Abstract
Cytokines are bioactive proteins produced by many different cells of the immune system. Due to their role in different inflammatory disease states and maintaining homeostasis, there is enormous clinical interest in the quantitation of cytokines. The typical standard methods for quantitation of cytokines are immunoassay-based techniques including enzyme-linked immusorbent assays (ELISA) and bead-based immunoassays read by either standard or modified flow cytometers. A review of recent developments in analytical methods for measurements of cytokine proteins is provided. This review briefly covers cytokine biology and the analysis challenges associated with measurement of these biomarker proteins for understanding both health and disease. New techniques applied to immunoassay-based assays are presented along with the uses of aptamers, electrochemistry, mass spectrometry, optical resonator-based methods. Methods used for elucidating the release of cytokines from single cells as well as in vivo collection methods are described.
Keywords: Cytokines, Immunoassay, Microfluidics, Optical Resonators, Aptamers, Review
1. Introduction
Cytokines and chemoattractant cytokines known as chemokines are highly localized soluble signaling proteins produced by many cells of the immune system (neutrophils, monocytes, macrophages, B-cells, and T-cells) to regulate immune responses [1–3]. These proteins are widespread through mammals and interestingly, recently discovered in invertebrates [4, 5]. There are several different families of cytokine proteins and the number of identified proteins continues to grow. For example the interleukin family is categorized in numerical order (up to IL-38) and other families include those describing functional activity such as the tumor necrosis family. Cytokines differ not only in their function, but also have a wide variety of molecular weight ranges from approximately 6 to 70 kDa.
Cytokines act as mediators and modulators within highly localized environments and regulate immunological responses, hematopoietic development, and cell-to-cell communication as well as host responses to infectious agents and inflammatory stimuli [6]. They interact with each other in complex ways that may be additive, synergistic or antagonistic, or may involve the induction of one cytokine by another. Cytokines are pleiotropic which refers to their ability to address multiple targets and physiological effects as shown in Figure 1. The term cytokine redundancy is associated with cytokine pleiotropy and is a common feature of some cytokines [7]. The physiological effects of cytokines often depend on the relative concentrations of several cytokines [8]. This illustrates the importance of recognizing that cytokines influence physiology via networks (Figure 1) [9, 10]. Furthermore, since cytokines work in networks, it is significantly important to be able to measure multiple cytokines in a single sample. Some examples of cytokine properties and analysis challenges combined with an overview of approaches aimed to meet these challenges is provided in Figure 2.
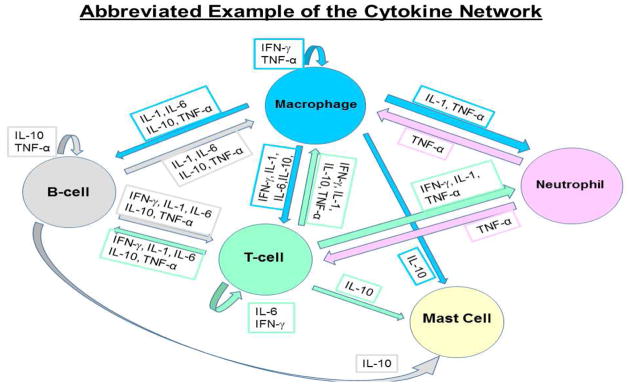
Figure 1.
An abbreviated version of the cytokine network highlighting the molecular communication between different cell types in the immune system.
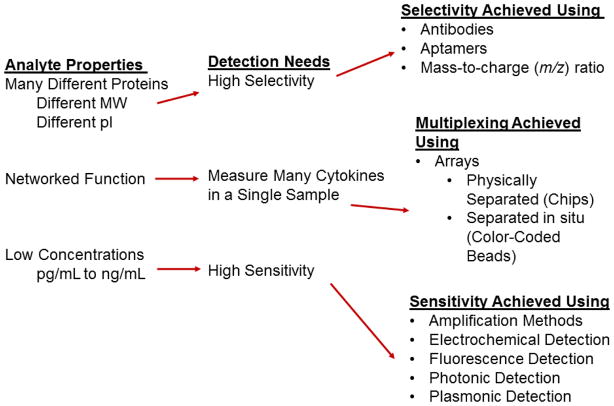
Figure 2.
Overview of cytokine properties, detection needs and the different methods that have been used to meet these detection challenges.
Cytokine production is often transient and tightly regulated. Due to the high biological activity of most cytokines, their homeostatic concentration in body fluids is low, e.g. picomolar concentrations [11]. However, if required, the concentration of cytokines can increase up to 1,000-fold. In healthy individuals, cytokines are either not detectable or present at pg/mL concentrations in body fluid or tissues. Elevated concentrations of cytokines indicate activation of cytokine pathways associated with inflammation or disease progression [12]. For this reason, cytokine measurements are important as these proteins are widely used as biomarkers to understand and predict disease progression and monitor the effects of treatment [13]. Since cytokines are biomarkers of inflammatory-based diseases, nearly every type of disease has involvement of cytokines as potential biomarkers. Some recent representative reviews for these different diseases and the roles for cytokines are provided here: Alloreactivity (rejection to clinical transplantation) [14, 15], Alzheimer’s [16], asthma [17], atherosclerosis [18], colon cancer [19], cancer [20, 21], depression [22], heart disease [23, 24], HIV [25], kidney injury [26], Parkinson’s disease [27], sepsis [28], and rheumatoid arthritis [29]. As a result, understanding of the cytokine orchestra and its regulation abnormalities in these diseases could ultimately lead to promising and specific treatments for patients [30, 31].
1.1. Cytokine Soluble Receptors
Cytokine receptors may be both membrane-bound at the cell surface and soluble. Soluble cytokine receptors regulate cytokine activity by functioning as either agonists or antagonists of cytokine signaling. Soluble receptors are common for many cytokines as part of the homeostatic process and prevents cytokine levels to approach what is known as a cytokine storm,” also known as hypercytokinemia, an immune reaction with highly elevated levels of various cytokines that can lead to death. A comprehensive, although dated, review of cytokine soluble receptors is available [32]. Soluble receptors for cytokines include: sIL-1-RI, sIL-1-RII, sIL-2-Rα, sIL-2-Rβ, sIL-4-R, sIL-6-Rα, sIL-6-Rβ, sIL-7-R, sIL-9-R, s-mpl, sLIF-R, sCNTF-R, sG-CSF-R, sECF-R, sTGF-β-R, sPDGF-R, sNGF-R, sTNF-RI, sTNF-RII, sIFNα, and sIFNγ. If the cytokine is bound to its soluble receptor in a biological sample it typically cannot be quantified with an immunoassay-based measurement. This is important biological information to know about these proteins since if there is a significant upregulation in these receptors, the absolute cytokine concentrations observed may be lower than anticipated.
2. Standard Cytokine Assays
Accurate and sensitive methods for the measurement and detection of cytokines are an obvious prerequisite for the study of cytokine biology, biochemistry, the possible involvement of these molecules in pathology, and establishing the potency of cytokine medicinal products intended for therapeutic use [33]. Therefore, cytokines are important biomarkers to monitor the effect of drugs influencing the immune system or inflammation [34]. Many standard assay procedures exist for measurement of cytokines from different mammalian systems and have been reviewed [35]. Most analysts are familiar with understanding the possible components within the sample matrix that may affect or interfere with the measurement as being critical to measurement success. Some of these issues and a brief discussion of the many common assays available for cytokine measurements are provided below.
2.1 Commercially Available Assays – Bioassay, ELISA, ELISPOT/ FluoroSpot, and PCR
The common methods for detecting cytokines have been reviewed and include bioassays [33, 34, 36, 37], ELISA methods, Enzyme Linked Immuno Spot Assays (ELISPOT) which allow detection of cytokines from single cells [38–40] and indirect methods to determine gene expression for cytokine production (PCR). The FluoroSpot assay is a modification of the ELISPOT assay that uses a bound fluorophore instead of an enzyme reaction. These assays are now available to measure two separate cytokines from the same cell by using different anti-cytokine antibodies. ELISPOT is similar to ELISA in that cytokines produced from individually sorted cells (typically performed on a cell sorter (flow cytometer) are determined [41]. One of the advantages of this approach is that low frequency cells producing cytokines can be quickly identified and sorted.
Bioassays determine active concentrations of cytokines by determining outcomes for biological processes that are driven by cytokines. These processes include cellular proliferation or production of other cytokines. While the length of time necessary to perform most bioassays is a concern, a significant advantage of these assays is their ability to discriminate active vs. inactive forms of cytokines [36, 37, 42]. A wide variety of commercially available cytokine bioassays can be sourced for humans and mice, but much fewer for rats and other species. However, when compared to the availability of ELISA kits and antibodies, there are far fewer bioassays available for cytokines.
ELISA platforms are widely available and used for quantifying cytokines. Most cytokine ELISA measurements typically require approximately 100 μL of sample. With the increased understanding that cytokines are part of an overall signaling network [9], there has been a greater interest in assays that allow for detection of more than one cytokine or are multiplexed in their analyte measurement capability. These multiplexed assays including protein arrays and bead-based immunoassays are discussed in a separate section below.
Polymerase chain reaction (PCR) methods for measuring the regulation of cytokine gene expression are also commonly used for studies involving cytokines. This is a mature technology for cytokine RNA expression measurements that more than two decades ago allowed for measurements from as few as 20 cells. With improved technology for handling picoliter sized volumes from advances in microfluidics [43, 44], instruments that can perform PCR on a single cell are available and have been used for single-cell studies of cytokine expression [45].
2.2. Multiplexed Based Immunoassays: Protein Arrays and Bead-Based
The physical release of soluble cytokines and other soluble released factors from cells has now been “omed” and is called the secretome [46]. Elucidating the secretome or the cytokine network as described above requires analysis tools that have multiplexing capability. Significant advances in miniaturization, lithography, and fluidics have paved the way for commercially available multiplexed assays for measuring cytokines. These assays are briefly described below to allow the focus of this review to be on new methods for measuring cytokines.
2.2.1. Bead-Based Immunoassay
Microsphere-based immunoassays that can be monitored with flow cytometry platforms have been developed allowing highly sensitive and multiplexed measurements of cytokines within low microliter volume samples [47–51]. These platforms incorporate sets of microspheres (~ 5 to 7 μm o.d.) identifiable by the concentration of fluorophores embedded into the beads that serve as the solid support for the immunoassay. The flow cytometry system is capable of discriminating the fluorophore intensity in the bead set as well as the mean fluorescence intensity (MFI) for the labeled detection antibody (Figure 3) [52]. With high affinity antibodies and rapid reaction kinetics in solution, microsphere-based multiplexed immunoassays for flow cytometry provide a more rapid, specific and reproducible analysis method that has a three to four logarithmic range of sensitivity compared with one to two logs for ELISAs [53].
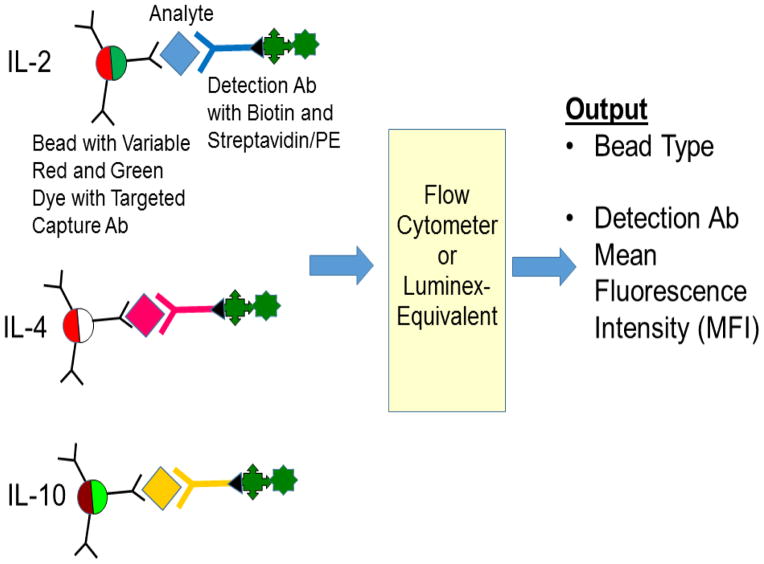
Figure 3.
Overview of bead-based immunoassays. Different color-coded beads with dyes that fluoresce either red or green are used. The instrument measures the bead color intensity and the mean fluorescence intensity of the labeled detection antibody which is typically labeled with a streptavidin/phycoerythrin (PE) conjugate.
2.2.2. Micropatterned Antibody Cytokine Arrays
Similar to the bead-based immunoassays, there has been a tremendous interest in development of multiplexed protein arrays. Antibodies have been patterned onto surfaces using photolithography and have retained their activity [54]. This has led to the development of commercial arrays used for cytokine detection that were able to detect 1–15 pg/mL of IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-12, IL-13, and TNF-α in 15 μL using a sandwich ELISA with Cy-5 streptavidin used as the marker vs. biotinylated detection antibodies [55]. The use of antibody microarrays and all the cytokines that have been detected with this technique has been extensively reviewed [56].
A comparison between the competitive assay vs. sandwich assay approach for five cytokines, IL-1β, MIP-1β, TGF-β1, TNF-α, and VEGF on a protein array was performed [57]. If possible, competitive assays would be preferred since washing steps as well as the assay time would be significantly reduced. It was found that a sandwich assay was preferred as there was less variability observed. This was contributed to many factors one of which includes the possibility of altered binding affinity for fluorescent-labeled cytokines.
Chip- or slide-based protein arrays for cytokines both commercially available and created in house by various researchers have seen wide application. These arrays have the capacity to measure a few cytokines to more than 30 in a sample. Using protein microarrays, cytokines have been detected from cell culture [58], wound fluids [59], dermal [60, 61] and pancreatic fluid [61] and to determine the effectiveness of antiviral therapies [62]. Zajac et al., described a protein microarray for measurement of IL-1β, IL-6, IL-8, IL-13, MIP-1β, and TNF-α using quantum dots [63]. Modified chips from a commercial vendor have been used for measuring 80 cytokines from tear fluid [64].
2.3. Challenges and Validation
There are challenges associated with measurements of cytokines using protein microarrays including quality control and necessary algorithms to transform the data into meaningful clinical results [65]. For example, Wu and Grainger used different additives as a means to stabilize the cytokine antibodies for human IL-1β, IL-4, and TNF-α spotted onto a microarray [66]. With their approach, the chips were stable for 1 month at 4°C. Additional concerns for multiplexed assays both on chips and bead-based have been raised regarding quality control, low precision between assays, and general lack of reference methods [67]. Others have modified the surfaces to prevent non-specific adsorption using nonfouling polymer brushes [68].
There has been a significant interest in the validation of these different types of assays. Different studies have reported different outcomes for these kits. A common observation is differences in absolute cytokine concentrations outputted for the same samples when multiplexed kits are compared to standard ELISA measurements [50, 69, 70]. Others have found issues between different kits with respect to the coefficient of variation and other figures of merit [71, 72].
In a large international validation study, 12 laboratories located in four countries compared many different immunoassay platforms for detection of human cytokines, IL-1β and IL-6 [73]. The different platforms included standard ELISA, Luminex bead-based assays (both commercial and in-house created) and electro-chemiluminescence assays. In general, the intra-laboratory comparisons were within expected ranges. There were large inter-laboratory variations among the different platforms. The Luminex assays had the lowest inter-laboratory reproducibility.
3. Precautions during data interpretation sample preparation, and quality control
Whiteside has written many thorough articles describing many potential concerns with different aspects of cytokine measurements [34, 74]. Typical points include issues of protease degradation as well as activation of cytokine release due to the collection and processing of samples. As proteins, cytokines can be degraded in biological samples due to the presence of proteases. Other analytical concerns include sample handling and storage stability especially with freeze-thaw cycles. As mentioned in Section 1, cytokines bound to their soluble receptor are typically not quantified using immunoassay measurements.
3.1. Sample Storage and other Sample Processing-Related Issues
With the increased use of biological therapeutics applied to diseases to affect an immune response, e.g., cancer, multiple sclerosis, and rheumatoid arthritis, there is tremendous interest in understanding how cytokine responses are modulated by these different treatments. Thus, it is important to be able to compare cytokine concentrations across multiple studies and from −80°C stored samples that may have been stored for years. Zhou et al., have provided an overview of the many issues related to collection of samples that will be quantified for cytokines as well as issues of storage since cytokines have a short half-life and freeze-thaw cycles should be avoided [75]. In a recent study, Butterfield et al., reported several technical issues that were observed for cytokine analyses of healthy sera and melanoma patients whose sera was measured within a few months of blood draw vs. samples that were stored at −80°C for five years [76]. In this work, they investigated the cytokines: IL-4, IL-6, IL-8, IL-10, TNF-α, IFN-γ, GM-CSF. Important observations included the following: 1) Interleukin-8, IL-8 (CXCL8) increased four- to six-fold in old patient samples. 2) MCP-1 (CCL2) increased four- to six-fold in new patient samples and over ten-fold in healthy donor samples. 3) IL-10 was variable in different samples. These observations were similar to those reported by de Jager, et al. [77]. In the work of de Jager, et al., the effects of anticoagulants, long term storage, and freeze-thaw cycles were investigated for human IL-1α, IL-β, IL-2, IL-4, IL-5, IL-6, IL-8 (CXCL8), IL-10, IL-12, IL-12, IL-15, IL-17, IL-18, TNF-α, and IFN-γ. The studies from Butterfield et al. and de Jager et al. also mentioned the instability of cytokines after incurring several freeze-thaw cycles.
Gu et al., asked the question about repeated measures of cytokines and whether in some cases these are reliable long-term biomarkers [78]. This is important as frequently only one measurement of different cytokines will be used with no comparative measure. They tested 41 cytokines using the Luminex xMap system and found that only 16 of these 41 cytokine biomarkers had sufficient sensitivity and reproducibility within sera samples. Others have also asked similar questions with respect to the reliability of these different markers. Clendenen tested the reliability of measurements from healthy and menopausal women two years apart and found among 31 cytokines, cytokine soluble receptors, or growth factors that 22 were a reliable measurement [79].
The different variables that affect assays for cytokines have been reviewed including issues regarding variability in standards [80, 81]. Other sample preparation problems have included cytokine removal during albumin depletion steps [82] and protein/antibody interference of mM alcohol concentrations [83].
4. Needs for New Assay Developments Beyond the Standard Techniques
Standard diagnostic measurements for cytokines require long incubation times and extensive workflows. The significant diagnostic importance of cytokines has yielded numerous transduction platforms for cytokine quantification. The rationale for creating specific sensors to cytokines is that such quantitative information about these relevant biomarkers would add significant value to clinicians to determine treatment course [84, 85]. The desire for point of care measurements is primarily based on obtaining rapid answers while significantly reducing the personnel necessary to perform accepted antibody-based measurements (ELISA or flow-cytometry-based assays). Several requirements are necessary for such devices to ultimately replace the accepted diagnostic measurement systems that have immunoassays as their base. The primary issue is that any sensor or method measure proteins at clinically-relevant concentrations in the presence of many potential interferrents from a biological sample. Another driving force is the desire to perhaps only run a few samples at a time rather than full plates with immunoassays.
