Abstract
Most of multi-atlas segmentation methods focus on the registration between the full-size volumes of the data set. Although the transformations obtained from these registrations may be accurate for the global field of view of the images, they may not be accurate for the local prostate region. This is because different magnetic resonance (MR) images have different fields of view and may have large anatomical variability around the prostate. To overcome this limitation, we proposed a two-stage prostate segmentation method based on a fully automatic multi-atlas framework, which includes the detection stage i.e. locating the prostate, and the segmentation stage i.e. extracting the prostate. The purpose of the first stage is to find a cuboid that contains the whole prostate as small cubage as possible. In this paper, the cuboid including the prostate is detected by registering atlas edge volumes to the target volume while an edge detection algorithm is applied to every slice in the volumes. At the second stage, the proposed method focuses on the registration in the region of the prostate vicinity, which can improve the accuracy of the prostate segmentation. We evaluated the proposed method on 12 patient MR volumes by performing a leave-one-out study. Dice similarity coefficient (DSC) and Hausdorff distance (HD) are used to quantify the difference between our method and the manual ground truth. The proposed method yielded a DSC of 83.4%±4.3%, and a HD of 9.3 mm±2.6 mm. The fully automated segmentation method can provide a useful tool in many prostate imaging applications.
Keywords: Magnetic resonance imaging (MRI), edge volume, prostate segmentation, multi-atlas, registration, adaptive threshold
1. INTRODUCTION
MR imaging of the prostate is widely used in clinical practice for prostate cancer diagnosis and therapy [1–7]. MRI has high spatial resolution and high soft-tissue contrast for visualizing the size, shape, and location of the prostate. Segmentation of the prostate plays an important role for clinical applications. However, an automated segmentation of prostate MR images is a challenging task due to the fact that the shape of the prostate varies significantly and it has inhomogeneous intensities in the prostate area of MR volumes [8].
Recently, atlas-based segmentation [9–12] has become a frequently used automatic method for prostate delineation. Klein et al. [9] proposed an automatic prostate segmentation method based on registering manually segmented atlas images. A localized mutual information based similarity measure is used in the registration stage. This method focused on the global registration between the whole volumes. However, due to local structure variation, a global registration strategy for label fusion may not achieve the most accurate delineation of the local region around the prostate. Ou et al. [10] proposed a multi-atlas-based automatic pipeline to segment the prostate MR images. Initial segmentation of the prostate was computed based on the atlas selection. Once the initial segmentation is obtained, their method focused on the initially segmented prostate region to get final results by simply re-running the atlas-to-target registration. There is no difference between the two-phase segmentation except for the target image size. It is not an efficient way and there is no much improvement by adding the second phase. In this paper, we introduce a two-stage framework. In the first stage, an edge volume is used as the target volume in the multi-atlas segmentation for fast detecting the location of the prostate. In the second stage, a smaller volume around the prostate is used as the target volume for accurate segmentation of the prostate.
2. METHODS
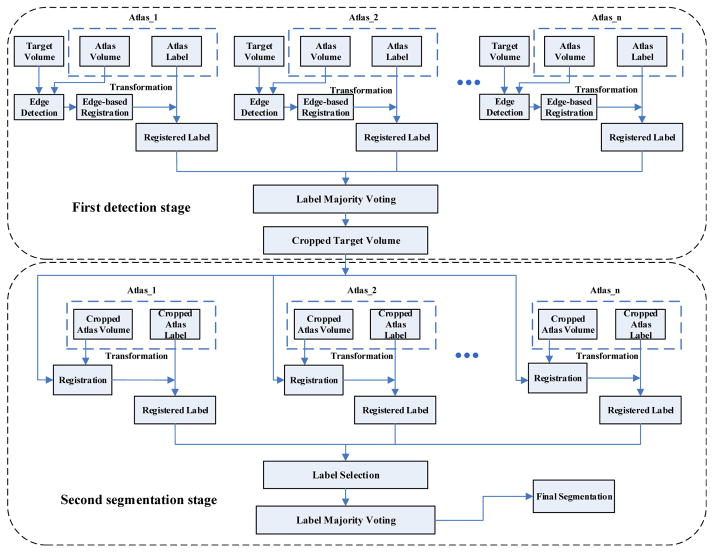
In this work, we are primarily interested in segmenting the prostate in a small region around the prostate. A two-stage prostate extraction framework is proposed, which includes the detection stage and the segmentation stage. For the detection stage, we first detect the location of the prostate in the MR volume. Instead of using intensity-based volume registration, an edge volume registration method is adopted to quickly determine the volume of interest (VOI) around the prostate. To obtain the edge volume, an edge detection algorithm is adopted on each slice of MR volumes. For the segmentation stage, only the prostate vicinity needs to be registered between the cropped atlas volume and the cropped target volume, which has a lower computational cost than registering the whole volumes. Fig. 1 shows the flowchart of the proposed method. In the following presentation, the main notations are listed in Table 1. In the next section, we will present the proposed detection and segmentation framework in details.
