Abstract
Transforming growth factor (TGF)-β is stored in the extracellular matrix as a latent complex with its prodomain. Activation of TGF-β1 requires the binding of αv integrin to an RGD sequence in the prodomain and exertion of force on this domain, which is held in the extracellular matrix by latent TGF-β binding proteins. Crystals of dimeric porcine proTGF-β1 reveal a ring-shaped complex, a novel fold for the prodomain, and show how the prodomain shields the growth factor from recognition by receptors and alters its conformation. Complex formation between αvβ6 integrin and the prodomain is insufficient for TGF-β1 release. Force-dependent activation requires unfastening of a ‘straitjacket’ that encircles each growth-factor monomer at a position that can be locked by a disulphide bond. Sequences of all 33 TGF-β family members indicate a similar prodomain fold. The structure provides insights into the regulation of a family of growth and differentiation factors of fundamental importance in morphogenesis and homeostasis.
The TGF-β family is key to specifying the body plan during metazoan development1,2. Members of this family, including nodal, activins, inhibins, bone morphogenetic proteins (BMPs) and growth differentiation factors (GDFs), specify the anterior/posterior and dorsal/ventral axes, endoderm, mesoderm and ectoderm, left–right asymmetry and details of individual organs. TGF-β1, TGF-β2 and TGF-β3 are important in development, wound healing, immune responses and tumour-cell growth and inhibition1,3.
Although TGF-β synthesis and expression of its receptors are widespread, activation is localized to sites where TGF-β is released from latency. TGF-β family members are synthesized with large amino-terminal prodomains, which are required for the proper folding and dimerization of the carboxy-terminal growth-factor domain4. Despite intracellular cleavage by furin, after secretion, noncovalent association persists between the dimeric growth-factor domain and pro-domain of TGF-β, and of an increasingly recognized number of other family members. The prodomain is sufficient to confer latency on some family members and it also targets many members for storage in the extracellular matrix, in complex with latent TGF binding proteins (LTBPs) or fibrillins5,6.
The prodomains of TGF-β1 and TGF-β3 contain an RGD motif that is recognized by αv integrins. Mice with the integrin-binding RGD motif mutated to RGE recapitulate all major phenotypes of TGF-β1-null mice, including multi-organ inflammation and defects in vasculogenesis, thus demonstrating the essential role of integrins in TGF-β activation7. Among αv integrins, the phenotypes of integrin β6-null and integrin β8-null mice demonstrate the particular importance of the αvβ6 and αvβ8 integrins for activation of TGF-β1 and TGF-β3 in vivo8,9.
Integrin binding alone is not sufficient for TGF-β activation. Activation by αvβ6 integrin requires incorporation of TGF-β1 into the extracellular matrix, by association with LTBP, and association of the β6 cytoplasmic domain with the actin cytoskeleton5,10,11. Furthermore, contractile force is necessary for TGF-β activation by myofibroblasts8. Thus, tensile force exerted by integrins across the LTBP–prodomain–TGF-β complex is hypothesized to change the conformation of the prodomain and to free TGF-β for receptor binding5,8. Here, we describe the structure of latent TGF-β, mechanisms for latency and integrin-dependent activation, and broad implications for the regulation of bioactivity in the TGF-β family.
Crystal structure
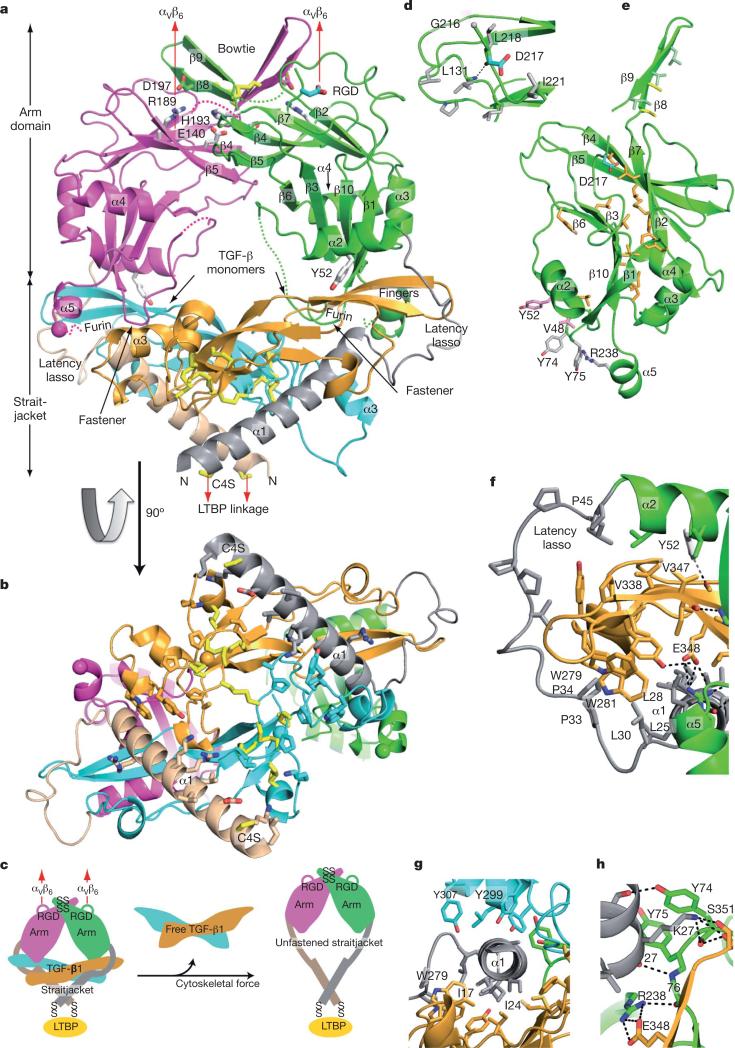
The structure of pro-TGF-β1 at 3.05Å (Fig. 1a–c and Supplementary Table 1) was solved using multi- and single-wavelength anomalous diffraction. Electron density maps (Supplementary Fig. 1) were improved by multi-crystal, multi-domain averaging over four monomers per asymmetric unit. In a ring-like shape, two prodomain arm domains connect at the elbows to crossed ‘forearms’ formed by the two growth-factor monomers and by prodomain ‘straitjacket’ elements that surround each growth-factor monomer (Fig. 1a–c and 4a). The centre of the ring contains solvent. The arms come together at the neck, where they are disulphide-linked in a bowtie, and RGD motifs locate to each shoulder (Fig. 1a). On the opposite side of the ring where the straitjacketed forearms cross, LTBP would be linked to straitjacket residue Cys 4, which is mutated to serine in the crystallization construct (Fig. 1a–c).
Figure 1. Architecture of proTGF-β1.