5. Immunoassay Improvements
Standard immunoassays for cytokines use detection antibodies that provide for colorimetric detection using absorbance-based plate readers. Knowing that cytokine basal levels are near or below pg/mL concentrations, there has been a great interest in extending the low limits of detection for cytokine immunoassays into the fg/mL or lower levels. Using I-labeling with multiphotons to increase sensitivity to fg/mL is one such example [86]. Different platforms have been proposed for these purposes including use of bioluminescence and immunoPCR methods that use the amplification of nucleic acids as a means to improve detection limits (Figure 4). The use of electrochemiluminescence as a means to decrease detection limits is discussed in the electrochemistry subsection of this review.
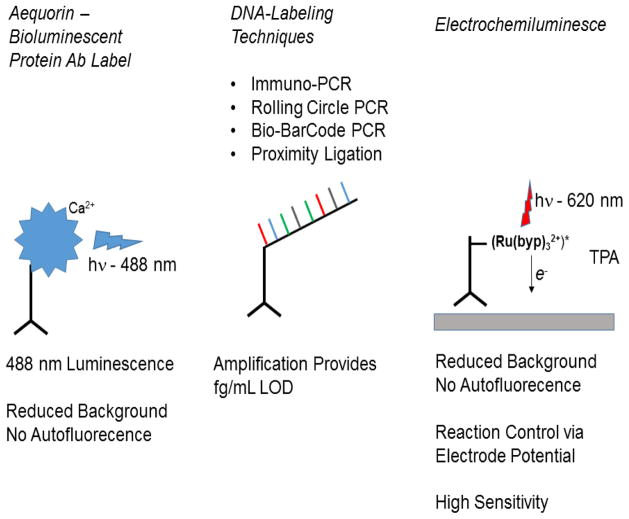
Figure 4.
Overview of immunoassay improvements. Note that aequorin requires calcium activation to achieve its photo-protein state. Additionally, several steps are included in the electrochemiluminescence. These steps are outlined in the text and in other review articles describing these techniques.
5.1. Bioluminescent Proteins
Bioluminenescent proteins are used as alternate labeling schemes since the light is produced from a biochemical reaction and there is no background from autofluorescence [87]. The photoprotein, aequorin, has been used for different cytokine mRNA analyses with PCR [88, 89]. Aequorin and derivatives have been engineered for use in cytokine immunoassays with reported detection limits for IL-1β, IL-6, and IL-8 in the low pg/mL range [90].
5.2. Immuno-PCR
The incorporation of DNA strands into an analysis has been widely used for amplification processes during detection of biomolecules and has been extensively reviewed [91]. There are many different variations of using the concepts of adding DNA and amplifying the signal using polymerase chain reaction (PCR) methods. Given the diversity of possibilities to both ligate the nucleic acid sequence to an antibody combined with the variety of transduction methods for nucleic acid amplification, it is not surprising that immuno-PCR methods have a wide variety of methods and schemes reported in the literature [92]. Compared to standard ELISA methods, the immuno-PCR methods have been reported to provide anywhere from 10–100,000 fold improvements in the limit of detection (LOD). In general, these assays appear to have ranges of LOD between 1000 to 100,000 molecules (~ 0.016 to 16 amol). For example, detection limits using an immuno-PCR method for TNF-α quantification has reported detection limits in the 1–10 fg/mL range. While many different variations of the immuno-PCR process have been summarized in the above-mentioned review article [91], the most common methods for measurement of protein antigens include coupling of single-stranded DNA to antibodies and performing a variety of amplification methods described below.
5.2.1. Rolling circle amplification
Rolling circle amplification (RCA) is a signal amplification technique that has been applied to cytokine analysis [93]. Like many other new techniques in cytokine assay development, RCA is aimed towards the capability of detecting cytokine proteins on a microarray-based chip device. In this method, the detection antibody to a specified cytokine antigen is biotinylated. Then, an additional antibody that binds to the biotin-labeled detection antibody and has a specific oligonucleotide primer is added to the detection well. Circular DNA that binds to this primer is then added with the appropriate nucleotides and DNA polymerase. This process initiates the rolling-circle amplification leading to a long DNA concatamer to which fluorescently-labeled complementary oligonucleotide probes will bind.
In the studies by Schweitzer and colleagues, 75 different cytokines were tested in the RCA platform [93]. A comprehensive comparison between the RCA platform vs. standard ELISA was provided. The assay sensitivity for the RCA vs. ELISA was either equivalent or actually higher (higher concentrations of the LOD as compared to ELISA). The same research group also used this assay to detect the different cytokine kinetics after LPS and TNF-α induction of Langerhans cells and dendritic cells.
5.2.2. BioBar Code
The bio-barcode amplification approach was originally conceived by Mirkin’s research group and uses the concept of immobilizing short oligonucleotides onto gold nanoparticles [94]. This approach allows for the amplification of signal to occur via oligonucleotide binding processes without the need to perform PCR. For cytokine detection, the method was modified to include the bio-barcode DNA into porous silica nanoparticles to which a capture antibody to the protein antigen has been immobilized. A second amine modified nanoparticle has a secondary antibody attached to it to allow for capture of the entire complex of the protein and the associated silica particle. This is then removed from the sample using a magnet and the resulting excess bio-barcode oligonucleotide strands are released to attach to complementary oligonucleotides on Au nanoparticles. This has been demonstrated for use with interleukin-2 in serum samples. A semi-log response was obtained in buffer solutions for IL-2, but became nonlinear when applied to human serum samples suggesting either antibody cross reactivity or other non-specific binding issues. Detection limits of 30 aM (10−18 M) or 0.45 fg/mL for IL-2 were reported [95, 96].
5.2.3. Proximity Ligation
In this PCR-based method, two separate antibodies are prepared with oligonucleotide chains [97]. When these antibodies bind to the antigen, this allows the oligonucleotide chains to be close enough together to perform a ligation and amplification procedure. This method has been used for detection of IL-2, IL-4, PDGF-BB, and VEGF. In their original paper describing this method for cytokine analysis, the authors demonstrated sensitivity down to approximately 10−18 M or 0.015 fg/mL for each of the proteins listed above in 1 μL samples [98]. This method has been improved by using dual fluorophore readouts [99] and by adding additional probes [100].
6 Antibody-Based Flow Assays
6.1. Immunochemistry-based Liquid Chromatographic Separations
In an attempt to overcome many of the limitations of standard immunoassays including: long preparation times (8–24 hrs), measurement of only one analyte at a time, and potential cross reactivity of antibodies, separations-based methods that could increase throughput as well as allow for more than one analyte to be quantified (multiplexing) have been described for many different proteins [101]. One reported immunochromatographic method for cytokines described the separation of granulocyte colony stimulating factor (GCSF) from its pegylated form [102]. Pegylated cytokines are commonly used in pharmaceutical-based immunotherapy as a means to increase half-life.
Schnek, et al. created an immunochromatographic method for IL-4 [11]. The detection limit was 50 pM for a 20 μL injection (750 pg/mL for ~ 15 kDa IL-4) of standards and 500 pM for IL-4 in cell culture medium. Liquid chromatography with different types of column chemistries was applied to separate cytokines. Separated cytokines were passed into a reactor to which fluorescently-labeled antibodies were added. Excess antibody was removed via a cytokine-immobilized column allowing only complexes of the cytokine bound to the fluorescently-labeled Ab to be quantified. The assay was influenced by solvent ionic strength as salt content of greater than 165 mM diminished the response. Reaction times were optimized to approximately a minute in the outflowing reactor. Interestingly, mobile phases that contained methanol linearly decreased the detection output for IL-4 from 0 to 80 v/v% methanol. At 80% (v/v) of methanol there was no response. Different retention behavior for the cytokines IL-2, IL-10, and IL-12 that could not be predicted based on cytokine molecular weight was reported.
Variants of interferon-α (IFN-α) have been used as biological therapy for various diseases including hepatitis B and C, melanoma, and leukemia. Using immobilized antibodies in a capillary column, Chaves, and Queiroz, immobilized antibodies against the variant, IFN-α2a, in a capillary tube to capture the therapeutic target. Captured protein was then quantified using LC-fluorescence methods. The detection range was 0.006 million international units (MIU)/mL (~ 120 ng/mL) to 3 MIU/mL International units are preferred as a means to standardize activity levels across manufacturers for this therapeutic product [103]. The World Health Organization states that 11,000 IU can be made using 220 ng of IFN-α.1
6.2. Capillary Electrophoresis/Immunochemistry-based Separations
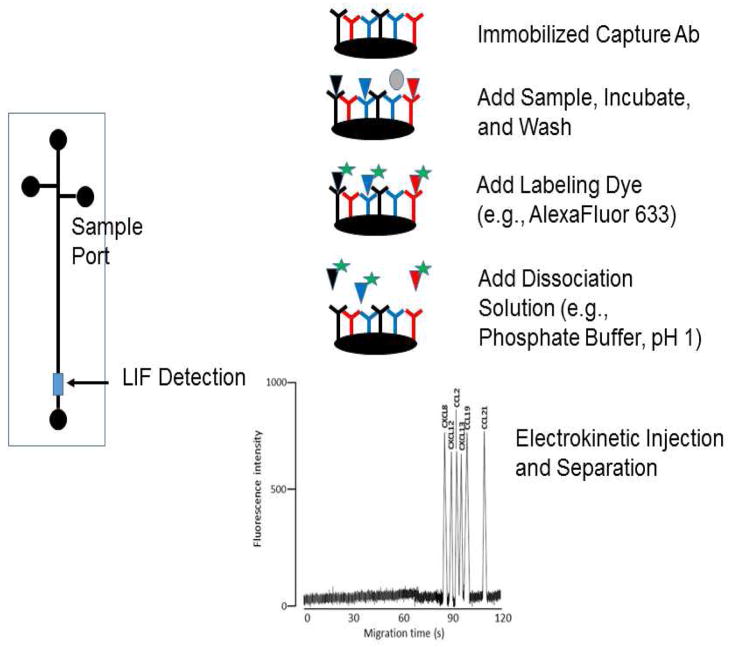
Capillary electrophoretic immunoaffinity chromatography has been described for cytokine measurements. In this technique, capture antibodies are immobilized into the fused silica capillary or onto a chip-based device (Figure 5). The cytokines are then labeled with a fluorescent dye. In the initial report of this work, cytokines are captured and then released via acidic wash separated using the capillary electrophoresis and detected using laser induced fluorescence (LIF) [104]. This work was then extended to using a recycling method with antibodies for 30 separate analytes immobilized onto glass beads. The reported detection limits were in the pg/mL range and a usage of these beads was reported to be 200 times [105]. Several different cytokines have been detected using immunoaffinity capillary electrophoretic approaches including the use of receptors as an alternative to antibodies [106–111].
Figure 5.
Capillary electrophoresis with immunoaffinity capture. The example electrophorogram is reprinted with permission from: Detection of cerebral spinal fluid-associated chemokines in birth traumatized premature babies by chip-based immunoaffinity CE, T.M. Phillips and E. Wellner, Electrophoresis. 2013 34(11):1530–8. John Wiley & Sons. doi: 10.1002/elps.201200634.
6.3. Antibody flow-based assays
A flow-based assay using fluorescent-labeled antibodies against IL-10 that were co-injected with samples to be quantified has been reported [112]. IL-10 was bound to an affinity support to capture the labeled-antibodies that are not bound to IL-10 from the sample. The LOD from cell media was reported to be 2.5 nM (45 ng/mL; ~18 kDa for IL-10 monomer) with a sample throughput of 20 samples/hr. The IL-10 assay performance was not affected by the presence of 50 nM IL-6, IL-8 or TNF-α. Similar approaches were performed for IL-8 with quantifiable levels of approximately 5 nM (40 ng/mL for ~ 8 kDa for IL-8) [113, 114].
Worsley et al described a lateral flow assay for IL-6 and TNF-α that was able to detect these two important cytokines into the 7–10 pg/mL range in plasma [115]. Using up-converting phosphor technology, IL-10 was detected at a targeted level of 100 pg/mL [116].
Qiu et al. described a flow-based immunoassay for IL-8 [117]. In their work, they created the necessary microfluidics for the cassette and used polymeric microbeads created by the Walt laboratory [118]. Polymeric beads with antibodies against VEGF and IL-8 were created and the assay was performed for detection of IL-8 at a concentration of 125 nM (~ 1 μg/mL assuming ~ 8 kDa for IL-8).
Cesaro-Tadic and colleagues were able to create a high-sensitivity chip for measurement of TNF-α. This fluorescence-based assay used Eu-based chelates for the detection step. Capture times for TNF-α had to be optimized to 12 minutes since 3 to 5 minutes did not provide appropriate assay sensitivity. Assay LOD was reported to be 20 pg/mL [119].
A microfluidic chip for interleukin-2 (IL-2), interleukin-4 (IL-4), and tumor necrosis factor-alpha (TNF-α) has been prepared that incorporates a fluorescent probe into a polymer material to demarcate the antibodies that have been immobilized to that chip [120]. This is similar in concept to the Luminex bead-based assay system. Detection limits for standard solutions were comparable to ELISA and Luminex assays in the 1–10 pg/mL range for these three cytokines.
6.4. Microfluidics and Lab on a Chip
McDevitt and colleagues created a total analysis system that was modified to be able to quantify bead-based immunoassays for c-reactive protein (CRP) and IL-6 [121]. This is part of an ongoing effort by this team to create programmable bio-chips (Figure 6) [122]. The applications of these lab-on-a-chip devices have included measuring different cytokines in salivary fluid. In these studies IL-1β, IL-6, and TNF-α were measured in different groups for studies of periodontal disease. The cytokines exhibited detection limits and ranges which were equivalent to those found for standard ELISA [123].
Figure 6.
Example of a programmable biochip. A) Capture antibody is immobilized to porous agarose. Detection antibodies are labeled with appropriate quantum dots to allow spectral discrimination. B) Agarose beads are driven into wells through a pressurized system. C) The design features of the chip are shown with the fluid lines. D) A modeling analysis of the overall pressures in the chip. Reprinted with permission from: JV Jokerst, J Chou, JP Camp, J Wong, A Lennart, AA Pollard, PN Floriano, N Christodoulides, GW Simmons, Y Zhou, MF Ali, JT McDevitt, Location of Biomarkers and Reagents within Agarose Beads of a Programmable Bionano-chip. Small. 2011, 7(5): 613–624. John Wiley & Sons, Inc. doi:10.1002/smll.201002089
Other microfluidic platforms for cytokine measurements have also been described. Using an electrowetting-based digital microfluidics platform, Sista and colleagues developed a total analysis system for IL-6 and insulin using magnetic beads [124]. The performance of the IL-6 assay seemed to be within the range typically observed for a standard ELISA measurement. The overall reaction time for the assay was 7 minutes. Other chip based methods have been described for IL-8 [125], IL-10 [126] plus IL-6 and TNF-α [127].
7. Aptamer Assays
Aptamers are single strands of either DNA or RNA oligonucleotides or nucleic acid ligands that can be used to bind different analytes with high specificity and affinity and are selected from large combinatorial libraries [128]. Their uses as molecular recognition agents and possible substitutes for antibodies in chemical analysis have been widely recognized [129–132]. The major advantages of aptamers are principally their stability and synthetic chemical creation rather than production from animals. This chemical stability allows apatmers to be reused numerous times. Aptamers are created by a process known as SELEX [133, 134]. There has been a recent emerging interest in using aptamers for various proteomics applications as selection agents [135, 136]. However, despite the enormous interest in these potential antibody substitutes, the number of aptamers that bind different cytokines is currently quite limited [137]. The vast majority of aptamer applications have involved using the aptamer as a substitute for an antibody in an immunoassay-type platform although a few interesting exceptions exist using electrochemical-based detection platforms and others including aptamer-labeled pores [138, 139]. Reported aptamers that bind cytokines include: interferon-γ (IFN-γ) [140]; interleukin-6 (IL-6) [141]; IL-17 [142]; IL-32 [143]; oncostatin M, a member of the IL-6 family [144]; platelet derived growth factor (PDGF) [145]; tumor necrosis factor-α [146, 147], and vascular endothelial growth factor (VEGF) [148]. Among these cytokines, new analytical chemistry assays or devices have been described using aptamers against IFN-γ, PDGF and VEGF.
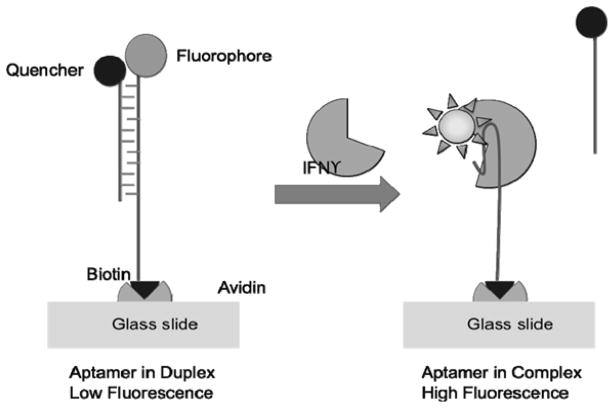
The Revzin group has published several papers describing various diagnostic methods for cytokine detection using a human IFN-γ aptamer [140, 149–151]. IFN-γ is an important inflammatory cytokine during host defense [152]. It is a heavily glycosylated protein with molecular weights reported between 15 and 25 kDa. They created different modifications of a previously published IFN-γ aptamer sequence [153]. One of the advantages of aptamers is the ability to use them as molecular beacons where a quencher of fluorescence is bound to a DNA strand that binds the aptamer as shown in Figure 7 allowing for measurement of fluorescence resonance energy transfer (FRET). The advantage of the molecular beacon approach can lead to single-step assays without the need for time consuming washing steps that are required for a standard immunoassay. The aptamer FRET sensor worked in culture media where the concentrations of other proteins would exceed that for IFN-γ by 100 to 10,000 fold. For the culture media, an approximate 20 to 30% loss in signal from serum possibly due to non-specific binding was observed. A linear range of 5 to 100 nM (~ 100 ng/mL = 5 nM assuming an approximate molecular weight of 20,000 Da) was observed for this sensor. The FRET based sensor from this group has been integrated into arrays and has been used to detect the amount of IFN-γ released from a single cell as 7.9 fg/cell/hr [149, 151].
Figure 7.
Aptamer-based FRET assay for IFN-γ. Reprinted with permission from: N Tuleuova; CN Jones; J Yan; E Ramanculov; Y Yokobayashi; A Revzin; Anal. Chem. 2010, 82, 1851–1857. DOI: 10.1021/ac9025237 Copyright © 2010 American Chemical Society
This group went on to create an electrochemical sensor by conjugating a methylene blue redox probe to the IFN-γ aptamer [140]. This aptamer was attached to a gold electrode. A significant finding was the need to optimize the density of the aptamer onto the gold electrode. A reported detection limit of 0.6 nM or 1 ng/mL IFN-γ was reported [135].
