barbatus(N = 3) and A. picea (N = 4). Relative gene expression was normalized with the geometric mean of 18 s rRNA and β-actin as the calibrator (* = p < 0.05;** = p < 0.01; *** = p < 0.001 levels of significance) and fold change was calculated as P. barbatus relative to A. picea was calculated as follows: 2Target(Pbar-Apic)/2Calibrator(Pbar-Apic)(Pbar = P. barbatus, Apic = A. picea). -1 was divided by values less than one to calculate negative relative basal expression. Significant up-regulation in P. barbatus and A. picea are colored in red and blue, respectively
Fig. 7.
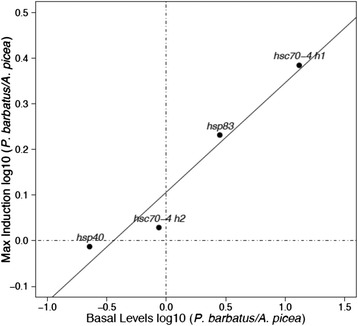
The positive relationship between the log ratios of basal expression levels (P. barbatus/A. picea) at rearing temperatures and max induction (β1 slope = 0.2398, r2 = 0.918, p < 0.05)
Discussion
Molecular characterization of Hymenopteran Hsps reveals functionally important divergence in identity, amino acid sequence, and regulation of chaperone proteins (Table 2, Figs. 1 and 2). Both hsp83 and hsc70-4 display Hymenoptera-specific gains and losses, resulting in unique sets of homologues. Although most codons exhibited purifying selection (Table 1), instances of positive selection along branches leading to and within the Hymenoptera (Figs. 1 and 2, left) suggest novel chaperoning activity [48]. This sequence divergence, coupled with cis-regulatory HSE distribution and expression patterns (Figs. 1 and 2, right; Figs. 4 and 5), suggests that although there is substantial conservation of ancestral inducibility, the HSR response in Hymenoptera has been additionally augmented by expansion and subfunctionalization of novel gene duplicates that are activated by thermal stress.
As in other taxonomic groups, cytoplasmic Hsps mainly mediate the HSR in Hymenoptera (Figs. 4 and 5), whereas mitochondrial and ER-localizing forms of Hsp70 [9, 49] and Hsp90 appear to play little role ([50, 51], but see [52]). The set of inducible Hsps identified likely interact with one another to protect and refold denatured proteins. Upon protein denaturation, Hsp40 delivers unfolded proteins to Hsp70, and the two together mediate refolding through cycles of substrate binding and release driven by ATP binding and hydrolysis [53]. Despite their interdependence, however, the extent of functional diversification of hsc70-4 and hsp40 differed substantially (Figs. 1, 2 and 3). Hsc70-4 showed the most dramatic variation, with the primary inducible member hsp70 in Drosophila completely lost in Hymenoptera, which instead induces two hsc70-4 paralogues that vary in both HSE configuration and fold-increase in response to heat stress (Figs. 2 and 4). Utilization of hsc70 in the stress response across the insects appears to be widespread, with HSEs present in most of the taxa sampled (Fig. 2). Hsc70-4 also contains gene duplications in other taxa, suggesting that this gene family has undergone multiple evolutionary gains, losses and functional shifts. For example, Culex quinquefasciatus has two paralogues, one of which is accompanied by cis-regulatory HSEs (Fig. 2), suggesting that one copy is heat-inducible and the other serves housekeeping functions.
For hsp83, we found two paralogues in bees and wasps, one with an ancestral contiguous arrangement of HSEs, and one with a derived split arrangement similar to that of Drosophila hsp70. This split arrangement reduces cooperative binding of HSF trimers, leading to lower basal expression and higher inducibility than in the more contiguous motif [14, 54, 55]. The presence of two differentially regulated paralogues may reflect novel functionalization in hsp83 to provide both basal and inducible Hsp expression. Foraging bees are known to super-heat thoracic muscles prior and during flight, which necessitates both constitutive and inducible chaperoning activity [32, 34]. Transcriptomic screens in Apis mellifera have found weak support for Hsp90 up-regulation in foraging relative to nurse bees, but more detailed and precise quantification of each paralogue will determine whether they have subfunctionalized into constitutive and inducible roles [56]. In contrast, except for the nuptial flight of males and queens, worker ants are flightless, which may explain the secondary loss of the ancestral paralogue but the retention of the more inducible form.