Arm, straitjacket and TGF-β1 monomer segments are coloured differently. a, b, Overall structure. Spheres mark the last residue visible in density in the prodomain and the first residue of the growth factor. Disordered segments are dashed. Red arrows show the directions of forces during activation by integrins. Key side chains are shown in stick representation, including Asp of the RGD motif in cyan and CED mutations in white. Disulphide bonds and the Cys 4 mutation to Ser are in yellow. c, Schematic of the structure and activation mechanism. SS, disulphide bonds. d, Hydrophobic residues near Asp 217 of the RGD motif. e, Arm domain. Side chains for the hydrophobic core are shown in gold (also marked in Fig. 2 and Supplementary Fig. 2), conserved α2-helix residues that interact with the growth factor are in pink, fastener residues are in silver and bowtie residues are in light green for aliphatics and yellow for Cys. f–h, Straitjacket and fastener details.
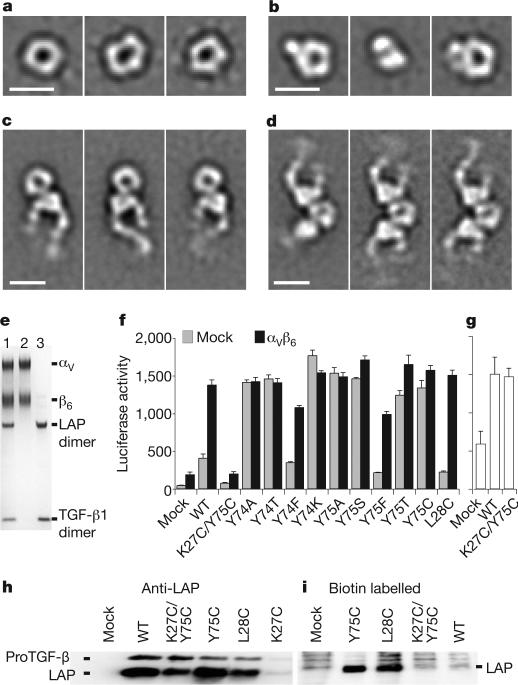
Figure 4. ProTGF-β1 complexes with LTBP and and αvβ6 integrin, and activation of TGF-β.
a–d, Representative negative-stain electron microscopy class averages of proTGF-β (a), the complex of proTGF-β with a fragment of LTBP1 containing TGF-βinding domain 3–EGF–EGF–TGF-βinding domain 4 (b) and complexes of proTGF-β1 with αvβ6 integrin, prepared with an excess of proTGF-β1 (c) or an excess of αvβ6 integrin (d). Scale bars, 100Å. e, Non-reducing SDS–PAGE of the complex peak from S200 gel filtration used for electron microscopy in d (lane 1), αvβ6 integrin (lane 2) and proTGF-β (lane 3). LAP, latency-associated protein (prodomain). f, Activation of TGF-β1. 293T cells stably transfected with αvβ6 integrin or a mock control were additionally transfected with the indicated wild-type (WT) or mutant proTGF-β1 constructs, or with empty vector (mock), and co-cultured with TGF-β indicator cells5. g, Material made by the indicated mutants in 293T cells was heated at 80 °C and assayed with indicator cells. Error bars in f and g show the s.e.m. of 3–9 samples from 1–3 representative experiments. h, i, Western blots of proTGF-β1 secreted by the indicated transfectants, using an antibody to the prodomain (h) or streptavidin to detect biotinylated cysteines (i).
The arm domain, residues 46–242, has a novel fold12 with unusual features. Its two anti-parallel, four-stranded β-sheets bear extensive hydrophobic faces but these overlap only partially in the hydrophobic core (Fig. 1a, e). The hydrophobic faces are extended by long meanders between the two sheets and burial by the α2, α3 and α4 helices.
β-strands β8 and β9 extend on the two-fold pseudo-symmetry axis to link the two arm domains in a bowtie at the neck (Fig. 1a). The bow is tied with reciprocal inter-prodomain disulphide bonds, Cys 194–Cys 196 and Cys 196–Cys 194, and by hydrophobic residues (Fig. 1a, e).
Arg 215 of the RGD motif locates to a disordered loop (residues 209–215) following the bowtie β9 strand. Partially ordered Gly 216 and Asp 217 of the RGD motif (Fig. 1a, d) begin the long, 12-residue meander across the hydrophobic face of the neck-proximal β-sheet that connects to β10 in the forearm-proximal β-sheet.
The straitjacket, residues 1–45, is formed by the α1 helix and the latency lasso (Fig. 1a–c and 2a). The latency lasso, an extended loop that connects the α1 and α2 helices, has little contact with the remainder of the prodomain while encircling the tip of each TGF-β monomer (Fig. 1a, b, f). Six proline residues and three aliphatic residues make hydrophobic contacts with an extensive array of growth-factor aromatic and aliphatic residues, and help these to stabilize the conformation of the latency lasso (Fig. 1f).
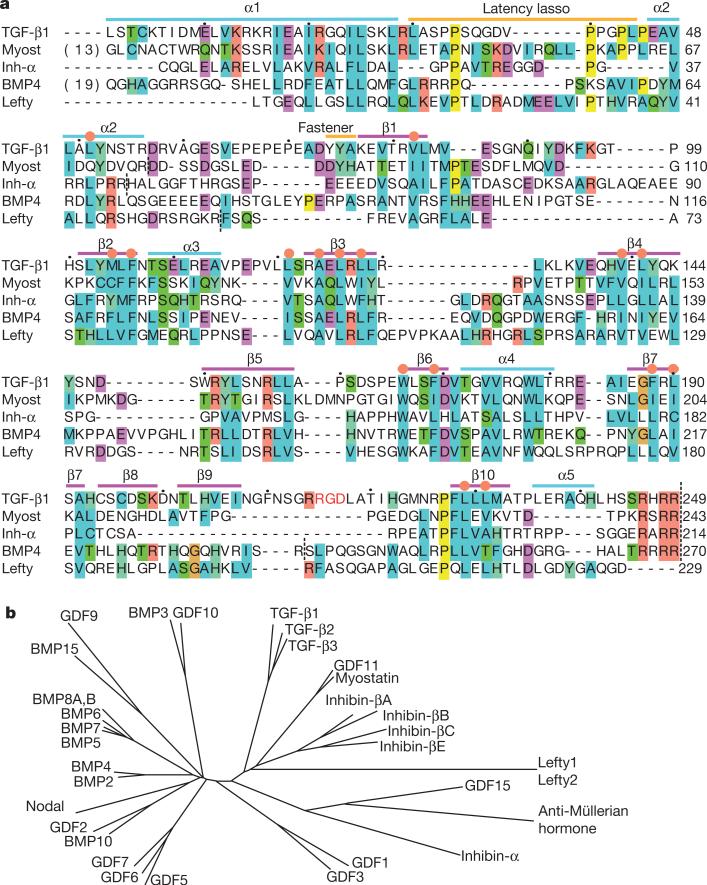
Figure 2. The TGF-β family.
a, Sequence alignment of five representative prodomains. Orange circles mark the core hydrophobic residues shown in Fig. 1e. Inh-α, inhibin-α; Myost, myostatin. Black dots over TGF-β1 sequence mark decadal residues. Vertical dashed lines mark cleavage sites. b, Phylogenetic tree of the TGF-β family, based on the alignment in Supplementary Fig. 2.