One of the advantages of using aptamers as compared to antibodies for specific target capture is their ability to be reused despite the use of harsh protein denaturing conditions. For the IFN-γ aptamer immobilized to a fluidic chip, 7 M urea was passed through during multiple catch and release cycles [149]. After four cycles of these harsh conditions, the aptamer was still able to capture IFN-γ.
Besides IFN-γ, aptamers to platelet derived growth factor (PDGF), particularly the “B” chain, PDGF-BB, have been widely reported and used in various detection schemes. One of the most sensitive methods that is available is what is termed the “proximity dependent DNA ligation assay” since it allows for amplification of the signal using PCR [97]. Using this method, Frederiksson and colleagues were able to detect as little as 40 zmol (10−21 mol) of PDGF-BB (MW ~ 24 kDa) in μL biological samples (~ 10 pg/mL in 1 μL sample) using a redesigned aptamer to PDGF-BB [145]. Using an aptamer against PDGF-BB and gold nanoparticles, Li et al., reported detection limits of 83 amol/L (1.9 fg/mL) and a linear range from 1 fmol/L to 100 pmol/L (24 fg/mL to 2.4 ng/mL)[154].
Weihong Tan’s research group has demonstrated the use of a molecular beacon approach with FRET measurements to discriminate between PDGF-BB and the variant PDGF-AA [155]. In this work, they modified an aptamer developed by Green, et al.[156]. They demonstrate different levels of fluorescence quenching between the different variants of PDGF, PDGF-BB, PDGF-AA and PDGF-AB with PDGF-BB quenching the FRET the most. Reported detection limits were in the 10 nM range for PDGF-BB (~ 240 ng/mL). This group has continued developing and improving their aptamer-based FRET detection by testing different fluorophore combinations and gold nanoparticles [157, 158].
An aptamer-based capillary electrophoresis separation that was capable of identifying PDGF-AA, PDGF-AB, PDGF-BB and their receptors has been reported by Le’s group[159]. Using the same aptamer for PDGF-BB that many others have used [156], this group was able to use the differential electrophoretic mobility differences between the aptamer bound to the PDGF-BB vs. the other forms as a means to quantify these proteins. The detection limits were in the low nM range for all three PDGF variants. Similarly, the PDGF receptor α was detected in the nM range.
The rolling circle amplification (RCA) approach has been used with the PDGF-BB aptamer [160]. Using a similar type of amplification approach called strand displacement amplification and a PDGF-BB aptamer, a detection limit of 0.9 pM has been reported [161]. Others have also reported on different variations of RCA with aptamers for cytokine detection [162, 163].
The company SomaLogic has focused on developing an extensive selection of aptamers to different biomarkers [130, 164]. The main driving force for designing these different targets is to allow for a rapid production of much-needed, reproducible, and stable affinity agents to new targets as they are discovered in biomedical research. Such reagents could then be kept in a large database of affinity reagents that would include both antibodies and aptamers for use in different proteomics applications [165]. There have been new aptamers made to different cytokine proteins with detection limits reported in the 10 fM range for IL-6 (1.4 pg/mL) and VEGF (0.27 pg/mL) [166]. While there have been some papers that highlight the work of this company, most of their work has been published in the patent literature for biomarkers of many different diseases especially different cancers. These aptamers developed by this company are designed to have slow dissociation off rates to facilitate their use in a variety of proteomics applications.
8. Mass Spectrometry
Cytokines are low abundance proteins with endogenous concentrations in the pg/mL range. Despite the wide spread use of mass spectrometry for qualitative proteomic studies, detection and quantitation of low abundance proteins is a significant challenge to using mass spectrometry as a means to quantify cytokines. Indeed there are many advantages to using mass spectrometry based approaches to measure proteins rather than immunoassays [167]. Since peptide fragments, rather than whole proteins, are used to confirm protein identity, there are significant challenges for low-abundance protein quantitation [168]. These challenges include differences in digestion processes due to different protein amino acid sequences and structure combined with efficiency of labeling techniques used for quantifying the peptides [169, 170]. While it is theoretically possible to detect cytokines from approximately 100 μL samples using mass spectrometry, typically detection suffers greatly from matrix interferences and/or suppression. For example, even with affinity capture, a series of interferon proteins spiked into serum could not be detected below 1 ng/mL using LC-ion trap MS, LC-Orbitrap MS or LC-FTMS [171]. Thus, antibody-capture techniques are commonly employed for quantifying low abundance proteins. Additionally, many studies with cytokines are performed in cell culture where concentrations are often much higher than found in a more tightly regulated in vivo environment. The measurement of the cytokine secretome from activated monocytes using label-free mass spectrometry approaches has been reported [172]. Others have also recently reported on the secretome of macrophages [173].
Many studies involving the detection of cytokines have focused on the use of specific cell types or sets to maximize the cytokine output. For example, a study that focused on mass spectrometry based detection of interleukin-2 (IL-2) from highly specialized T-cells used a variety of chromatographic, mass spectrometric (Orbitrap), and bioinformatics approaches to weed out IL-2’s signature peptides from the complex sample [174]. That is, in the spectral searches any peptide fragments not related to proteins derived from T-cells were removed from the search. Note that concentrations of IL-2 were in the low ng/mL range that was easily detected by the standard ELISA approach with an LOD of 7 pg/mL.
Insulin-like growth factor-1 (IGF-1) is a growth hormone that is implicated in different disease states and is believed to also be an important component to balance during exercise [175, 176]. Using a triple-quadrupole MS/MS method, Bredehöft and co-workers developed a mass spectrometry assay that could be used for doping control of IGF-1 and many of its synthetic analogs which are designed for faster clearance [177]. The LLOD for IGF-1 and the analogs were reported to be in the 20 to 50 ng/mL range. The significant advantage of the mass spectrometry method as compared to the standard radioimmunoassay detection method is that the MS method allows for exact typing of the manufacturer source of synthetic analogs by its ability to provide molecular weight information about the peptide products formed during the collisionally-induced dissociation (CID) event in the triple quadrupole.
A commercially-available click chemistry technique that placed an azide group onto the proteins of interest that can be captured with a special column has been used within cell cultures to capture different synthesized proteins including cytokines. Using this technique combined with stable isotope labeling (SILAC) in different types of cultured cells using various conditions (normal, starvation and lipopolysaccharide, LPS) different cytokines were identified [178]. Under normal conditions only IL-6 and IL-11 could be identified at ng/mL concentrations. With starvation conditions, more cytokines and chemokines were measured including IL-6, IL-11, CXCL1, CXCL5 and TGF-β2. When LPS was given, 12 cytokines were identified from a macrophage culture.
A common method to quantify cytokines and other low-abundance proteins is to use antibodies to capture either the entire protein or specific peptide fragments. Affinity capture has been used for IFN-γ, IL-4, TNF-α using MALDI and 30 to 40 ng per spot [179]. The different isoforms of monocyte chemoattractant protein-4/CCL13 have been identified using immunoaffinity capture [180]. Additionally, matrix effects were noted in a SELDI-TOF experiment attempting to measure the chemokine, CCL18 [181].
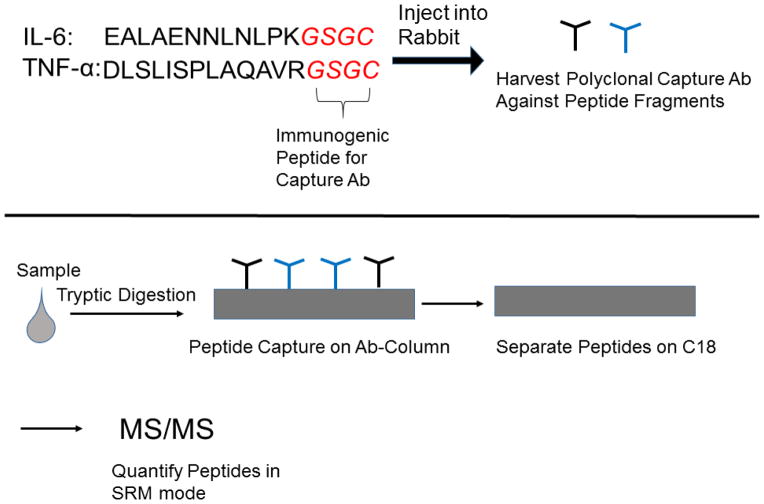
A method involving quantitative labeling and immuno-capture of peptides is SISCAPA (Stable Isotope Standards and Capture by Anti-Peptide Antibodies) was developed and tested using LC-MS in SRM with standards of the cytokines, IL-6 and TNF-α (Figure 8) [182]. This technique has been used to measure IL-33 in plasma in the range of 1.5–5000 ng/mL[183]. This work has been extended to use with a series of cytokines with reports of average detection limits around 10 ng/mL [184].
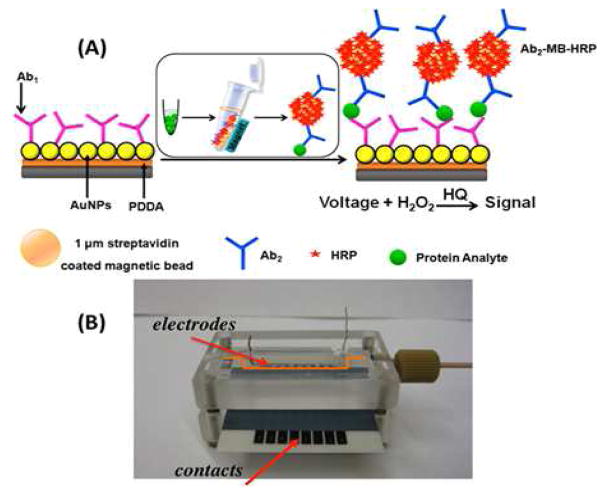
Figure 8.
Example of a multiplexed electrochemical-based immunoassay. The magnetic beads are coated with horseradish peroxidase (HRP) that reacts with hydrogen peroxide and hydroquinone (HQ) to give a product that is measured at −0.3V vs. Ag/AgCl. The bottom picture (B) shows the multiplexed channels. Reprinted with permission from: R Malhotra; V Patel; BV Chikkaveeraiah; BS Munge; SC Cheong; RB Zain; MT Abraham; DK Dey; JS Gutkind; JF Rusling; Anal. Chem. 2012, 84, 6249–6255. DOI: 10.1021/ac301392g Copyright © 2012 American Chemical Society
8.1. Mass Cytometry
Fluorescence spectroscopic methods for detection are limited by the number of available fluorophores and the knowledge that many fluorophores show spectral overlaps. Keeping this in mind, there has been increasing interest in using selective mass tags to allow for multiplexed mass spectrometric detection of various analytes. With appropriate (m/z) resolution of the mass analyzer, mass spectrometry allows for a highly multiplexed analysis of targeted analytes when a mass tag is placed onto different solutes.
Recently, there has been interest in the use of lanthanide or other metal tags for immunoassays [185, 186]. These assays involve conjugating polymeric materials with different lanthanides to antibodies providing a unique mass spectral signature for immunoassays. These assays have been used for different immunoassays including a demonstration for human-derived platelet-derived growth factor (hPDGF) and its isoform (hPDGF-AA) using a terbium-labeled antibody combined with inductively coupled plasma-MS detection [187]. The concept has been expanded to measure a variety of different cellular components related to immune responses [188].
9. Electrochemical Based Methods
Electrochemical-based methods for protein detection have been used for different immunoassays for quite some time. The primary advantage for electrochemical methods is the inexpensive equipment necessary for the measurement and the high sensitivity associated with particularly amperometric-based measurements. The use of electrochemical techniques combined with aptamer sensing has been reviewed [189]. Electrochemistry-based techniques for cancer biomarker detection have been reviewed [20].
9.1. Amperometric-Based Protein Measurements
Electrochemical measurements using labeled antibodies that produce electroactive substrates that can be measured using amperometric detection have been of interest for decades and have been extensively reviewed [190–193]. With the advent of magnetic beads that can be immobilized with antibodies and moved around easily on microfluidic systems, electrochemical methods for protein detection have become more popular.
While many groups have described the creation of electrochemical immunoassays for proteins, only a few have focused on measurements of cytokines. In particular the Rusling group has published many papers describing different variants of electrochemical immunoassay for measurements of cytokines and other proteins related to dysfunction caused by cancer [194]. Different materials have been used including carbon nanotubes in combination with electrochemiluminescence gave detection limits of 0.25 pg/mL for IL-6 in serum [195]. More recently nanostructured arrays have been created with the use of horseradish-peroxidase labeled antibodies for highly sensitive detection of IL-6, IL-8, VEGF and VEGF-C which resulted in detection limits in the 5 to 50 fg/mL range (Figure 9) [196].
Figure 9.
Schematic illustrating the Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) approach. The targeted peptides from the cytokines are listed in black. The peptide sequence denoted in red (GSGC) is added to allow for a more immunogenic response during antibody production. Polyclonal antibodies are retrieved and immobilized for affinity chromatography capture followed by RPLC-separation and MS/MS detection.
Other groups have reported electrochemical assays for single cytokines. Using glucose oxidase and ferrocene as a mediator, TNF-α, was detected in the range of 5 pg/mL to 10 ng/mL in spiked serum samples [197]. TNF-α has been detected using alkaline-phosphatase labeled nanospheres in serum down to the 10 pg/mL level [198].
9.2. Electrochemiluminescence
Electrochemiluminescence or electrogenerated chemiluminescence occurs when a chemiluminescence reaction is created with indirect chemical reporters that produce an excited state via an electrochemical reaction [199]. For immunoassays, the common reagents for this reaction are either phenantroline-ruthenium(II) tris(bipyridyl), Ru(bpy)32+ complexes, with tripropylamine (TPA) or luminol. Typically, a gold electrode is used to oxidize Ru2+ to Ru3+ followed by the subsequent reduction of Ru3+ by tripropylamine (TPA) resulting in chemiluminescence. The theory and applications of electrochemiluminescence for use in bioanalysis have been reviewed [200–202].
Electrochemiluminescence has been implemented for different cytokine immunoassays resulting in often one or two orders of magnitude reduction in LOD down to the 1 pg/mL level [203]. Commercial immunoassays based on this technology are available from Meso Scale Discovery (MSD) [204]. Comparisons between the MSD assays and flow cytometry bead-based assays from Becton Dickinson (BD) for an array of cytokines showed nearly equivalent lower limits of quantitation (LLOQ) between the two platforms [204]. The Luminex platform, which is a different bead-based immunoassay, has also been compared to the MesoScale Discovery platform and like the BD platform appears to be comparable [205].
More recent applications of electrochemiluminescence have incorporated various nano-based or chip-based strategies. The measurement of VEGF has been reported at the single-bead level using electrochemiluminescence methods [206]. Using a tumor necrosis factor antibody, the combination of quantum dots with electrochemiluminescence was reported with antibody protein detection limits of 7 pg/mL [207]. Carbon nanotubes have been employed in electrochemiluminescence detection schemes for IL-6 down to 0.25 pg/mL [195]. Microfluidic-based devices using electrochemiluminescence have allowed for IL-6 to be detected at the 10 fg/mL level in serum [208].
9.3. Other Reports using Electrochemical Methods
Using an electrochemical approach with a hairpin aptamer towards IFN-γ, Zhang et al., reported subnanomolar detection of IFN-γ [209]. Interleukin-6 (IL-6) has been quantified using a capacitive-based biosensor [210]. TNF-α has been measured down to 57 fM using electrochemical impedance spectroscopy [211]. Bettazzi and colleagues used a magnetic bead based assay to measure TNF-α [212]
10. Optical Methods
Optical sensors have been widely employed for bioanalytical detection methods. Due to the wide variety of available optical methods for detection, it is challenging to try to sort through these different techniques as they relate to cytokine measurements. In general, it is possible to break these methods down into bead-based methods where beads are optically encoded to allow for multiplexed signal analysis, label-free methods that are used with various types of plasmonic resonance/microcavities shifts, single-molecule techniques, and general fluorescence techniques. A comprehensive overview of optical sensors can be found in a recent book edited by Ligler and Taitt [213]. A recent review describing optical biosensors for unlabeled targets provides a comprehensive table for detection capability of many different types of optical sensors [214].
10.1. Optically Encoded Beads
Walt’s group has described 3.1 μm fluorescently encoded polymeric beads that were used for cytokine detection in human saliva [118]. In their work, the encoded beads were combined with a fluorescent microscope for array analysis of ten cytokines (VEGF, EFG, IP-10/CXCL10, IL-8/CXCL8, MCP-1/CCL2, TIMP-1, RANTES/CCL5, MIP-1β/CCL4, Eotaxin-2 and IL-6) from 100 μL of sample with a total assay time of 2.5 hrs. While the array detection limits were higher than ELISA measurements used for comparison, they were sufficient to allow for quantitation of cytokines in patient samples. Detection limits in pM ranged from 8 to 469 pM for the different cytokines.
Theilacker et al., describe the use of 1 μm beads for a multiplexed assay where the beads were externally labeled with different intensity of select Alexa 488, Alexa 594 and Atto 680 dyes [215]. They measured human IL-6, IL-8, prostate specific antigen (PSA), and TNF-α into the pg/mL range. However, interestingly they noted significant problems with the TNF-α assay with non-specific interactions between the TNF-α capture antibody and the IL-6 and IL-8 detection antibodies.
10.2. Molecular Imprints – With Fluorescence Sensors
Molecular imprinting involves taking a specific target termed a template and then building a scaffold surrounding it typically using polymers [216]. The main goal is to impart molecular recognition capability based upon shape. There has been wide use of this approach for small molecular templates particularly in the separations field for sample preparation or separation of enantiomers. With the wide use and successful application of this technique for low molecular weight solutes (< 1500 Da), there has been an interest in using proteins for templates in order to create artificial receptors [217]. Artificial receptors created in different materials would be expected to have far more chemical stability than biological receptors.
Sol-gels provide an improvement over the standard use of polymers for molecular imprinting. The chemical aspects such as porosity, structure control and morphology can be better achieved using sol-gel chemistry leading to many uses of these materials in chemical analysis [218]. This has led to wide use of these materials for sensing capabilities [219, 220] Bright’s group developed a xerogel sensor for human interleukin-1α with an integrated luminescent reporter [221]. The sensor had less than a 2 min response time, was stable to multiple cycles and could detect IL-1α with an LOD of 2 pg/mL in spiked samples. The sensor was also stable for 4 months at room temperature.
Despite the great interest in molecular imprints for protein sensing, the progress towards developing sensors that would meet clinical analysis needs has been limited. However, there is a recent review describing the challenges of using molecular imprint technology for protein detection[222]. Some have described how imprinting proteins can be challenging as the protein itself may actually be too large to fit and the issue become one of binding sites being accessible [223]. To overcome this potential problem, the possibility of epitope mapping for imprinted materials has been described [224, 225]. There are many citations to these papers, but none appear to be towards cytokines.
