Abstract
Background
Species or clades may retain or shift their environmental niche space over evolutionary time. Understanding these processes offers insights into the environmental processes fuelling lineage diversification and might also provide information on past range dynamics of ecosystems. However, little is known about the relative contributions of niche conservatism versus niche divergence to species diversification in the tropics. Here, we examined broad-scale patterns of niche evolution within a Pliocene–Pleistocene clade of epiphytic Bulbophyllum orchids (30 spp.) whose collective distribution covers the northwest and eastern forest ecosystems of Madagascar.
Results
Using species occurrence data, ecological niche models, and multivariate analyses of contributing variables, we identified a three-state niche distribution character for the entire clade, coinciding with three major forest biomes viz. phytogeographical provinces in Madagascar: A, Northwest ‘Sambirano’; B, ‘Eastern Lowlands’; and C, ‘Central Highlands’. A time-calibrated phylogeny and Bayesian models of niche evolution were then used to detect general trends in the direction of niche change over the clade’s history (≤5.3 Ma). We found highest transitions rates between lowlands (A and B) and (mostly from B) into the highland (C), with extremely low rates out of the latter. Lowland-to-highland transitions occurred frequently during the Quaternary, suggesting that climate-induced vegetational shifts promoted niche transitions and ecological speciation at this time.
Conclusions
Our results reveal that niche transitions occurred frequently and asymmetrically within this Madagascan orchid clade, and in particular over Quaternary time scales. Intrinsic features germane to Bulbophyllum (e.g., high dispersal ability, drought tolerance, multiple photosynthetic pathways) as well as extrinsic factors (ecological, historical) likely interacted to generate the niche transition patterns observed. In sum, our results support the emerging idea of dramatic environmental and climatic fluctuations in Madagascar during the recent geological past, which overturns the long-held paradigm of long-term stability in tropical forest settings. The generality of the patterns and timings reported here awaits the availability of additional comparative studies in other Madagascan endemics.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-016-0586-3) contains supplementary material, which is available to authorized users.
Keywords: Macroecological niche, Niche shift vs. niche conservatism, Ancestral character reconstruction, Orchidaceae
Background
Whether species or clades retain or shift their environmental niche space over time is a key question relevant to our understanding of speciation and large-scale patterns of diversification [1–7]. At the species level, the ability of populations to colonize and spread into new (e.g., climatic) niches supports a role for ecological speciation in which divergent natural selection on ecologically important traits promotes population isolation through adaptation to new environments [1, 8–10]. By contrast, niche conservatism is the inability of populations to adapt to new environmental conditions and but nonetheless may facilitate allopatric speciation by maintaining ancestral niches and niche-related traits over time [1, 2, 7]. Moreover, at the phylogenetic level, geographical patterns of niche shifts versus niche conservatism can offer insights into the environmental processes fuelling lineage diversification and may also provide important information on the range dynamics of species and ecosystems over geological time (e.g., [7, 9, 11–13]). However, few phylogenetic-dating studies have explicitly addressed this latter issue in tropical biota, and those that did, mainly focussed on animals (e.g., [13, 14] but see [15–17] for plant examples).
Madagascar is one of the world’s richest biodiversity hotspots [18], with about 3.2 % and 2.8 % of the estimated global total for number of species of vascular plants and vertebrates, respectively [19, 20]. This rich tropical species diversity is commonly attributed to the island’s long-term geological isolation from Africa (<150 million years ago, Ma) and India (<90 Ma), a high geological and topographical complexity, and regionally pronounced and locally steep environmental (e.g., climatic, elevational) gradients [21–23]. Originally, the forest ecosystems and climates of Madagascar, like elsewhere in the tropics, were believed to have been relatively stable through most of their Cenozoic history, thus allowing the built-up of high species diversity (e.g., [24–26]; but see [7, 27–29]). However, Madagascar’s pollen and vertebrate subfossil record is scarce (e.g., [30, 31]), which makes it difficult to formulate a priori hypotheses about the impact of past vegetation-climate changes on the niche evolution of local biota. Nonetheless, in agreement with the few palaeo-data available (e.g., [32–35]), there is growing evidence from phylogeographical studies of forest-dwelling plants and animals (e.g., [36–38]) that dramatic vegetational shifts took place in Madagascar during the climatic fluctuations of the Quaternary (≤2.6 Ma). Furthermore, using palaeoclimatic modelling, Rakotoarinivo et al. [29] have shown that Quaternary changes in vegetation-precipitation relationships in Madagascar exerted a strong influence on plant species richness, as exemplified by palms (Arecaceae) of the island’s eastern rainforest biomes. It is feasible, therefore, that Madagascan forest ecosystems underwent repeated range expansion/contraction cycles due to Quaternary palaeoclimatic change, which in turn promoted phylogenetic niche shifts along steep environmental gradients [21–23, 39–42]. An alternative, mutually non-exclusive hypothesis is that niche conservatism had likewise a role in facilitating speciation and lineage divergence through the maintenance of long-term allopatry in proposed glacial refuge locations [21], notably habitat isolation caused by aridification of low-elevation/montane river catchments [23, 29].
Molecular phylogenetic tools are increasingly used to examine patterns and processes of diversification in plant and animal groups from Madagascar [23, 38, 43–47]. However, to our knowledge, no study to date has specifically examined niche evolution within a Madagascan clade using phylogenetic reconstruction and ecological modelling as integrative tools of the emergent field of ‘phyloclimatic analysis’ (e.g., [11, 13, 48] and references therein; but see [47]). For example, if niche shifts play an important role in phylogenetic history, we would expect that species niches have non-overlapping geographical distributions and are phylogenetically over-dispersed (i.e., mixed among clades); conversely, under phylogenetic niche conservatism, species sharing the same niche will be phylogenetically clustered [1, 4, 5, 10, 47, 49, 50]. Another prediction of the niche shift hypothesis is that the distribution of niche variables across a phylogenetic tree is not significantly different from a random distribution, while under niche conservatism one would expect a strong tendency of related species to resemble each other in such variables more than if taken at random from the tree [3, 15, 51, 52].
Here, we adopted a phyloclimatic analysis approach to determine broad-scale patterns of niche evolution during the diversification of a well-defined clade of the pantropic genus BulbophyllumThouars (Orchidaceae, Epidendroideae) from Madagascar and adjacent islands. With about 2200 to 2400 described species, Bulbophyllum is one of the largest genera within the orchid family, and has a major center of diversity in Madagascar (>210 spp.), where species mainly occur as epiphytes in a wide range of rainforest habitats [53–56]. Based on molecular phylogenetic evidence [56–59], Madagascan Bulbophyllum forms a monophyletic group of Late Miocene age (c. 10.5 Ma) with two major lineages: a species-rich core clade (‘A–B’) mainly distributed in eastern rainforest of mid-to-high elevation (c. 800–1300 m), and a species-poor clade (‘C’) that is eco-geographically more varied (Fig. 1, see below). That said, new species of Bulbophyllum continue to be discovered in Madagascar and the Mascarenes ([60, 61]; G. A. Fischer, unpubl. data) but are rare and threatened with extinction, as typical for other rainforest taxa of these islands (e.g., [29]).
Fig. 1.

Members of Madagascan Bulbophyllum clade C in their different forest habitats. (a) B. complanatum from the seasonally dry region of Northwest Madagascar, (b) B. elliotii from the humid rainforest of the eastern coast, (c) B. occultum from the mid-to-high elevation humid rainforest of the eastern slopes. Photos by G. A. Fischer
In this study, we used a time-calibrated species-level phylogeny of Madagascan Bulbophyllum ‘clade C’ (30 spp.), previously constructed from plastid and nuclear markers [59], as a framework to examine patterns and processes of niche evolution during the clade’s Plio-/Pleistocene diversification (c. ≤ 5.3 Ma). In general, we used a broad-scale, macroecological perspective at the clade-level rather than looking at the niche space of individual species, as the majority of Bulbophyllum species have small to very small ranges, so that sample sizes would be too low to implement such an approach for most species (e.g., [62, 63]). Accordingly, based on georeferenced occurrence data and bioclimatic/elevation variables of the entire clade, we first identified and characterized the major partitions of the dataset in terms of their (supposedly) realized niche space (sensu Soberón [64, 65] using, respectively, fuzzy clustering/ordination analyses in conjunction with ecological niche models (ENMs) [66] and predicted niche occupancy (PNO) profiles [11]. Second, we traced transitions between these realized niches across the tree and used a Bayesian modelling framework to test for a general trend in the direction of niche change through time. Finally, we assessed the extent of phylogenetic signal in variables of the clade’s niches. Together, our results reveal that niche transitions occurred frequently and asymmetrically within this Madagascan orchid clade, and in particular over Quaternary time scales. This supports the emerging idea that climate-induced vegetational shifts in the recent geological past gave rise to current eco-geographical patterns of biota in this tropical biodiversity hotspot.
Methods
Study system and phylogenetic framework
Taxonomically, Madagascan Bulbophyllum ‘clade C’ comprises members of sects. Bifalcula, Humblotiorchis, and Calamaria with altogether 32 recognized species ([56]; G. A. Fischer, B. Gravendeel, J. Hermans, A. Sieder, M. Kiehn, J. Andriantiana, P. J. Cribb, unpubl. data; see (Table 1). These leaf-succulent epiphytes (or rarely lithophytes) are mainly restricted to Northwest/East Madagascar (27 spp.) and/or adjacent islands (Mascarenes: La Réunion/Mauritius, Comores, Seychelles; 3 spp.), with two found in Madagascar and/or East Africa (B. humblotii, B. malawiense; see [59]). Preferential habitats range from seasonally dry to coastal/littoral and humid evergreen forests at various altitudes (0–1800 m) ([56, 58, 67]; G. A. Fischer, B. Gravendeel, J. Hermans, A. Sieder, M. Kiehn, J. Andriantiana, P. J. Cribb, unpubl. data). As typical for Bulbophyllum, most clade C species are adapted to insect (usually fly) mediated outcrossing (e.g., [68, 69]), excepting eight species (indicated in Table 1) that are polymorphic for both outcrossing and selfing variants [70, 71]. This intra-specific mating type polymorphism, however, is not correlated with the macroecological niche states and their contributing variables reported herein (A. Gamisch, unpubl. data), and thus will not be considered further.
Table 1.