In contrast to hsc70 and hsp83, hsp40 was much more conserved. There was a single gene copy per taxon in which most sites were under purifying selection, suggesting that their co-chaperoning activity has been retained across insects. In particular, the conserved J domain of Hsp40 stimulates the ATPase domain of Hsp70 proteins. Across the insect hsp40 phylogeny, HSE configuration remained conserved for all but D. melanogaster, whose primary motif was further from the transcriptional start site (Fig. 3). Although the Hsp40 gene family is one of the most diverse molecular chaperones, we captured the paralogue that participates in the HSR because it was significantly up-regulated in response to heat stress. Interestingly, hsp40 in P. barbatus peaked in up-regulation at a less extreme temperature than did the other Hsp proteins (Fig. 4d). Early expression of hsp40 may enhance chaperoning activity by binding to existing and accumulating pools of hsc70 and also by providing crosstalk with Hsp90-mediated chaperoning [57].
Comparisons of two ant species that experience very different thermal ranges revealed correlated shifts in both the basal expression and inducibility of Hsps that reflect the higher and more frequent thermal stress expected in extreme habitats (Fig. 7, Additional file 7: Figure S7). Workers of the harvester ant P. barbatus forage in extreme desert heat [58, 59] and may be more reliant on both constitutive and inducible mechanisms to cope with thermal stress than workers of A. picea, which are more temperature sensitive and occur in thermally buffered mesic deciduous forest [47, 60]. The gene expression responses of P. barbatus and A. picea are consistent with previous work comparing two hot-desert ant species of Cataglyphis with the cool woodland ant Formica polyctena [33]. In that study, HSP70 (hsc70-4) basal expression and induction were higher in Cataglyphis, although alternative paralogues were not fully distinguished. Although Hsp chaperoning activity expends energy (ATP), there may not be trade-offs between continual and maximum induction of Hsps because investment in the HSR is less costly than the loss of biochemical activity from protein denaturation [4, 61, 62]. In addition, the HSR in P. barbatus in this study was shifted upward by ~5–7 °C (Fig. 4), suggesting underlying differences in overall proteome stability that permit P. barbatus to tolerate significantly higher temperatures than A. picea (Additional file 7: Figure S7).
Conclusions
Our study represents the most comprehensive survey to date of Hsp sequence and cis-regulatory evolution for insects. Hymenoptera have unique Hsp evolutionary histories shaped by gains, losses, and changes in cis-regulation. Based on the presence of conserved cis-regulatory elements (HSEs), we reliably predicted the heat inducible Hsps that are critical for mounting the HSR in ants, suggesting that the ancestral inducibility has been retained. We uncovered greater diversity in the number, arrangement, and location of cis-regulatory HSEs in Hymenoptera for two major Hsp genes (hsp83 and hsc70-4), suggesting that the HSR is mediated through changes in cis-regulation. Furthermore, Hsp expression patterns were associated with the thermal limits of two ant species that inhabit different thermal environments. Collectively, our analyses suggest Hsp sequence and expression patterns may reflect the forces of selection acting on thermal tolerance in ants and other social Hymenoptera.
Methods
Phylogenetic reconstruction
To reconstruct the evolutionary relationships of heat shock proteins, we identified orthologous Hsps in 17 insect species representing five insect orders using the well-characterized Hsps of Drosophila melanogaster as a reference (Additional file 8: Table S1). Reciprocal best BLAST (blastp) searches (e-value < 1E-10, and top bit score) were used to identify annotated orthologues of the known D. melanogaster paralogues with an ant-specific genome database (http://antgenomes.org/, [63]) as well as with the NCBI non-redundant protein and nucleotide databases (Additional file 8: Table S1). To find unannotated sequences, we queried D. melanogaster orthologues with tblastn to each insect species’ genome. To identify additional homologues not found with BLAST, we employed a similar search with Hmmer 3.0 [64]. We used Drosophila melanogaster transcripts to search (hmmsearch) each individual genome and identified orthologues based on e-value < 1E-10 and top bit score. HMMER searches recovered nine additional copies from two genes (gp93 and hsp70) for Culex quinquefasciatus. Identified nucleotide sequences were translation-aligned with MAFFT using default settings [65] to identify homologous codons for subsequent selection analyses and the resultant alignment was translated for phylogenetic reconstruction [66]. We reconstructed gene relationships of homologous Hsps with PhyML [66, 67], and bootstrap support was estimated for 1000 replicate searches utilizing an amino acid substitution model inferred from Prottest3 [68]. Similar phylogenetic relationships were recovered with a Bayesian analysis using MrBayes [66, 69].
Tests of selection
Selection at the protein-coding level was quantified as the ratio of the nonsynonymous substitution rate to the synonymous substitution rate (ω = dN/dS); ω > 1 indicates positive selection, whereas ω < 1 indicates purifying selection, and ω = 1 is indicates neutral evolution [70]. For each homologous Hsp, we tested for selection at individual codons as well as across the phylogeny using the HyPhy package [71] on the web-interface Datamonkey (http://www.datamonkey.org).