10.3. Surface Plasmon Resonance
Surface plasmon resonance requires a system with polarized light passing through a prism or chip with a coated metal that acts like a mirror [226]. At a specific angle (the resonance angle), the light will excite surface plasmons. Surface plasmons arise from the interaction of free electrons in the metal with the polarized light resulting in a dip in the light intensity resulting in the phenomenon called surface plasmon resonance (SPR). The resonance angle is dependent on the optical properties (refractive indices) on both sides of the metal. When molecules bind to the metal surface or to objects immobilized to the metal surface such as antibodies, the resonance angle changes allowing detection of binding events. For this reason, SPR has been widely used as a means for obtaining label-free information about the kinetics of various biological-based binding events (proteins/antibodies, ligands/receptors). For example, Chou et al., measured IL-6 from cell culture using an SPR assay with the Biacore 3000 instrument that had approximate detection limits of 1 ng/mL [227].
The group of Karl Booksh was the first to report an SPR-based sensor for measurement of cytokines in physiologically-relevant samples [228]. A common challenge with SPR-based sensors is the issue of overcoming signals produced via non-specific binding events onto the sensor. This is especially challenging for SPR sensors used in samples that contain biological materials. They report measuring IL-1β, IL-6, and TNF-α in cell culture media samples down to the ~ 1 ng/mL range with maximum sensor response times reached in approximately 10 minutes. Detection limits were improved 3-fold over the use of standard CM-dextrans by using an N- hydroxysuccinimide ester of 16-mercaptohexadecanoic acid (NHS-MHA) to coat the gold chip.
Other groups have reported the use of SPR. IL-6 has been measured from cell culture media. To reduce non-specific interactions non-specific binding, a mixed self-assembled monolayer of mercaptoundecanoic acid (MUA) and mercaptohexanol (MCH) was coated onto the chip [227]. By adding gold nanorods, detection limits for TNF-α were reduced by 40-fold in an SPR imaging study [229]. Using gold nanoparticles and an approach to reduce non-specific binding, TNF-α was detected down to 54.9 pg/mL in spiked human serum [230]. To determine the different TH1 responses from cells circulating in the blood, Rice and colleagues used SPR imaging [231]. Bustos measured growth factors secreted from cells using SPR [232]. Using unique protein-engineered albumin binding protein, interferon-γ (IFN-γ) has been measured to the 0.2 nM level using SPR [233].
10.4 Nanoparticles and LSPR
Nanoparticles are known to exhibit localized surface plasmon resonance effects which have become of significant interest to exploit for use in sensing applications [234]. Bolduc and Masson have recently reviewed the advances in SPR sensors as well as localized surface plasmon resonance (LSPR) devices [235]. Typically these gold nanoparticles exhibit similar surface plasmon effects as typically observed in a standard gold thin film in common instruments such as the Biacore. By lining gold nanoparticles with attached IL-1β antibodies onto an optical fiber, a sensor was created with detection limits of 22 pg/mL with a detection range of 32 to 250 pg/mL, which is comparable to a standard ELISA [236]. However, it should be noted that 200 μL of sample was used for analysis which is twice as much as that used in a standard ELISA. Using an LSPR approach, the release of TNF-α has been detected being secreted from cells derived from clinical blood samples [237]. An LSPR sensor has been used to measure recombinant IL-6 produced from E.Coli [238]. A fiber optic with gold nanoparticles was used to measure TNF-α from synovial fluid with detection limits reported of 34 pg/mL [239]. This group also measured IL-1β in a separate study down to 21 pg/mL for the LOD [236]. Jeong and colleagues developed a fiber optic LSPR sensor for detection of IFN-γ and prostate specific antigen with detection limits for IFN-γ in the low pg/mL range [240].
10.5. Single Molecule Detection
Despite significant efforts in immunology towards characterizing, classifying and understanding cytokine regulation, there still remain significant needs towards understanding how cytokines are released, regulated and secreted by immune cells [241]. Lacy and Snow clearly point out in a recent review that in many cases, the understanding of these control mechanisms for cytokines remain a “significant ‘black box’ in immunology.” Cytokines can be released via trafficking as well as granulocyte (vesicle) release. Given the low concentrations of cytokines and the research significance to elucidate cell-to-cell interactions, the use of single-molecule detection (SMD) strategies towards cytokine measurements has a role to play in answering questions about cell-to-cell and other cytokine-mediated interactions.
To apply SMD techniques to the study of any molecule requires a significant confinement in volume to be able to have the sensitivity to make the measurement [242]. The use of noble metal nanoparticles has been applied to the SMD of cytokines. Noble metals do not suffer some of the problems of photobleaching associated with fluorophores and blinking associated with quantum dots. The Xu research group at Old Dominion has described their major progress towards creating stable noble metal nanoparticles that have been called single molecule nanoparticle optical nanobiosensors (SMNOBS) that have been used for sensing TNF-α [243]. This work demonstrated the ability to track the on-off kinetics of TNF-α onto the Ag nanoparticles. A concentration range of 1 to 200 ng/mL was used for TNF-α with 50 nM of the antibody-conjugated Ag nanoparticles.
The approach using TNF-α and SMNOBS was further investigated to determine TNF-α binding and internalization into L929 cells [244]. The internalization processes of TNF-α binding onto a single cell followed by the apoptotic response was followed using the SMNOBS and an approach called far-field photostable optical nanoscopy (PHOTON). The statistics for the numbers of TNF-α/receptor complexes necessary to achieve apoptosis was measured in more than 20 separate single cells along with the time to achieve apoptosis was measured. It was determined that a threshold number of complexes with TNF-α on these cells was necessary to initiate apoptosis.
10.6. Microring Resonators
Bailey’s group has reviewed the various platforms for label-free sensing devices including plasmonic, photonic, and mechanical [245]. An example of the readout for whispering-gallery mode is shown in Figure 10. Luchansky et al., provide the theoretical framework behind their microring sensors [246]. Luchansky and Bailey report measuring IL-2 (15.5 kDa) down to 100 pg/mL (6.5 pM) by using a sandwich assay to increase the total mass bound [247]. IL-2 was measured in Jurkat T-cells from culture. The assay takes approximately 45 minutes to perform. The group also demonstrated similar effectiveness of measurement for IL-8 in vitro. The use of a sandwich assay improves the LOD because IL-2 by itself (50 ng/mL) gives a resonance shift, Δpm, of ~ 20. Addition of the detection antibody causes another resonance shift of Δpm of ~ 40. In their work, they have 24 active sensing rings on their microresonator of which 15 are used for actual sensing and the other 9 are used as controls for nonspecific binding events, bulk changes to the refractive index and possible thermal drift.
Figure 10.
Example of whispering gallery mode. Glass beads that exhibit a resonance are immobilized with capture antibodies. As the beads come into resonance with the incident wavelength, the increase in intensity can be observed. Analyte addition to the beads causes this resonance wavelength to shift. The change in the wavelength is proportional to analyte concentration. The photograph of the beads in resonance are reprinted with permission from: HA Huckabay, SM Wildgen, RC Dunn, Label-free detection of ovarian cancer biomarkers using whispering gallery mode imaging. Biosensors and Bioelectronics, Volume 45, 2013, 223 – 229. © Elsevier. dx.doi.org/10.1016/j.bios.2013.01.072.
Using whispering-gallery mode which amplifies the number of times a target molecule is interrogated, Armani and colleagues created a sensing device for IL-2 measurements in serum. This device has a calibration range of 5 × 10−18 M to 1 × 10−6 M [248]. A simulation of fluid flows needed in a microfluidic chip to obtain optimal performance with a similar integrated photonic device for IL-6 has been described [249]. Dunn and colleagues have measured different cytokines using whispering-gallery mode in microfluidic devices [250, 251].
10.6. New Platforms
Ruckstuhl, et al., describe a single use optical cell for immunoassays which only requires approximately 15 minutes of sample preparation time based on an optical technical called supercritical angle fluorescence [252]. This technique allows for a one step sandwich immunoassay of IL-2 in an in vitro solution. Even though fluorescent molecules are homogenous in the solution (as labeled detection antibodies), the technique requires fluorescence to be emitted above the critical angle of the interface with the aqueous sample. Detection limits for this assay based on 3σ of the S/N was 4.5 pg/mL for IL-2. Carter et al., report the use of Arrayed Imaging Reflectometry to detect multiple cytokines [253]. A fiber-optic biosensor dip probe with diode laser excitation and a charged-coupled device spectrometer and functions on a technique of sandwich immunoassay was used to measure IL-6. Detection limit were low pM and an estimated IL-6 concentration in a patient with lupus was 5.9 ± 0.6 pM. Some samples had concentrations of IL-6 too low to detect [254, 255].
Choi et al., described a label-free measurement of TNF-α using their nanostructured photonic crystal device containing a low refractive index UV-cured polymer and a high refractive index layer of TiO2 [256, 257]. Different designs with different fluidic geometries were investigated. Each design had different reported strengths and weaknesses with respect to equilibration time, instrument performance and sample volume needs. Detection limits for TNF-α in phosphate buffered saline solutions were reported in the range of 16.7 to 48.4 ng/mL.
Adalsteinsson and colleagues described the use of ZnO nanorods as a means to increase the detection limits of antibody arrays for cytokine detection [258]. Measuring IL-18 and TNF-α in standards and urine samples, the detection limits for these two cytokines using an Alexa-488 labeled detection antibody were in the low fg/mL range. The mechanism for fluorescence enhancement using the ZnO nanorods relative to more standard polymeric platforms such as PMMA, polystyrene (PS), or PS-b-PVP is still being elucidated. The ZnO nanorods appear to behave as efficient evanescent waveguides enabling fluorescence enhancement.
11. Detecting Cytokines Released from Cells and Single Cells
The immune system is highly plastic and many different cell types (T-cells and macrophages) within this system can be activated to different polarization states. To be able to detect these polarization states, there has been a significant interest in capturing these cells followed by analysis of both their cytokine release as well as their release rates. While flow cytometry is a powerful analytical tool to determine the types of inflammatory cells by identification of cellular markers, direct flow cytometry of a single cell cannot be used to determine the cytokine release from these cells.
Enzyme-linked immuno spot assays (ELISPOT) have been used to detect cytokine proteins released from activated cells [13, 38–40]. There is currently a significant interest in the chemical analysis of single cells [259–261]. The ELISPOT is not able to provide protein production information from a single cell. The current multiplexing capability for ELISPOT is two separate cytokines. Several new platforms have been designed to measure cytokine output from captured immune cells. Lakowicz has improved on the ELISPOT assay by including plasmonic nanoparticles to allow for real-time detection of TNF-α and IFN-γ from mouse macrophage [262]
Zhu et al., report a side-by-side array for T-cell capture (CD4 and CD8) followed by measurement of cytokine detection of IFN-γ and IL-2 in the ng/mL range from human blood [263]. They followed up on this work by coating their wells with poly (ethylene glycol) microwells and still had detection capability in the ng/mL range from T-cells evoked to release cytokines [264].
Using microengraving technology to capture a small number of cells, the cytokine production of IFN-γ, IL-2, IL-6, IL-17, and TNG-α were determined from different types of cells [265]. Wells that were 1.25 × 105 μm3 (50 μm on each side) were created to capture different inflammatory cells. Cells were captured and then subjected to various treatments to elicit cytokine release and determine relative release rates of cytokines per second. With this work they were able to get frequency histograms of cytokine production from different labeled cells. This work has now been translated for use in clinical medicine by measuring alterations in cytokine production from human CD8+ cells [266]. The technique has been used in other studies to determine production of cytokines from different T-cells [267]. An aptamer to PDGF was completely redesigned to work in biological media and was attached to a cell membrane to detect PDGF after cellular release [268].
12. In Vivo Methods
The significance of cytokine signaling within mammalian systems has led to much interest in the ability to capture cytokine signaling events within mammalian systems. The majority of the published papers related to unraveling the cytokine network use microdialysis sampling.
12.1 Direct Imaging Methods
12.1.1. Radiolabeled Cytokines
Determining levels of local inflammation in human subjects is of significant clinical interest and different imaging techniques have been described [269]. Cytokines are known to be upregulated during inflammation. Attempts to non-invasively image this protein upregulation in experimental animals and humans has been made possible through the use of radiolabeled cytokines using different methods [270, 271]. The advantage of radiolabeled probes or radiolabeled pharmaceuticals is that these allow for the visualization and quantitation of physiological processes in vivo [272]. However, despite the critical clinical need for targeting specific cytokines related to inflammatory responses, there have been observed challenges in the field with creating and implementing these imaging procedures. For some cytokines, adverse physiological effects were noted after administration even at low doses of 10 ng/kg such as IL-1. However, IL-1 receptor antagonist (IL-1ra) did not cause the adverse effects that IL-1 did [273]. Other issues with cytokines are the non-specific adsorption that can be observed. Even though there are some problems, there still are different groups reporting work with labeled cytokines [274, 275].
Despite the exquisite selectivity of having labelled-cytokine proteins, the imaging of inflammation states has shifted to using F-labeled deoxy-glucose techniques (18F-DG).
12.1.2. Nanoparticles
The use of nanoparticles in medicine as contrast agents has seen wide application space. The chemical nature of nanoparticles allows them to be used imaging techniques involving either light adsorption, emission or magnetic properties. Targeting the imaging of cytokines using nanoparticle technology is an emerging field. The Wang and Kotov group have been making steady progress towards developing imaging agents for TNF-α in arthritis applications [276, 277]. In particular, the group has made extensive use of photoacoustic imaging because it is becoming widely used in diagnostics since it combines advantages of light and ultrasound.
IL-13 peptides are useful in identifying glial tumors since these tumors have a specific receptor for the peptide. In order to image the binding interaction using an MR instrument, the contrast provided by a standard contrast agent such as gadolinium (Gd) has to be optimized. The use of metallofullerenes has allowed for more specific uses of Gd [278].
12.2 Microdialysis Sampling of Cytokines
Microdialysis sampling has been well-described in the literature for different applications in neuroscience and pharmaceutics [279, 280]. The technique involves using catheters or probes with semi-permeable membranes and microliter-per-minute perfusion flow rates to sample from living tissues. Microdialysis sampling of cytokines has been reviewed [281, 282].
A limitation of the microdialysis sampling technique for cytokine localized collection and subsequent quantitation is the relatively large volume of sample required. ELISAs typically require 100 μL of sample for the detection of a single cytokine. Collection of this volume at a moderate microdialysis sampling flow rate of 1 μL/min would require nearly a two hour sampling time which would result in low temporal resolution. Such poor temporal resolution would be unacceptable for very acute immune responses. Thus, when using microdialysis sampling for in vivo protein collection, there is a severe tradeoff between temporal resolution and the need for high extraction efficiency. This problem is exacerbated by the knowledge that simultaneous measurement of multiple cytokines reveals more about the underlying biological processes than a single measurement. A standard ELISA method can only analyze one single cytokine per well. The ability to measure multiple cytokines simultaneously is extremely important in a variety of physiological conditions because the concentration fluctuations of one cytokine often induce changes in other networked cytokines. For these reasons, many microdialysis samples from both laboratory animals and humans have used either the multiplexed-bead based assay approach for quantitation or microarrays due to the low microliter sample volume needs combined with multiplexing capability [283–285].
The application of microdialysis is now emerging as potentially the most successful approach for clinical in vivo studies in both healthy and diseased subjects to recover targeted cytokine molecules from the sites of action in tissue. Commercially available microdialysis probes with a 100 kDa MWCO membrane are commonly used for in vivo microdialysis of cytokines. However, the extraction efficiency across these membranes is quite low, e.g., 3% at 1.0 μL/min for a 10 mm probe. Other researchers create their own dialysis probes in-house using 3,000 kDa MWCO membranes. Given the wide use of the microdialysis sampling technique for studies in the brain and other tissues [286–288], it is likely continued use of it in the clinic for cytokine collection will remain.
13. Summary and Conclusions
Cytokines are important biomarkers that are measured in a variety of clinical contexts. Many of the new methods described for cytokine measurement aim to greatly reduce the time necessary for ELISA development. Additionally, limited sample volumes (microliters) in some cases drives the development of methods capable of measuring cytokines in a few microliters. With the exception of the electrochemical-based methods, the amplification methods (e.g., immune-PCR), and a few of the optical methods, nearly all the new techniques for cytokine measurement have either equivalent or higher detection limits that standard ELISA (~10–1000 pg/mL range).
Cytokine measurements with different techniques is a highly active field of study and the enormous number of citations to cytokine measurements was far higher than we originally anticipated. We apologize to authors whose important work we may have not cited. Finding many of these papers in the published literature was actually challenging. For example, persons who measure IL-6 or TNF-α may not refer to these as cytokines in their manuscripts. Additionally, performing a substance identifier search on these particular protein names in SciFinder Scholar with a cross-reference to “analytical study” does not provide any citations. Therefore, we were left with searching authors that we knew, their citations, combined with judiciously-chosen search strings. Nonetheless, a significant body of work remained to be reviewed and this literature continues to grow.
Highlights.
Cytokines are important biomarkers related to immune responses
Chemical analysis is challenging due to pg/mL concentrations
Different chemical methods to analyze cytokines are reviewed.
Acknowledgments
We acknowledge the NIH for support of different projects that have required cytokine analysis.
Biographies
 Julie Stenken has a B.S. in Chemistry from the University of Akron and a PhD from the University of Kansas with an emphasis on microdialysis sampling research. During the 1994–1995 academic year, she was a J. William Fulbright Fellow at the Department of Clinical Pharmacology at the Karolinska Institute in Stockholm, Sweden. Her first academic appointment was at Rensselaer Polytechnic Institute. In 2007, she become the 21st Century Chair in Proteomics at the University of Arkansas. Her research interests are in the areas of bioanalytical chemistry, biomaterials, in vivo measurements, macrophage polarization and microdialysis sampling.
Julie Stenken has a B.S. in Chemistry from the University of Akron and a PhD from the University of Kansas with an emphasis on microdialysis sampling research. During the 1994–1995 academic year, she was a J. William Fulbright Fellow at the Department of Clinical Pharmacology at the Karolinska Institute in Stockholm, Sweden. Her first academic appointment was at Rensselaer Polytechnic Institute. In 2007, she become the 21st Century Chair in Proteomics at the University of Arkansas. Her research interests are in the areas of bioanalytical chemistry, biomaterials, in vivo measurements, macrophage polarization and microdialysis sampling.
 Andreas Poschenrieder from Regensburg, Germany, has studied chemistry at the Universities of Regensburg, Bath, Arkansas and Munich. His B.S. research studies included whole-cell biosensors and the properties of biomimetic membranes for biomaterial development. Forhis masters program and the EU Atlantis Program, he worked in Dr. Stenken’s group, USA, to obtain a Transatlantic Dual Bachelor Degree in Chemistry and improved bioanalytical methods for the in-vivo detection of cytokines. In 2013, he started his PhD at the TechnischeUniversiätMünchen. Within the IGSSE, sponsored by the Excellence Initiative, his current research focuses on the design of CXCR4 ligands for medical applications.