Niche-cluster membership of 604 data point localities itemized per species of Bulbophyllum clade C (sects. Bifalcula, Calamaria, Humblotiorchis), and outgroups, used as tip-states for the reconstruction of ancestral niche states A, B, and C (see Fig. 4). Note, two clade C species are not included in the present study (i.e., B. cryptostachium Schltr., B. sp. nov. ‘F’)
| Niche states | Coding | |||
|---|---|---|---|---|
| Section/species | A | B | C | |
| Bifalcula | ||||
| B. capuronii Bosser | 0 | 4 | 0 | B |
| B. complanatum a H.Perrier | 11 | 0 | 0 | A |
| B. implexum Jum. & H.Perrier | 12 | 0 | 0 | A |
| B. minutum Thouars | 0 | 12 | 0 | B |
| B. sp. nov. A | 0 | 3 | 0 | B |
| B. sp. nov. B | 0 | 0 | 1 | C |
| Calamaria | ||||
| B. bicoloratum a Schltr. | 3 | 10 | 43 | C |
| B. cirrhoglossum H.Perrier | 0 | 4 | 0 | B |
| B. elliotii Rolfe | 1 | 19 | 0 | B |
| B. erectum a Thouars | 21 | 48 | 0 | AB |
| B. hildebrandtii Rchb.f. | 17 | 0 | 14 | AC |
| B. histrionicum G.A.Fischer & P.J.Cribb | 20 | 0 | 0 | A |
| B. incurvum Thouars | 0 | 0 | 5 | C |
| B. lecouflei Bosser | 2 | 3 | 0 | AB |
| B. luteobracteatum Jum. & H.Perrier | 0 | 0 | 4 | C |
| B. malawiense B.Morris | 0 | 0 | 2 | C |
| B. obtusatum a (Jum. & H.Perrier) Schltr. | 1 | 0 | 9 | C |
| B. occultum a Thouars | 1 | 61 | 86 | BC |
| B. pervillei Rolfe | 0 | 18 | 0 | B |
| B. pusillum a (H.Perrier) G.A.Fischer & P.J.Cribb | 2 | 2 | 35 | C |
| B. quadrifarium a Rolfe | 0 | 11 | 1 | B |
| B. rubrum Jum. & H.Perrier | 9 | 0 | 0 | A |
| B. ruginosum H.Perrier | 0 | 0 | 3 | C |
| B. senghasii G.A.Fischer & A.Sieder | 1 | 0 | 0 | A |
| B. sp. nov. C | 0 | 7 | 0 | B |
| B. sp. nov. D | 0 | 0 | 1 | C |
| B. sp. nov. E1 | 12 | 0 | 0 | A |
| B. sp. nov. E2 | 1 | 0 | 0 | A |
| B. trifarium Rolfe | 8 | 0 | 2 | A |
| Humblotiorchis | ||||
| B. humblotii a Rolfe | 4 | 35 | 6 | B |
| Alcistachys | ||||
| B. variegatum Thouars | 0 | 12 | 3 | B |
| Inversiflorum | ||||
| B. cardiobulbum Bosser | 0 | 0 | 10 | C |
| Kainochilus | ||||
| B. horizontale Bosser | 0 | 0 | 4 | C |
aClade C species polymorphic for (outcrossing/selfing) mating type [71]
The present study capitalizes on a nearly complete molecular phylogeny of clade C (comprised of 30 spp. plus one outgroup species each of related sects. Alcistachys, Kainochilus, and Inversiflorum; Table 1) as previously reconstructed from an eight-gene dataset of five plastid and three nuclear markers [59]. Phylogenetic analyses, using maximum parsimony (MP) (Paup* v. 4.0b10, [72]) and Bayesian inference (BI) (Mrbayes v. 3.1.2, [73]), were conducted on the combined plastid/nuclear dataset (6505 bp in total). Bayesian posterior probability (PP > 0.95) and parsimony bootstrap percentage (BP ≥ 85) were used as estimates of strong clade support. The trees retrieved identified sects. Bifalcula (6 spp.) and Humblotiorchis (1 sp.) as successive sister taxa to sect. Calamaria (23 spp.). An estimate of absolute age for each node was obtained from a relaxed molecular clock analysis in Beast v. 1.6.1 [74] using multiple fossils and geological events for calibration (for details see [59]). The resulting maximum clade credibility (MCC) chronogram is used here to represent the clade C phylogeny and to trace the history of niche shifts across this phylogeny (see below).
Identification and characterization of macroecological niches
The fundamental niche space of species or higher taxa can be separated into variables of biotic interactions and resource-consumer dynamics (‘Eltonian niche’) or into variables of broad-scale environmental conditions (‘Grinnellian niche’) [64]. The interplay between ‘Eltonian’ processes (e.g., competitive exclusion), ‘Grinnellian’ factors (e.g., climate) and others (e.g., dispersal limitation, demographic constraints), defines the realized niche [64]. For the purpose of this study we rely on a reasonable approximation of the realized niche based on climatic and elevation variables of a comprehensive locality dataset for all 30 clade C species (plus outgroups) [65, 75]. This database was assembled from two main sources: (1) own field data gathered during expeditions in Madagascar (G.A.F. and A. Sieder, Vienna) and La Réunion (A.G.); and (2) voucher specimen data (from the period 1850–2010) obtained from herbaria (G, K, MO, NEU, P, TAN, W, WU). Specimen localities without coordinates were geotagged using an internal Gazetteer of Kew Royal Botanic Gardens, topographical maps of Madagascar (1:100.000), and GoogleEarth™. We discarded duplicate presence records per species as well as implausible locality data (e.g., outside a 1 km radius of the Gazetteer locality and/or those of likely misidentified species accessions). In total, the final dataset comprised 604 georeferenced point localities (coordinates available on request), with the number per species ranging from 1 to 148 (mean ± SD, 18.84 ± 28.71).
For each locality in the database, we extracted altitude and 19 bioclimatic (‘bioclim’) data layers for current conditions (~1950–2000) at 30 arc-seconds resolution from the WorldClim database v. 1.4 ([76]; http://www.worldclim.org), using the R-package Raster v. 1.9-70 [77]. To investigate how these 604 data points group in multi-dimensional environmental space into a specific number of different clusters, we used fuzzy C-means (FCM) clustering [78–81]. Compared to conventional (‘hard’) clustering methods, FCM clustering has the advantage that it allows gradual memberships to clusters measured as degrees (0, 1) [82]. This ‘degree of belonging (or fuzziness)’ is defined by a membership coefficient (m), which quantifies a data point’s membership to all the given clusters. FCM clustering is usually regarded more ‘natural’ than ‘hard’ clustering, especially when based on continuous (e.g., environmental, climatic) variables as used here [79, 81, 82].
The 604 data points were fuzzy clustered on the basis of their Euclidian distances calculated from the environmental variables (altitude, bioclim 1–19) using the vegdist function of the R package Vegan v. 2.0-3 [83]. The resultant distance matrix was multi-dimensionally scaled using the cmdscale command in R (i.e., the matrix was scaled such that the distances between the data points were approximately equal to their Euclidian distance). We employed seven different fuzzy validity indices (detailed in [Additional file 1]) to determine the number of clusters existing in the dataset using the functions cmeans and fclustIndex of the R package e1071 v. 1.6 [84]. The number of clusters (K) was set to vary from 2 to 15, each combined with a fuzziness scheme at ten steps (m = 1.1, 1.2 … 2.0), resulting in a total of 140 combinations. Each cmeans run converged before the maximum of 200 iterations was reached. Overall, this FCM analysis identified three clusters (hereafter termed ‘A’, ‘B’, and ‘C’) to which samples were assigned according to their maximum m value (see Results).
We compared the environmental space between the three FCM clusters using principal component analysis (PCA) in R (prcomp command). The PCA was performed on the matrix of variables (altitude, bioclim 1–19) and the component scores of each data point were projected in two dimensions. For each cluster pair, we assessed significance between the first two components (PC1, PC2) using Mann–Whitney U tests. This non-parametric approach was used because PC1 did not fulfil the parametric assumption of normal distribution based on Kolmogorov-Smirnov tests (data not shown).
ENMs were built independently for each FCM cluster using Maxent v. 3.3.3e [66], a maximum entropy method frequently employed for species distribution modelling (SDM) (e.g., [85, 86]). Based on the locality data assigned to each cluster, ENMs were developed using the 19 bioclim data layers plus altitude (see above) to visualize the geographic space and overlap of these clusters, and to compare their ranges with known phytogeographical zones in Madagascar. For each cluster, model performance was assessed using 100 bootstrap replicates (70 % training, 30 % testing). The area under the ‘Receiver Operating Characteristic (ROC) Curve’ (AUC; [87, 88]) was averaged across replicates to determine if models should be removed (AUC < 0.7). Maxent outputs of continuous probabilities for habitat suitability were converted to presence/absence predictions using a logistic threshold maximizing the absolute value of training sensitivity plus specificity (e.g., [89]).
Using the approach of Evans et al. [11], we further characterized each FCM cluster by predicted niche occupancy (PNO) profiles for particular environmental variables using the R package Phyloclim v. 0.8.1 [90]. As input, this program uses Maxent raw data (i.e., cumulative probabilities of habitat suitability) for creating ‘unit area histograms of suitability’, which illustrate the predicted occupancy of an environmental variable by a given set of locality data (in this case of clusters A, B, and C). As predictive variables we selected six that received relatively high loadings in the PCA (see Results).
Ancestral niche reconstructions and testing alternative models of niche transition
We used the submodule Multistate of BayesTraits v. 1.0 ([91]; http://www.evolution.rdg.ac.uk/BayesTraits.html) to reconstruct ancestral niche states under a continuous-time Markov model of discrete character evolution. For each clade C (and outgroup) species, we coded niche state according to the majority of localities per species falling within a certain FCM cluster (hereafter referred to as ‘niche’), and as’polymorphic‘if at least one third of those localities fell into a different niche (see Table 1, showing near-equal species frequencies across niches: A: 9.5, B: 11.5, C: 12). Probabilities of transitions were calculated together with the probabilities of niche states for all internal nodes of the last 5000 post-burn-in trees from the BI analysis of clade C (with outgroup taxa pruned prior to analysis; [58]), which thus accounts for uncertainty in the phylogeny’s branch lengths and topology. The Markov chain Monte Carlo (MCMC) analyses were run for 5,050,000 generations, with a ratedev parameter of 250 (obtained from initial test runs), a reversible-jump hyperprior with an exponential prior (mean seeded from a uniform distribution on the interval 0 to 30), and a burn-in of 50,000 generations. The reconstructed ancestral niche states were plotted onto the Beast-derived (MCC) chronogram of clade C. Phylogenetic niche shifts were quantified by comparing the most probable states between ancestral and descendant nodes [16, 17, 92].
We used a modelling framework within BayesTraits to estimate niche transition rates (q) and to detect any general trend in the direction of niche change (see Table 2). Specifically, we compared a full (six-parameter) model that allowed transitions to vary freely to (1) each of six models with zero uni-directional transitions in all pairwise combinations of states (i.e., qAB = 0; qAC = 0; … qCB = 0); and (2) one model with zero bi-directional transitions from C (i.e., qCB = 0, qCA = 0). Note, this latter model was guided by our ancestral niche reconstructions (see Results). We used Tracer v. 1.5 [93] to compute the marginal likelihoods of the full and constrained models. For model comparison, we used the Bayes Factor (BF) ‘test statistic’ of 2 log(marginal likelihood [unconstrained model]) – log(marginal likelihood [constrained model]) [93–95]. Evidence against the constrained model (i.e., the null hypothesis, H0) was considered to be ‘positive’ (BF value = 2–6), ‘strong’ (6–10), or ‘very strong’ (>10).