Figure 1.
The flowchart of the proposed method. The first stage (top) is for the detection of prostate location, while the second stage (bottom) is for the segmentation of the prostate boundaries.
Table 1.
Notations used in the paper.
| V | Target volume | VC | Cropped target volume | |
| A(I, L) | Atlas, I is atlas volume, L is atlas label | AC(IC, LC) | Cropped atlas | |
| E | Edge volume of V | TC(VC, IC) | Transformation from IC to VC | |
| T(V, I) | Transformation from I to V | LCR | Registered cropped label | |
| L̂ | Final label |
|
The mutual information between the ith registered cropped volume and VC | |
| LR | Registered atlas label |
2.1 Prostate detection
At the first stage, multi-atlas segmentation framework [13, 14] is applied on the edge volume to obtain the VOI of the prostate. Given a target MR volume V, and a set of atlases A(I, L). We are looking for a labeled volume that contains a rough segmentation of the prostate for obtaining the location of the prostate.
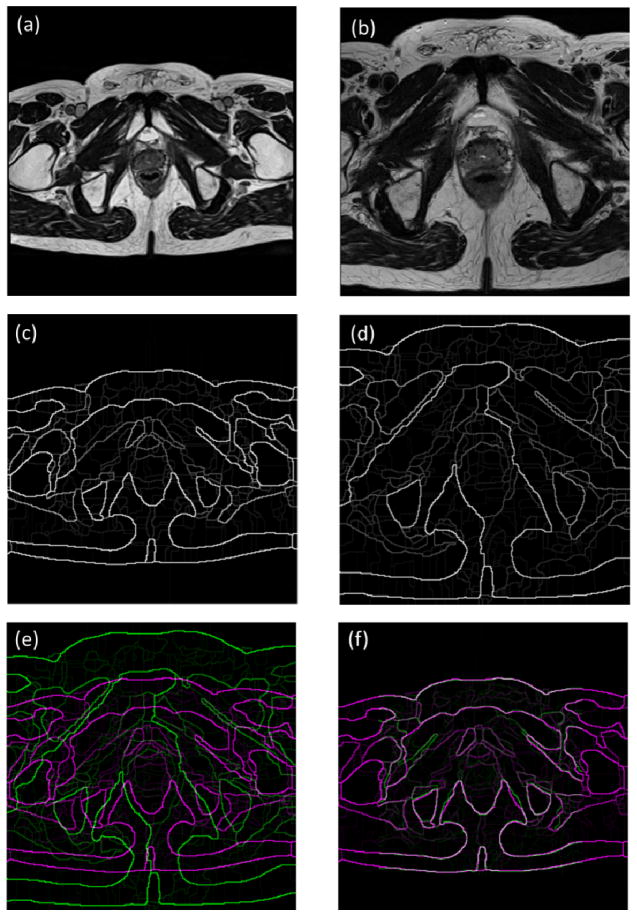
First, we obtain an edge volume of an atlas volume Ii represented as E(Ii), by applying an edge detection method [15] on the MR data in a slice-by-slice manner. The output of the edge detection algorithm is an intensity map. The intensity value ranges from 0 to 1. ‘0’ presents no edge at the current voxel, while ‘1’ represents a definite edge voxel. Similarly, the edge volume of the target volume V is obtained and represented as E(V). Edge detection removes redundant information in the MR images, which makes volumes easier to be registered. Therefore, edge volume registration can be more robust than intensity-based volume registration [3, 16] that can be trapped at local minima. Figure 2 shows the demonstration of the edge maps of MR images.
Figure 2.
The demonstration of the edge maps of MR images. (a) The target image. (b) The atlas image. (c) The edge map of the target image. (d) The edge map of the atlas image. (e) Two edge maps are superposed before registration. (f) Two edge maps are superposed after registration. Gray areas correspond to areas that have similar intensities, while magenta and green areas show places where one image is brighter than the other.