Andreas Poschenrieder from Regensburg, Germany, has studied chemistry at the Universities of Regensburg, Bath, Arkansas and Munich. His B.S. research studies included whole-cell biosensors and the properties of biomimetic membranes for biomaterial development. Forhis masters program and the EU Atlantis Program, he worked in Dr. Stenken’s group, USA, to obtain a Transatlantic Dual Bachelor Degree in Chemistry and improved bioanalytical methods for the in-vivo detection of cytokines. In 2013, he started his PhD at the TechnischeUniversiätMünchen. Within the IGSSE, sponsored by the Excellence Initiative, his current research focuses on the design of CXCR4 ligands for medical applications.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meager T. The Molecular Biology of Cytokines. New York: John Wiley & Sons; 1998. [Google Scholar]
- 2.Thompson AW, Lotze MT. The Cytokine Handbook. 4. Amsterdam: Academic Press; 2003. [Google Scholar]
- 3.Dinarello CA. Historical insights into cytokines. Eur J Immunol. 2007;37:S34–S45. doi: 10.1002/eji.200737772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beschin A, Bilej M, Torreele E, De BP. On the existence of cytokines in invertebrates. Cell Mol Life Sci. 2001;58:801–14. doi: 10.1007/PL00000901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beschin A, Bilej M, Magez S, Lucas R, de BP. Functional convergence of invertebrate and vertebrate cytokine-like molecules based on a similar lectin-like activity. Springer-Verlag; 2004. pp. 145–63. [DOI] [PubMed] [Google Scholar]
- 6.Lefkowitz DL, Lefkowitz SS. Macrophage-Neutrophil Interaction: Paradigm for Chronic Inflammation Revisited. Immunology and Cell Biology. 2001;79:502–6. doi: 10.1046/j.1440-1711.2001.01020.x. [DOI] [PubMed] [Google Scholar]
- 7.Gordon S, Fraser I, Nath D, Hughes D, Clarke S. Macrophages in tissues and in vitro. Curr Opin Immunol. 1992;4:25–32. doi: 10.1016/0952-7915(92)90119-y. [DOI] [PubMed] [Google Scholar]
- 8.Wakita T, Shintani F, Yagi G, Asai M, Nozawa S. Combination of inflammatory cytokines increases nitrite and nitrate levels in the paraventricular nucleus of conscious rats. Brain Res. 2001;905:12–20. doi: 10.1016/s0006-8993(01)02346-0. [DOI] [PubMed] [Google Scholar]
- 9.Balkwill FR, Burke F. The cytokine network. Immunol Today. 1989;10:299–304. doi: 10.1016/0167-5699(89)90085-6. [DOI] [PubMed] [Google Scholar]
- 10.Schmitz ML, Weber A, Roxlau T, Gaestel M, Kracht M. Signal integration, crosstalk mechanisms and networks in the function of inflammatory cytokines. Biochim Biophys Acta, Mol Cell Res. 2011;1813:2165–75. doi: 10.1016/j.bbamcr.2011.06.019. [DOI] [PubMed] [Google Scholar]
- 11.Schenk T, Irth H, Marko-Varga G, Edholm LE, Tjaden UR, van dGJ. Potential of on-line micro-LC immunochemical detection in the bioanalysis of cytokines. J Pharm Biomed Anal. 2001;26:975–85. doi: 10.1016/s0731-7085(01)00464-2. [DOI] [PubMed] [Google Scholar]
- 12.Dinarello CA. Proinflammatory cytokines. Chest. 2000;118:503–8. doi: 10.1378/chest.118.2.503. [DOI] [PubMed] [Google Scholar]
- 13.Bienvenu J, Monneret G, Fabien N, Revillard JP. The clinical usefulness of the measurement of cytokines. Clinical Chemistry and Laboratory Medicine. 2000;38:267–85. doi: 10.1515/CCLM.2000.040. [DOI] [PubMed] [Google Scholar]
- 14.Brunet M. Cytokines as predictive biomarkers of alloreactivity. Clin Chim Acta. 2012;413:1354–8. doi: 10.1016/j.cca.2012.04.014. [DOI] [PubMed] [Google Scholar]
- 15.Levine JE, Paczesny S, Sarantopoulos S. Clinical Applications for Biomarkers of Acute and Chronic Graft-versus-Host Disease. Biol Blood Marrow Transplant. 2012;18:S116–S24. doi: 10.1016/j.bbmt.2011.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velluto L, Iarlori C, Gambi D, Reale M. Cytokines and Alzheimer’s disease. Science Publishers, Inc; 2011. pp. 329–42. [Google Scholar]
- 17.Bhokta NR, Woodruff PG. Human asthma phenotypes: from the clinic, to cytokines, and back again. Immunol Rev. 2011;242:220–32. doi: 10.1111/j.1600-065X.2011.01032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gui T, Shimokado A, Sun Y, Akasaka T, Muragaki Y. Diverse roles of macrophages in atherosclerosis: from inflammatory biology to biomarker discovery. Mediators Inflammation. 2012;693083:14. doi: 10.1155/2012/693083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long TM, Raufman J-P. The diagnostic and prognostic role of cytokines in colon cancer. Gastrointest Cancer: Targets Ther. 2011;1:27–39. [Google Scholar]
- 20.Chikkaveeraiah BV, Bhirde AA, Morgan NY, Eden HS, Chen X. Electrochemical Immunosensors for Detection of Cancer Protein Biomarkers. ACS Nano. 2012;6:6546–61. doi: 10.1021/nn3023969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rusling JF, Kumar CV, Gutkind JS, Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst (Cambridge, U K) 2010;135:2496–511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lotrich F. Inflammatory cytokines, growth factors, and depression. Curr Pharm Des. 2012;18:5920–35. doi: 10.2174/138161212803523680. [DOI] [PubMed] [Google Scholar]
- 23.Rose NR. Critical Cytokine Pathways to Cardiac Inflammation. J Interferon Cytokine Res. 2011;31:705–10. doi: 10.1089/jir.2011.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gullestad L, Ueland T, Vinge LE, Finsen A, Yndestad A, Aukrust P. Inflammatory Cytokines in Heart Failure: Mediators and Markers. Cardiology. 2012;122:23–35. doi: 10.1159/000338166. [DOI] [PubMed] [Google Scholar]
- 25.Catalfamo M, Le SC, Lane HC. The role of cytokines in the pathogenesis and treatment of HIV infection. Cytokine Growth Factor Rev. 2012;23:207–14. doi: 10.1016/j.cytogfr.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee DW, Faubel S, Edelstein CL. Cytokines in acute kidney injury (AKI) Clin Nephrol. 2011;76:165–73. doi: 10.5414/cn106921. [DOI] [PubMed] [Google Scholar]
- 27.Iarlori C, Gambi D, Reale M. Parkinson’s disease and cytokines. Science Publishers, Inc; 2011. pp. 343–55. [Google Scholar]
- 28.Reinhart K, Bauer M, Riedemann NC, Hartog CS. New approaches to sepsis: molecular diagnostics and biomarkers. Clin Microbiol Rev. 2012;25:609–34. doi: 10.1128/CMR.00016-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burska A, Boissinot M, Ponchel F. Cytokines as biomarkers in rheumatoid arthritis. Mediators Inflamm. 2014;2014:545493. doi: 10.1155/2014/545493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schett G, Elewaut D, McInnes IB, Dayer J-M, Neurath MF. How cytokine networks fuel inflammation: Toward a cytokine-based disease taxonomy. Nat Med. 2013;19:822–4. doi: 10.1038/nm.3260. [DOI] [PubMed] [Google Scholar]
- 31.Striz I, Brabcova E, Kolesar L, Sekerkova A. Cytokine networking of innate immunity cells: a potential target of therapy. Clin Sci. 2014;126:593–612. doi: 10.1042/CS20130497. [DOI] [PubMed] [Google Scholar]
- 32.Rose-John S, Heinrich PC. Soluble receptors for cytokines and growth factors: generation and biological function. Biochem J. 1994;300:281–90. doi: 10.1042/bj3000281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorpe R, Wadhwa M, Bird CR, Mire-Sluis AR. Detection and measurement of cytokines. Blood Rev. 1992;6:133–48. doi: 10.1016/0268-960x(92)90025-l. [DOI] [PubMed] [Google Scholar]
- 34.Whiteside TL. Assays for cytokines. In: Thompson AW, Lotze MT, editors. Cytokine Handbook. 4. Amsterdam: Academic Press; 2003. pp. 1375–96. 6 plates. [Google Scholar]
- 35.Sachdeva N, Asthana D. Cytokine quantitation: technologies and applications. Front Biosci. 2007;12:4682–95. doi: 10.2741/2418. [DOI] [PubMed] [Google Scholar]
- 36.Mire-Sluis AR, Page L, Thorpe R. Quantitative cell line based bioassays for human cytokines. J Immunol Methods. 1995;187:191–9. doi: 10.1016/0022-1759(95)00220-1. [DOI] [PubMed] [Google Scholar]
- 37.House RV. Cytokine bioassays: an overview. Dev Biol Stand. 1999;97:13–9. [PubMed] [Google Scholar]
- 38.Favre N, Bordmann G, Rudin W. Comparison of cytokine measurements using ELISA, ELISPOT and semi-quantitative RT-PCR. J Immunol Methods. 1997;204:57–66. doi: 10.1016/s0022-1759(97)00033-1. [DOI] [PubMed] [Google Scholar]
- 39.Ekerfelt C, Ernerudh J, Jenmalm MC. Detection of spontaneous and antigen-induced human interleukin-4 responses in vitro: comparison of ELISPOT, a novel ELISA and real-time RT-PCR. J Immunol Methods. 2002;260:55–67. doi: 10.1016/s0022-1759(01)00520-8. [DOI] [PubMed] [Google Scholar]
- 40.Atochina O, Mylvaganam R, Akselband Y, McGrath P. Comparison of results using the gel microdrop cytokine secretion assay with ELISPOT and intracellular cytokine staining assay. Cytokine. 2004;27:120–8. doi: 10.1016/j.cyto.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Kalyuzhny AE. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2012. Handbook of ELISPOT: Methods and Protocols. [Google Scholar]
- 42.Meager A. Measurement of cytokines by bioassays: Theory and application. Methods. 2006;38:237–52. doi: 10.1016/j.ymeth.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 43.Marcus JS, Anderson WF, Quake SR. Microfluidic Single-Cell mRNA Isolation and Analysis. Anal Chem. 2006;78:3084–9. doi: 10.1021/ac0519460. [DOI] [PubMed] [Google Scholar]
- 44.Marcus JS, Anderson WF, Quake SR. Parallel Picoliter RT-PCR Assays Using Microfluidics. Anal Chem. 2006;78:956–8. doi: 10.1021/ac0513865. [DOI] [PubMed] [Google Scholar]
- 45.Diercks A, Kostner H, Ozinsky A. Resolving cell population heterogeneity: real-time PCR for simultaneous multiplexed gene detection in multiple single-cell samples. PLoS One. 2009;4 doi: 10.1371/journal.pone.0006326. No pp. given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skalnikova H, Motlik J, Gadher SJ, Kovarova H. Mapping of the secretome of primary isolates of mammalian cells, stem cells and derived cell lines. Proteomics. 2011;11:691–708. doi: 10.1002/pmic.201000402. [DOI] [PubMed] [Google Scholar]
- 47.Carson RT, Vignali DAA. Simultaneous quantitation of 15 cytokines using a multiplexed flow cytometric assay. J Immunol Methods. 1999;227:41–52. doi: 10.1016/s0022-1759(99)00069-1. [DOI] [PubMed] [Google Scholar]
- 48.Earley MC, Vogt RF, Jr, Shapiro HM, Mandy FF, Kellar KL, Bellisario R, et al. Report from a workshop on multianalyte microsphere assays. Cytometry. 2002;50:239–42. doi: 10.1002/cyto.10140. [DOI] [PubMed] [Google Scholar]
- 49.Vignali DA. Multiplexed particle-based flow cytometric assays. J Immunol Methods. 2000;243:243–55. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- 50.Khan SS, Smith MS, Reda D, Suffredini AF, McCoy JP., Jr Multiplex bead array for detection of soluble cytokines: comparisons of sensitivity and quantitative values among kits from multiple manufacturers. Cytometry, Part B. 2004;61B:35–9. doi: 10.1002/cyto.b.20021. [DOI] [PubMed] [Google Scholar]
- 51.Morgan E, Varro R, Sepulveda H, Ember JA, Apgar J, Wilson J, et al. Cytometric bead array: a multiplexed assay platform with applications in various areas of biology. Clin Immunol. 2004;110:252–66. doi: 10.1016/j.clim.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 52.Kellar KL, Iannone MA. Multiplexed microsphere-based flow cytometric assays. Exp Hematol. 2002;30:1227–37. doi: 10.1016/s0301-472x(02)00922-0. [DOI] [PubMed] [Google Scholar]
- 53.Oliver KG, Kettman JR, Fulton RJ. Multiplexed analysis of human cytokines by use of the FlowMetrix system. Clin Chem. 1998;44:2057–60. [PubMed] [Google Scholar]
- 54.Fodor SPA, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–73. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 55.Pawlak M, Schick E, Bopp MA, Schneider MJ, Oroszlan P, Ehrat M. Zeptosens’ protein microarrays: a novel high performance microarray platform for low abundance protein analysis. Proteomics. 2002;2:383–93. doi: 10.1002/1615-9861(200204)2:4<383::AID-PROT383>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 56.Mustafa SA, Hoheisel JD, Alhamdani MSS. Secretome profiling with antibody microarrays. Mol BioSyst. 2011;7:1795–801. doi: 10.1039/c1mb05071k. [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Nath N, Reichert WM. Parallel comparison of sandwich and direct label assay protocols on cytokine detection protein arrays. Anal Chem. 2003;75:5274–81. [Google Scholar]
- 58.Li Y, Schutte RJ, Abu-Shakra A, Reichert WM. Protein array method for assessing in vitro biomaterial-induced cytokine expression. Biomaterials. 2005;26:1081–5. doi: 10.1016/j.biomaterials.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 59.Schutte RJ, Xie L, Klitzman B, Reichert WM. In vivo cytokine-associated responses to biomaterials. Biomaterials. 2009;30:160–8. doi: 10.1016/j.biomaterials.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Averbeck M, Beilharz S, Bauer M, Gebhardt C, Hartmann A, Hochleitner K, et al. In situ profiling and quantification of cytokines released during ultraviolet B-induced inflammation by combining dermal microdialysis and protein microarrays. Exp Dermatol. 2006;15:447–54. doi: 10.1111/j.0906-6705.2006.00429.x. [DOI] [PubMed] [Google Scholar]
- 61.Paulo JA, Lee LS, Wu B, Banks PA, Steen H, Conwell DL. Cytokine profiling of pancreatic fluid using the ePFT collection method in tandem with a multiplexed microarray assay. J Immunol Methods. 2011;369:98–107. doi: 10.1016/j.jim.2011.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Relucio KI, Beernink HT, Chen D, Israelski DM, Kim R, Holodniy M. Proteomic Analysis of Serum Cytokine Levels in Response to Highly Active Antiretroviral Therapy (HAART) J Proteome Res. 2005;4:227–31. doi: 10.1021/pr049930y. [DOI] [PubMed] [Google Scholar]
- 63.Zajac A, Song D, Qian W, Zhukov T. Protein microarrays and quantum dot probes for early cancer detection. Colloids Surf, B. 2007;58:309–14. doi: 10.1016/j.colsurfb.2007.02.019. [DOI] [PubMed] [Google Scholar]
- 64.Sack RA, Conradi L, Krumholz D, Beaton A, Sathe S, Morris C. Membrane array characterization of 80 chemokines, cytokines, and growth factors in open- and closed-eye tears: angiogenin and other defense system constituents. Invest Ophthalmol Vis Sci. 2005;46:1228–38. doi: 10.1167/iovs.04-0760. [DOI] [PubMed] [Google Scholar]
- 65.Master SR, Bierl C, Kricka LJ. Diagnostic challenges for multiplexed protein microarrays. Drug Discovery Today. 2006;11:1007–11. doi: 10.1016/j.drudis.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 66.Wu P, Grainger DW. Comparison of Hydroxylated Print Additives on Antibody Microarray Performance. J Proteome Res. 2006;5:2956–65. doi: 10.1021/pr060217d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ellington AA, Kullo IJ, Bailey KR, Klee GG. Antibody-based protein multiplex platforms: technical and operational challenges. Clin Chem. 2010;56:186–93. doi: 10.1373/clinchem.2009.127514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hucknall A, Kim D-H, Rangarajan S, Hill RT, Reichert WM, Chilkoti A. Simple fabrication of antibody microarrays on nonfouling polymer brushes with femtomolar sensitivity for protein analytes in serum and blood. Adv Mater. 2009;21:1968–71. doi: 10.1002/adma.200803125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Young S-H, Antonini JM, Roberts JR, Erdely AD, Zeidler-Erdely PC. Performance evaluation of cytometric bead assays for the measurement of lung cytokines in two rodent models. J Immunol Methods. 2008;331:59–68. doi: 10.1016/j.jim.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 70.Fu Q, Zhu J, Van Eyk JE. Comparison of multiplex immunoassay platforms. Clin Chem. 2010;56:314–8. doi: 10.1373/clinchem.2009.135087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bastarache JA, Koyama T, Wickersham NE, Mitchell DB, Mernaugh RL, Ware LB. Accuracy and reproducibility of a multiplex immunoassay platform: A validation study. J Immunol Methods. 2011;367:33–9. doi: 10.1016/j.jim.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siawaya JFD, Roberts T, Babb C, Black G, Golakai HJ, Stanley K, et al. An evaluation of commercial fluorescent bead-based Luminex cytokine assays. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002535. No pp. given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fichorova RN, Richardson-Harman N, Alfano M, Belec L, Carbonneil C, Chen S, et al. Biological and technical variables affecting immunoassay recovery of cytokines from human serum and simulated vaginal fluid: a multicenter study. Anal Chem. 2008;80:4741–51. doi: 10.1021/ac702628q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Whiteside TL. Cytokines and cytokine measurements in a clinical laboratory. Clin Diagn Lab Immunol. 1994;1:257–60. doi: 10.1128/cdli.1.3.257-260.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou X, Fragala MS, McElhaney JE, Kuchel GA. Conceptual and methodological issues relevant to cytokine and inflammatory marker measurements in clinical research. Curr Opin Clin Nutr Metab Care. 2010;13:541–7. doi: 10.1097/MCO.0b013e32833cf3bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Butterfield LH, Potter DM, Kirkwood JM. Multiplex serum biomarker assessments: technical and biostatistical issues. J Transl Med. 2011;9:173. doi: 10.1186/1479-5876-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.de Jager W, Bourcier K, Rijkers GT, Prakken BJ, Seyfert-Margolis V. Prerequisites for cytokine measurements in clinical trials with multiplex immunoassays. BMC Immunol. 2009;10 doi: 10.1186/1471-2172-10-52. No pp. given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gu Y, Zeleniuch-Jacquotte A, Linkov F, Koenig KL, Liu M, Velikokhatnaya L, et al. Reproducibility of serum cytokines and growth factors. Cytokine. 2009;45:44–9. doi: 10.1016/j.cyto.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clendenen TV, Arslan AA, Lokshin AE, Idahl A, Hallmans G, Koenig KL, et al. Temporal reliability of cytokines and growth factors in EDTA plasma. BMC Res Notes. 2010;3 doi: 10.1186/1756-0500-3-302. No pp. given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ledur A, Fitting C, David B, Hamberger C, Cavaillon J-M. Variable estimates of cytokine levels produced by commercial ELISA kits: results using international cytokine standards. J Immunol Methods. 1995;186:171–9. doi: 10.1016/0022-1759(95)00184-c. [DOI] [PubMed] [Google Scholar]