Table 2.
Results of testing alternative models of niche transition in BayesTraits using logarithmic Bayes Factor (BF) comparisons
| Model | Marginal likelihood | Standard error | 2(log BF) | q AB | q AC | q BA | q BC | q CA | q CB |
|---|---|---|---|---|---|---|---|---|---|
| No constraint | −34.74 | +/− 0.017 | - | 77.03 | 68.17 | 68.99 | 71.50 | 41.74 | 53.28 |
| q AB = 0 | −37.73 | +/− 0.020 | 5.93 | 0 | 57.61 | 64.94 | 79.81 | 38.02 | 63.57 |
| q AC = 0 | −36.64 | +/− 0.019 | 3.53 | 74.17 | 0 | 67.35 | 81.99 | 31.09 | 53.85 |
| q BA = 0 | −37.37 | +/− 0.020 | 5.12 | 75.50 | 76.98 | 0 | 70.84 | 63.44 | 59.58 |
| q BC = 0 | −38.20 | +/− 0.023 | 5.85 | 78.44 | 83.30 | 74.54 | 0 | 42.40 | 42.62 |
| q CA = 0 | −34.38 | +/− 0.017 | −0.75 | 75.47 | 62.09 | 72.48 | 71.12 | 0 | 51.80 |
| q CB = 0 | −35.21 | +/− 0.017 | 0.80 | 80.33 | 65.13 | 69.27 | 66.25 | 40.37 | 0 |
| q CB = 0, qCA = 0 | −35.00 | +/− 0.017 | 0.46 | 77.64 | 59.20 | 70.45 | 63.67 | 0 | 0 |
Phylogenetic signal in environmental variables
To provide a complementary perspective on the clade’s niche evolution, we used Blomberg’s K [51] to test for correlations between environmental variables and phylogenetic relatedness. In general, this test statistic is used to assess phylogenetic signal by asking whether a given phylogeny (topology and branch lengths) better fits a set of quantitative tip data than expected for data randomly permuted across the tips of the tree [15, 51, 96]. Values of K range from zero to infinity, where K < 1 indicates weak phylogenetic signal, and K > 1 strong signal, implying that close relatives are, respectively, more or less divergent for traits than expected under a Brownian model (K = 1; [51]). First, based on the locality data, we calculated the centroid value of each principal component (PC1, PC2) per FCM cluster, and also the median values of environmental variables (altitude, bioclim 1–19) for each clade C species. Those ‘trait’ values were then used as input data of the R package Picante v. 1.5-2 [97] for calculating values of K for each trait over 100 trees from the BI analysis (derived from [59]). Significance of K was assessed by comparing observed patterns of the variance of independent contrasts of the trait to a null model by shuffling taxa across the tips of the phylogeny (999 randomized data sets).
Results
Macroecological niche identification and characterization
Our results of the FCM cluster analysis offered more than one optimal solution for partitioning the 604 occurrence data of Madagascan Bulbophyllum clade C (plus outgroups) in environmental space (Additional file 1). In detail, of the seven validity measures employed, two (i.e., Dunn’s separation and Xie-Beni indices) suggested clustering of the samples into three groups (K = 3, with m = 1.1 or 2.0), while two others (Bezdek’s partition coefficient and Bezdek’s partition entropy) favoured K = 4 with m = 1.1. By contrast, the remaining indices suggested K = 12, 13, or 14 as best partitions (bold and underlined in [Additional file 1]). However, we favoured K = 3 (with m = 1.1) as suggested by the majority of validity indices (four out of seven) when considering their next to optimal values (bold in [Additional file 1]). Accordingly, the 604 locality data were assigned to three clusters depending on the maximum m value per sample (i.e., A: 125, B: 249, C: 229).
Figure 2a shows the two-dimensional PCA plot of the 604 samples as assigned to their respective FCM clusters (i.e., A, B, or C). Samples of clusters C and B mainly differed along the first component (PC1, accounting for 52 % of the total variance), which was positively correlated mainly with altitude, measures of temperature (e.g., bio6, bio10), and precipitation (bio12) (Additional file 2). By contrast, samples of A and B were largely separated along PC2 (27 %), which was mainly positively correlated with temperature seasonality (bio4) but negatively with both precipitation seasonality (bio15) and isothermality (bio15); (Additional file 2). As a consequence, clusters A and C differed along both PC axes. However, all pairwise comparisons between clusters revealed significant differences for each of the two axes tested (Mann–Whitney U tests, all P < 0.001).
Fig. 2.
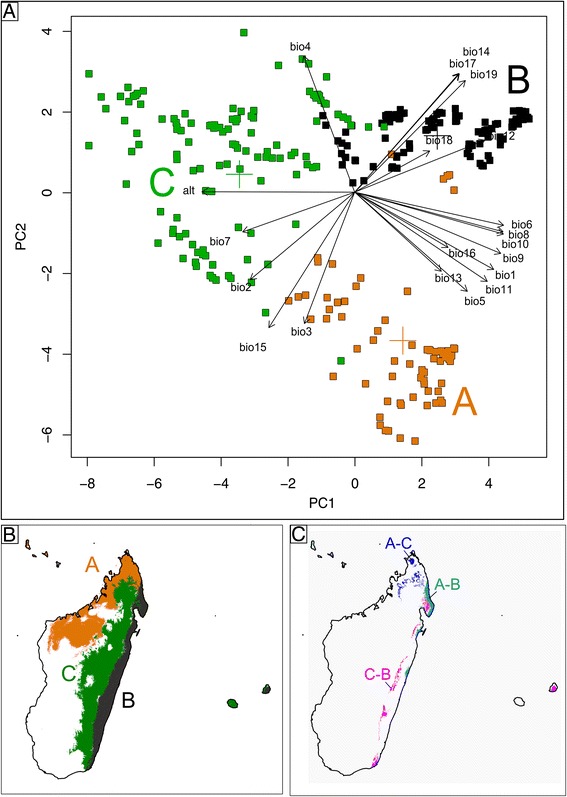
Macroecological clusters viz. niches (A, B, C) of Bulbophyllum clade C from Madagascar and adjacent islands (Comores, La Réunion, Mauritius). a Principal component analysis (PCA) biplot of 604 occurrence data (squared symbols) of clade C species (plus three outgroup species) based on 19 bioclimatic variables (bio 1–19) and altitude (alt). Each data point is coloured according to its hard cluster membership as defined by fuzzy C-means (FCM) clustering. Coloured crosses indicate centroids. (Note: the factor loadings do not correspond to the x or y axes, see [Additional file 2] for factor loadings). b Binary ecological niche models (ENMs) for clusters A, B, and C using a maximum training sensitivity plus specificity logistic threshold. c Range overlap between the ENMs of clusters (A–B, A–C, C–B) based on the same threshold
Spatial-climatic niche characterisation
The ENMs generated for clusters A, B, and C had high AUC values (range: 0.978–0.992), indicating good predictive performance. Based on these models, the predicted distributions of these clusters in Madagascar (Fig. 2b) are largely parapatric with little spatial overlap (c. 1.6–4.6 %; Fig. 2c). Considering further the PNO profiles of each cluster for selected environmental variables (Fig. 3), as well as phytogeographic provinces in Madagascar [20, 98–101], the predicted distributions of clusters A, B, and C (hereafter ‘niches’) can be characterized as follows. Niche A occupies the seasonally dry region of Northwest Madagascar at low-to-mid elevations (0–800 m) (Fig. 2b), and is characterized mainly by constantly high temperatures, coupled with low annual precipitation that markedly varies across seasons (Fig. 3). Phytogeographically, niche A largely coincides with the ‘Sambirano’ rainforest domain [20, 98], which receives heavy seasonal rainfall due to the influence of both easterly trade winds and Indian monsoon currents, but also extends into the western seasonally dry forest at about the same elevation [20, 98–101]. By contrast, niche B coincides with the ‘Eastern Lowlands’ (Fig. 2b) of coastal (littoral) forest, intersected by marshland, and humid rainforest at low-to-mid elevation (c. 0–800 m). When compared to niches A and C, niche B is intermediate in both annual temperature and temperature seasonality; however, it receives the highest annual precipitation, which is continually supplied throughout the year (Fig. 3). Finally, niche C largely coincides with the eastern slopes and plateau of the ‘Central Highlands’ (800–1200 m) (Fig. 2b) characterized by, respectively, mid-to-high elevation humid rainforest and sclerophyllous forest/mountain (‘Philippia’) scrubland [100]. In terms of annual temperature regime, niche C is the coldest and most variable one; it receives similar levels of annual precipitation as niche A but at a more constant rate throughout the year (Fig. 3).
Fig. 3.
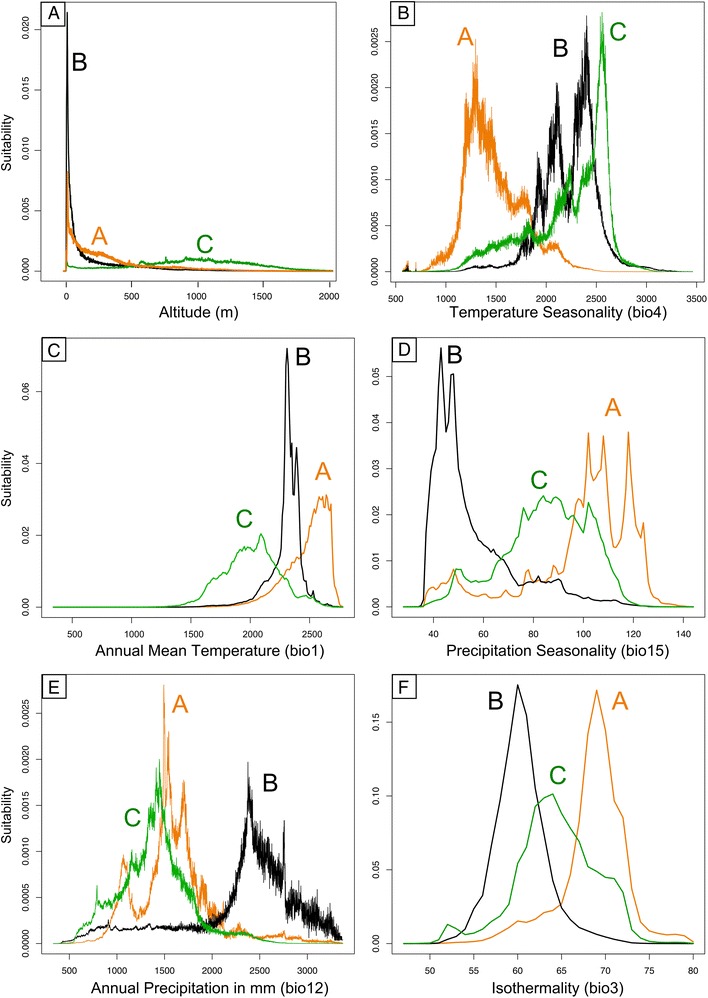
Predicted niche occupancy (PNO) profiles of niche clusters A, B, and C of Madagascan Bulbophyllum clade C (see Fig. 2): for (a) altitude, (b) temperature seasonality (bio4), (c) annual mean temperature (bio1), (d) precipitation seasonality (bio15), (e) annual precipitation (bio12), and (f) isothermality (bio3)
Ancestral niche reconstructions and patterns of niche transition
The Bayesian MCMC reconstruction of ancestral niche states (A, B, C) along the chronogram of clade C (Fig. 4) placed its crown node (c. 5.3 Ma) in either A or B with near equal probabilities. This analysis further revealed frequent niche transitions within this clade, especially in more recent geological times. If we consider only transitions associated with well-supported sister species or subclades (>0.95 PP), then niche shifts may have occurred up to 21 times independently, and all but one of those fall into the Quaternary (≤2.6 Ma; Fig. 4). Altogether 11 transitions appear on internal nodes, including eight from A to B or vice versa, but only one each from A and B to C, and one from C to B. The 10 remaining transitions are located on terminal tip branches (associated with speciation events), including six (mostly from B to C), three from A to B or vice versa, and only one from C to B. Clearly, these inferences about the number of independent transitions and reversals have to be treated with some caution because ancestral states at several nodes appeared equivocal (Fig. 4). Nonetheless, these results qualitatively suggest frequent niche transitions between A and B and almost irreversible transitions from mainly B to C.