Second, a transformation T (rotation, translation, and scaling) from E(Ii) to E(V) is calculated by minimizing the mean of squared error (MSE) between E(V) and registered E(Ii). After registration, the registered atlas label LR is calculated by applying the obtained transformation to the corresponding atlas label L. Similarly, the other registered atlas labels of atlas database can be obtained.
Finally, the majority voting is used to get the initial segmentation of the prostate. To make sure that the initial segmentation can cover the whole prostate, we first dilate the prostate mask obtained from the label majority voting. Then the bounding box of the dilated mask is used to crop the target volume for the second segmentation stage.
2.2 Prostate segmentation
Once obtain the cropped volume VC, the similar multi-atlas framework is used for the second segmentation stage. The difference between two stages is that an affine transformation (rotation, translation, scaling, and shear) and a mutual information [17] similarity metric are adopted at the second stage. First, we use affine transformation TC(VC, IC) for registering the cropped atlases volume IC to the cropped target volume VC, while mutual information is used as the similarity metric. As affine transformation and mutual information metric are only performed on cropped volume, it can alleviate the computational problem significantly. Note that the atlas volumes are also cropped based on their corresponding label volumes, which is presented as AC(IC, LC).
Second, for a given target, some certain atlases in the database may be more appropriate as the candidates than others. Combining only these atlases is likely to produce a better segmentation than ones that produced from the full atlas database. This consideration provides a motivation for the selection of atlases that are appropriate for a given target volume. In our work, the mutual information between the target volume and registered atlas volume is used to guide the selection of atlases. In the final atlas fusion process, we use an adaptive threshold method to pick up the appropriate registered labels for label majority voting.
The registered atlas label LCR whose mutual information between the corresponding registered cropped volume ICR and the cropped target volume VC is greater than the median value of mutual information of other 11 cropped atlas volumes (there are 12 volumes for the leave-one-out experiment) will be selected as a candidate. Therefore, the i-th registered label will be selected if it satisfies the following condition:
| (1) |
Finally, we use the selected labels to obtain the final segmentation L̂ by applying majority voting.
2.3 Evaluation
The leave-one-out cross validation method was adopted in our experiments for all MR volumes within the dataset. In each experiment, one MR volume was excluded from the dataset and was considered as the target volume, while other 11 volumes were used as the atlases. Our segmentation method was evaluated by two quantitative metrics, which are dice similarity coefficient (DSC) and Hausdorff distance (HD) [18, 19]. The DSC was calculated using:
| (2) |
where |A| is the number of voxels in the manual segmentation and |B| is the number of voxels in the segmentation of algorithm. To compute the HD, a distance from a voxel x to a surface Y is first defined as:
| (3) |
Then the HD between two surfaces X and Y is calculated by:
| (4) |
3. EXPERIMENTS AND RESULTS
The proposed automatic segmentation method has been implemented using MATLAB on a computer with 3.4 GHz CPU and 128 GB memory. The volume size varies from 320×320×23 to 320×320×61. The execution time of segmenting each MR volume is about 4 minutes.
3.1 Qualitative evaluation results
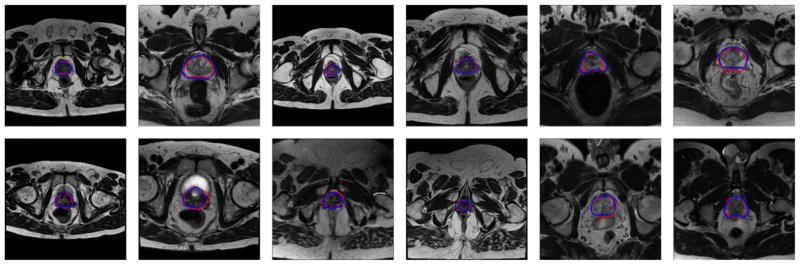
Quantitative evaluation was obtained by comparing the automatically generated segmentations with the manual segmentations provided by an experienced radiologist. The qualitative results from 12 prostate volumes are shown in Fig. 3. Blue curves are the manual segmented ground truth by the radiologist, while the red curves are segmentation obtained from the proposed method. The proposed method yields satisfactory segmentation results for all the 12 volumes.
Figure 3.
The qualitative results of 12 prostate volumes. The red curves are the prostate contours obtained by the proposed method, while the blue curves are the contours obtained by manually labeling.