We identified individual codon sites for positive selection using Single-Likelihood Ancestor Counting (SLAC), Random Effects Likelihood (REL), and Fixed Effects Likelihood [72]. SLAC calculates the number of observed and expected dN and dS rates and conservatively estimates ω using a recommended cutoff of p = 0.1 [72]. The REL method is an extension of analyses in PAML [70] that allows dN and dS to vary across sites and uses a Bayes factor (>50) to assess selection [72]. FEL estimates dN and dS from the codon substitution model and implements a likelihood ratio to evaluate significance using a recommended cutoff of p = 0.1 [72].
In addition to testing for selection at sites along the gene, we tested for changes in selective pressures across the reconstructed amino acid phylogeny, which might indicate evolutionary shifts in gene function. Episodic diversifying selection was assessed using branch-REL and MEME [73, 74]: branch-site REL detects episodic diversifying selection for individual lineages [73], whereas MEME is an extension of FEL that detects episodic diversifying evolution by allowing ω to vary across branches and sites [74].
Identification of genomic structure and cis-regulatory Heat Shock Elements (HSE)
Identification of genomic structure and cis-regulatory HSEs was performed for Hsps that were detectable by qPCR (for methods, see Quantitative real time PCR). We mapped transcripts to their respective genomic regions in Geneious Pro 6.1 [75] and identified exons and introns, making further manual alignments by hand when necessary. The transcriptional start site (TSS) was predicted using Neural Network Promoter Predictor (NNPP) [76]. Previous chip-seq experiments in D. melanogaster revealed that HSF binds primarily to Hsp promoters within 1250 bps of the TSS [77]; sequences were trimmed to this length and locally aligned to identify orthologous HSEs.
To identify cis-regulatory HSEs, we followed a modified search procedure adapted from Tian et al. [17]. Promoter sequences were searched for the putative HSE motif (head conformation = nGAAnnTTCnnGAAn or tail conformation = nTTCnnGAAnnTTCn) [78], allowing for a two base-pair mismatch from the preferred sequence [66]. HSE motifs were then manually screened and annotated by the number and type of subunit occupying the distal end (subunits beginning with ‘nGAAn’ or ‘nTTCn’ refer to the head or tail conformation, respectively). Mismatches occurring at critical sites for HSF binding (G in 2nd position of head conformation, C in 4th position of tail conformation) [79] were discarded, unless motifs were interior to a HSE with three or more subunits, known as a gapped HSE [17].
A final screen was employed to quantify the binding strength of each HSE subunit. Briefly, a WebLogos [80] was generated for head and tail types recovered from the search. Bit scores for the preferred base at each of the five possible positions in a subunit were summed; the match between the individual subunits and the preferred subunit was expressed as the ratio of the summed observed bit score over the preferred bit score. Subunits with scores less than 0.5 were discarded unless flanked with subunits with scores greater than 0.5, again indicating a ‘gapped’ HSE. 253 out of 1753 total HSEs were retained after screening (Additional file 9: Table S2).
Field collections and lab rearing
Hsp induction was quantified in workers sampled from lab-acclimated colonies of Pogonomyrmex barbatus and Aphaenogaster picea. Three Pogonomyrmex barbatus colonies were reared from queens collected with permission following a mating flight at the Welder Wildlife Foundation in Sinton Co., Texas (28.10837 °N 97.42265 °W) in June 2007. Colonies were maintained in an environmental room at the University of Vermont, Department of Biology, with a 12:12 light dark light cycle at 30 °C in 17 × 12 × 6 cm plastic nest boxes provided with three 16 × 150 mm disposable glass test tubes in which water was supplied behind a cotton stopper as a nest site. Each week, colonies were fed two mealworms (Tribolium molitor) and an ad libitum seed mixture composed of oat bran, wheat germ, millet, thistle seeds, and quinoa.
Eight colonies of A. picea were collected with permission from the University of Vermont in May and June 2012 from black spruce forest adjacent to Molly Bog (44.508611°N, 72.702222°W), located near Stowe, Vermont. Entire live colonies containing 500–1000 workers, brood, and queen were excavated from the leaf litter. Colonies were maintained for 1 month in the laboratory at 25 °C +/− 1 °C with 12 h light/dark cycles in a 7 x 3 ¼ x 1 ¾ inch plastic nest box covered with red cellophane and connected to an open plastic foraging arena filled with ~1 cm sand and lined with Insect-a-slip (BioQuip) to prevent escape. 1–3 cotton-plugged water tubes (16 × 150 mm) were provided in the nest box for each colony to maintain humidity. Approximately 200 μl of 20 % honey water and one bisected mealworm were provided in each foraging arena every 3 days.