Fig. 4.
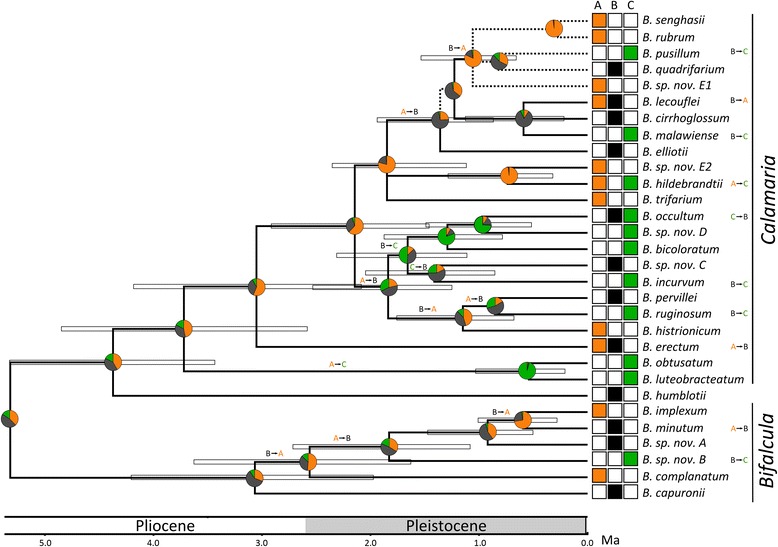
Niche shifts of Madagascan Bulbophyllum clade C. BayesTraits MCMC reconstruction of ancestral niche states (A, B, C) plotted onto a Beast-derived chronogram of Madagascan Bulbophyllum clade C with branch lengths proportional to time (million of years ago, Ma; modified from [59]). Boxes at tips correspond to the character state coding (see Table 1), while pie charts at nodes represent probabilities for ancestral states. Niche shifts and their directionality are indicated at internal branches or next to taxon names if associated with tip branches. Stippled lines indicate phylogenetic posterior probability (PP) support below 0.95. Horizontal bars indicate 95 % highest posterior density (HPD) for node ages
Estimates of transition rates in BayesTraits (Table 2) confirmed this partly asymmetrical niche change within clade C. According to the unconstrained (full) model, transitions were most probable between niches A and B, followed by those from either B to C or from A to C, whereas reversals from C were rendered highly unlikely (see also Fig. 5). Note that this analysis assigned high probability to A to C transitions, despite being observed only two times along the phylogeny (Fig. 4; see above). When we compared the fit of the full model against each of the seven models with asymmetrical transitions (Table 2), there was ‘positive’ to ‘strong’ evidence against all four models with zero (uni-directional) transitions from A or B (BF = 3.53–5.93). However, none of the remaining models, constraining either uni- or bi-directional transitions from C, provided a worse fit than the full model (BF = −0.75–0.80) (Table 2).
Fig. 5.
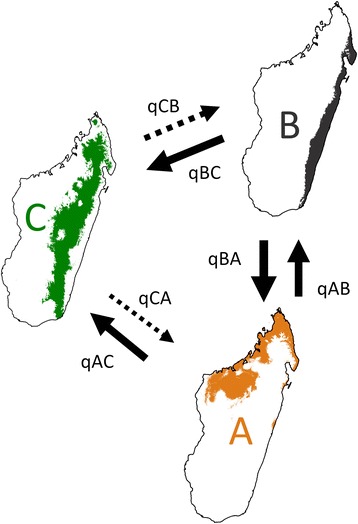
Transition probabilities between niche states (A, B, C). The size of the arrows is proportional to the pairwise transition rates (q AB, q BA, q AC, q CA, q CB, q BC) estimated by BayesTraits, whereas the dashed lines indicate transition rates not significantly different from zero (see Table 2)
Phylogenetic signal
We found no phylogenetic signal of Blomberg’s K for the two principal components (PC1, PC2) and their constituent variables (altitude, bioclim 1–19) (all K < 1; Table 3). Moreover, for PC2 and all variables, the observed distribution of values across the tips of the phylogeny was not significantly different from a random distribution in most of the 100 BI trees analyzed (average 97 %; median P = 0.098–0.749). By contrast, for PC1, the percentage of trees rejecting the null hypothesis of a random signal was relatively high (84 %; median P = 0.03), indicating an even lesser amount of phylogenetic clustering than would be expected by chance (i.e., under a Brownian model of evolution).
Table 3.
Tests for phylogenetic signal of Blomberg’s K for the centroid values of principal environmental components (PC1, PC2) per cluster (A, B, C), and the median values of contributing variables (altitude, bioclim 1–19) per species, over 100 Bayesian trees of Bulbophyllum clade C (derived from [59])
| Variable | Median K | Median P | % of trees not rejecting H0 (P > 0.05)a |
|---|---|---|---|
| PC1 | 0.443 | 0.030 | 16 |
| PC2 | 0.328 | 0.295 | 100 |
| Altitude | 0.354 | 0.297 | 99 |
| Bio1 = Annual Mean Temperature | 0.366 | 0.145 | 93 |
| Bio2 = Mean Diurnal Range | 0.399 | 0.098 | 93 |
| Bio3 = Isothermality | 0.395 | 0.101 | 93 |
| Bio4 = Temperature Seasonality | 0.319 | 0.340 | 100 |
| Bio5 = Max Temperature of Warmest Month | 0.383 | 0.121 | 87 |
| Bio6 = Min Temperature of Coldest Month | 0.355 | 0.218 | 96 |
| Bio7 = Temperature Annual Range | 0.371 | 0.120 | 87 |
| Bio8 = Mean Temperature of Wettest Quarter | 0.366 | 0.220 | 99 |
| Bio9 = Mean Temperature of Driest Quarter | 0.353 | 0.230 | 98 |
| Bio10 = Mean Temperature of Warmest Quarter | 0.377 | 0.170 | 97 |
| Bio11 = Mean Temperature of Coldest Quarter | 0.363 | 0.154 | 92 |
| Bio12 = Annual Precipitation | 0.420 | 0.198 | 100 |
| Bio13 = Precipitation of Wettest Month | 0.343 | 0.231 | 100 |
| Bio14 = Precipitation of Driest Month | 0.331 | 0.370 | 100 |
| Bio15 = Precipitation Seasonality | 0.331 | 0.319 | 100 |
| Bio16 = Precipitation of Wettest Quarter | 0.364 | 0.236 | 99 |
| Bio17 = Precipitation of Driest Quarter | 0.335 | 0.372 | 100 |
| Bio18 = Precipitation of Warmest Quarter | 0.277 | 0.749 | 100 |
| Bio19 = Precipitation of Coldest Quarter | 0.338 | 0.305 | 100 |
aPercentages of trees not rejecting the null hypothesis of random signal for given variable
Discussion
Although tropical environments comprise numerous biodiversity hotspots [18], it is precisely in such environments where least is known about the patterns and processes that drive lineage diversification and niche evolution (e.g., [23, 102]). This is the first study to integrate time-calibrated phylogeny, ancestral niche reconstructions, and hypothesis testing of niche transitions to explore the relative importance of niche conservatism versus niche divergence in the evolutionary history of a plant lineage from the tropical biodiversity hotspot of Madagascar. Based on the evidence from Bulbophyllum orchids, our results reveal an exceptionally high number of broad-scale macroecological niche shifts during the Plio-Pleistocene diversification of ‘clade C’ (30 spp.) in general, and over Quaternary time scales, in particular. Furthermore, this pattern is accentuated by partly asymmetrical niche transitions with little evidence for reversals from highland to lowland areas. Although there are limitations to the methods used (e.g., niche classification based on macroenvironmental conditions only [47] and at the clade level rather than for individual species; equivocal character states at several nodes of the phylogeny), our broad-scale patterns and estimates provide novel insights that can be used for more explicit hypothesis testing to understand historical and ecological factors driving plant diversification in Madagascar.
Niche differentiation and lability within Madagascan Bulbophyllum clade C
Although all clade C species occur in tropical forest biomes, our FCM clustering in conjunction with the PCA (Fig. 2a) and our ENM results (Fig. 2b) revealed the presence of three macroecological niches (A, Northwest ‘Sambirano’; B, ‘Eastern Lowlands’; and C, ‘Central Highlands’), with ‘parapatric’ but de facto non-overlapping ranges (Fig. 2c) thereby largely coinciding with major provinces for the phytogeography of Madagascar [20, 98–101]. Interestingly, 26 of the 30 species included in this study can be assigned unequivocally to one of these three niches (Table 1, Fig. 4), suggesting that (broad-scale) niche conservatism prevails at the intra-specific level. Conversely, those species featuring multiple niche states (i.e., B. erectum, B. hildebrandtii, B. lecouflei, B. occultum) should be ideal candidates for fine-scale studies of incipient ecological speciation.
Our ancestral state reconstructions (Fig. 4) revealed significant evolutionary lability of niches (A, B, C) within clade C. Nonetheless, in several cases, niche conservatism has possibly been important in facilitating species divergence through the maintenance of long-term allopatry (e.g., regarding the sister-species B. occultum/B. bicoloratum; A. Gamisch and U. Jaros, unpubl. data; see also Fig. 4). However, across the entire clade, bioclimatic niche conservatism outweighs respective niche shifts by a ratio of only 2:1, which is considerably lower than the ratio previously reported (25:1) for vascular plants at a global scale [5]. In fact, the total number of 21 transitions inferred for clade C is exceptionally high when compared to other, similar-sized plant lineages that have been identified as undergoing bioclimatic niche shifts over Late Tertiary/Quaternary time scales. As for two examples, six biome shifts were reconstructed within a clade of African Coccinia [16], and just only two within a clade of South American Leucocoryne [103]. Clearly, these inferences about the number of independent transitions and reversals in Bulbophyllum clade C have to be treated with some caution due to the ambiguity in ancestral character reconstruction. In general, this difficulty to estimate ancestral states with confidence can have many different causes (e.g., taxonomic misclassification, incomplete taxon sampling, phylogenetic uncertainty, inappropriate state categorization, high transition rates among states; [104–106]). In the present case, we suspect this uncertainty is most likely caused by the high rates of transition among niche states ([107–109], M. Pagel, pers. comm.), as also inferred from the results of our BayesTraits analyses (Fig. 5, Table 2; see below). However, the statistical nature of this inference needs to be explored in more detail using simulations.