A highly hydrophobic face of the amphipathic α1 helix, bearing isoleucine and leucine residues, interacts with Trp 279 and Trp 281 and with aliphatic side chains on one growth-factor monomer (Fig. 1f, g). The tryptophan residues are further covered by lasso residues Leu 30, Pro 33 and Pro 34 (Fig. 1f). Notably, the α1 helix is buried deeply in an interface between the two growth-factor monomers (Fig. 1a, b, g). The interface on the second monomer includes three tyrosine residues (Fig. 1g).
Together with the straitjacket, the arm domain completes the encirclement of each growth-factor monomer. The α2 helix buries Val 338 and Val 347 of the TGF-β finger (Fig. 1f). The α5 helix projects from the base of the arm domain (Fig. 1a and e) and in it, prodomain residue Arg 238 forms a salt bridge to Glu 348 in TGF-β1 (Fig. 1f, h). These two residues are invariant in proTGF-β1, proTGF-β2 and proTGF-β3 (Supplementary Fig. 2).
The straitjacket is fastened to arm residues 74–76 (Fig. 1a, h). A backbone hydrogen bond between the nitrogen of Ala 76 and the oxygen of Lys 27 caps the C-terminal end of the α1 helix (Fig. 1h). Moreover, the carbonyl oxygen of Ala 76 forms a hydrogen bond to Arg 238 in the α5 helix (Fig. 1h). Lys 27 is a key fastener residue. Its side chain forms a π-cation bond to the side chain of Tyr 74, a hydrogen bond to the backbone of Tyr 74, and hydrogen bonds to the backbone and sidechain of Ser 351 (Fig. 1h). Van der Waals contacts between the bulky side chains of the fastener residues Lys 27, Tyr 74 and Tyr 75 also secure the straitjacket. Notably, Lys 27, Tyr 74 and Tyr 75 are invariant among TGF-β1, TGF-β2 and TGF-β3 (Supplementary Fig. 2). Fastening is reinforced by backbone hydrogen bonds between arm β1-strand residues 77 and 78 and growth-factor β-fingers, which join the prodomain and the growth factor in a super-β-sheet (Fig. 1a).
The TGF-β dimer forms the forearms, although TGF-β monomers have also been described as hand-like13–15 (Fig. 3). Each monomer has no hydrophobic core, aside from the cystine knot motif in which one disulphide passes between two polypeptide segments bridged by two other disulphides.
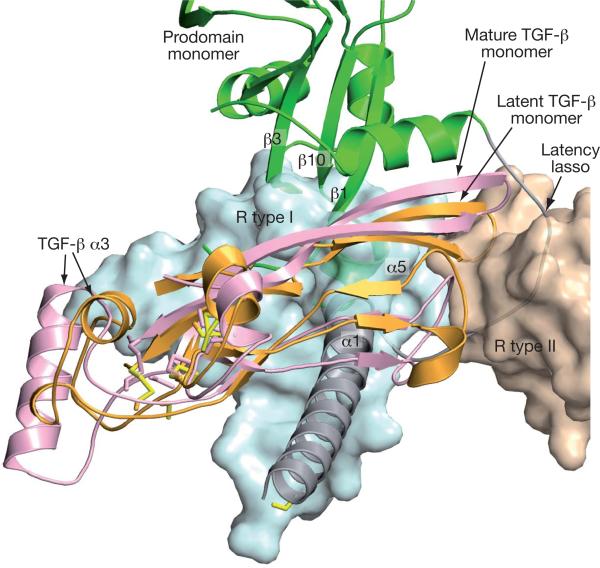
Figure 3. Shielding from receptor binding.
ProTGF-β1 and TGF-β1 in complex with its receptors (R type I and II) (ref. 15) were superimposed on the TGF-β dimers. For clarity, only one monomer of each is shown. The receptors are shown as transparent molecular surfaces. Elements of the prodomain that clash with the receptors are labelled.
Prodomain-bound TGF-β1 differs markedly from previous TGF-β structures in both the orientation between monomers and the position of elements within monomers (Fig. 3 and Supplementary Fig. 3). The Cα root-mean-squared deviations over all 112 residues are 7Å, and 2Å over the most similar 85 residues. The largest differences are imposed by the prodomain α1 helix. It occupies a similar position to the growth-factor α3 helix in mature TGF-β1 (Supplementary Fig. 3). Intercalation of the prodomain α1 helix between the growth-factor monomers reduces the total area buried between monomers from 850Å2 to 335Å2. The large conformational changes in TGF-β1 are driven by an intimate interaction between the growth-factor and prodomain dimers, which buries a total area of 2,440Å2.
Implications for biosynthesis
Folding and secretion of active TGF-β1 and activin A requires the co-expression of their prodomains4, whereas the TGF-β1 prodomain can be biosynthesized in the absence of the growth-factor domain16. These findings suggest that the C-terminal growth-factor domain folds either concomitantly with, or subsequently to, the N-terminal prodomain. The kinetics of biosynthesis of TGF-β1, activin and anti-Müllerian hormone are slow and for anti-Müllerian hormone, folding of the growth-factor domain is rate-limiting17. Regions of the prodomain that may be particularly important in templating the folding of the growth-factor domain include the β1 strand that forms a supersheet with the TGF-β fingers and the α1 and α2 helices, which pack against extensive hydrophobic interfaces on opposite sides of the growth-factor fingers (Fig. 1a, f, g). Residues Ile 17, Ile 24, Leu 25 and Leu 28 in the α1-helix interface, and Leu 30 in the lasso interface (Fig. 1f, g), have been specifically identified as important for TGF-β1 association18. The embrace of the fingers of each growth-factor monomer may complement the correct formation of the cystine knot and inter-monomer disulphide bonds in TGF-β. The structure of proTGF-β1 makes these disulphides accessible to disulphide isomerases during biosynthesis (Fig. 1b).
A definitive assignment of which growth-factor and prodomain monomers derive from the same polypeptide chain is not possible because of intracellular cleavage by furin and lack of density for residues 243–249. However, cleavage is incomplete and the small amount of uncleaved proTGF-β that is present in protein preparations co-crystallizes with cleaved proTGF-β (Supplementary Fig. 1d), indicating that there is no major conformational change after cleavage. A long prodomain–growth-factor connection through the centre of the ring, spanning ~50Å, would require substantial conformational change and would limit access to furin. Therefore, we have assigned the shorter ~30Å connection, between the C terminus of the prodomain and the N terminus of the growth factor, located on the same side of the ring (for example, the magenta and gold spheres in Fig. 1a, b).
This assignment indicates a swap in the growth-factor monomer that each prodomain monomer embraces, with the intimate interactions described above occurring between prodomains and growth-factor domains that are present on different polypeptide chains in the precursor in the endoplasmic reticulum (for example, the gold and green domains in Fig. 1). Thus, the surface area buried between the growth-factor domains and prodomains of different precursor monomers (900Å2) is substantially larger than that within the same precursor monomer (370Å2) and adds substantially to the inter-monomer interfaces of the growth factor (330Å2) and prodomain (600Å2). Swapping may be important in regulating the proportion of growth-factor heterodimers, which have unique functions in settings such as dorsoventral patterning19.