Thermal tolerance assays
Acute upper thermal limits in both species were determined by quantifying a LT50 temperature, defined as the temperature at which a one-hour exposure produced 50 % worker mortality after 3 days of recovery using the dose.p function in the MASS package within R (version 3.2.0) [81]. Ants were exposed to six different temperature regimes (30, 35, 40, , 42, 45, 46 °C for P. barbatus and 25, 30, 35, 36.5, 38.5 40 °C for A. picea). Temperature treatments were applied by confining 10–13 nest-mate workers together in a 5 mL screw-cap glass vial and submerging the vial in a pre-set Thermo Neslab EX17 heating water bath for 1 h. Temperature inside the vials was monitored with a temperature probe inserted in an empty 5 mL glass vial submerged in the water bath simultaneously. After the application of temperature treatment, ten ants were allowed to recover for survival counts in a 16 × 150 mm cotton-plugged water tube. For each treatment, three ants per colony from four of the eight A. picea colonies and the three P. barbatus colonies were flash frozen and stored at −80 °C for gene expression analyses.
Quantitative real time PCR
RNA was isolated from flash-frozen ants with RNAzol (Molecular Research Center, Inc., USA) and then purified with the RNeazy micro kit (QIAGEN, USA) for downstream gene expression quantification. Flash-frozen ants from each temperature treatment were pooled and homogenized in a Bullet Blender (Next Advance Inc., USA) homogenizer at top speed (10) with 1.4 mm zirconium silicate grinding beads (Quackenbush Co., Inc., USA) and 500 uL of RNAzol buffer (Molecular Research Center, Inc., USA) for 3 min. Following the manufacturer’s instructions for RNAzol, RNA samples were resuspended in 100 uL of water and subsequently purified with Rneasy micro kit with DNAse I (Qiagen, USA) treatment on the micro column to remove genomic DNA contamination. RNA was quantified with Nanodrop spectrophotometry; all sample 260/280 ratios were between 2.0–2.2, indicating acceptable RNA quality. mRNA was reverse transcribed into cDNA with High Capacity cDNA Reverse Transcription Kit (ABI, USA).
To detect specific heat shock proteins, primers were designed for a whole suite of genes for each gene family (Table 1, Additional file 10: Table S3). Table 1 highlights (in *) working primer sets. Quantitative PCR was performed on an ABI StepOnePlus Real-Time PCR system. Reactions were performed in 20 μl volumes with 2 ng of template cDNA, 500 nM total primer, and 10 μl of Power SYBR® Green Master Mix (Life Technologies, USA). Cycling conditions consisted of an initial 95 °C incubation for 2 min and then 40 cycles of 95 °C for 15 s, with 55 °C annealing and extension for 60 s. Following amplification, melt curve analyses confirmed the presence of a single amplicon. All gene products from preliminary experiments were sequenced for verification of specific gene amplification.
Gene expression fold changes were calculated relative to rearing temperatures using the ΔΔ CT method [82] after empirically determining ~100 % primer efficiencies for each primer set (Additional file 10: Table S3). The set of housekeeping genes for normalization was determined with Normfinder [83], which calculated the relative stability of four housekeeping genes (18 s rRNA, GAPDH, β-actin, and Ef1β) and selected the most stable genes across samples. For A. picea, 18 s rRNA and β-actin were most stable (0.20 stability), whereas 18 s rRNA and GAPDH (0.25 stability) were the most stable for P. barbatus. For cross-species comparisons, 18 s rRNA and β-actin were the most stable (0.05 stability). Differences in HSP up-regulation across temperature treatments were determined with a one-way Analysis of Variance (ANOVA) in which fold expression values were log10 transformed to meet assumptions of normality. Significant up-regulation relative to controls was identified with post hoc Tukey tests.
Availability of supporting data
The data sets supporting the results of this article are included in the Dryad Digital repository (https://datadryad.org/resource/doi:10.5061/dryad.8vj6j) [66], within the article and its additional files.
Acknowledgements
This work was supported by the National Science Foundation, Division of Environmental Biology (1136644). We thank Brent L. Lockwood, John Stanton-Geddes, Federico Osorio-Lopez, Yainna Hernàiz-Hernàndez, Michael Herrmann, and Katie Miller for discussions and manuscript suggestions. We also thank three anonymous reviewers for their comments and suggestions, which have substantially improved our manuscript.