In view of the high number of niche shifts within Madagascan Bulbophyllum clade C, it is not unexpected that we found no phylogenetic signal of Blomberg’s K for principal environmental components (PC1, PC2) and all their contributing variables (Table 3). Such lack of phylogenetic signal in ecological traits is generally interpreted as being indicative of high rates of niche evolution (viz. niche-related trait lability) among closely related species that rapidly diversified to fill new and/or empty niches [51, 110]. This also accords with altogether ten niche transitions associated with tip branches of the clade C phylogeny (Fig. 4), suggesting that niche shifts had a significant role in recent species diversification (see below).
But what factors then could have promoted these frequent niche transitions? One possible explanation is that these tropical orchids possess a suite of characteristics that make them better suited for such transitions than other plant groups. For instance, Bulbophyllum species, like other orchids, possess dust-like and wind dispersed seeds [111–113]. This high dispersal ability may facilitate both habitat selection and stabilizing selection on phenotypic (e.g., niche-related) traits [52, 114, 115]. Additionally, Bulbophyllum produces pseudobulbs with water-storage tissue, which indicates tolerance to drought stress [67, 116, 117]. Notably, too, this genus is highly variable in photosynthetic assimilation pathways, with species exhibiting C3, CAM (Crassulacean Acid Metabolism), and/or C3-CAM intermediate pathways [118–120]. Although little is known about this physiological variation in Madagascan Bulbophyllum (for which only C3 photosynthesis has been reported so far [121]), CAM is generally believed to assist species in occupying a wider range of habitats, including both semiarid and (sub) tropical environments [117, 120, 122, 123]. Together, these general characteristics may help explain the evolutionary versatility of clade C species to exploit new environmental niches and adapt to novel selective regimes (e.g., [124]). Although the species-specific (intrinsic) traits involved remain unknown, they likely interacted in a complex way with both ecological (abiotic and/or biotic) as well as historical (extrinsic) factors to generate the remarkable pattern of niche shifts exhibited by this group (see below).
Directionality of niche transitions
Given the high number of niche shifts and the lack of a phylogenetic signal for environmental variables within Bulbophyllum clade C, one may conjecture that niches varied at random along the phylogeny [7, 16]. This, however, was not observed as we detected several trends in the direction, frequency, and timing of particular niche shifts in this orchid group. Both ancestral niche reconstructions (Fig. 4) and BayesTraits analyses (Fig. 5, Table 2) concurred in that niche shifts within clade C mainly took place from niche A to B, and in the opposite direction, with either niche or, as observed, predominantly B acting as ancestral source for transitions into C. Remarkably, however, reversals back from the dryer and colder high-elevation niche C to the warmer lowlands (A or B) were hardly observed (Fig. 4), and also deemed improbable by the hypothesis testing framework (Fig. 5, Table 2).
The PCA results (Fig. 2a) offer some tentative explanation for the above transition patterns in suggesting differential constraints along the major environmental trajectories (PC1, PC2) linking the three niches. Accordingly, shifts between the lowland niches A and B would need to overcome constraints only along PC2, which may have required adaptive responses to differences in seasonality (temperature, precipitation) and isothermality. By contrast, shifts from B to C mainly proceed along PC1, implying adaptations to overall temperature regime, precipitation, as well as higher elevation. Finally, shifts between A and C would require adaptations along both trajectories (PC1, PC2), and likely therefore are the most severely constrained. In fact, transitions from A to C are rarely observed across the phylogeny (Fig. 4), despite being rendered highly probable by BayesTraits (Fig. 5, Table 2).
However, apart from these different ecological constraints, historical factors may have had a role in shaping the direction and frequency of niche transitions. For instance, Madagascan pollen fossil records, even though scarce, suggest that the primary forest vegetation of the ‘Central Highlands’ was more widespread at lower elevation during the colder (glacial) periods of the Late Quaternary, while receding towards the upper slopes of mountains during warmer inter-/postglacials [32–35]. More recently, this ‘Quaternary vegetational shift’ hypothesis has gained support from population genetic studies of forest-dependent rodent species from northern Madagascar [37, 38]. For Bulbophyllum, there is likewise preliminary evidence from species-specific ENMs for the Last Glacial Maximum (LGM; c. 21,000 year BP) that suitable habitat of high-elevation forest-dwellers (e.g., B. occultum) expanded downwards at that time (A. Gamisch, unpubl. data). It is feasible, therefore, that during the Quaternary climate-induced vegetational shifts (in both elevation and latitude) promoted niche shifts in Bulbophyllum not only between lowlands (A and B) but also from lowland (B) to highland (C) areas. In addition, our results show that 90 % of all speciation events within clade C (27/30) fall into the Quaternary, and when associated with niche shifts, they mostly occur from B to C (five out of ten; Fig. 4). This trend in the timing and frequency of low-to-highland transitions further supports the hypothesis that Quaternary vegetational shifts had an influential role not only in promoting niche shifts per se but also in driving ecological speciation in Madagascan Bulbophyllum.
An outstanding question is why reversals back to lowland niches, i.e., from C to A or B, were hardly observed (Fig. 4) and modelled with zero probability (Table 2). Given the high frequency of B to C transitions, and lesser abiotic (i.e., climatic) constraints along this trajectory relative to those from A to C (see above), one would have expected C to B reversals to occur at least to some extent. Of course, this asymmetry might simply reflect insufficient time elapsed for relevant physiological traits to (re-)evolve in response to warmer and wetter lowland (B) conditions. On the other hand, we may exclude the possibility that reversals are limited by higher extinction rates in niche C, because the time-calibrated phylogeny of the entire clade (Fig. 4) best fits a constant-rates model of diversification without extinction [59]. In other words, niche C is probably not a mere biogeographical ‘sink’ where extinction exceeds colonization. However, with recourse to the ‘Quaternary vegetational shift’ hypothesis, a viable historical scenario is that, associated with inter-/postglacial change to a warmer climate, the upward retreat of the lower limits of the Central Highland forest vegetation severed habitat connections to the Eastern Lowland forest [32, 35, 38]. In the case of Bulbophyllum, this hypothesis implies that, in addition to climate and topography, historical factors likewise imposed constraints on niche shifts from C to B. However, it is also possible that any asymmetry in niche shifts in this plant group from highland to lowland forests may have more to do with biotic interactions (e.g., competition with well-adapted residents; lack of suitable pollinators) than it does with physiological adaptation to lowland climates and/or the fragmentation of past migration routes (see also [125]).
Clearly, a full understanding why clade C species exhibit frequent yet irreversible transitions to higher elevation habitats (niche C) over geologically recent (Quaternary) time scales would require fine-scaled analyses. This, for instance, may involve sister-species or species populations distributed on either side of the B–C niche boundary (e.g., B. pervillei/B. ruginosum), and including genetic, phenotypic, ecological, and distributional (ENM) data for present and past (LGM) climate conditions to test for asymmetric migration and dispersal rates. It also needs to be explored in more ecological detail which biotic factors may have a major role in disconnecting the fundamental and realized niches of these tropical orchids, especially due to the role of competitors and/or pollinators. Nevertheless, the present results concur with phylogenetic reconstructions in other plant and animal taxa from tropical and extra-tropical regions, showing a general trend of niche transitions from lower to higher elevation habitats (e.g., [52, 86, 126–130]), whereas well-supported cases of reverse shifts seem to be rare (but see [39]). Whether this reflects actual patterns or bias in data collection remains unclear, and is worth addressing in greater depth (see [131] for the reminiscent case of mainland-island transitions).
Conclusions
On this broad macroecological scale, our findings highlight an unusually high incidence of niche transitions in an epiphytic orchid lineage from Madagascar, which seems to overturn the long-held paradigm of long-term stability in tropical forest ecosystems [24–26]; but see [7, 27–29]. Over the entire Plio-/Pleistocene history of Bulbophyllum clade C, the highest transitions rates were between the two lowland rainforest niches, Northwest ‘Sambirano’ (A) and ‘Eastern Lowlands’ (B), and into the mixed-forest/scrubland niche of the ‘Central Highlands’ (C), with extremely low rates out of the latter. Intrinsic features germane to this orchid group (e.g., high dispersal ability, drought tolerance, multiple photosynthetic pathways) as well as extrinsic factors (ecological, historical) likely interacted to generate this partly asymmetrical niche transition pattern. Our results also tend to suggest that Quaternary vegetational shifts had an important role in promoting niche transitions in general, and shaping recent speciation via Eastern Lowland-to-Central Highland shifts in particular. The generality of these patterns and timings awaits the availability of additional comparative studies in other Madagascan plant endemics, which might also shed light on the Central Highland biome as a possible ‘end of the colonization road’. Finally, this work clearly demonstrates the need for species-level sampling and molecular ecological analyses of Bulbophyllum clade C to better understand the relative importance of historical and contemporary factors driving speciation in this orchid group.
Availability of supporting data
The data sets supporting the results of this article are included within the article (and its Additional files 1 and 2). The phylogenetic data sets [59] supporting the results of this article are available in the Dryad Digital Repository [doi:10.5061/dryad.35935].
Acknowledgment
We like to thank members of the Botanical Garden of Vienna University (Michael Kiehn, Anton Sieder), the Parc Botanique et Zoologique de Tsimbazaza (Solo Rapanarivo, Jacky Andriantiana), and the Université de la Réunion (Thierry Pailler) for general support and help with fieldwork in Madagascar and La Réunion; the Département des Eaux et Fôrets (Madagascar) and the Parc National de La Réunion for issuing collecting permits; the curators of herbaria (G, K, MO, NEU, P, TAN, W, and WU) for the loan of specimens; and Elisabeth Egger and her team for cultivation of Bulbophyllum at Salzburg Botanical Garden (HBS). We extend special thanks to H. Peter Linder (University of Zürich) and Hanno Schäfer (Technical University Munich) for stimulating discussions on this research topic, to Mark Pagel (University of Reading) for help in interpreting the niche transition rates, and two anonymous reviewers for very insightful comments on earlier versions of this manuscript. This study was funded by the FWF (Austrian Science Fund) grants P20726-B03 (H.P.C.) and P17124-B0 (G.A.F.) and by the Stiftungs- und Förderungsgesellschaft der Paris-Lodron-Universität Salzburg.