Complex with LTBP and activation
In the large latent complex, a single LTBP molecule is disulphide-bonded to two proTGF-β monomers. We confirmed this unusual 1:2 stoichiometry by multi-angle light-scattering mass measurements of the complex with an LTBP fragment. The LTBP-crosslinking Cys 4 residues in each monomer (serines in our construct) are 40Å apart (Fig. 1b). Their linkage to LTBP will reinforce the straitjacket by fastening together the forearms (Fig. 1c). The large 40Å separation may represent a mechanism for preventing disulphide linkage between the two Cys 4 residues. In contrast, the two cognate cysteines in TGF-βinding domain 3 of LTBP become linked to one another in the absence of complex formation. These cysteines are surface-exposed and surrounded by acidic residues that interact with proTGF-β (refs 20, 21). In agreement with this, the straitjacket a1-helices bear basic residues (Fig. 1b). Moreover, between the two prodomain a1-helices, a concave growth-factor surface that bears numerous hydrophobic residues, including proline and disulphide-linked cysteine, is available for interaction with LTBP (Fig. 1b).
Negative-stain electron microscopy class averages of proTGF-β are in excellent agreement with our crystal structure (Fig. 4a and Supplementary Fig. 4). A complex with an LTBP fragment that contains TGF-β-binding domains 3 and 4 and two intervening EGF domains shows no major conformational change in the proTGF-β moiety (Fig. 4b). An additional density corresponding to the LTBP fragment is present on the periphery of the ring, as expected from our crystal structure, and causes the ring to lie at an angle to the substrate in some class averages (Fig. 4b, middle panel and Supplementary Fig. 4c).
The RGD motifs in the shoulders of proTGF-β are highly accessible for integrin binding (Fig. 1a). In contrast to many RGD motifs that are present in extended loops, the position of Asp 217 is stabilized by burial of Leu 218 and by a 218–131 backbone hydrogen bond (Fig. 1d). Exposed hydrophobic side chains that are nearby on the body of the arm domain (Fig. 1d) may increase affinity for integrins.
The integrin αvβ6 ectodomain, with a C-terminal clasp, formed complexes with proTGF-β1 in Ca2+ and Mg2+ that could be isolated by gel filtration, demonstrating unusually tight binding for an integrin (Fig. 4c–e and Supplementary Fig. 4a). Different input ratios of proTGF-β1 and αvβ6 integrin yielded 1:1 (Fig. 4c) and 1:2 (Fig. 4d, e) complexes. Binding to ligand stabilized extension of the integrin legs and the open conformation of the αvβ6 headpiece (Fig. 4c, d and Supplementary Fig. 4d, e). Integrins were bound to proTGF-β at the interface between a large density, corresponding to the αv β-propeller domain, and a small density, corresponding to the β6 βI domain, with their legs extending away from proTGF-β. The spacing between the binding sites on the ring seen by electron microscopy (40–50Å) was appropriate for that between the two RGDs in the crystal structure (45Å). No major conformational change in proTGF-β1 was apparent, even with two integrins bound (Fig. 4d), and SDS–polyacrylamide gel electrophoresis (SDS–PAGE) confirmed the presence of TGF-β1 in the complex (Fig. 4e). These biochemical and structural studies demonstrate that integrin binding to proTGF-β1 is not sufficient for release of TGF-β1, consistent with previous cell-biological assays5,8,10. The requirements of (1) attachment of proTGF-β through LTBP to the extracellular matrix; (2) integrin attachment to the cytoskeleton and (3) cellular contraction indicate that the generation of tensile force across proTGF-β1 is required for activation of TGF-β5,8,10,11.
The structure enables the overall mechanism of TGF-β1 activation by applied force to be readily predicted (Fig. 1c). Tensile force applied to the RGD motifs in the shoulders by αv integrins attached to the actin cytoskeleton will be resisted at the opposite end of the ring, where the Cys 4 residues in the straitjacket are disulphide-linked to LTBP, which is tightly associated with the extracellular matrix. Pulling force will be applied in the directions shown by red arrows in Fig. 1a.
The direction of the pulling force and fold topology strongly influence the unfolding pathway and resistance to force23. β-Sheet proteins are the most force-resistant and thus the arm domain will be the most force-resistant portion of the prodomain. Pulling against the RGD motif will be transmitted through the long meander to the β10 strand. Force transmitted from the Cys 4 residues through the straitjacket will be resisted by the β1 strand. The β1 and β10 strands are each parallel to applied force and adjacent in a β-sheet (Fig. 1) and are thus in the most force-resistant structural geometry known, the hydrogen-bond clamp23. By contrast, the topologies and geometries of the α-helices and the long latency lasso of the straitjacket are ill-suited to resist force. Force on Cys 4 will apply leverage to the C-terminal end of the α1 helix and weaken interactions with fastener residues. After unfastening, the long latency lasso, which has no stabilizing hydrogen-bond interactions, will be easily elongated and straightened by the applied tensile force. Thus, freed by opening of its straitjacket, TGF-β will be released from the prodomain and activated for receptor binding (Fig. 1c).
The prodomain not only holds TGF-β in a markedly different conformation from when it is free or bound to receptors, it also blocks receptor access completely. TGF-β family members are recognized by two type I receptors and two type II receptors that surround the growth-factor dimer (Supplementary Fig. 3e)15. Binding of the type II receptor to the finger-tips of the growth factor is blocked by the latency lasso, and binding of the type I receptor to the body of the growth-factor domain is blocked by the prodomain α1 helix, α5 helix, the fastener and the ends of β-strands 1, 3 and 10 (Fig. 3). Although straitjacket removal might be sufficient to allow binding of type II receptors, type I receptor interactions overlap with so many interactions between TGF-β and the arm domain (Fig. 3) that complete release from the prodomain would be required for receptor binding. The structure thus shows that integrins could not expose TGF-β sufficiently for receptor activation if it remained bound to the prodo-main, and that other explanations should be sought for the greater activity of integrin-activated TGF-β on neighbouring cells than on distant cells10.
To test the importance of unfastening in TGF-β activation, key residues were mutated. Non-conservative substitutions of the fastener residues Tyr 74 and Tyr 75 resulted in spontaneous, non-integrin-dependent TGF-β activation (Fig. 4f). Among the different amino acids to which Tyr 74 and Tyr 75 were mutated, only phenylalanine was not activating. As a control, mutation of nearby Leu 28, in a hydrophobic interface with TGF-β, was not activating (Fig. 4f). These results are consistent with the importance to fastening of π-bonding and of van der Waals interactions of the aromatic tyrosine side chains.