Abbreviations
- ANOVA
Analysis of Variance
- Hsp
heat shock protein
- HSR
heat shock response
- HSE
heat shock element
- HSF
heat shock factor
- qPCR
quantitative polymerase chain reaction
Additional files
Maximum likelihood phylogeny of Hsp60 (mitochondrial form) for 17 species of insects (rooted on A. pisum) using a JTT amino acid substitution model and 1000 bootstraps replicates. (DOCX 233 kb)
Maximum likelihood phylogeny of hsc70-3 (BIP) for 17 species of insects (rooted on A. pisum) using a JTT amino acid substitution model and 1000 bootstrap replicates. (DOCX 230 kb)
Maximum likelihood phylogeny of hsc70-5 for 17 species of insects (rooted on A. pisum) using a JTT amino acid substitution model and 1000 bootstrap replicates. (DOCX 240 kb)
Local alignment of the genomic region of orthologous hsp83 from 17 insect species spanning 5 insect Orders. (DOCX 89 kb)
Local alignment of the genomic region of orthologous hsc70-4 from 17 insect species spanning 5 insect Orders. (DOCX 231 kb)
Local alignment of the genomic region of orthologous hsp40 from 17 insect species spanning 5 insect Orders. (DOCX 97 kb)
Percent survival (+/1 SD) of Aphaenogaster picea and P. barbatus (right panel) from heat shock treatments at different temperature treatments. (DOCX 79 kb)
Nucleotide sequences used to characterize the molecular evolution of heat shock proteins. (DOCX 138 kb)
Sequence annotations (position, length, arrangement) of cis-regulatory HSEs for each HSP gene and across all species screened. (DOCX 177 kb)
Primer sets for qPCR including housekeeping and heat shock genes. (DOCX 79 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
ADN, SHC, NJG conceived and designed the study. ADN conducted the phylogenetic reconstruction, tests for selection, thermal tolerance assays, qPCR, and wrote the manuscript. SHC and NJG assisted with the statistical analyses and writing of the manuscript. All three authors have read and approved the submitted manuscript.
Contributor Information
Andrew D. Nguyen, Email: anbe642@gmail.com
Nicholas J. Gotelli, Email: ngotelli@uvm.edu
Sara Helms Cahan, Email: scahan@uvm.edu.
References
- 1.Lindquist S, Craig EA. The Heat-Shock Proteins. Annu Rev Genet. 1988;22:631–77. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 2.Feder ME, Hofmann GE. Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol. 1999;61:243–82. doi: 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- 3.Kültz D. Evolution of the cellular stress proteome: from monophyletic origin to ubiquitous function. J Exp Biol. 2003;206:3119–24. doi: 10.1242/jeb.00549. [DOI] [PubMed] [Google Scholar]
- 4.Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci. 2011;108:680–5. doi: 10.1073/pnas.1017570108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–32. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- 6.Ashburner M, Bonner JJ. The induction of gene activity in drosophila by heat shock. Cell. 1979;17:241–54. doi: 10.1016/0092-8674(79)90150-8. [DOI] [PubMed] [Google Scholar]
- 7.Richter K, Haslbeck M, Buchner J. The Heat Shock Response: Life on the Verge of Death. Mol Cell. 2010;40:253–66. doi: 10.1016/j.molcel.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 8.Nei M, Rooney AP. Concerted and Birth-and-Death Evolution of Multigene Families*. Annu Rev Genet. 2005;39:121–52. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krenek S, Schlegel M, Berendonk TU. Convergent evolution of heat-inducibility during subfunctionalization of the Hsp70 gene family. BMC Evol Biol. 2013;13:49. doi: 10.1186/1471-2148-13-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fink AL. Chaperone-Mediated Protein Folding. Physiol Rev. 1999;79:425–49. doi: 10.1152/physrev.1999.79.2.425. [DOI] [PubMed] [Google Scholar]
- 11.Johnson JL. Evolution and function of diverse Hsp90 homologs and cochaperone proteins. Biochimica et Biophysica Acta (BBA) - Molecular Cell Research. 2012;1823:607–13. doi: 10.1016/j.bbamcr.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 12.Morris JP, Thatje S, Hauton C. The use of stress-70 proteins in physiology: a re-appraisal. Mol Ecol. 2013;22:1494–502. doi: 10.1111/mec.12216. [DOI] [PubMed] [Google Scholar]
- 13.Bonner JJ, Ballou C, Fackenthal DL. Interactions between DNA-bound trimers of the yeast heat shock factor. Mol Cell Biol. 1994;14:501–8. doi: 10.1128/MCB.14.1.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fernandes M, Xiao H, Lis JT. Binding of heat shock factor to and transcriptional activation of heat shock genes in Drosophila. Nucleic Acids Res. 1995;23:4799–804. doi: 10.1093/nar/23.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santoro N, Johansson N, Thiele DJ. Heat Shock Element Architecture Is an Important Determinant in the Temperature and Transactivation Domain Requirements for Heat Shock Transcription Factor. Mol Cell Biol. 1998;18:6340–52. doi: 10.1128/MCB.18.11.6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bettencourt BR, Feder ME. Hsp70 Duplication in the Drosophila melanogaster Species Group: How and When Did Two Become Five? Mol Biol Evol. 2001;18:1272–82. doi: 10.1093/oxfordjournals.molbev.a003912. [DOI] [PubMed] [Google Scholar]