Additional files
Fuzzy C-means (FCM) clustering analysis of 604 locality data points of Bulbophyllum clade C (and outgroup taxa). (DOC 209 kb)
Rankings of loadings for the first two principal components (PC1, PC2) of 604 Bulbophyllum clade C (plus outgroup) locality values of 20 environmental variables (altitude, bioclim 1–19). (DOC 40 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AG, HPC and GAF conceived and designed the experiments. AG performed the experiments. AG and HPC analysed the data. AG and HPC drafted the manuscript. All authors read and approved the final manuscript.
Contributor Information
Alexander Gamisch, Email: alexandergamisch@gmx.at.
Gunter Alexander Fischer, Email: gfischer@kfbg.org.
Hans Peter Comes, Email: Hans-Peter.Comes@sbg.ac.at.
References
- 1.Wiens JJ. Speciation and ecology revisited: phylogenetic niche conservatism and the origin of species. Evolution. 2004;58:193–197. doi: 10.1111/j.0014-3820.2004.tb01586.x. [DOI] [PubMed] [Google Scholar]
- 2.Wiens JJ, Graham CH. Niche conservatism: integrating evolution, ecology, and conservation biology. Annu Rev Ecol Evol Syst. 2005;36:519–539. doi: 10.1146/annurev.ecolsys.36.102803.095431. [DOI] [Google Scholar]
- 3.Losos JB. Phylogenetic niche conservatism, phylogenetic signal, and the relationship between phylogenetic relatedness and ecological similarity among species. Ecol Lett. 2008;11:995–1007. doi: 10.1111/j.1461-0248.2008.01229.x. [DOI] [PubMed] [Google Scholar]
- 4.Pearman PB, Guisan A, Broennimann O, Randin CF. Niche dynamics in space and time. Trends Ecol Evol. 2008;23:149–158. doi: 10.1016/j.tree.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Crisp MD, Arroyo MTK, Cook LG, Gandolfo MA, Jordan GJ, McGlone MS, et al. Phylogenetic biome conservatism on a global scale. Nature. 2009;458:754–756. doi: 10.1038/nature07764. [DOI] [PubMed] [Google Scholar]
- 6.Pyron RA, Burbrink FT. Lineage diversification in a widespread species: roles for niche divergence and conservatism in the Common Kingsnake, Lampropeltis getula. Mol Ecol. 2009;18:3443–3457. doi: 10.1111/j.1365-294X.2009.04292.x. [DOI] [PubMed] [Google Scholar]
- 7.Wiens JJ, Ackerly DD, Allen AP, Anacker BL, Buckley LB, Cornell HV, et al. Niche conservatism as an emerging principle in ecology and conservation biology. Ecol Lett. 2010;13:1310–1324. doi: 10.1111/j.1461-0248.2010.01515.x. [DOI] [PubMed] [Google Scholar]
- 8.Coyne JA, Orr HA. Speciation. Sunderland: Sinauer Associates; 2004. [Google Scholar]
- 9.Martínez-Cabrera HI, Schlichting CD, Silander JA, Jones CS. Low levels of climate niche conservatism may explain clade diversity patterns in the South African genus Pelargonium (Geraniaceae) Am J Bot. 2012;99:954–960. doi: 10.3732/ajb.1100600. [DOI] [PubMed] [Google Scholar]
- 10.Hua X, Wiens JJ. How does climate influence speciation? Am Nat. 2013;182:1–12. doi: 10.1086/670690. [DOI] [PubMed] [Google Scholar]
- 11.Evans MEK, Smith SA, Flynn RS, Donoghue MJ. Climate, niche evolution, and diversification of the’bird-cage evening primroses’ (Oenothera, sections Anogra and Kleinia) Am Nat. 2009;173:225–240. doi: 10.1086/595757. [DOI] [PubMed] [Google Scholar]
- 12.Ahmadzadeh F, Flecks M, Carretero MA, Böhme W, Ilgaz C, Engler JO, et al. Rapid lizard radiation lacking niche conservatism: ecological diversification within a complex landscape. J Biogeogr. 2013;40:1807–1818. doi: 10.1111/jbi.12121. [DOI] [Google Scholar]
- 13.Nyári ÁS, Reddy S. Comparative phyloclimatic analysis and evolution of ecological niches in the scimitar babblers (Aves: Timaliidae: Pomatorhinus) PLoS ONE. 2013;8:e55629. doi: 10.1371/journal.pone.0055629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wiens JJ, Graham CH, Moen DS, Smith SA, Reeder TW. Evolutionary and ecological causes of the latitudinal diversity gradient in hylid frogs: treefrog trees unearth the roots of high tropical diversity. Am Nat. 2006;168:579–596. doi: 10.1086/507882. [DOI] [PubMed] [Google Scholar]
- 15.Couvreur TLP, Porter-Morgan H, Wieringa JJ, Chatrou LW. Little ecological divergence associated with speciation in two African rain forest tree genera. BMC Evol Biol. 2011;11:296. doi: 10.1186/1471-2148-11-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holstein N, Renner SS. A dated phylogeny and collection records reveal repeated biome shifts in the African genus Coccinia (Cucurbitaceae) BMC Evol Biol. 2011;11:28. doi: 10.1186/1471-2148-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De-Nova JA, Medina R, Montero JC, Weeks A, Rosell JA, Olson ME, Eguiarte LE, Magallón S. Insights into the historical construction of species-rich Mesoamerican seasonally dry tropical forests: the diversification of Bursera (Burseraceae, Sapindales) New Phytol. 2012;193:276–287. doi: 10.1111/j.1469-8137.2011.03909.x. [DOI] [PubMed] [Google Scholar]
- 18.Myers N, Mittermeier R, Mittermeier GC, DaFonseca GAB, Kent J. Biodiversity hotspots for conservation priorities. Nature. 2000;403:853–858. doi: 10.1038/35002501. [DOI] [PubMed] [Google Scholar]
- 19.de Wit MJ. Madagascar: heads it’s a continent, tails it’s an island. Annu Rev Earth Planet Sci. 2003;31:213–248. doi: 10.1146/annurev.earth.31.100901.141337. [DOI] [Google Scholar]
- 20.Yoder AD, Nowak MD. Has vicariance or dispersal been the predominant biogeographic force in Madagascar? Only time will tell. Annu Rev Ecol Evol Syst. 2006;37:405–31. doi: 10.1146/annurev.ecolsys.37.091305.110239. [DOI] [Google Scholar]
- 21.Wilmé L, Goodman SM, Ganzhorn JU. Biogeographic evolution of Madagascar’s microendemic biota. Science. 2006;312:1063–1065. doi: 10.1126/science.1122806. [DOI] [PubMed] [Google Scholar]
- 22.Pearson RG, Raxworthy CJ. The evolution of local endemism in Madagascar: watershed versus climatic gradient hypotheses evaluated by null biogeographic models. Evolution. 2009;63:959–967. doi: 10.1111/j.1558-5646.2008.00596.x. [DOI] [PubMed] [Google Scholar]
- 23.Vences M, Wollenberg KC, Vieites DR, Lees DC. Madagascar as a model region of species diversification. Trends Ecol Evol. 2009;24:456–465. doi: 10.1016/j.tree.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Richards PW. The Tropical Rain Forest: An Ecological Study. Cambridge: Cambridge Univ. Press; 1952. [Google Scholar]
- 25.Fisher AG. Latitudinal variations in organic diversity. Evolution. 1960;14:64–81. doi: 10.2307/2405923. [DOI] [Google Scholar]
- 26.Ruddiman WF. Earth’s Climate: Past and Future. 2. New York: W.H. Freeman and Company; 2001. [Google Scholar]
- 27.Weir JT. Divergent timing and patterns of species accumulation in lowland and highland neotropical birds. Evolution. 2006;60:842–855. doi: 10.1111/j.0014-3820.2006.tb01161.x. [DOI] [PubMed] [Google Scholar]
- 28.Mittelbach GG, Schemske DW, Cornell HV, Allen AP, Brown JM, Bush MB, et al. Evolution of the latitudinal diversity gradient: speciation, extinction, and biogeography. Ecol Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 29.Rakotoarinivo M, Blach-Overgaard A, Baker WJ, Dransfield J, Moat J, Svenning J-C. Palaeo-precipitation is a major determinant of palm species richness patterns across Madagascar: a tropical biodiversity hotspot. Proc R Soc B. 2013;280:20123048. doi: 10.1098/rspb.2012.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burney DA, James HF, Grady F, Rafamantanantsoa JG, Ramilisonina, Wright HT, et al. Environmental change, extinction, and human activity: evidence from caves in NW Madagascar. J Biogeogr. 1997;24:755–767. doi: 10.1046/j.1365-2699.1997.00146.x. [DOI] [Google Scholar]
- 31.Goodman SM, Wilmé L. Inventaires de la faune et de la flore du nord de Madagascar dans la region Loky-Manambato, Analamerana et Andavakoera. Recherches pour le Developpement, serie Sciences biologiques. 2006;23:1–238. [Google Scholar]
- 32.Burney DA. Climate change and fire ecology as factors in the Quaternary biogeography of Madagascar. In: Lourencxo WR, editor. Biogeographie de Madagascar. Paris: Editions de l’ORSTOM; 1996. pp. 49–58. [Google Scholar]
- 33.Gasse F, Van Campo E. Late Quaternary environmental changes from a pollen and diatom record in the southern tropics (Lake Tritrivakely, Madagascar) Palaeogeogr Palaeoclimatol Palaeoecol. 2001;167:287–308. doi: 10.1016/S0031-0182(00)00242-X. [DOI] [Google Scholar]
- 34.Farrera I, Harrison SP, Prentice IC, Ramstein G, Guiot J, Bartlein PJ, et al. Tropical climates at the Last Glacial Maximum: a new synthesis of terrestrial palaeoclimate data. I. Vegetation, lake-levels and geochemistry. Clim Dynam. 1999;15:823–856. doi: 10.1007/s003820050317. [DOI] [Google Scholar]
- 35.Burney DA, Burney LP, Godfrey LR, Jungers WL, Goodman SM, Wright HT, et al. A chronology for late prehistoric Madagascar. J Hum Evol. 2004;47:25–63. doi: 10.1016/j.jhevol.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 36.Adrianoelina O, Rakotondraoelina H, Ramamonjisoa L, Maley J, Danthu P, Bouvet J. Genetic diversity of Dalbergia monticola (Fabaceae) an endangered tree species in the fragmented oriental forest of Madagascar. Biodivers Conserv. 2006;15:1109–1128. doi: 10.1007/s10531-004-2178-6. [DOI] [Google Scholar]
- 37.Rakotoarisoa JE, Raheriarisena M, Goodman SM. A phylogeographic study of the endemic rodent Eliurus carletoni (Rodentia: Nesomyinae) in an ecological transition zone of northern Madagascar. J Hered. 2012;104:23–35. doi: 10.1093/jhered/ess083. [DOI] [PubMed] [Google Scholar]
- 38.Rakotoarisoa JE, Raheriarisena M, Goodman SM. Late Quaternary climatic vegetational shifts in an ecological transition zone of northern Madagascar: insights from genetic analyses of two endemic rodent species. J Evol Biol. 2013;26:1019–1034. doi: 10.1111/jeb.12116. [DOI] [PubMed] [Google Scholar]