- 81.Aziz N, Nishanian P, Mitsuyasu R, Detels R, Fahey JL. Variables that affect assays for plasma cytokines and soluble activation markers. Clin Diagn Lab Immunol. 1999;6:89–95. doi: 10.1128/cdli.6.1.89-95.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Granger J, Siddiqui J, Copeland S, Remick D. Albumin depletion of human plasma also removes low abundance proteins including the cytokines. Proteomics. 2005;5:4713–8. doi: 10.1002/pmic.200401331. [DOI] [PubMed] [Google Scholar]
- 83.von Maltzan K, Pruett SB. ELISA assays and alcohol: increasing carbon chain length can interfere with detection of cytokines. Alcohol. 2011;45:1–9. doi: 10.1016/j.alcohol.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Braunwald E. Biomarkers in heart failure. N Engl J Med. 2008;358:2148–59. doi: 10.1056/NEJMra0800239. [DOI] [PubMed] [Google Scholar]
- 85.Bozza FA, Salluh JI, Japiassu AM, Soares M, Assis EF, Gomes RN, et al. Cytokine profiles as markers of disease severity in sepsis: a multiplex analysis. Crit Care. 2007;11:R49. doi: 10.1186/cc5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Drukier AK, Ossetrova N, Schors E, Brown LR, Tomaszewski J, Sainsbury R, et al. Ultra-Sensitive Immunoassays Using Multi-Photon-Detection in Diagnostic Proteomics of Blood. J Proteome Res. 2005;4:2375–8. doi: 10.1021/pr050320n. [DOI] [PubMed] [Google Scholar]
- 87.Daunert S, Deo SK, editors. Photoproteins in Bioanalysis. Wiley-VCH Verlag GmbH & Co. KGaA; 2006. [Google Scholar]
- 88.Xiao L, Yang C, Nelson CO, Holloway BP, Udhayakumar V, Lal AA. Quantitation of RT-PCR amplified cytokine mRNA by aequorin-based bioluminescence immunoassay 1. J Immunol Methods. 1996;199:139–47. doi: 10.1016/s0022-1759(96)00174-3. [DOI] [PubMed] [Google Scholar]
- 89.Actor JK, Kuffner T, Dezzutti CS, Hunter RL, McNicholl JM. A flash-type bioluminescent immunoassay that is more sensitive than radioimaging: quantitative detection of cytokine cDNA in activated and resting human cells. J Immunol Methods. 1998;211:65–77. doi: 10.1016/s0022-1759(97)00190-7. [DOI] [PubMed] [Google Scholar]
- 90.Scott D, Dikici E, Ensor M, Daunert S. Bioluminescence and its impact on bioanalysis. Annu Rev Anal Chem. 2011;4:297–319. doi: 10.1146/annurev-anchem-061010-113855. [DOI] [PubMed] [Google Scholar]
- 91.Adler M, Wacker R, Niemeyer CM. Sensitivity by combination: immuno-PCR and related technologies. Analyst (Cambridge, U K) 2008;133:702–18. doi: 10.1039/b718587c. [DOI] [PubMed] [Google Scholar]
- 92.Potuckova L, Franko F, Bambouskova M, Draber P. Rapid and sensitive detection of cytokines using functionalized gold nanoparticle-based immuno-PCR, comparison with immuno-PCR and ELISA. J Immunol Methods. 2011;371:38–47. doi: 10.1016/j.jim.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 93.Schweitzer B, Roberts S, Grimwade B, Shao W, Wang M, Fu Q, et al. Multiplexed protein profiling on microarrays by rolling-circle amplification. Nat Biotechnol. 2002;20:359–65. doi: 10.1038/nbt0402-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nam J-M, Thaxton CS, Mirkin CA. Nanoparticle-based bio-bar codes for the ultrasensitive detection of proteins. Science (Washington, DC, U S) 2003;301:1884–6. doi: 10.1126/science.1088755. [DOI] [PubMed] [Google Scholar]
- 95.Nam J-M, Wise AR, Groves JT. Colorimetric Bio-Barcode Amplification Assay for Cytokines. Anal Chem. 2005;77:6985–8. doi: 10.1021/ac0513764. [DOI] [PubMed] [Google Scholar]
- 96.Nam J-M, Jang K-J, Groves JT. Detection of proteins using a colorimetric bio-barcode assay. Nat Protoc. 2007;2:1438–44. doi: 10.1038/nprot.2007.201. [DOI] [PubMed] [Google Scholar]
- 97.Gustafsdottir SM, Schallmeiner E, Fredriksson S, Gullberg M, Soederberg O, Jarvius M, et al. Proximity ligation assays for sensitive and specific protein analyses. Anal Biochem. 2005;345:2–9. doi: 10.1016/j.ab.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 98.Gullberg M, Gustafsdottir SM, Schallmeiner E, Jarvius J, Bjarnegard M, Betsholtz C, et al. Cytokine detection by antibody-based proximity ligation. Proc Natl Acad Sci U S A. 2004;101:8420–4. doi: 10.1073/pnas.0400552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hammond M, Nong RY, Ericsson O, Pardali K, Landegren U. Profiling cellular protein complexes by proximity ligation with dual tag microarray readout. PLoS One. 2012;7:e40405. doi: 10.1371/journal.pone.0040405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schallmeiner E, Oksanen E, Ericsson O, Spangberg L, Eriksson S, Stenman U-H, et al. Sensitive protein detection via triple-binder proximity ligation assays. Nat Methods. 2007;4:135–7. doi: 10.1038/nmeth974. [DOI] [PubMed] [Google Scholar]
- 101.Weller MG. Immunochromatographic techniques - a critical review. Fresenius’ J Anal Chem. 2000;366:635–45. doi: 10.1007/s002160051558. [DOI] [PubMed] [Google Scholar]
- 102.Miller KJ, Herman AC. Affinity Chromatography with Immunochemical Detection Applied to the Analysis of Human Methionyl Granulocyte Colony Stimulating Factor in Serum. Anal Chem. 1996;68:3077–82. doi: 10.1021/ac960201e. [DOI] [PubMed] [Google Scholar]
- 103.Mire-Sluis AR, Das RG, Zoon K, Padilla A, Thorpe R, Meager A. The biological properties, assay, and standardization of interferon-alpha: a need for a WHO collaborative study. J Interferon Cytokine Res. 1996;16:637–43. doi: 10.1089/jir.1996.16.637. [DOI] [PubMed] [Google Scholar]
- 104.Phillips TM, Dickens BF. Analysis of recombinant cytokines in human body fluids by immunoaffinity capillary electrophoresis. Electrophoresis. 1998;19:2991–6. doi: 10.1002/elps.1150191632. [DOI] [PubMed] [Google Scholar]
- 105.Phillips TM. Multi-analyte analysis of biological fluids with a recycling immunoaffinity column array. J Biochem Biophys Methods. 2001;49:253–62. doi: 10.1016/s0165-022x(01)00202-0. [DOI] [PubMed] [Google Scholar]
- 106.Castle PE, Phillips TM, Hildesheim A, Herrero R, Bratti MC, Rodriguez AC, et al. Immune Profiling of Plasma and Cervical Secretions Using Recycling Immunoaffinity Chromatography. Cancer Epidemiol, Biomarkers Prev. 2003;12:1449–56. [PubMed] [Google Scholar]
- 107.Phillips TM. Rapid analysis of inflammatory cytokines in cerebrospinal fluid using chip-based immunoaffinity electrophoresis. Electrophoresis. 2004;25:1652–9. doi: 10.1002/elps.200305873. [DOI] [PubMed] [Google Scholar]
- 108.Kuchen S, Robbins R, Sims GP, Sheng C, Phillips TM, Lipsky PE, et al. Essential Role of IL-21 in B Cell Activation, Expansion, and Plasma Cell Generation during CD4+ T Cell-B Cell Collaboration. J Immunol. 2007;179:5886–96. doi: 10.4049/jimmunol.179.9.5886. [DOI] [PubMed] [Google Scholar]
- 109.Summy-Long JY, Hu S, Long A, Phillips TM. Interleukin-1β release in the supraoptic nucleus area during osmotic stimulation requires neural function. J Neuroendocrinol. 2008;20:1224–32. doi: 10.1111/j.1365-2826.2008.01783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kalish H, Phillips TM. Application of immunoaffinity capillary electrophoresis to the measurements of secreted cytokines by cultured astrocytes. J Sep Sci. 2009;32:1605–12. doi: 10.1002/jssc.200900047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Phillips TM, Kalish H, Wellner E. Receptor affinity CE for measuring bioactive inflammatory cytokines in human skin biopsies. Electrophoresis. 2009;30:3947–54. doi: 10.1002/elps.200900311. [DOI] [PubMed] [Google Scholar]
- 112.Kjellstrom S, Emneus J, Marko-Varga G. Flow immunochemical bio-recognition detection for the determination of interleukin-10 in cell samples. J Immunol Methods. 2000;246:119–30. doi: 10.1016/s0022-1759(00)00307-0. [DOI] [PubMed] [Google Scholar]
- 113.Kjellstrom S, Burestedt E, Emneus J, Marko-Varga G. Determination of interleukin-8 by on-line non-competitive flow immunoassay and restricted access trace enrichment. Int J Bio-Chromatogr. 2000;5:17–30. [Google Scholar]
- 114.Burestedt E, Kjellstrom S, Emneus J, Marko-Varga G. Development of an offline noncompetitive flow immunoassay for the determination of interleukin-8 in cell samples. Anal Biochem. 2000;279:46–54. doi: 10.1006/abio.1999.4442. [DOI] [PubMed] [Google Scholar]
- 115.Worsley GJ, Attree SL, Noble JE, Horgan AM. Rapid duplex immunoassay for wound biomarkers at the point-of-care. Biosens Bioelectron. 2012;34:215–20. doi: 10.1016/j.bios.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 116.Corstjens PLAM, de DCJ, van dP-vSJJ, Wiesmeijer KC, Riuttamaeki T, van MKE, et al. Lateral flow assay for simultaneous detection of cellular- and humoral immune responses. Clin Biochem. 2011;44:1241–6. doi: 10.1016/j.clinbiochem.2011.06.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Qiu X, Thompson JA, Chen Z, Liu C, Chen D, Ramprasad S, et al. Finger-actuated, self-contained immunoassay cassettes. Biomed Microdevices. 2009;11:1175–86. doi: 10.1007/s10544-009-9334-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Blicharz TM, Siqueira WL, Helmerhorst EJ, Oppenheim FG, Wexler PJ, Little FF, et al. Fiber-Optic Microsphere-Based Antibody Array for the Analysis of Inflammatory Cytokines in Saliva. Anal Chem. 2009;81:2106–14. doi: 10.1021/ac802181j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cesaro-Tadic S, Dernick G, Juncker D, Buurman G, Kropshofer H, Michel B, et al. High-sensitivity miniaturized immunoassays for tumor necrosis factor α using microfluidic systems. Lab Chip. 2004;4:563–9. doi: 10.1039/b408964b. [DOI] [PubMed] [Google Scholar]
- 120.Appleyard DC, Chapin SC, Doyle PS. Multiplexed protein quantification with barcoded hydrogel microparticles. Anal Chem. 2011;83:193–9. doi: 10.1021/ac1022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Christodoulides N, Tran M, Floriano PN, Rodriguez M, Goodey A, Ali M, et al. A microchip-based multianalyte assay system for the assessment of cardiac risk. Anal Chem. 2002;74:3030–6. doi: 10.1021/ac011150a. [DOI] [PubMed] [Google Scholar]
- 122.Jokerst JV, Jacobson JW, Bhagwandin BD, Floriano PN, Christodoulides N, McDevitt JT. Programmable Nano-Bio-Chip Sensors: Analytical Meets Clinical. Anal Chem (Washington, DC, U S) 2010;82:1571–9. doi: 10.1021/ac901743u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Christodoulides N, Floriano PN, Miller CS, Ebersole JL, Mohanty S, Dharshan P, et al. Lab-on-a-chip methods of point-of-care measurements of salivary biomarkers of periodontitis. Ann N Y Acad Sci. 2007;1098:411–28. doi: 10.1196/annals.1384.035. [DOI] [PubMed] [Google Scholar]
- 124.Sista RS, Eckhardt AE, Srinivasan V, Pollack MG, Palanki S, Pamula VK. Heterogeneous immunoassays using magnetic beads on a digital microfluidic platform. Lab Chip. 2008;8:2188–96. doi: 10.1039/b807855f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Thompson JA, Du X, Grogan JM, Schrlau MG, Bau HH. Polymeric microbead arrays for microfluidic applications. J Micromech Microeng. 2010;20:115017/1–/8. [Google Scholar]
- 126.Konry T, Dominguez-Villar M, Baecher-Allan C, Hafler DA, Yarmush ML. Droplet-based microfluidic platforms for single T cell secretion analysis of IL-10 cytokine. Biosens Bioelectron. 2011;26:2707–10. doi: 10.1016/j.bios.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Abe K, Hashimoto Y, Yatsushiro S, Yamamura S, Bando M, Hiroshima Y, et al. Simultaneous immunoassay analysis of plasma IL-6 and TNF-α on a microchip. PLoS One. 2013;8:e53620. doi: 10.1371/journal.pone.0053620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature (London) 1990;346:818–22. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 129.McGown LB, Joseph MJ, Pitner JB, vonk GP, Linn CP. The nucleic acid ligand. A new tool for molecular recognition. Anal Chem. 1995;67:663A–8A. doi: 10.1021/ac00117a002. [DOI] [PubMed] [Google Scholar]
- 130.Brody EN, Gold L. Aptamers as therapeutic and diagnostic agents. Rev Mol Biotechnol. 2000;74:5–13. doi: 10.1016/s1389-0352(99)00004-5. [DOI] [PubMed] [Google Scholar]
- 131.Famulok M, Mayer G. Aptamer Modules as Sensors and Detectors. Acc Chem Res. 2011;44:1349–58. doi: 10.1021/ar2000293. [DOI] [PubMed] [Google Scholar]
- 132.Iliuk AB, Hu L, Tao WA. Aptamer in Bioanalytical Applications. Anal Chem. 2011;83:4440–52. doi: 10.1021/ac201057w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nery AA, Wrenger C, Ulrich H. Recognition of biomarkers and cell-specific molecular signatures: aptamers as capture agents. J Sep Sci. 2009;32:1523–30. doi: 10.1002/jssc.200800695. [DOI] [PubMed] [Google Scholar]
- 134.Fang X, Tan W. Aptamers Generated from Cell-SELEX for Molecular Medicine: A Chemical Biology Approach. Acc Chem Res. 2010;43:48–57. doi: 10.1021/ar900101s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zichi D, Eaton B, Singer B, Gold L. Proteomics and diagnostics: Let’s Get Specific, again. Curr Opin Chem Biol. 2008;12:78–85. doi: 10.1016/j.cbpa.2008.01.016. [DOI] [PubMed] [Google Scholar]
- 136.Gold L, Ayers D, Bertino J, Bock C, Bock A, Brody EN, et al. Aptamer-based multiplexed proteomic technology for biomarker discovery. PLoS One. 2010;5:e15004. doi: 10.1371/journal.pone.0015004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Guthrie JW, Hamula CLA, Zhang H, Le XC. Assays for cytokines using aptamers. Methods (San Diego, CA, U S) 2006;38:324–30. doi: 10.1016/j.ymeth.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 138.Wu Z-S, Zhou H, Zhang S, Shen G, Yu R. Electrochemical Aptameric Recognition System for a Sensitive Protein Assay Based on Specific Target Binding-Induced Rolling Circle Amplification. Anal Chem. 2010;82:2282–9. doi: 10.1021/ac902400n. [DOI] [PubMed] [Google Scholar]
- 139.Platt M, Willmott GR, Lee GU. Resistive Pulse Sensing of Analyte-Induced Multicomponent Rod Aggregation Using Tunable Pores. Small. 2012;8:2436–44. S/1–S/4. doi: 10.1002/smll.201200058. [DOI] [PubMed] [Google Scholar]
- 140.Liu Y, Tuleouva N, Ramanculov E, Revzin A. Aptamer-Based Electrochemical Biosensor for Interferon Gamma Detection. Anal Chem (Washington, DC, U S) 2010;82:8131–6. doi: 10.1021/ac101409t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kim M, Jeoung Y-H, Lee SJ, Choi I, Pyun K-H, Lee Y. In vitro selection of DNA aptamers binding to the recombinant human interleukin-6. Mol Cells. 1995;5:555–62. [Google Scholar]
- 142.Ishiguro A, Akiyama T, Adachi H, Inoue J-i, Nakamura Y. Therapeutic potential of anti-interleukin-17A aptamer: suppression of interleukin-17A signaling and attenuation of autoimmunity in two mouse models. Arthritis Rheum. 2011;63:455–66. doi: 10.1002/art.30108. [DOI] [PubMed] [Google Scholar]
- 143.Kim S, Kim J-H, Yoon S, Kim K-S, Yoon M-Y, Yoon D-Y, et al. Generation of antagonistic RNA aptamers specific to proinflammatory cytokine interleukin-32. Bull Korean Chem Soc. 2010;31:3561–6. [Google Scholar]
- 144.Rhodes A, Deakin A, Spaull J, Coomber B, Aitken A, Life P, et al. The generation and characterization of antagonist RNA aptamers to human oncostatin M. J Biol Chem. 2000;275:28555–61. doi: 10.1074/jbc.M002981200. [DOI] [PubMed] [Google Scholar]
- 145.Fredriksson S, Gullberg M, Jarvius J, Olsson C, Pietras K, Gustafsdottir SM, et al. Protein detection using proximity-dependent DNA ligation assays. Nat Biotechnol. 2002;20:473–7. doi: 10.1038/nbt0502-473. [DOI] [PubMed] [Google Scholar]
- 146.Yan X, Gao X, Zhang Z. Isolation and characterization of 2′-amino-modified RNA aptamers for human TNFα. Genomics, Proteomics Bioinf. 2004;2:32–42. doi: 10.1016/S1672-0229(04)02005-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Orava EW, Jarvik N, Shek YL, Sidhu SS, Gariepy J. A Short DNA Aptamer That Recognizes TNFα and Blocks Its Activity in Vitro. ACS Chem Biol. doi: 10.1021/cb3003557. Ahead of Print. [DOI] [PubMed] [Google Scholar]