3.2 Quantitative evaluation results
In Table 2, we list the DSC and HD calculated from the leave-one-out cross validation of the proposed method on 12 MR image volumes. Our fully automatic segmentation approach yielded a DSC of 83.4%± 4.3%. For the 12 volumes, the HD is 9.3mm±2.6mm, which indicates the proposed method can detect prostate tissue with a relatively low error.
Table 2.
Quantitative results.
| #Volume | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Avg. | Std. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DSC(%) | 82.3 | 85.1 | 83.0 | 91.0 | 75.2 | 78.5 | 82.5 | 79.2 | 84.6 | 85.3 | 87.2 | 86.4 | 83.4 | 4.3 |
| HD(mm) | 10.1 | 7.4 | 6.9 | 5.2 | 12.7 | 12.5 | 7.4 | 13.3 | 8.4 | 7.5 | 9.2 | 11.2 | 9.3 | 2.6 |
3.3 Discussion
The main task of the first stage is to find the location and the approximate size of the prostate. Therefore, a conservative segmentation is obtained to make sure that the initial segmentation is inside the prostate region. In this work, the first stage yields an average DSC of 67% on the full-size volume, which is good enough for detecting the prostate but is not good for the final segmentation. Based on the results of the fully automatic stage, the DSC of the second stage on the cropped volume has been increased to 83%, which demonstrates the necessity of the second stage.
4. CONCLUSION AND FUTURE WORK
We proposed an automatic two-stage method to segment the prostate from MR volumes. To the best of our knowledge, this is the first study to use edge volumes to register MR volumes for multi-atlas-based prostate segmentation. Based on the low computational cost and robust edge volume registration, a volume of interest (VOI) contains the prostate can be obtained. Once the prostate VOI is obtained, the top best registered atlases are chosen for the label fusion by measuring mutual information in the vicinity of the prostate. The experimental results show that the proposed method can be used to aid in the delineation of the prostate from MR volumes, which could be useful in a number of clinical applications.
Table 3.
Comparison between our label fusion method and the MV and STAPLE methods [20].
| #Volume | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | Avg. | Std. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MV | 73.3 | 62.1 | 79.4 | 88.1 | 53.8 | 53.7 | 81.3 | 73.4 | 80.2 | 83.7 | 69.3 | 69.5 | 72.3 | 11.2 |
| STAPLE | 80.2 | 79.6 | 84.0 | 90.1 | 66.7 | 72.9 | 84.5 | 78.9 | 83.7 | 85.1 | 77.2 | 80.8 | 80.3 | 6.1 |
| Ours | 82.3 | 85.1 | 83.0 | 91.0 | 75.2 | 78.5 | 82.5 | 79.2 | 84.6 | 85.3 | 87.2 | 86.4 | 83.4 | 4.3 |
Acknowledgments
This research is supported in part by NIH grants R21CA176684, R01CA156775 and P50CA128301, Georgia Cancer Coalition Distinguished Clinicians and Scientists Award, and the Center for Systems Imaging (CSI) of Emory University School of Medicine.
References
- 1.Wu Qiu JY, Ukwatta Eranga, Sun Yue, Rajchl Martin, Fenster Aaron. Prostate Segmentation: An Efficient Convex Optimization Approach With Axial Symmetry Using 3-D TRUS and MR Images. IEEE Transactions on Medical Imaging. 2014;33(4):947–960. doi: 10.1109/TMI.2014.2300694. [DOI] [PubMed] [Google Scholar]
- 2.Hu YP, Ahmed HU, Taylor Z, Allen C, Emberton M, Hawkes D, Barratt D. MR to ultrasound registration for image-guided prostate interventions. Medical Image Analysis. 2012;16(3):687–703. doi: 10.1016/j.media.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 3.Fei B, Lee Z, Boll DT, Duerk JL, Lewin JS, Wilson DL. Image registration and fusion for interventional MRI guided thermal ablation of the prostate cancer. MICCAI. 2003:364–372. doi: 10.1109/TMI.2003.809078. [DOI] [PubMed] [Google Scholar]
- 4.Fei B, Duerk JL, Boll DT, Lewin JS, Wilson DL. Slice-to-volume registration and its potential application to interventional MRI-guided radio-frequency thermal ablation of prostate cancer. IEEE Trans Med Imaging. 2003;22(4):515–525. doi: 10.1109/TMI.2003.809078. [DOI] [PubMed] [Google Scholar]