- 39.Knox EB, Palmer JD. Chloroplast DNA variation and the recent radiation of the giant senecios (Asteraceae) on the tall mountains of eastern Africa. Proc Natl Acad Sci U S A. 1995;92:10349–10353. doi: 10.1073/pnas.92.22.10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Graham CH, Ron SR, Santos JC, Schneider CJ, Moritz C. Integrating phylogenetics and environmental niche models to explore speciation mechanisms in dendrobatid frogs. Evolution. 2004;58:1781–1793. doi: 10.1111/j.0014-3820.2004.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 41.Hall JP. Montane speciation patterns in Ithomiola butterflies (Lepidoptera: Rhiodinidae): are they consistently moving up in the world? Proc R Soc B. 2005;272:2457–2466. doi: 10.1098/rspb.2005.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buerki S, Devey DS, Callmander MW, Phillipson PB, Forest F. Spatio-temporal history of the endemic genera of Madagascar. Bot J Linn Soc. 2013;171:304–329. doi: 10.1111/boj.12008. [DOI] [Google Scholar]
- 43.Janssen T, Bystriakova N, Rakotondrainibe F, Coomes D, Labatel J-N, Schneider H. Neoendemism in Madagascan scaly tree ferns results from recent, coincident diversification bursts. Evolution. 2008;62:1876–1889. doi: 10.1111/j.1558-5646.2008.00408.x. [DOI] [PubMed] [Google Scholar]
- 44.Strijk JS, Noyes RD, Strasberg D, Cruaud C, Chase MW, Abbott RJ, et al. In and out of Madagascar: dispersal to peripheral islands, insular speciation and diversification of Indian Ocean daisy trees (Psiadia, Asteraceae) PLoS ONE. 2012;7:e42932. doi: 10.1371/journal.pone.0042932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hong-Wa C, Besnard G. Species limits and diversification in the Madagascar olive (Noronhia, Oleaceae) Bot J Linn Soc. 2014;174:141–161. doi: 10.1111/boj.12112. [DOI] [Google Scholar]
- 46.Miraldo A, Hanski IA. Competitive release leads to range expansion and rampant speciation in Malagasy dung beetles. Syst Biol. 2014;63:480–92. doi: 10.1093/sysbio/syu011. [DOI] [PubMed] [Google Scholar]
- 47.Evans M, Aubriot X, Hearn D, Lanciaux M, Lavergne S, Cruaud C, et al. Insights on the evolution of plant succulence from a remarkable radiation in Madagascar (Euphorbia) Syst Biol. 2014;63:698–711. doi: 10.1093/sysbio/syu035. [DOI] [PubMed] [Google Scholar]
- 48.Jakob SS, Heibl C, Rödder D, Blattner FR. Population demography influences climatic niche evolution: evidence from diploid American Hordeum species (Poaceae) Mol Ecol. 2010;19:1423–1438. doi: 10.1111/j.1365-294X.2010.04582.x. [DOI] [PubMed] [Google Scholar]
- 49.Moritz C, Patton JL, Schneider CJ, Smith TB. Diversification of rainforest faunas: an integrated molecular approach. Annu Rev Ecol Syst. 2000;31:533–563. doi: 10.1146/annurev.ecolsys.31.1.533. [DOI] [Google Scholar]
- 50.Kozak KH, Wiens JJ. Climatic zonation drives latitudinal variation in speciation mechanisms. Proc R Soc B. 2007;274:2995–3003. doi: 10.1098/rspb.2007.1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blomberg SP, Garland T, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 52.Boucher FC, Thuiller W, Roquet C, Douzet R, Aubert S, Alvarez N, et al. Reconstructing the origins of high-alpine niches and cushion life form in the genus Androsace s.l. (Primulaceae) Evolution. 2012;66:1255–1268. doi: 10.1111/j.1558-5646.2011.01483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vermeulen JJ. Orchid Monographs 2: A Taxonomic Revision of the Continental African Bulbophyllinae. Leiden: E.J. Brill; 1987. [Google Scholar]
- 54.Brieger FG, Maatsch R, Senghas K. Rudolf Schlechter: Die Orchideen. 3. Berlin and Hamburg: Verlag Paul Parey; 1992. [Google Scholar]
- 55.Sieder A, Rainer H, Kiehn M. CITES checklist for Bulbophyllum and allied taxa (Orchidaceae). Botanical Garden of the University of Vienna. 2007. www.cites.org/common/com/nc/tax_ref/Bulbophyllum.pdf. Accessed 22 February 2013
- 56.Pridgeon AM, Cribb PJ, Chase MW, Rasmussen N. Genera Orchidacearum Vol. 6. Epidendroideae (Part 3) Oxford: Oxford University Press; 2014. [Google Scholar]
- 57.Fischer GA. Evolution of the orchid genus Bulbophyllum in Madagascar. PhD thesis. Austria: University of Vienna; 2007.
- 58.Fischer GA, Gravendeel B, Sieder A, Andriantiana J, Heiselmayer P, Cribb PJ, et al. Evolution of resupination of Madagascan species of Bulbophyllum (Orchidaceae) Mol Phylogenet Evol. 2007;45:358–376. doi: 10.1016/j.ympev.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 59.Gamisch A, Fischer GA, Comes HP. Multiple independent origins of auto-pollination in tropical orchids (Bulbophyllum) in light of the hypothesis of selfing as an evolutionary dead end. BMC Evol Biol. 2015;15:192. doi: 10.1186/s12862-015-0471-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fischer GA, Sieder A, Hermans J, Andriantiana J, Kiehn M, Cribb PJ. Description of four new species from Madagascar. Novon. 2009;19:344–352. doi: 10.3417/2007141. [DOI] [Google Scholar]
- 61.Bernet P, Castillon JB. Orchidaceae des îles Mascareignes: commentaires sur Bulbophyllum densum et Bulbophyllum pendulum sur l’île Maurice et description d’un Bulbophyllum nouveau de La Réunion; nom nouveau pour Habenaria chloroleuca. Richardiana. 2012;14:13–24. [Google Scholar]
- 62.Frey MA. The relative importance of geography and ecology in species diversification: evidence from a tropical marine intertidal snail (Nerita) J Biogeogr. 2010;37:1515–1528. [Google Scholar]
- 63.Normand S, Ricklefs RE, Skov F, Bladt J, Tackenberg O, Svenning JC. Postglacial migration supplements climate in determining plant species ranges in Europe. Proc R Soc B. 2011;278:3644–3653. doi: 10.1098/rspb.2010.2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Soberón J. Grinnellian and Eltonian niches and geographic distributions of species. Ecol Lett. 2007;10:1115–1123. doi: 10.1111/j.1461-0248.2007.01107.x. [DOI] [PubMed] [Google Scholar]
- 65.Soberón J, Nakamura M. Niches and distributional areas: concepts, methods and assumptions. Proc Natl Acad Sci U S A. 2009;106:19644–19650. doi: 10.1073/pnas.0901637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecol Model. 2006;190:231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 67.Cribb P, Hermans J. A Field Guide to the Orchids of Madagascar. Kew: Royal Botanic Gardens; 2009. [Google Scholar]
- 68.Humeau L, Micheneau C, Jacquemyn H, Gauvin-Bialecki A, Fournel J, Pailler T. Sapromyiophily in the native orchid, Bulbophyllum variegatum, on Réunion (Mascarene Archipelago, Indian Ocean) J Trop Ecol. 2011;27:591–599. doi: 10.1017/S0266467411000411. [DOI] [Google Scholar]
- 69.Chen LJ, Zhang GQ, Li LQ, Zhang YT, Rao WH. Development and maintenance of a cross-mixed mating system in the orchid Bulbophyllum orientale. J Phylogen Evol Biol. 2014;2:124. [Google Scholar]
- 70.Gamisch A, Staedler YM, Schönenberger J, Fischer GA, Comes HP. Histological and micro-CT evidence of stigmatic rostellum receptivity promoting auto-pollination in the Madagascan orchid Bulbophyllum bicoloratum. PLoS ONE. 2013;8:e72688. doi: 10.1371/journal.pone.0072688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gamisch A, Fischer GA, Comes HP. Recurrent polymorphic mating type variation in Madagascan Bulbophyllum species exemplifies high incidence of auto-pollination in tropical orchids. Bot J Linn Soc. 2014;175:242–258. doi: 10.1111/boj.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Swofford DL . PAUP*: Phylogenetic Analysis using Parsimony (* and other methods) version 4.0b10. Sunderland: Sinauer Associates; 2001. [Google Scholar]
- 73.Ronquist F, Huelsenbeck JP. MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 74.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hof C, Rahbek C, Araujo MB. Phylogenetic signals in the climatic niches of the world’s amphibians. Ecography. 2010;33:242–250. [Google Scholar]
- 76.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int J Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 77.Hijmans RJ, van Etten J. raster: Geographic analysis and modeling with raster data. R package version 1.9-70. 2012. http://CRAN.R-project.org/package=raster. Accessed 27 February 2012.
- 78.Bezdek JC. Pattern Recognition with Fuzzy Objective Function Algorithms. New York: Plenum Press; 1981. [Google Scholar]
- 79.Rezaee MR, Lelieveldt BPF, Reiber JHC. A new cluster validity index for the fuzzy c-mean. Pattern Recogn Lett. 1998;19:237–246. doi: 10.1016/S0167-8655(97)00168-2. [DOI] [Google Scholar]
- 80.Recknagel F. Ecological Informatics. Understanding Ecology by Biologically-Inspired Computation. Heidelberg: Springer; 2003. [Google Scholar]
- 81.Markovic D, Freyhof J, Wolter C. Where are all the fish: potential of biogeographical maps to project current and future distribution patterns of freshwater species. PLoS ONE. 2012;7:e40530. doi: 10.1371/journal.pone.0040530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bora DJ, Gupta AK. A comparative study between fuzzy clustering algorithm and hard clustering algorithm. Int J Comp Trends Tech. 2014;10:108–113. doi: 10.14445/22312803/IJCTT-V10P119. [DOI] [Google Scholar]
- 83.Oksanen J, Blanchet GF, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. vegan: Community Ecology Package. R package version 2.0-3. 2012. http://CRAN.R-project.org/package=vegan. Accessed 23 March 2012.
- 84.Dimitriadou E, Hornik K, Leisch F, Meyer D, Weingessel A. e1071: Misc Functions of the Department of Statistics (e1071), TU Wien. R package version 1.6-0. 2012. http://cran.r-project.org/web/packages/e1071/index.html. Accessed 24 March 2012.