- 17.Tian S, Haney RA, Feder ME. Phylogeny Disambiguates the Evolution of Heat-Shock cis-Regulatory Elements in Drosophila. PLoS One. 2010;5:e10669. doi: 10.1371/journal.pone.0010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Astakhova LN, Zatsepina OG, Przhiboro AA, Evgen’ev MB, Garbuz DG. Novel arrangement and comparative analysis of hsp90 family genes in three thermotolerant species of Stratiomyidae (Diptera) Insect Mol Biol. 2013;22:284–96. doi: 10.1111/imb.12020. [DOI] [PubMed] [Google Scholar]
- 19.Krebs RA, Feder ME. Natural Variation in the Expression of the Heat-Shock Protein HSP70 in a Population of Drosophila melanogaster and Its Correlation with Tolerance of Ecologically Relevant Thermal Stress. Evolution. 1997;51:173–9. doi: 10.2307/2410970. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann AA, Sørensen JG, Loeschcke V. Adaptation of Drosophila to temperature extremes: bringing together quantitative and molecular approaches. J Therm Biol. 2003;28:175–216. doi: 10.1016/S0306-4565(02)00057-8. [DOI] [Google Scholar]
- 21.Dong Y, Miller LP, Sanders JG, Somero GN. Heat-Shock Protein 70 (Hsp70) Expression in Four Limpets of the Genus Lottia: Interspecific Variation in Constitutive and Inducible Synthesis Correlates With in situ Exposure to Heat Stress. Biol Bull. 2008;215:173–81. doi: 10.2307/25470698. [DOI] [PubMed] [Google Scholar]
- 22.Carmel J, Rashkovetsky E, Nevo E, Korol A. Differential Expression of Small Heat Shock Protein Genes Hsp23 and Hsp40, and heat shock gene Hsr-omega in Fruit Flies (Drosophila melanogaster) along a Microclimatic Gradient. J Hered. 2011;102:593–603. doi: 10.1093/jhered/esr027. [DOI] [PubMed] [Google Scholar]
- 23.Buchman AR, Berg P. Comparison of intron-dependent and intron-independent gene expression. Mol Cell Biol. 1988;8:4395–405. doi: 10.1128/MCB.8.10.4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nott A, Meislin SH, Moore MJ. A quantitative analysis of intron effects on mammalian gene expression. RNA. 2003;9:607–17. doi: 10.1261/rna.5250403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Hir H, Nott A, Moore MJ. How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci. 2003;28:215–20. doi: 10.1016/S0968-0004(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 26.Dunn RR, Agosti D, Andersen AN, Arnan X, Bruhl CA, Cerdá X, et al. Climatic drivers of hemispheric asymmetry in global patterns of ant species richness. Ecol Lett. 2009;12:324–33. doi: 10.1111/j.1461-0248.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- 27.Sanders NJ, Lessard J-P, Fitzpatrick MC, Dunn RR. Temperature, but not productivity or geometry, predicts elevational diversity gradients in ants across spatial grains. Glob Ecol Biogeogr. 2007;16:640–9. doi: 10.1111/j.1466-8238.2007.00316.x. [DOI] [Google Scholar]
- 28.Munyai TC, Foord SH. Temporal Patterns of Ant Diversity across a Mountain with Climatically Contrasting Aspects in the Tropics of Africa. PLoS One. 2015;10:e0122035. doi: 10.1371/journal.pone.0122035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaspari M, Clay NA, Lucas J, Yanoviak SP, Kay A. Thermal adaptation generates a diversity of thermal limits in a rainforest ant community. Glob Change Biol. 2015;21:1092–102. doi: 10.1111/gcb.12750. [DOI] [PubMed] [Google Scholar]
- 30.Traniello JFA, Fujita MS, Bowen RV. Ant foraging behavior: ambient temperature influences prey selection. Behav Ecol Sociobiol. 1984;15:65–8. doi: 10.1007/BF00310217. [DOI] [Google Scholar]
- 31.Penick CA, Tschinkel WR. Thermoregulatory brood transport in the fire ant, Solenopsis invicta. Insect Soc. 2008;55:176–82. doi: 10.1007/s00040-008-0987-4. [DOI] [Google Scholar]
- 32.Heinrich B. Mechanisms of Body-Temperature Regulation in Honeybees, Apis Mellifera: I. Regulation of Head Temperature. J Exp Biol. 1980;85:61–72. [Google Scholar]
- 33.Gehring WJ, Wehner R. Heat shock protein synthesis and thermotolerance in Cataglyphis, an ant from the Sahara desert. PNAS. 1995;92:2994–8. doi: 10.1073/pnas.92.7.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elekonich MM. Extreme thermotolerance and behavioral induction of 70-kDa heat shock proteins and their encoding genes in honey bees. Cell Stress Chaperones. 2009;14:219–26. doi: 10.1007/s12192-008-0063-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu PJ, Xiao JH, Xia QY, Murphy B, Huang DW. Apis mellifera has two isoforms of cytoplasmic HSP90. Insect Mol Biol. 2010;19:593–7. doi: 10.1111/j.1365-2583.2009.00923.x. [DOI] [PubMed] [Google Scholar]