In the fastener, the Cα carbons of Lys 27 and Tyr 75 are only 4.1Å apart, permissive for disulphide bond formation in a K27C/Y75C mutant, as confirmed by free-cysteine labelling of the Y75C mutant but not the K27C/Y75C mutant (Fig. 4h, i). The K27C mutation greatly reduced expression (Fig. 4h). Similarly, a K27A mutation greatly reduces expression, and also releases free TGF-β1 (ref. 18). The Y75C mutant was constitutively active (Fig. 4f). The K27C/Y75C double mutation rescued expression compared to K27C, prevented the spontaneous release of TGF-β that was seen with Y75C and, compared to wild type, made proTGF-β completely resistant to integrin-αvβ6-dependent activation (Fig. 4f, h). Denaturants such as heat can unfold proteins by pathways distinct from applied force23. Heat released comparable amounts of active TGF-β from wild type and the K27C/Y75C mutant (Fig. 4g). Thus, a disulphide bond can fasten the strait-jacket permanently and prevent integrin-dependent activation. These results support the hypothesis that tensile force applied to the prodo-main by integrins can release TGF-β, and emphasise the importance of straitjacket unfastening in integrin-dependent activation.
Mutations in disease
Camurati–Engelmann disease (CED), which is characterized by thickening of the shafts of the long bones with pain in muscle and bone, is caused by mutations in the prodomain of TGF-β1 that increase its release18,24. Among CED mutations, Y52H disrupts an α2-helix residue that cradles the TGF-β fingers (Fig. 1a, f). The charge-reversal E140K and H193D mutations disrupt a pH-regulated salt bridge between Glu 140 and His 193 in the dimerization interface of the prodomain (Fig. 1a). ‘Hotspot’ residue Arg 189 is substantially buried: it forms a cation-π bond with Tyr 142 and salt bridges across the dimer interface with bowtie residue Asp 197 (Fig. 1a). Moreover, CED mutations in Cys 194 and Cys 196 demonstrate the importance of the bowtie disulphide bonds.
Implications for the large TGF-β family
The TGF-β family consists of 33 members (Fig. 2b)2. Although growth-factor domains are highly conserved, prodomains vary in length from 169 to 433 residues, and are variously described as unrelated in sequence or low in homology. However, alignment shows that all prodomains have a similar fold (Fig. 2a and Supplementary Fig. 2). Deeply buried hydrophobic residues in core secondary-structure elements of the arm domain, that is, the α2 helix and β-strands 1–3, 6, 7 and 10, are conserved in all members (gold side chains in Fig. 1e and orange circles in Fig. 2 and Supplementary Fig. 2).
Most family members also contain clear sequence signatures for the amphipathic C-terminal portion of the a1 helix that inserts intimately between the two growth-factor monomers (Fig. 2 and Supplementary Fig. 2). A similar insertion in inhibin-α and inhibin-βA has been demonstrated by mapping disruptive mutations to the equivalents of Ile 24 and Leu 28 in TGF-β (Fig. 1f, g)25. Many family members also contain proline-rich latency lasso loops with lengths that are compatible with encirclement of the growth-factor β-finger (Fig. 2 and Supplementary Fig. 2). Thus, a prodomain structure similar to that of proTGF-β, including a portion of the straitjacket, is widespread in the TGF-β family. However, the low sequence identity and many insertions and deletions indicate substantial specializations.
Differences in prodomain dimerization among family members are indicated by variations in cysteine positions. The bowtie (β-strands 8 and 9) and its disulphides are specializations. Inhibin-α and -β subunits have cysteines in similar positions, whereas other family members either have cysteine residues in the β7 strand or lack cysteines altogether in this region (Fig. 2 and Supplementary Fig. 2).
The interface between the two arm domains in the β4 and β5 strands is modest in size and lacks hydrophobic and conserved residues. GDF1 and GDF15 specifically lack the β4 and β5 strands, which are adjacent in sequence and structure, on the edge of a β-sheet (Fig. 1a and Supplementary Fig. 2). Therefore, arm-domain dimerization seems to be variable or absent in some family members.
The close relatives of TGF-β, myostatin and GDF11, which are also latent, show conservation of the fastener residues Lys 27 and Tyr 75 (Fig. 2a and Supplementary Fig. 2). Myostatin regulates muscle mass and is stored in the extracellular matrix, bound to LTBP3. Release of myostatin and GDF11 from latency requires cleavage of the prodo-main between Arg 75 and Asp 76 by BMP1/tolloid metalloproteinases (reviewed in ref. 26). This cleavage is between the α2 helix and the fastener (Fig. 2a). Thus at least two different methods of unfastening the straitjacket, force and proteolysis, can release family members from latency.
An increasingly large number of TGF-β family members are recognized to remain associated with their prodomains after secretion, including BMP4, BMP7, BMP10, GDF2, GDF5 and GDF8 (ref. 27). Furthermore, many of these prodomains bind with high affinity to fibrillin-1 and fibrillin-2. Targeting by the prodomain to the extra-cellular matrix may be of wide importance in regulating bioactivity in the TGF-β family6. Moreover, binding to LTBPs or fibrillins seems to strengthen the prodomain–growth-factor complex6. Thus, although only a limited number of TGF-β family members are latent as prodomain–growth-factor complexes, the concept of latency may extend to other members when their physiologically relevant complexes with LTBPs and fibrillins are considered.
The signalling range of BMP4 in vivo is increased by extracellular cleavage of the prodomain by furin-like proteases at a second site upstream of the prodomain–growth-factor cleavage site28. Notably, the second site is in the disordered loop bearing the arginine of RGD in TGF-β1 (Fig. 2a). Loss of the central β10 strand between the two cleavage sites results in loss of binding of the BMP4 prodomain to its growth factor27.
The prodomain of Nodal, which binds to Cripto, targets Nodal for cleavage by proteases secreted by neighbouring cells29. Anti-Müllerian hormone is secreted largely uncleaved and association with the pro-domain greatly potentiates its activity in vivo30. Lefty protein, which is involved in establishing bilateral asymmetry, is not cleaved between the arm and growth-factor domains, and is cleaved instead between the α2 helix and the fastener31 (Fig. 2). Notably, release of the strait-jacket should be sufficient to enable access of type II receptors to growth-factor domains.
Concluding perspective
We have described the structure of latent TGF-β1 and a force-dependent mechanism for its activation by αv integrins. It is notable that so many members of the TGF-β family associate with fibrillins or with LTBPs, which co-assemble in the elastic fibres of connective tissues6. Forces acting on elastic fibres would extend fibrillins and LTBPs, and we speculate that this could weaken their association with TGF-β family members, enabling release and activation. It is thus possible that force-dependent regulation of TGF-β family activation could extend beyond integrin-dependent mechanisms and could be important in a wide variety of contexts, including regulation of bone and tissue growth. Although prodomains in the TGF-β family are diverse, their sequences are highly conserved between species2. Further studies are required to address the diversity of mechanisms by which prodomains regulate latency and activation in the TGF-β family.