- 85.Phillips SJ, Dudık M. Modeling of species distributions with Maxent: new extensions and a comprehensive evaluation. Ecography. 2008;31:161–175. doi: 10.1111/j.0906-7590.2008.5203.x. [DOI] [Google Scholar]
- 86.Qi XS, Chen C, Comes HP, Sakaguchi S, Liu YH, Tanaka N, et al. Molecular data and ecological niche modelling reveal a highly dynamic evolutionary history of the East Asian Tertiary relict Cercidiphyllum (Cercidiphyllaceae) New Phytol. 2012;196:617–630. doi: 10.1111/j.1469-8137.2012.04242.x. [DOI] [PubMed] [Google Scholar]
- 87.Fielding AH, Bell JF. A review of methods for the assessment of prediction errors in conservation presence/absence models. Environ Conserv. 1997;24:38–49. doi: 10.1017/S0376892997000088. [DOI] [Google Scholar]
- 88.Fawcett T. An introduction to ROC analysis. Pattern Recogn Lett. 2006;27:861–874. doi: 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- 89.Hu J, Jiang Z. Climate change hastens the conservation urgency of an endangered ungulate. PLoS ONE. 2011;6:e22873. doi: 10.1371/journal.pone.0022873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Heibl C. phyloclim: Integrating phylogenetics and climatic niche modelling. 2011. http://cran.r-project.org/web/packages/phyloclim/index.html. Accessed 23 May 2012.
- 91.Pagel M, Meade A, Barker D. Bayesian estimation of ancestral character states on phylogenies. Syst Biol. 2004;53:673–684. doi: 10.1080/10635150490522232. [DOI] [PubMed] [Google Scholar]
- 92.Heibl C, Renner SS. Distribution models and a dated phylogeny for Chilean Oxalis species reveal occupation of new habitats by different lineages, not rapid adaptive radiation. Syst Biol. 2012;61:823–834. doi: 10.1093/sysbio/sys034. [DOI] [PubMed] [Google Scholar]
- 93.Rambaut A, Suchard MA, Xie D, Drummond AJ. Tracer v1.5. 2013. [http://tree.bio.ed.ac.uk/software/tracer/]
- 94.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc. 1995;90:773–795. doi: 10.1080/01621459.1995.10476572. [DOI] [Google Scholar]
- 95.Suchard MA, Weiss RE, Sinsheimer JS. Bayesian selection of continuous time Markov chain evolutionary models. Mol Biol Evol. 2001;18:1001–1013. doi: 10.1093/oxfordjournals.molbev.a003872. [DOI] [PubMed] [Google Scholar]
- 96.Abellán P, Ribera I. Geographic location and phylogeny are the main determinants of the size of the geographical range in aquatic beetles. BMC Evol Biol. 2011;11:344. doi: 10.1186/1471-2148-11-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 98.Humbert H. Les territoires phytogeographiques de Madagascar, Leur cartographie. Année Biol. 1955;31:195–204. [Google Scholar]
- 99.Straka H. Über Moore und Torf auf Madagaskar und den Maskarenen. Erdkunde. 1960;14:81–97. [Google Scholar]
- 100.Du Puy DJ, Moat J. A refined classification of the primary vegetation of Madagascar based on the underlying geology: using GIS to map its distribution and to assess its conservation status. In: Lourenço WR, editor. Proceedings of the International Symposium on the Biogeography of Madagascar. Paris: Editions de l’ORSTOM; 1996. pp. 205–218. [Google Scholar]
- 101.Dransfield J, Rakotoarinivo M. The biogeography of Madagascar palms. In: Bramwell D, Caujapé-Castells J, editors. The Biology of Island Floras. Cambridge: Cambridge University Press; 2011. pp. 179–196. [Google Scholar]
- 102.Gonzalez-Voyer A, Padial JM, Castroviejo-Fisher S, de la Riva I, Vilà C. Correlates of species richness in the largest Neotropical amphibian radiation. J Evol Biol. 2011;24:931–942. doi: 10.1111/j.1420-9101.2011.02243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jara-Arancio P, Arroyo MTK, Guerrero PC, Hinojosa LF, Arancio G, Méndez M. Phylogenetic perspectives on biome shifts in Leucocoryne (Alliaceae) in relation to climatic niche evolution in western South America. J Biogeogr. 2014;41:328–338. doi: 10.1111/jbi.12186. [DOI] [Google Scholar]
- 104.Salisbury BA, Kim J. Ancestral state estimation and taxon sampling density. Syst Biol. 2001;50:557–564. doi: 10.1080/106351501750435103. [DOI] [PubMed] [Google Scholar]
- 105.Revell LJ. Ancestral character estimation under the threshold model from quantitative genetics. Evolution. 2014;68:743–759. doi: 10.1111/evo.12300. [DOI] [PubMed] [Google Scholar]
- 106.Duchêne S, Lanfear R. Phylogenetic uncertainty can bias the number of evolutionary transitions estimated from ancestral state reconstruction methods. J Exp Zool (Mol Dev Evol) 2015;324:517–524. doi: 10.1002/jez.b.22638. [DOI] [PubMed] [Google Scholar]
- 107.Schluter D, Price TD, Mooers AØ, Ludwig D. Likelihood of ancestor states in adaptive radiation. Evolution. 1997;51:1699–1711. doi: 10.2307/2410994. [DOI] [PubMed] [Google Scholar]
- 108.Cunningham CW. Some limitations of ancestral character-state reconstruction when testing evolutionary hypotheses. Syst Biol. 1999;48:665–674. doi: 10.1080/106351599260238. [DOI] [Google Scholar]
- 109.Paradis E. Analysis of Phylogenetics and Evolution using R. New York: Springer; 2006. [Google Scholar]
- 110.Kamilar JM, Cooper N. Phylogenetic signal in primate behaviour, ecology and life history. Phil Trans R Soc B. 2013;368:20120341. doi: 10.1098/rstb.2012.0341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ridley HN. The Dispersal of Plants Throughout the World. Reeve & Co: Ashford; 1930. [Google Scholar]
- 112.Arditti J, Ghani AKA. Numerical and physical properties of orchid seeds and their biological implications. Tansley Review No. 110. New Phytol. 2000;145:367–421. doi: 10.1046/j.1469-8137.2000.00587.x. [DOI] [PubMed] [Google Scholar]
- 113.Jersáková J, Malinová T. Spatial aspects of seed dispersal and seedling recruitment in orchids. New Phytol. 2007;176:235–237. doi: 10.1111/j.1469-8137.2007.02224.x. [DOI] [PubMed] [Google Scholar]
- 114.Donoghue MJ. A phylogenetic perspective on the distribution of plant diversity. Proc Natl Acad Sci U S A. 2008;105:11549–11555. doi: 10.1073/pnas.0801962105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Crisp MD, Cook LG. Phylogenetic niche conservatism: what are the underlying evolutionary and ecological causes? New Phytol. 2012;196:681–694. doi: 10.1111/j.1469-8137.2012.04298.x. [DOI] [PubMed] [Google Scholar]
- 116.Johansson DR. Ecology of vascular epiphytes in West African rain forests. Acta Phytogeogr Suec. 1974;59:1–136. [Google Scholar]
- 117.Silvera K, Santiago LS, Cushman JC, Winter K. Crassulacean acid metabolism and epiphytism linked to adaptive radiations in the Orchidaceae. Plant Physiol. 2009;149:1838–1847. doi: 10.1104/pp.108.132555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Arditti J. Aspects of physiology of orchids. Adv Bot Res. 1969;7:421–655. doi: 10.1016/S0065-2296(08)60091-9. [DOI] [Google Scholar]
- 119.Winter K, Wallace BJ, Stocker GC, Roksandic Z. Crassulacean acid metabolism in Australian vascular epiphytes and some related species. Oecologia. 1983;57:129–141. doi: 10.1007/BF00379570. [DOI] [PubMed] [Google Scholar]
- 120.Earnshaw MJ, Winter K, Ziegler H, Stichler W, Cruttwell NEG, Kerenga K, et al. Altitudinal changes in the incidence of crassulacean acid metabolism in vascular epiphytes and related life forms in Papua New Guinea. Oecologia. 1987;73:566–572. doi: 10.1007/BF00379417. [DOI] [PubMed] [Google Scholar]
- 121.Kluge M, Brulfert J, Rauh W, Ravelomanana D, Ziegler H. Ecophysiological studies on the vegetation of Madagascar: a δ13C and δD survey for incidence of crassulacean acid metabolism (CAM) among orchids from montane forests and succulents from the xerophytic thorn-Bush. Isot Environ Health Stud. 1995;31:191–210. doi: 10.1080/10256019508234018. [DOI] [Google Scholar]
- 122.Lüttge U. Ability of crassulacean acid metabolism plants to overcome interacting stresses in tropical environments. AoB Plants. 2010;2010:plq005. doi: 10.1093/aobpla/plq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Silvestro D, Zizka G, Schulte K. Disentangling the effects of key innovations on the diversification of Bromelioideae (Bromeliaceae) Evolution. 2013;68:163–175. doi: 10.1111/evo.12236. [DOI] [PubMed] [Google Scholar]
- 124.Bone RE, Smith JAC, Arrigo N, Buerki S. A macro-ecological perspective on crassulacean acid metabolism (CAM) photosynthesis evolution in Afro-Madagascan drylands: Eulophiinae orchids as a case study. New Phytol. 2015;doi:10.1111/nph.13572. [DOI] [PubMed]
- 125.Donoghue MJ, Edwards EJ. Biome shifts and niche evolution in plants. Annu Rev Ecol Evol Syst. 2014;45:547–572. doi: 10.1146/annurev-ecolsys-120213-091905. [DOI] [Google Scholar]
- 126.Monasterio M, Sarmiento L. Adaptive radiation of Espeletia in the cold Andean tropics. Trends Ecol Evol. 1991;6:387–391. doi: 10.1016/0169-5347(91)90159-U. [DOI] [PubMed] [Google Scholar]
- 127.Kropf M, Kadereit JW, Comes HP. Differential cycles of range contraction and expansion in European high mountain plants during the Late Quaternary: insights from Pritzelago alpina (L.) O. Kuntze (Brassicaceae) Mol Ecol. 2003;12:931–949. doi: 10.1046/j.1365-294X.2003.01781.x. [DOI] [PubMed] [Google Scholar]
- 128.Kadereit JW, Griebeler EM, Comes HP. Quaternary diversification in European alpine plants: pattern and process. Phil Trans R Soc B. 2004;359:265–274. doi: 10.1098/rstb.2003.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Brumfield RT, Edwards SV. Evolution into and out of the Andes: a Bayesian analysis of historical diversification in Thamnophilus antshrikes. Evolution. 2007;61:346–367. doi: 10.1111/j.1558-5646.2007.00039.x. [DOI] [PubMed] [Google Scholar]
- 130.Nakazato T, Warren DL, Moyle LC. Ecological and geographic modes of species divergence in wild tomatoes. Am J Bot. 2010;97:680–93. doi: 10.3732/ajb.0900216. [DOI] [PubMed] [Google Scholar]
- 131.Bellemain E, Ricklefs RE. Are islands the end of the colonization road? Trends Ecol Evol. 2008;23:461–468. doi: 10.1016/j.tree.2008.05.001. [DOI] [PubMed] [Google Scholar]