- 36.Wang H, Li K, Zhu J-Y, Fang Q, Ye G-Y, Wang H, et al. Cloning and expression pattern of heat shock protein genes from the endoparasitoid wasp, Pteromalus puparum in response to environmental stresses. Arch Insect Biochem Physiol. 2012;79:247–63. doi: 10.1002/arch.21013. [DOI] [PubMed] [Google Scholar]
- 37.Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, et al. Genomic Comparison of the Ants Camponotus floridanus and Harpegnathos saltator. Science. 2010;329:1068–71. doi: 10.1126/science.1192428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nygaard S, Zhang G, Schiøtt M, Li C, Wurm Y, Hu H, et al. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 2011;21:1339–48. doi: 10.1101/gr.121392.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith CD, Zimin A, Holt C, Abouheif E, Benton R, Cash E, et al. Draft genome of the globally widespread and invasive Argentine ant (Linepithema humile) PNAS. 2011;108:5673–8. doi: 10.1073/pnas.1008617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith CR, Smith CD, Robertson HM, Helmkampf M, Zimin A, Yandell M, et al. Draft genome of the red harvester ant Pogonomyrmex barbatus. PNAS. 2011;108:5667–72. doi: 10.1073/pnas.1007901108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suen G, Teiling C, Li L, Holt C, Abouheif E, Bornberg-Bauer E, et al. The Genome Sequence of the Leaf-Cutter Ant Atta cephalotes Reveals Insights into Its Obligate Symbiotic Lifestyle. PLoS Genet. 2011;7:e1002007. doi: 10.1371/journal.pgen.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, et al. The genome of the fire ant Solenopsis invicta. PNAS. 2011;108:5679–84. doi: 10.1073/pnas.1009690108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.The Honeybee Genome Sequencing Consortium Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–49. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadd BM, Barribeau SM, Bloch G, de Graaf DC, Dearden P, Elsik CG, et al. The genomes of two key bumblebee species with primitive eusocial organization. Genome Biol. 2015;16:76. doi: 10.1186/s13059-015-0623-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Werren JH, Richards S, Desjardins CA, Niehuis O, Gadau J, Colbourne JK. Functional and Evolutionary Insights from the Genomes of Three Parasitoid Nasonia Species. Science. 2010;327:343–8. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnson RA. Habitat segregation based on soil texture and body size in the seed-harvester ants Pogonomyrmex rugosus and P. barbatus. Ecol Entomol. 2000;25:403–12. doi: 10.1046/j.1365-2311.2000.00286.x. [DOI] [Google Scholar]
- 47.Lubertazzi D: The Biology and Natural History of Aphaenogaster rudis. Psyche: A Journal of Entomology 2012, 2012
- 48.Carretero-Paulet L, Albert VA, Fares MA. Molecular Evolutionary Mechanisms Driving Functional Diversification of the HSP90A Family of Heat Shock Proteins in Eukaryotes. Mol Biol Evol. 2013;30:2035–43. doi: 10.1093/molbev/mst113. [DOI] [PubMed] [Google Scholar]
- 49.Gross TL, Myles KM, Adelman ZN. Identification and Characterization of Heat Shock 70 Genes in Aedes aegypti (Diptera: Culicidae) J Med Entomol. 2009;46:496–504. doi: 10.1603/033.046.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sørensen JG, Nielsen MM, Kruhøffer M, Justesen J, Loeschcke V. Full genome gene expression analysis of the heat stress response in Drosophila melanogaster. Cell Stress Chaperones. 2005;10:312–28. doi: 10.1379/CSC-128R1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bettencourt BR, Hogan CC, Nimali M, Drohan BW. Inducible and constitutive heat shock gene expression responds to modification of Hsp70 copy number in Drosophila melanogaster but does not compensate for loss of thermotolerance in Hsp70 null flies. BMC Biol. 2008;6:5. doi: 10.1186/1741-7007-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clark MS, Thorne MAS, Toullec J-Y, Meng Y, Guan LL, Peck LS, et al. Antarctic Krill 454 Pyrosequencing Reveals Chaperone and Stress Transcriptome. PLoS One. 2011;6:e15919. doi: 10.1371/journal.pone.0015919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu X-B, Shao Y-M, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–70. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xiao H, Lis JT. Heat shock and developmental regulation of the Drosophila melanogaster hsp83 gene. Mol Cell Biol. 1989;9:1746–53. doi: 10.1128/MCB.9.4.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao H, Perisic O, Lis JT. Cooperative binding of drosophila heat shock factor to arrays of a conserved 5 bp unit. Cell. 1991;64:585–93. doi: 10.1016/0092-8674(91)90242-Q. [DOI] [PubMed] [Google Scholar]