- 148.Ng EWM, Adamis AP. Anti-VEGF aptamer (pegaptanib) therapy for ocular vascular diseases. Ann N Y Acad Sci. 2006;1082:151–71. doi: 10.1196/annals.1348.062. [DOI] [PubMed] [Google Scholar]
- 149.Tuleuova N, Revzin A. Micropatterning of Aptamer Beacons to Create Cytokine-Sensing Surfaces. Cell Mol Bioeng. 2010;3:337–44. doi: 10.1007/s12195-010-0148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tuleuova N, Jones CN, Yan J, Ramanculov E, Yokobayashi Y, Revzin A. Development of an Aptamer Beacon for Detection of Interferon-gamma. Anal Chem. 2010;82:1851–7. doi: 10.1021/ac9025237. [DOI] [PubMed] [Google Scholar]
- 151.Liu Y, Yan J, Howland MC, Kwa T, Revzin A. Micropatterned Aptasensors for Continuous Monitoring of Cytokine Release from Human Leukocytes. Anal Chem. 2011;83:8286–92. doi: 10.1021/ac202117g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Pestka S, Krause CD, Walter MR. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- 153.Balasubramanian V, Nguyen LT, Balasubramanian SV, Ramanathan M. Interferon-γ-inhibitory oligodeoxynucleotides alter the conformation of interferon-γ. Mol Pharmacol. 1998;53:926–32. [PubMed] [Google Scholar]
- 154.Li Y-Y, Zhang C, Li B-S, Zhao L-F, Li X-b, Yang W-J, et al. Ultrasensitive densitometry detection of cytokines with nanoparticle-modified aptamers. Clin Chem (Washington, DC, U S) 2007;53:1061–6. doi: 10.1373/clinchem.2006.082271. [DOI] [PubMed] [Google Scholar]
- 155.Fang X, Sen A, Vicens M, Tan W. Synthetic DNA aptamers to detect protein molecular variants in a high-throughput fluorescence quenching assay. Chem BioChem. 2003;4:829–34. doi: 10.1002/cbic.200300615. [DOI] [PubMed] [Google Scholar]
- 156.Green LS, Jellinek D, Jenison R, Oestman A, Heldin C-H, Janjic N. Inhibitory DNA Ligands to Platelet-Derived Growth Factor B-Chain. Biochemistry. 1996;35:14413–24. doi: 10.1021/bi961544+. [DOI] [PubMed] [Google Scholar]
- 157.Vicens MC, Sen A, Vanderlaan A, Drake TJ, Tan W. Investigation of molecular beacon aptamer-based bioassay for platelet-derived growth factor detection. Chem BioChem. 2005;6:900–7. doi: 10.1002/cbic.200400308. [DOI] [PubMed] [Google Scholar]
- 158.Huang C-C, Huang Y-F, Cao Z, Tan W, Chang H-T. Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal Chem. 2005;77:5735–41. doi: 10.1021/ac050957q. [DOI] [PubMed] [Google Scholar]
- 159.Zhang H, Li X-F, Le XC. Differentiation and detection of PDGF isomers and their receptors by tunable aptamer capillary electrophoresis. Anal Chem. 2009;81:7795–800. doi: 10.1021/ac901471w. [DOI] [PubMed] [Google Scholar]
- 160.Csordas A, Gerdon AE, Adams JD, Qian J, Oh SS, Xiao Y, et al. Detection of proteins in serum by micromagnetic aptamer PCR (MAP) technol. Angew Chem, Int Ed. 2010;49:355–8. S/1–S/6. doi: 10.1002/anie.200904846. [DOI] [PubMed] [Google Scholar]
- 161.Zhang Z-z, Zhang C-y. Highly Sensitive Detection of Protein with Aptamer-Based Target-Triggering Two-Stage Amplification. Anal Chem (Washington, DC, U S) 2012;84:1623–9. doi: 10.1021/ac2029002. [DOI] [PubMed] [Google Scholar]
- 162.Yang L, Fung CW, Cho EJ, Ellington AD. Real-Time Rolling Circle Amplification for Protein Detection. Anal Chem (Washington, DC, U S) 2007;79:3320–9. doi: 10.1021/ac062186b. [DOI] [PubMed] [Google Scholar]
- 163.Lee J, Icoz K, Roberts A, Ellington AD, Savran CA. Diffractometric Detection of Proteins using Microbead-Based Rolling Circle Amplification. Anal Chem. 2010;82:197–202. doi: 10.1021/ac901716d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gold L, Walker JJ, Wilcox SK, Williams S. Advances in human proteomics at high scale with the SOMAscan proteomics platform. New Biotechnol. 2012;29:543–9. doi: 10.1016/j.nbt.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 165.Gloriam DE, Orchard S, Bertinetti D, Bjorling E, Bongcam-Rudloff E, Borrebaeck CAK, et al. A community standard format for the representation of protein affinity reagents. Mol Cell Proteomics. 2010;9:1–10. doi: 10.1074/mcp.M900185-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bock C, Coleman M, Collins B, Davis J, Foulds G, Gold L, et al. Photoaptamer arrays applied to multiplexed proteomic analysis. Proteomics. 2004;4:609–18. doi: 10.1002/pmic.200300631. [DOI] [PubMed] [Google Scholar]
- 167.Hoofnagle AN, Wener MH. The fundamental flaws of immunoassays and potential solutions using tandem mass spectrometry. J Immunol Methods. 2009;347:3–11. doi: 10.1016/j.jim.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Parker CE, Pearson TW, Anderson NL, Borchers CH. Mass-spectrometry-based clinical proteomics - a review and prospective. Analyst (Cambridge, U K) 2010;135:1830–8. doi: 10.1039/c0an00105h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Elliott MH, Smith DS, Parker CE, Borchers C. Current trends in quantitative proteomics. J Mass Spectrom. 2009;44:1637–60. doi: 10.1002/jms.1692. [DOI] [PubMed] [Google Scholar]
- 170.Carr SA, Anderson L. Protein quantitation through targeted mass spectrometry: the way out of biomarker purgatory? Clin Chem (Washington, DC, U S) 2008;54:1749–52. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Izrael-Tomasevic A, Phu L, Phung QT, Lill JR, Arnott D. Targeting Interferon Alpha Subtypes in Serum: A Comparison of Analytical Approaches to the Detection and Quantitation of Proteins in Complex Biological Matrices. J Proteome Res. 2009;8:3132–40. doi: 10.1021/pr900076q. [DOI] [PubMed] [Google Scholar]
- 172.Groessl M, Luksch H, Roesen-Wolff A, Shevchenko A, Gentzel M. Profiling of the human monocytic cell secretome by quantitative label-free mass spectrometry identifies stimulus-specific cytokines and proinflammatory proteins. Proteomics. 2012;12:2833–42. doi: 10.1002/pmic.201200108. [DOI] [PubMed] [Google Scholar]
- 173.Meissner F, Scheltema RA, Mollenkopf H-J, Mann M. Direct proteomic quantification of the secretome of activated immune cells. Science (Washington, DC, U S) 2013;340:475–8. doi: 10.1126/science.1232578. [DOI] [PubMed] [Google Scholar]
- 174.Finoulst I, Vink P, Rovers E, Pieterse M, Pinkse M, Bos E, et al. Identification of low abundant secreted proteins and peptides from primary culture supernatants of human T-cells. J Proteomics. 2011;75:23–33. doi: 10.1016/j.jprot.2011.03.034. [DOI] [PubMed] [Google Scholar]
- 175.Firth SM, Baxter RC. Cellular actions of the insulin-like growth factor binding proteins. Endocr Rev. 2002;23:824–54. doi: 10.1210/er.2001-0033. [DOI] [PubMed] [Google Scholar]
- 176.AE, Nemet D. Exercise training, physical fitness and the growth hormone-insulin-like growth factor-1 axis and cytokine balance. 2010;55:128–40. doi: 10.1159/000321977. [DOI] [PubMed] [Google Scholar]
- 177.Bredehoft M, Schanzer W, Thevis M. Quantification of human insulin-like growth factor-1 and qualitative detection of its analogues in plasma using liquid chromatography/electrospray ionisation tandem mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:477–85. doi: 10.1002/rcm.3388. [DOI] [PubMed] [Google Scholar]
- 178.Eichelbaum K, Winter M, Diaz MB, Herzig S, Krijgsveld J. Selective enrichment of newly synthesized proteins for quantitative secretome analysis. Nat Biotechnol. 2012;30:984–90. doi: 10.1038/nbt.2356. [DOI] [PubMed] [Google Scholar]
- 179.Boyle MDP, Hess JL, Nuara AA, Buller RM. Application of immunoproteomics to rapid cytokine detection. Methods (San Diego, CA, U S) 2006;38:342–50. doi: 10.1016/j.ymeth.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 180.Rossi L, Moharram R, Martin BM, White RL, Panelli MC. Detection of human MCP-4/CCL13 isoforms by SELDI immunoaffinity capture. J Transl Med. 2006;4 doi: 10.1186/1479-5876-4-5. No pp. given. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.van Breemen MJ, Bleijlevens B, de Koster CG, Aerts JMFG. Limitations in quantitation of the biomarker CCL18 in Gaucher disease blood samples by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry. Biochim Biophys Acta, Proteins Proteomics. 2006;1764:1626–32. doi: 10.1016/j.bbapap.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 182.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using stable isotope standards and capture by anti-peptide antibodies (SISCAPA) J Proteome Res. 2004;3:235–44. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 183.Kuhn E, Addona T, Keshishian H, Burgess M, Mani DR, Lee RT, et al. Developing multiplexed assays for troponin I and interleukin-33 in plasma by peptide immunoaffinity enrichment and targeted mass spectrometry. Clin Chem (Washington, DC, U S) 2009;55:1108–17. doi: 10.1373/clinchem.2009.123935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kuhn E, Whiteaker JR, Mani DR, Jackson AM, Zhao L, Pope ME, et al. Interlaboratory evaluation of automated, multiplexed peptide immunoaffinity enrichment coupled to multiple reaction monitoring mass spectrometry for quantifying proteins in plasma. Mol Cell Proteomics. 2012;11:M111.013854, 14. doi: 10.1074/mcp.M111.013854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang C, Wu F, Zhang Y, Wang X, Zhang X. A novel combination of immunoreaction and ICP-MS as a hyphenated technique for the determination of thyroid-stimulating hormone (TSH) in human serum. J Anal At Spectrom. 2001;16:1393–6. [Google Scholar]
- 186.Giesen C, Waentig L, Panne U, Jakubowski N. History of inductively coupled plasma mass spectrometry-based immunoassays. Spectrochim Acta, Part B. 2012;76:27–39. [Google Scholar]
- 187.Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S. Highly multiparametric analysis by mass cytometry. J Immunol Methods. 2010;361:1–20. doi: 10.1016/j.jim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 188.Bendall SC, Simonds EF, Qiu P, Amir E-aD, Krutzik PO, Finck R, et al. Single-Cell Mass Cytometry of Differential Immune and Drug Responses Across a Human Hematopoietic Continuum. Science. 2011;332:687–96. doi: 10.1126/science.1198704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Li B, Ellington AD. Electrochemical techniques as powerful readout methods for aptamer-based biosensors. RSC Biomol Sci. 2012;26:211–41. [Google Scholar]
- 190.Rishpon J, Buchner V. Electrochemical antibody-based sensors. Elsevier Science B.V; 2005. pp. 329–73. [Google Scholar]
- 191.Wang J. Nanoparticle-based electrochemical bioassays of proteins. Electroanalysis. 2007;19:769–76. [Google Scholar]
- 192.Li X-M, Yang X-Y, Zhang S-S. Electrochemical enzyme immunoassay using model labels. TrAC, Trends Anal Chem. 2008;27:543–53. [Google Scholar]
- 193.Kuramitz H. Magnetic microbead-based electrochemical immunoassays. Anal Bioanal Chem. 2009;394:61–9. doi: 10.1007/s00216-009-2650-y. [DOI] [PubMed] [Google Scholar]
- 194.Rusling JF, Kumar CV, Gutkind JS, Patel V. Measurement of biomarker proteins for point-of-care early detection and monitoring of cancer. Analyst (Cambridge, U K) 2010;135:2496–511. doi: 10.1039/c0an00204f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sardesai NP, Barron JC, Rusling JF. Carbon Nanotube Microwell Array for Sensitive Electrochemiluminescent Detection of Cancer Biomarker Proteins. Anal Chem (Washington, DC, U S) 2011;83:6698–703. doi: 10.1021/ac201292q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Malhotra R, Patel V, Chikkaveeraiah BV, Munge BS, Cheong SC, Zain RB, et al. Ultrasensitive Detection of Cancer Biomarkers in the Clinic by Use of a Nanostructured Microfluidic Array. Anal Chem (Washington, DC, U S) 2012;84:6249–55. doi: 10.1021/ac301392g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Wang J, Liu G, Engelhard MH, Lin Y. Sensitive Immunoassay of a Biomarker Tumor Necrosis Factor-α Based on Poly(guanine)-Functionalized Silica Nanoparticle Label. Anal Chem. 2006;78:6974–9. doi: 10.1021/ac060809f. [DOI] [PubMed] [Google Scholar]
- 198.Yin Z, Liu Y, Jiang L-P, Zhu J-J. Electrochemical immunosensor of tumor necrosis factor α based on alkaline phosphatase functionalized nanospheres. Biosens Bioelectron. 2011;26:1890–4. doi: 10.1016/j.bios.2010.03.025. [DOI] [PubMed] [Google Scholar]
- 199.Blackburn GF, Shah HP, Kenten JH, Leland J, Kamin RA, Link J, et al. Electrochemiluminescence detection for development of immunoassays and DNA probe assays for clinical diagnostics. Clin Chem (Winston-Salem, N C) 1991;37:1534–9. [PubMed] [Google Scholar]
- 200.Miao W. Electrogenerated Chemiluminescence and Its Biorelated Applications. Chem Rev (Washington, DC, U S) 2008;108:2506–53. doi: 10.1021/cr068083a. [DOI] [PubMed] [Google Scholar]
- 201.Rhyne PW, Wong OT, Zhang YJ, Weiner RS. Electrochemiluminescence in bioanalysis. Bioanalysis. 2009;1:919–35. doi: 10.4155/bio.09.80. [DOI] [PubMed] [Google Scholar]
- 202.Muzyka K. Current trends in the development of the electrochemiluminescent immunosensors. Biosens Bioelectron. 2014;54:393–407. doi: 10.1016/j.bios.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 203.Sennikov SV, Krysov SV, Injelevskaya TV, Silkov AN, Grishina LV, Kozlov VA. Quantitative analysis of human immunoregulatory cytokines by electrochemiluminescence method. J Immunol Methods. 2003;275:81–8. doi: 10.1016/s0022-1759(03)00007-3. [DOI] [PubMed] [Google Scholar]
- 204.Dabitao D, Margolick JB, Lopez J, Bream JH. Multiplex measurement of proinflammatory cytokines in human serum: Comparison of the Meso Scale Discovery electrochemiluminescence assay and the Cytometric Bead Array. J Immunol Methods. 2011;372:71–7. doi: 10.1016/j.jim.2011.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Chowdhury F, Williams A, Johnson P. Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling. J Immunol Methods. 2009;340:55–64. doi: 10.1016/j.jim.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 206.Deiss F, La Fratta CN, Symer M, Blicharz TM, Sojic N, Walt DR. Multiplexed Sandwich Immunoassays Using Electrochemiluminescence Imaging Resolved at the Single Bead Level. J Am Chem Soc. 2009;131:6088–9. doi: 10.1021/ja901876z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Yuan L, Hua X, Wu Y, Pan X, Liu S. Polymer-Functionalized Silica Nanosphere Labels for Ultrasensitive Detection of Tumor Necrosis Factor-alpha. Anal Chem (Washington, DC, U S) 2011;83:6800–9. doi: 10.1021/ac201558w. [DOI] [PubMed] [Google Scholar]
- 208.Sardesai NP, Kadimisetty K, Faria R, Rusling JF. A microfluidic electrochemiluminescent device for detecting cancer biomarker proteins. Anal Bioanal Chem. 2013;405:3831–8. doi: 10.1007/s00216-012-6656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhang H, Jiang B, Xiang Y, Chai Y, Yuan R. Label-free and amplified electrochemical detection of cytokine based on hairpin aptamer and catalytic DNAzyme. Analyst. 2012;137:1020–3. doi: 10.1039/c2an15962g. [DOI] [PubMed] [Google Scholar]
- 210.Berggren C, Bjarnason B, Johansson G. An immunological interleukine-6 capacitive biosensor using perturbation with a potentiostatic step. Biosens Bioelectron. 1998;13:1061–8. doi: 10.1016/s0956-5663(98)00058-x. [DOI] [PubMed] [Google Scholar]
- 211.Pui TS, Kongsuphol P, Arya SK, Bansal T. Detection of tumor necrosis factor (TNF-α) in cell culture medium with label free electrochemical impedance spectroscopy. Sens Actuators, B. 2013;181:494–500. [Google Scholar]
- 212.Bettazzi F, Enayati L, Sanchez IC, Motaghed R, Mascini M, Palchetti I. Electrochemical bioassay for the detection of TNF-α using magnetic beads and disposable screen-printed array of electrodes. Bioanalysis. 2013;5:11–9. doi: 10.4155/bio.12.293. [DOI] [PubMed] [Google Scholar]
- 213.Ligler FS, Taitt CR, editors. Optical Biosensors: Today and Tomorrow. 2008. [Google Scholar]
- 214.Fan X, White IM, Shopova SI, Zhu H, Suter JD, Sun Y. Sensitive optical biosensors for unlabeled targets: A review. Anal Chim Acta. 2008;620:8–26. doi: 10.1016/j.aca.2008.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Theilacker N, Roller EE, Barbee KD, Franzreb M, Huang X. Multiplexed protein analysis using encoded antibody-conjugated microbeads. J R Soc Interface. 2011;8:1104–13. doi: 10.1098/rsif.2010.0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kryscio DR, Peppas NA. Critical review and perspective of macromolecularly imprinted polymers. Acta Biomater. 2012;8:461–73. doi: 10.1016/j.actbio.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 217.Takeuchi T, Hishiya T. Molecular imprinting of proteins emerging as a tool for protein recognition. Org Biomol Chem. 2008;6:2459–67. doi: 10.1039/b715737c. [DOI] [PubMed] [Google Scholar]
- 218.Walcarius A, Collinson MM. Analytical chemistry with silica sol-gels: traditional routes to new materials for chemical analysis. Annu Rev Anal Chem. 2009;2:121–43. doi: 10.1146/annurev-anchem-060908-155139. [DOI] [PubMed] [Google Scholar]
- 219.Mujahid A, Lieberzeit PA, Dickert FL. Chemical sensors based on molecularly imprinted sol-gel materials. Materials. 2010;3:2196–217. [Google Scholar]