METHODS
ProTGF-β1
The porcine proTGF-β1 construct with the rat serum albumin leader sequence (MKWVTFLLLLFISGSAFS), followed by eight histidine residues, a streptavidin-binding peptide (TTGWRGGHVVELAGELEQLRARLEHH PQGQREP)32 and a HRV-3C protease site (LEVLFQGP) was amplified from pcDNA-GS-TGF-β1 (ref. 33). Porcine proTGF-β1 with the C4S mutation was amplified from the latter construct. The C4S mutation increases proTGF-β1 expression33 and avoids inappropriate disulphide bond formation34. No crystals were obtained with this construct. One or two N-linked sites were deleted in the N147Q and N107Q/N147Q constructs, which were expressed similarly to wild type35. The best crystals were obtained with N147Q; the N107Q/N147Q mutant yielded needles that could not be optimized. CHO-Lec 3.2.8.1 cells were transfected by electroporation and cultured with 10 μg ml−1 puromycin. Clones were screened for expression using a sandwich enzyme-linked immunosorbent assay (ELISA) with a capture antibody to prodomain-1 (R&D Systems) and a biotinylated detection antibody to the His tag (Qiagen). The clone with highest expression of proTGF-β1 (~2 mg l−1) was expanded and cultured in roller bottles with J/J medium and 5% fetal bovine serum (FBS). Supernatants were collected every 5 d, clarified by centrifugation, concentrated tenfold with tangential flow filtration (Vivaflow 200, Sartorius Stedim), diluted fivefold with 10 mM Tris-HCl, 0.14 M NaCl (TBS, pH 8.0), then concentrated fivefold. Material was adjusted to 0.2 M NaCl and purified using Ni-NTA agarose (Qiagen) (25 ml per 5 l of culture supernatant), then washed with three column volumes of 0.6 M NaCl, 0.01 M Tris (pH 8.0) and eluted with 0.25 M imidazole in TBS. Material was adjusted to pH 7.4, applied to Strep-tactin agarose (IBA) (3 ml per 5 l of culture supernatant) and washed with TBS (pH 7.4). Then 4 ml of recombinant His-tagged HRV-3C protease (Novagen, 100 U mg−1, 1 mg ml−1), diluted 20-fold in TBS (pH 7.4) with 10% glycerol, was applied to the column, which was held at 4 °C for 16 h. The flow-through with two column volumes of TBS (pH 7.4), containing untagged proTGF-β1, was concentrated to 1 ml and applied to MonoQ and Superdex S200 columns connected in series and equilibrated with TBS (pH 7.5). Purified proTGF-β1 was concentrated to about 15 mg ml−1 in 10 mM Tris (pH 7.5), 75 mM NaCl for crystal screening in 96-well Greiner micro-plates (100 nl hanging-drop vapour diffusion format) using a mosquito crystallization robot (Molecular Dimensions) at 20 °C. Hits were optimized in 24-well plates using hanging-drop vapour diffusion. However, better-diffracting crystals could only be obtained from sitting drops containing equal volumes of 12–15 μl protein and well solution under the optimized conditions of 6.5–7.5% PEG 3500, 17–18% isopropanol and 0.1 M sodium citrate (pH 5.6), with the addition of 4–5% glycerol to slow crystal growth and improve crystal size and shape. Maximum single-crystal dimensions reached 450 μm × 150 μm × 40 μm. Before cooling the crystals to 100 K in liquid nitrogen, three rounds of increases in PEG 3350 concentration (12 h for each increase of 8% per cycle) were carried out in the mother liquor36. The final PEG 3350 concentration of about 31% was sufficient for cryoprotection. Crystals are summarized in Supplementary Table 1. There are two complexes per asymmetric unit, with a Matthews coefficient of 2.9Å3 Da−1, giving a solvent content of 57.8%.
To prepare Se-Met proTGF-β1, cells were washed with PBS (pH 7.4), supplemented with 1% FBS, then incubated for 8 h with methionine-free α-MEM (SAFC Biosciences) supplemented with 50 mg l−1 L-Se-Met (Sigma) and 10% dialyzed FBS. After replacement with the same medium, cells were cultured for 4 d. Se-Met proTGF-β1, at a yield of 1.5 mg l−1, was purified and crystallized identically to native proTGF-β1. Furthermore, a heavy-atom derivative was obtained by soaking crystals in mother liquor containing 0.4 mM HgBr2 for 4 h.
Structure determination and refinement
Native Se multiple-wavelength anomalous dispersion (MAD) and single-wavelength anomalous dispersion (SAD) Hg derivative data were collected at 100 K at beamline 23-ID, then processed using HKL2000 (ref. 37) and XDS38. Statistics are in Supplementary Table 1. Initial experimental phases were determined independently using Se-MAD and Hg-SAD, with 19 out of 24 Se sites and 14 Hg sites in the asymmetric unit located using PHENIX39. Electron density maps from Se-Met phasing, calculated after fourfold non-crystallographic symmetry (NCS) averaging, clearly defined the orientation of each monomer. The mature TGF-β1 homodimer was easily docked into the map using model 1KLC with MOLREP40 in CCP4 (ref. 41). The prodo-main was built into the map manually. A crude model of proTGF-β1 was obtained after rigid-body refinement by PHENIX, with both domains as one rigid body. The same model was docked into Hg-SAD density for the two homodimers, using MOLREP.
To improve the phases and extend them to higher resolution, multi-crystal averaging (two crystals in total: Se-MAD and Hg derivative), multi-domain averaging (with separate masks for each prodomain and TGF-β monomer) and solvent flattening and histogram matching were performed using DMMULTI42 from the CCP4 suite. The mask for each domain was calculated by NCSMASK in CCP4, and NCS matrices for each domain between molecules and crystals were computed by LSQKAB in CCP4. Rigid-body refinements were carried out by PHENIX for each lattice, on the basis of the averaged maps. The new models for each lattice were then used to calculate a set of new NCS matrices for the next cycle of DMMULTI. These steps were cycled twice.
Model building in COOT43 was based on multi-crystal, multi-domain averaged electron density maps and 2Fo – Fc maps. NCS restraints and translation-libration-screw (TLS) groups were used in refinement with PHENIX. The sequence-to-structure register was confirmed using Se anomalous maps. The multi-crystal, multi-domain averaging was repeated using the refined structure at an Rfree of about 33% and no major differences were found. Two residues from the 3C protease site remain at the N terminus after cleavage. The structures include residues 0–62, 70–208, 216–241, 250–361 and one N-acetylglucosamine (NAG) residue (chain A); residues 1–62, 70–208, 216–242, 250–299, 310-361 and two NAG residues (chain B); residues 1–62, 68–208, 216–241, 250–361 and three NAG residues (chain C) and residues 0–62, 69–208, 216–242, 250–361 and two NAG residues (chain D). Validation and Ramachandran statistics used MOLPROBITY44. All structure figures were generated using Pymol (DeLano Scientific).
In the asymmetric unit of the crystal, a second pro-TGF dimer extends each two-stranded β-ribbon to form a four-stranded, inter-dimer super-β-sheet in which Leu 203 forms a hydrophobic lattice contact. In its absence, Leu 203 may mediate hydrophobic interactions within the bowtie.