- 56.Whitfield CW, Cziko A-M, Robinson GE. Gene Expression Profiles in the Brain Predict Behavior in Individual Honey Bees. Science. 2003;302:296–9. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- 57.Brychzy A, Rein T, Winklhofer KF, Hartl FU, Young JC, Obermann WMJ. Cofactor Tpr2 combines two TPR domains and a J domain to regulate the Hsp70/Hsp90 chaperone system. EMBO J. 2003;22:3613–23. doi: 10.1093/emboj/cdg362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gordon DM. The rewards of restraint in the collective regulation of foraging by harvester ant colonies. Nature. 2013;498:91–3. doi: 10.1038/nature12137. [DOI] [PubMed] [Google Scholar]
- 59.Helms KR, Vinson B. Surface activity of native ants co-occurring with the red imported fire ant, Solenopsis invicta (Hymenoptera: Formicidae) Southwestern Entomologist. 2005;30:223–37. [Google Scholar]
- 60.Pelini SL, Diamond SE, MacLean H, Ellison AM, Gotelli NJ, Sanders NJ, et al. Common garden experiments reveal uncommon responses across temperatures, locations, and species of ants. Ecol Evol. 2012;2:3009–15. doi: 10.1002/ece3.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krebs RA, Loeschcke V. Costs and Benefits of Activation of the Heat-Shock Response in Drosophila melanogaster. Funct Ecol. 1994;8:730–7. doi: 10.2307/2390232. [DOI] [Google Scholar]
- 62.Sørensen JG, Kristensen TN, Loeschcke V. The evolutionary and ecological role of heat shock proteins. Ecol Lett. 2003;6:1025–37. doi: 10.1046/j.1461-0248.2003.00528.x. [DOI] [Google Scholar]
- 63.Wurm Y, Uva P, Ricci F, Wang J, Jemielity S, Iseli C, et al. Fourmidable: a database for ant genomics. BMC Genomics. 2009;10:5. doi: 10.1186/1471-2164-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mistry J, Finn RD, Eddy SR, Bateman A, Punta M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucl Acids Res. 2013;41:e121–1. doi: 10.1093/nar/gkt263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl Acids Res. 2002;30:3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nguyen A, Gotelli NJ, Helms CS. Data from: the evolution of heat shock protein sequences, cis-regulatory elements, and expression profiles in the eusocial Hymenoptera. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 68.Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–5. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice Across a Large Model Space. Syst Biol. 2012;61:539–42. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–73. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 71.Pond SLK, Frost SDW. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics. 2005;21:2531–3. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 72.Pond SLK, Frost SDW. Not So Different After All: A Comparison of Methods for Detecting Amino Acid Sites Under Selection. Mol Biol Evol. 2005;22:1208–22. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- 73.Pond SLK, Murrell B, Fourment M, Frost SDW, Delport W, Scheffler K. A Random Effects Branch-Site Model for Detecting Episodic Diversifying Selection. Mol Biol Evol. 2011;28:3033–43. doi: 10.1093/molbev/msr125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Kosakovsky Pond SL. Detecting Individual Sites Subject to Episodic Diversifying Selection. PLoS Genet. 2012;8:e1002764. doi: 10.1371/journal.pgen.1002764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput Chem. 2001;26:51–6. doi: 10.1016/S0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 77.Gonsalves SE, Moses AM, Razak Z, Robert F, Westwood JT. Whole-Genome Analysis Reveals That Active Heat Shock Factor Binding Sites Are Mostly Associated with Non-Heat Shock Genes in Drosophila melanogaster. PLoS One. 2011;6:e15934. doi: 10.1371/journal.pone.0015934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Perisic O, Xiao H, Lis JT. Stable binding of Drosophila heat shock factor to head-to-head and tail-to-tail repeats of a conserved 5 bp recognition unit. Cell. 1989;59:797–806. doi: 10.1016/0092-8674(89)90603-X. [DOI] [PubMed] [Google Scholar]
- 79.Fernandes M, Xiao H, Lis JT. Fine structure analyses of the Drosophila and Saccharomyces heat shock factor--heat shock element interactions. Nucleic Acids Res. 1994;22:167–73. doi: 10.1093/nar/22.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.R Development Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria. 2010. [Google Scholar]
- 82.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2 − ΔΔCT Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 83.Andersen CL, Jensen JL, Ørntoft TF. Normalization of Real-Time Quantitative Reverse Transcription-PCR Data: A Model-Based Variance Estimation Approach to Identify Genes Suited for Normalization, Applied to Bladder and Colon Cancer Data Sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]