- 5.Litjens G, Debats O, Barentsz J, Karssemeijer N, Huisman H. Computer-aided detection of prostate cancer in MRI. IEEE transactions on medical imaging. 2014;33(5):1083–1092. doi: 10.1109/TMI.2014.2303821. [DOI] [PubMed] [Google Scholar]
- 6.Toth R, Madabhushi A. Multifeature Landmark-Free Active Appearance Models: Application to Prostate MRI Segmentation. IEEE Transactions on Medical Imaging. 2012;31(8):1638–1650. doi: 10.1109/TMI.2012.2201498. [DOI] [PubMed] [Google Scholar]
- 7.Schulz J, Skrovseth SO, Tommeras VK, Marienhagen K, Godtliebsen F. A semiautomatic tool for prostate segmentation in radiotherapy treatment planning. BMC Med Imaging. 2014;14:4. doi: 10.1186/1471-2342-14-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yuan J, Qiu W, Ukwatta E, Rajchl M, Sun Y, Fenster A. An efficient convex optimization approach to 3D prostate MRI segmentation with generic star shape prior. Prostate MR Image Segmentation Challenge, MICCAI. 2012:1–8. [Google Scholar]
- 9.Klein S, van der Heide UA, Lips IM, van Vulpen M, Staring M, Pluim JPW. Automatic segmentation of the prostate in 3D MR images by atlas matching using localized mutual information. Medical Physics. 2008;35(4):1407–1417. doi: 10.1118/1.2842076. [DOI] [PubMed] [Google Scholar]
- 10.Ou Y, Doshi J, Erus G, Davatzikos C. Multi-atlas segmentation of the prostate: A zooming process with robust registration and atlas selection. Medical Image Computing and Computer Assisted Intervention (MICCAI) Grand Challenge: Prostate MR Image Segmentation. 2012;7:1–7. [Google Scholar]
- 11.Nouranian S, Mahdavi SS, Spadinger I, Morris WJ, Salcudean SE, Abolmaesumi P. An Automatic Multi-atlas Segmentation of the Prostate in Transrectal Ultrasound Images Using Pairwise Atlas Shape Similarity. MICCAI. 2013:173–180. doi: 10.1007/978-3-642-40763-5_22. [DOI] [PubMed] [Google Scholar]
- 12.Wang HZ, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Transactions on Pattern Analysis and Machine Intelligence. 2013;35(3):611–623. doi: 10.1109/TPAMI.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langerak TR, Berendsen FF, Van der Heide UA, Kotte A, Pluim JPW. Multiatlas-based segmentation with preregistration atlas selection. Medical Physics. 2013;40(9):8. doi: 10.1118/1.4816654. [DOI] [PubMed] [Google Scholar]
- 14.Lötjönen JM, Wolz R, Koikkalainen JR, Thurfjell L, Waldemar G, Soininen H, Rueckert D. Fast and robust multi-atlas segmentation of brain magnetic resonance images. Neuroimage. 2010;49(3):2352–2365. doi: 10.1016/j.neuroimage.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 15.Dollár P, Zitnick CL. Structured forests for fast edge detection. IEEE International Conference on Computer Vision (ICCV); 2013. pp. 1841–1848. [Google Scholar]
- 16.Fei B, Wang H, Muzic RF, Jr, Flask C, Wilson DL, Duerk JL, Feyes DK, Oleinick NL. Deformable and rigid registration of MRI and microPET images for photodynamic therapy of cancer in mice. Med Phys. 2006;33(3):753–60. doi: 10.1118/1.2163831. [DOI] [PubMed] [Google Scholar]
- 17.Viola P, Wells WM., III Alignment by maximization of mutual information. International journal of computer vision. 1997;24(2):137–154. [Google Scholar]
- 18.Garnier C, Bellanger JJ, Wu K, Shu HZ, Costet N, Mathieu R, de Crevoisier R, Coatrieux JL. Prostate Segmentation in HIFU Therapy. IEEE Transactions on Medical Imaging. 2011;30(3):792–803. doi: 10.1109/TMI.2010.2095465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fei B, Yang X, Nye JA, Aarsvold JN, Raghunath N, Cervo M, Stark R, Meltzer CC, Votaw JR. MR/PET quantification tools: registration, segmentation, classification, and MR-based attenuation correction. Med Phys. 2012;39(10):6443–6454. doi: 10.1118/1.4754796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warfield SK, Zou KH, Wells WM. Simultaneous truth and performance level estimation (STAPLE): An algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging. 2004;23(7):903–921. doi: 10.1109/TMI.2004.828354. [DOI] [PMC free article] [PubMed] [Google Scholar]