- 220.Holthoff EL, Bright FV. Molecularly templated materials in chemical sensing. Anal Chim Acta. 2007;594:147–61. doi: 10.1016/j.aca.2007.05.044. [DOI] [PubMed] [Google Scholar]
- 221.Tao Z, Tehan EC, Bukowski RM, Tang Y, Shughart EL, Holthoff WG, et al. Templated xerogels as platforms for biomolecule-less biomolecule sensors. Anal Chim Acta. 2006;564:59–65. doi: 10.1016/j.aca.2006.01.076. [DOI] [PubMed] [Google Scholar]
- 222.Whitcombe MJ, Chianella I, Larcombe L, Piletsky SA, Noble J, Porter R, et al. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem Soc Rev. 2011;40:1547–71. doi: 10.1039/c0cs00049c. [DOI] [PubMed] [Google Scholar]
- 223.Ge Y, Turner APF. Too large to fit? Recent developments in macromolecular imprinting. Trends Biotechnol. 2008;26:218–24. doi: 10.1016/j.tibtech.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 224.Rachkov A, Minoura N. Towards molecularly imprinted polymers selective to peptides and proteins. The epitope approach. Biochim Biophys Acta, Protein Struct Mol Enzymol. 2001;1544:255–66. doi: 10.1016/s0167-4838(00)00226-0. [DOI] [PubMed] [Google Scholar]
- 225.Nishino H, Huang C-S, Shea KJ. Selective protein capture by epitope imprinting. Angew Chem, Int Ed. 2006;45:2392–6. doi: 10.1002/anie.200503760. [DOI] [PubMed] [Google Scholar]
- 226.Schasfoort RBM, Tudos AJ. Handbook of Surface Plasmon Resonance. Cambridge, UK: Royal Society of Chemistry; 2008. [Google Scholar]
- 227.Chou T-H, Chuang C-Y, Wu C-M. Quantification of Interleukin-6 in cell culture medium using surface plasmon resonance biosensors. Cytokine. 2010;51:107–11. doi: 10.1016/j.cyto.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 228.Battaglia TM, Masson J-F, Sierks MR, Beaudoin SP, Rogers J, Foster KN, et al. Quantification of cytokines involved in wound healing using surface plasmon resonance. Anal Chem. 2005;77:7016–23. doi: 10.1021/ac050568w. [DOI] [PubMed] [Google Scholar]
- 229.Law W-C, Yong K-T, Baev A, Prasad PN. Sensitivity Improved Surface Plasmon Resonance Biosensor for Cancer Biomarker Detection Based on Plasmonic Enhancement. ACS Nano. 2011;5:4858–64. doi: 10.1021/nn2009485. [DOI] [PubMed] [Google Scholar]
- 230.Martinez-Perdiguero J, Retolaza A, Bujanda L, Merino S. Surface plasmon resonance immunoassay for the detection of the TNFα biomarker in human serum. Talanta. 2014;119:492–7. doi: 10.1016/j.talanta.2013.11.063. [DOI] [PubMed] [Google Scholar]
- 231.Rice JM, Stern LJ, Guignon EF, Lawrence DA, Lynes MA. Antigen-specific T cell phenotyping microarrays using grating coupled surface plasmon resonance imaging and surface plasmon coupled emission. Biosens Bioelectron. 2012;31:264–9. doi: 10.1016/j.bios.2011.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Bustos RH, Suesca E, Millan D, Gonzalez JM, Fontanilla MR. Real-time quantification of proteins secreted by artificial connective tissue made from uni- or multidirectional collagen I scaffolds and oral mucosa fibroblasts. Anal Chem (Washington, DC, U S) 2014;86:2421–8. doi: 10.1021/ac4033164. [DOI] [PubMed] [Google Scholar]
- 233.Sipova H, Sevcu V, Kuchar M, Ahmad JN, Mikulecky P, Osicka R, et al. Surface plasmon resonance biosensor based on engineered proteins for direct detection of interferon-gamma in diluted blood plasma. Sens Actuators, B. 2012;174:306–11. [Google Scholar]
- 234.Haes AJ, Van DRP. A unified view of propagating and localized surface plasmon resonance biosensors. Anal Bioanal Chem. 2004;379:920–30. doi: 10.1007/s00216-004-2708-9. [DOI] [PubMed] [Google Scholar]
- 235.Bolduc OR, Masson J-F. Advances in Surface Plasmon Resonance Sensing with Nanoparticles and Thin Films: Nanomaterials, Surface Chemistry, and Hybrid Plasmonic Techniques. Anal Chem. 2011;83:8057–62. doi: 10.1021/ac2012976. [DOI] [PubMed] [Google Scholar]
- 236.Chiang C-Y, Hsieh M-L, Huang K-W, Chau L-K, Chang C-M, Lyu S-R. Fiber-optic particle plasmon resonance sensor for detection of interleukin-1β in synovial fluids. Biosens Bioelectron. 2010;26:1036–42. doi: 10.1016/j.bios.2010.08.047. [DOI] [PubMed] [Google Scholar]
- 237.Oh B-R, Huang N-T, Chen W, Seo JH, Chen P, Cornell TT, et al. Integrated Nanoplasmonic Sensing for Cellular Functional Immunoanalysis Using Human Blood. ACS Nano. 2014;8:2667–76. doi: 10.1021/nn406370u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Shin Y-B, Lee J-M, Park M-R, Kim M-G, Chung BH, Pyo H-B, et al. Analysis of recombinant protein expression using localized surface plasmon resonance (LSPR) Biosens Bioelectron. 2007;22:2301–7. doi: 10.1016/j.bios.2006.12.028. [DOI] [PubMed] [Google Scholar]
- 239.Huang Y-C, Chiang C-Y, Li C-H, Chang T-C, Chiang C-S, Chau L-K, et al. Quantification of tumor necrosis factor-α and matrix metalloproteinases-3 in synovial fluid by a fiber-optic particle plasmon resonance sensor. Analyst (Cambridge, U K) 2013;138:4599–606. doi: 10.1039/c3an00276d. [DOI] [PubMed] [Google Scholar]
- 240.Jeong H-H, Erdene N, Park J-H, Jeong D-H, Lee H-Y, Lee S-K. Real-time label-free immunoassay of interferon-gamma and prostate-specific antigen using a Fiber-Optic Localized Surface Plasmon Resonance sensor. Biosens Bioelectron. 2013;39:346–51. doi: 10.1016/j.bios.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 241.Lacy P, Stow JL. Cytokine release from innate immune cells: association with diverse membrane trafficking pathways. Blood. 2011;118:9–18. doi: 10.1182/blood-2010-08-265892. [DOI] [PubMed] [Google Scholar]
- 242.Leake MC. The physics of life: one molecule at a time. Philos Trans R Soc, B. 2013;368:20120248/1–/9. doi: 10.1098/rstb.2012.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Huang T, Nallathamby PD, Xu X-HN. Photostable single-molecule nanoparticle optical biosensors for real-time sensing of single cytokine molecules and their binding reactions. J Am Chem Soc. 2008;130:17095–105. doi: 10.1021/ja8068853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Huang T, Browning LM, Xu X-HN. Far-field photostable optical nanoscopy (PHOTON) for real-time super-resolution single-molecular imaging of signaling pathways of single live cells. Nanoscale. 2012;4:2797–812. doi: 10.1039/c2nr11739h. [DOI] [PubMed] [Google Scholar]
- 245.Qavi AJ, Washburn AL, Byeon J-Y, Bailey RC. Label-free technologies for quantitative multiparameter biological analysis. Anal Bioanal Chem. 2009;394:121–35. doi: 10.1007/s00216-009-2637-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Luchansky MS, Washburn AL, Martin TA, Iqbal M, Gunn LC, Bailey RC. Characterization of the evanescent field profile and bound mass sensitivity of a label-free silicon photonic microring resonator biosensing platform. Biosens Bioelectron. 2010;26:1283–91. doi: 10.1016/j.bios.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Luchansky MS, Bailey RC. Silicon Photonic Microring Resonators for Quantitative Cytokine Detection and T-Cell Secretion Analysis. Anal Chem. 2010;82:1975–81. doi: 10.1021/ac902725q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Armani AM, Kulkarni RP, Fraser SE, Flagan RC, Vahala KJ. Label-free, single-molecule detection with optical microcavities. Science. 2007;5839:783–7. doi: 10.1126/science.1145002. [DOI] [PubMed] [Google Scholar]
- 249.Armani AM, Fraser SE. Label-free detection of cytokines using optical microcavities. Proc SPIE. 2008;6862:68620C/1–C/11. [Google Scholar]
- 250.Huckabay HA, Dunn RC. Whispering gallery mode imaging for the multiplexed detection of biomarkers. Sens Actuators, B. 2011;160:1262–7. [Google Scholar]
- 251.Huckabay HA, Wildgen SM, Dunn RC. Label-free detection of ovarian cancer biomarkers using whispering gallery mode imaging. Biosens Bioelectron. 2013;45:223–9. doi: 10.1016/j.bios.2013.01.072. [DOI] [PubMed] [Google Scholar]
- 252.Ruckstuhl T, Winterflood CM, Seeger S. Supercritical angle fluorescence immunoassay platform. Anal Chem. 2011;83:2345–50. doi: 10.1021/ac1032758. [DOI] [PubMed] [Google Scholar]
- 253.Carter JA, Mehta SD, Mungillo MV, Striemer CC, Miller BL. Analysis of inflammatory biomarkers by Arrayed Imaging Reflectometry. Biosens Bioelectron. 2011;26:3944–8. doi: 10.1016/j.bios.2011.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Kapoor R, Wang C-W. Highly specific detection of interleukin-6 (IL-6) protein using combination tapered fiber-optic biosensor dip-probe. Biosens Bioelectron. 2009;24:2696–701. doi: 10.1016/j.bios.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 255.Wang CW, Manne U, Reddy VB, Oelschlager DK, Katkoori VR, Grizzle WE, et al. Development of combination tapered fiber-optic biosensor dip probe for quantitative estimation of interleukin-6 in serum samples. J Biomed Opt. 2010;15:067005/1–/7. doi: 10.1117/1.3523368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Choi CJ, Belobraydich AR, Chan LL, Mathias PC, Cunningham BT. Comparison of label-free biosensing in microplate, microfluidic, and spot-based affinity capture assays. Anal Biochem. 2010;405:1–10. doi: 10.1016/j.ab.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 257.Mathias PC, Ganesh N, Cunningham BT. Application of Photonic Crystal Enhanced Fluorescence to a Cytokine Immunoassay. Anal Chem (Washington, DC, U S) 2008;80:9013–20. doi: 10.1021/ac801377k. [DOI] [PubMed] [Google Scholar]
- 258.Adalsteinsson V, Parajuli O, Kepics S, Gupta A, Reeves WB, Hahm J-i. Ultrasensitive Detection of Cytokines Enabled by Nanoscale ZnO Arrays. Anal Chem. 2008;80:6594–601. doi: 10.1021/ac800747q. [DOI] [PubMed] [Google Scholar]
- 259.Turner EH, Cohen D, Pugsley HR, Gonzalez GD, Whitmore CD, Zhu C, et al. Chemical cytometry: the chemical analysis of single cells. Anal Bioanal Chem. 2008;390:223–6. doi: 10.1007/s00216-007-1665-5. [DOI] [PubMed] [Google Scholar]
- 260.Borland LM, Kottegoda S, Phillips KS, Allbritton NL. Chemical analysis of single cells. Annu Rev Anal Chem. 2008;1:191–227. doi: 10.1146/annurev.anchem.1.031207.113100. [DOI] [PubMed] [Google Scholar]
- 261.Lin Y, Trouillon R, Safina G, Ewing AG. Chemical Analysis of Single Cells. Anal Chem. 2011;83:4369–92. doi: 10.1021/ac2009838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Szmacinski H, Toshchakov V, Piao W, Lakowicz JR. Imaging of Protein Secretion from a Single Cell Using Plasmonic Substrates. Bionanoscience. 2013;3:30–6. doi: 10.1007/s12668-013-0076-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Zhu H, Stybayeva G, Macal M, Ramanculov E, George MD, Dandekar S, et al. A microdevice for multiplexed detection of T-cell-secreted cytokines. Lab Chip. 2008;8:2197–205. doi: 10.1039/b810244a. [DOI] [PubMed] [Google Scholar]
- 264.Zhu H, Stybayeva G, Silangcruz J, Yan J, Ramanculov E, Dandekar S, et al. Detecting Cytokine Release from Single T-cells. Anal Chem (Washington, DC, U S) 2009;81:8150–6. doi: 10.1021/ac901390j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Han Q, Bradshaw EM, Nilsson B, Hafler DA, Love JC. Multidimensional analysis of the frequencies and rates of cytokine secretion from single cells by quantitative microengraving. Lab Chip. 2010;10:1391–400. doi: 10.1039/b926849a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Varadarajan N, Julg B, Yamanaka YJ, Chen H, Ogunniyi AO, McAndrew E, et al. A high-throughput single-cell analysis of human CD8+ T cell functions reveals discordance for cytokine secretion and cytolysis. J Clin Invest. 2011;121:4322–31. doi: 10.1172/JCI58653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Han Q, Bagheri N, Bradshaw EM, Hafler DA, Lauffenburger DA, Love JC. Polyfunctional responses by human T cells result from sequential release of cytokines. Proc Natl Acad Sci U S A. 2012;109:1607–12. S/1–S/12. doi: 10.1073/pnas.1117194109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Zhao W-A, Schafer S, Choi J-H, Yamanaka YJ, Lombardi ML, Bose S, et al. Cell-surface sensors for real-time probing of cellular environments. Nat Nanotechnol. 2011;6:524–31. doi: 10.1038/nnano.2011.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wu C, Li F, Niu G, Chen X. PET imaging of inflammation biomarkers. Theranostics. 2013;3:448–66. 19. doi: 10.7150/thno.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Signore A, Procaccini E, Annovazzi A, Chianelli M, van dLC, Mire-Sluis A. The developing role of cytokines for imaging inflammation and infection. Cytokine. 2000;12:1445–54. doi: 10.1006/cyto.2000.0746. [DOI] [PubMed] [Google Scholar]
- 271.Mather SJ, Chianelli M. Radiolabelled cytokines. Q J Nucl Med. 1996;40:290–300. [PubMed] [Google Scholar]
- 272.Signore A, Mather SJ, Piaggio G, Malviya G, Dierckx RA. Molecular imaging of inflammation/infection: Nuclear medicine and optical imaging agents and methods. Chem Rev. 2010;110:3112–45. doi: 10.1021/cr900351r. [DOI] [PubMed] [Google Scholar]
- 273.van der Laken CJ, Boerman OC, Oyen WJG, van de Ven MTP, van der Meer JWM, Corstens FHM. Imaging of infection in rabbits with radioiodinated interleukin-1 (α and β), its receptor antagonist and a chemotactic peptide: a comparative study. Eur J Nucl Med. 1998;25:347–52. doi: 10.1007/s002590050231. [DOI] [PubMed] [Google Scholar]
- 274.Liu Z, Barber C, Wan L, Liu S, Hui MM, Furenlid LR, et al. SPECT imaging of inflammatory response in ischemic-reperfused rat hearts using a 99mTc-labeled dual-domain cytokine ligand. J Nucl Med. 2013;54:2139–45. doi: 10.2967/jnumed.113.123497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Cawthorne C, Prenant C, Smigova A, Julyan P, Maroy R, Herholz K, et al. Biodistribution, pharmacokinetics and metabolism of interleukin-1 receptor antagonist (IL-1RA) using [18F]-IL1RA and PET imaging in rats. Br J Pharmacol. 2011;162:659–72. doi: 10.1111/j.1476-5381.2010.01068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Chamberland DL, Agarwal A, Kotov N, Fowlkes JB, Carson PL, Wang X. Photoacoustic tomography of joints aided by an etanercept-conjugated gold nanoparticle contrast agent-an ex vivo preliminary rat study. Nanotechnology. 2008;19:095101/1–/7. doi: 10.1088/0957-4484/19/9/095101. [DOI] [PubMed] [Google Scholar]
- 277.Agarwal A, Shao X, Rajian JR, Zhang H, Chamberland DL, Kotov NA, et al. Dual-mode imaging with radiolabeled gold nanorods. J Biomed Opt. 2011;16:051307/1–/7. doi: 10.1117/1.3580277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Fillmore HL, Shultz MD, Henderson SC, Cooper P, Broaddus WC, Chen ZJ, et al. Conjugation of functionalized gadolinium metallofullerenes with IL-13 peptides for targeting and imaging glial tumors. Nanomedicine (London, U K) 2011;6:449–58. doi: 10.2217/nnm.10.134. [DOI] [PubMed] [Google Scholar]
- 279.Westerink BHC, Cremers TIFH. Handbook of Microdialysis Sampling: Methods, Applications, and Clinical Aspects. Amsterdam: Academic Press; 2007. [Google Scholar]
- 280.Müller M. Microdialysis in Drug Development. In: Crommelin DJA, Lipper RA, editors. AAPS Advances in Pharmaceutical Sciences Series. New York: AAPS Springer; 2013. [Google Scholar]
- 281.Ao X, Stenken JA. Microdialysis sampling of cytokines. Methods. 2006;38:331–41. doi: 10.1016/j.ymeth.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 282.Clough GF, Stenken JA, Church MK. High Molecular Weight Targets and Treatments Using Microdialysis. In: Mueller M, editor. Microdialysis in Drug Development. New York: Springer; 2013. pp. 243–68. [Google Scholar]
- 283.Vasicek TW, Jackson MR, Poseno TM, Stenken JA. In Vivo Microdialysis Sampling of Cytokines from Rat Hippocampus: Comparison of Cannula Implantation Procedures. ACS Chem Neurosci. 2013;4:737–46. doi: 10.1021/cn400025m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Clough GF, Jackson CL, Lee JJ, Jamal SC, Church MK. What can microdialysis tell us about the temporal and spatial generation of cytokines in allergen-induced responses in human skin in vivo? J Invest Dermatol. 2007;127:2799–806. doi: 10.1038/sj.jid.5700930. [DOI] [PubMed] [Google Scholar]
- 285.Helmy A, Carpenter KLH, Menon DK, Pickard JD, Hutchinson PJA. The cytokine response to human traumatic brain injury: temporal profiles and evidence for cerebral parenchymal production. J Cereb Blood Flow Metab. 2011;31:658–70. doi: 10.1038/jcbfm.2010.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Clough GF. Microdialysis of large molecules. AAPS Journal. 2005;7:E686–E92. doi: 10.1208/aapsj070369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Helmy A, Carpenter Keri LH, Skepper Jeremy N, Kirkpatrick Peter J, Pickard John D, Hutchinson Peter J. Microdialysis of cytokines: methodological considerations, scanning electron microscopy, and determination of relative recovery. Journal of Neurotrauma. 2009;26:549–61. doi: 10.1089/neu.2008.0719. [DOI] [PubMed] [Google Scholar]
- 288.Hillman J, Aneman O, Anderson C, Sjogren F, Saberg C, Mellergard P. A microdialysis technique for routine measurement of macromolecules in the injured human brain. Neurosurgery. 2005;56:1264–8. doi: 10.1227/01.neu.0000159711.93592.8d. discussion 8–70. [DOI] [PubMed] [Google Scholar]