Mutagenesis
Wild-type human proTGF-β1 was inserted into the pEF1-puro plasmid. Site-directed mutagenesis was performed using QuikChange (Stratagene). All mutations were confirmed by DNA sequencing.
Free cysteine labelling and prodomain detection
HEK-293T cells were transfected using Polyfect reagent (Qiagen) according to the manufacturer's instructions, using 2 μg of proTGF-β1 cDNA per 6-cm dish of cells at 70–80% confluency. The cells were then cultured in FreeStyle serum-free medium (Invitrogen) for 3 d. Supernatant was reacted with 450 μM biotin-BMCC (1-biotinamido-4-(4′-(maleimidoethylcyclohexane)-carboxamido)butane) (Pierce) for 60 min at 22° C, followed by the addition of 40 mM N-ethylmaleimide. ProTGF-β1 was immunoprecipitated with 1.5 μg anti-human-prodomain-1 (LAP-1) antibody (R&D Systems) and Protein A Sepharose beads (GE Healthcare) at 4 °C for 2 h, then subjected to reducing SDS 10% PAGE. After transfer to polyvinylidene difluoride membranes (Millipore), biotin was detected using streptavidin-horseradish peroxidase with the ECL-plus western blotting kit (GE Healthcare). Total proTGF-β1 was similarly detected on a separate blot using biotinylated human prodomain-1 (LAP-1) antibody.
TGF-β1 activation assay
Transformed mink lung epithelial cells (TMLCs) stably transfected with a luciferase construct under plasminogen activator inhibitor promoter 1 (ref. 5) were provided by D. Rifkin (New York University). HEK-293T cell transfectants stably expressing αv and β6 were selected with puromycin and G418. Clones expressing high levels of integrin αvβ6 were selected by immunofluorescent flow cytometry using an anti-β6 antibody. Cells stably transfected with empty vector were used as a control. These cells were subsequently transiently transfected with human wild-type or mutant proTGF-β1, using lipofecta-mine with 0.4 μg plasmid DNA per well in a 48-well plate. After 16–24 h, each well was used to seed 3 wells of a 96-well plate with about 15,000 cells, which were co-cultured with 15,000 TMLCs in 100 ml DMEM with 0.1% BSA for 16–24 h. TGF-β1-induced luciferase activity in cell lysates was measured using the luciferase assay system (Promega). To assess heat-releasable TGF-β1, cells were transfected as above except that polyfect was used in 6-well plates. After 2 d, cells were collected and heated in 150 μl of DMEM with 0.1% BSA at 80 °C for 10 min. TGF-β1 activity in 50 °l aliquots was measured using the luciferase assay with TMLCs.
Negative-stain electron microscopy
A large latent-complex fragment was isolated from supernatants of 293T cells transiently co-transfected with native human proTGF-β and a human LTBP fragment containing the same N-terminal tag as was used above on proTGF-β. The LTBP fragment contained the TB3 and TB4 domains and two intervening EGF-like domains (residues Thr 1333–Asn 1578, immature numbering). Multi-angle light scattering gave an Mr of 119,400, compared to a calculated Mr of 120,400 for a 2:1 proTGF-β:LTBP fragment (2 × 46,400 for proTGF-β, including 2 × 7,500 for three high-mannose N-linked sites, plus 27,600 for the LTBP fragment). The αvβ6 ectodomain was expressed using C-terminal α-helical coiled-coils and tags; purified, then subjected to negative-stain electron microscopy as previously described45. Purified proTGF-β1 or proTGF-β1 in complex with an LTBP fragment (20 μg), proTGF-β1 (30 μg) in molar excess over clasped αvβ6 (20 μg), or clasped αvβ6 (60 μg) in excess over proTGF-β (10 μg) were subjected to Superdex S200 chromatography in TBS (pH 7.5) with 1 mM Ca2+ and 1 mM Mg2+. Peak fractions corresponding to the purified proteins or complexes were subjected to negative-stain electron microscopy. Particle selection, alignment, classification and averaging were conducted as previously described46.
Supplementary Material
Acknowledgements
We thank P. Sun for porcine TGF-β1 cDNA, K. Koli for human TGF-β1 and LTBP1 cDNAs, D. Rifkin for human LTBP1 cDNA and transformed mink lung epithelial cells, and the staff of the Advanced Photon Source General Medical Sciences and National Cancer Institute (APS GM/CA-CAT) beamline 23-ID.
Footnotes
Author Contributions M.S. cloned, expressed and purified proTGF-β1, crystallized the protein, collected and processed X-ray data, refined and analysed the structure, designed and performed biochemical assays and wrote the paper. J.Z. collected and processed X-ray data, refined and analysed the structure and performed electron microscopy studies. R.W. designed and performed TGF-β1 assays. X.C. performed electron microscopy studies. L.M. processed X-ray data. T.W. provided electron microscopy supervision. T.A.S. designed and supervised the project, refined and analysed the structure and wrote the paper.
Author Information X-ray structures have been deposited in the Protein Data Bank under the accession number 3RJR.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Wu MY, Hill CS. TGF-β superfamily signaling in embryonic development and homeostasis. Dev. Cell. 2009;16:329–343. doi: 10.1016/j.devcel.2009.02.012. [DOI] [PubMed] [Google Scholar]
- 2.Derynck R, Miyazono K. In: The TGF-β Family. Derynck R, Miyazono K, editors. Cold Spring Harbor Laboratory Press; 2008. pp. 29–43. Ch. 2. [Google Scholar]
- 3.Blobe GC, Schiemann WP, Lodish HF. Role of transforming growth factor β in human disease. N. Engl. J. Med. 2000;342:1350–1358. doi: 10.1056/NEJM200005043421807. [DOI] [PubMed] [Google Scholar]
- 4.Gray AM, Mason AJ. Requirement for activin A and transforming growth factor-β 1 pro-regions in homodimer assembly. Science. 1990;247:1328–1330. doi: 10.1126/science.2315700. [DOI] [PubMed] [Google Scholar]
- 5.Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin αVβ6-mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J. Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ramirez F, Sakai LY. Biogenesis and function of fibrillin assemblies. Cell Tissue Res. 2010;339:71–82. doi: 10.1007/s00441-009-0822-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Z, et al. Absence of integrin-mediated TGFβ1 activation in vivo recapitulates the phenotype of TGFβ1-null mice. J. Cell Biol. 2007;176:787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wipff PJ, Hinz B. Integrins and the activation of latent transforming growth factor β1 — an intimate relationship. Eur. J. Cell Biol. 2008;87:601–615. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 9.Aluwihare P, et al. Mice that lack activity of αVβ6- and αVβ8-integrins reproduce the abnormalities of Tgfβ1- and Tgfβ3-null mice. J. Cell Sci. 2009;122:227–232. doi: 10.1242/jcs.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger JS, et al. The integrin αVβ6 binds and activates latent TGF β1: A mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 11.Yoshinaga K, et al. Perturbation of transforming growth factor (TGF)-β1 association with latent TGF-β binding protein yields inflammation and tumors. Proc. Natl Acad. Sci. USA. 2008;105:18758–18763. doi: 10.1073/pnas.0805411105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Daopin S, Piez KA, Ogawa Y, Davies DR. Crystal structure of transforming growth factor-β2: an unusual fold for the superfamily. Science. 1992;257:369–373. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 14.Schlunegger MP, Grutter MG. An unusual feature revealed by the crystal structure at 2.2 Å resolution of human transforming growth factor-β2. Nature. 1992;358:430–434. doi: 10.1038/358430a0. [DOI] [PubMed] [Google Scholar]
- 15.Radaev S, et al. Ternary complex of transforming growth factor-β1 revealsisoform-specific ligand recognition and receptor recruitment in the superfamily. J. Biol. Chem. 2010;285:14806–14814. doi: 10.1074/jbc.M109.079921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentry LE, Nash BW. The pro domain of pre-pro-transforming growth factor β1 when independently expressed is a functional binding protein for the mature growth factor. Biochemistry. 1990;29:6851–6857. doi: 10.1021/bi00481a014. [DOI] [PubMed] [Google Scholar]
- 17.Belville C, et al. Mutations of the anti-Müllerian hormone gene in patients with persistent Müllerian duct syndrome: biosynthesis, secretion, and processing of the abnormal proteins and analysis using a three-dimensional model. Mol. Endocrinol. 2004;18:708–721. doi: 10.1210/me.2003-0358. [DOI] [PubMed] [Google Scholar]
- 18.Walton KL, et al. Two distinct regions of latency-associated peptide coordinate stability of the latent transforming growth factor-β1 complex. J. Biol. Chem. 2010;285:17029–17037. doi: 10.1074/jbc.M110.110288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Little SC, Mullins MC. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nature Cell Biol. 2009;11:637–643. doi: 10.1038/ncb1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lack J, et al. Solution structure of the third TB domain from LTBP1 provides insight into assembly of the large latent complex that sequesters latent TGF-β. J. Mol. Biol. 2003;334:281–291. doi: 10.1016/j.jmb.2003.09.053. [DOI] [PubMed] [Google Scholar]
- 21.Chen Y, et al. Amino acid requirements for formation of the TGF-β-latent TGF-β binding protein complexes. J. Mol. Biol. 2005;345:175–186. doi: 10.1016/j.jmb.2004.10.039. [DOI] [PubMed] [Google Scholar]
- 22.Luo B-H, Carman CV, Springer TA. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 2007;25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forman JR, Clarke J. Mechanical unfolding of proteins: insights into biology, structure and folding. Curr. Opin. Struct. Biol. 2007;17:58–66. doi: 10.1016/j.sbi.2007.01.006. [DOI] [PubMed] [Google Scholar]
- 24.Janssens K, et al. Camurati-Engelmann disease: review of the clinical, radiological, and molecular data of 24 families and implications for diagnosis and treatment. J. Med. Genet. 2005;43:1–11. doi: 10.1136/jmg.2005.033522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walton KL, et al. A common biosynthetic pathway governs the dimerization and secretion of inhibin and related transforming growth factor β (TGFβ) ligands. J. Biol. Chem. 2009;284:9311–9320. doi: 10.1074/jbc.M808763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson SB, Goldberg AL, Whitman M. Identification of a novel pool of extracellular pro-myostatin in skeletal muscle. J. Biol. Chem. 2008;283:7027–7035. doi: 10.1074/jbc.M706678200. [DOI] [PubMed] [Google Scholar]
- 27.Sengle G, et al. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J. Biol. Chem. 2008;283:13874–13888. doi: 10.1074/jbc.M707820200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cui Y, et al. The activity and signaling range of mature BMP-4 is regulated by sequential cleavage at two sites within the prodomain of the precursor. Genes Dev. 2001;15:2797–2802. doi: 10.1101/gad.940001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanchet MH, et al. Cripto recruits Furin and PACE4 and controls Nodal trafficking during proteolytic maturation. EMBO J. 2008;27:2580–2591. doi: 10.1038/emboj.2008.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson CA, et al. Müllerian inhibiting substance requires its N-terminal domain for maintenance of biological activity, a novel finding within the transforming growth factor-β superfamily. Mol. Endocrinol. 1993;7:247–257. doi: 10.1210/mend.7.2.8469238. [DOI] [PubMed] [Google Scholar]
- 31.Ulloa L, et al. Lefty proteins exhibit unique processing and activate the MAPK pathway. J. Biol. Chem. 2001;276:21387–21396. doi: 10.1074/jbc.M006933200. [DOI] [PubMed] [Google Scholar]
- 32.Keefe AD, Wilson DS, Seelig B, Szostak JW. One-step purification of recombinant proteins using a nanomolar-affinity streptavidin-binding peptide, the SBP-Tag. Protein Expr. Purif. 2001;23:440–446. doi: 10.1006/prep.2001.1515. [DOI] [PubMed] [Google Scholar]
- 33.Zou Z, Sun PD. Overexpression of human transforming growth factor-β1 using a recombinant CHO cell expression system. Protein Expr. Purif. 2004;37:265–272. doi: 10.1016/j.pep.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Gentry LE, et al. Type 1 transforming growth factor beta: amplified expression and secretion of mature and precursor polypeptides in Chinese hamster ovary cells. Mol. Cell. Biol. 1987;7:3418–3427. doi: 10.1128/mcb.7.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunner AM, et al. Site-directed mutagenesis of glycosylation sites in the transforming growth factor-beta 1 (TGF beta 1) and TGF beta 2 (414) precursors and of cysteine residues within mature TGF beta 1: effects on secretion and bioactivity. Mol. Endocrinol. 1992;6:1691–1700. doi: 10.1210/mend.6.10.1448117. [DOI] [PubMed] [Google Scholar]
- 36.Heras B, Martin JL. Post-crystallization treatments for improving diffraction quality of protein crystals. Acta Crystallogr. D. 2005;61:1173–1180. doi: 10.1107/S0907444905019451. [DOI] [PubMed] [Google Scholar]
- 37.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W. In: International Tables for Crystallography, Vol. F: Crystallography of Biological Macromolecules. Rossmann MG, Arnold EV, editors. Springer; 2001. pp. 730–734. Ch. 25.2.9 XDS. [Google Scholar]
- 39.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 40.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. D. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 41.Bailey S. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 42.Cowtan K. Recent developments in classical density modification. Acta Crystallogr. D. 2010;66:470–478. doi: 10.1107/S090744490903947X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 44.Davis IW, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takagi J, Petre BM, Walz T, Springer TA. Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell. 2002;110:599–611. doi: 10.1016/s0092-8674(02)00935-2. [DOI] [PubMed] [Google Scholar]
- 46.Chen X, et al. Requirement of open headpiece conformation for activation of leukocyte integrin αXβ2. Proc. Natl Acad. Sci. USA. 2010;107:14727–14732. doi: 10.1073/pnas.1008663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.