Abstract
In the field of food and nutrition, complex natural products (NPs) are typically obtained from cells/tissues of diverse organisms such as plants, mushrooms, and animals. Among them, edible fruits, grains, and vegetables represent most of the human diet. Because of an important dietary dependence, the comprehensive metabolomic analysis of dietary NPs, performed holistically via the assessment of as many metabolites as possible, constitutes a fundamental building block for understanding the human diet. Both mass spectrometry (MS) and nuclear magnetic resonance (NMR) are important complementary analytic techniques, covering a wide range of metabolites at different concentrations. Particularly, 1-dimensional 1H-NMR offers an unbiased overview of all metabolites present in a sample without prior knowledge of its composition, thereby leading to an untargeted analysis. In the past decade, NMR-based metabolomics in plant and food analyses has evolved considerably. The scope of the present review, covering literature of the past 5 y, is to address the relevance of 1H-NMR–based metabolomics in food plant studies, including a comparison with MS-based techniques. Major applications of NMR-based metabolomics for the quality control of dietary NPs and assessment of their nutritional values are presented.
Keywords: metabolomics, nuclear magnetic resonance, food plants, natural products, mass spectrometry, quality control, nutritional value, food safety
Introduction
Natural products (NPs)7 comprise all chemicals or metabolites produced by a living organism, constituting an organism’s metabolome. In food analysis and nutrition, NPs are complex metabolic mixtures that originate from various sources such as microorganisms, plants, animals, and humans. Analyses dedicated to the comprehensive characterization of complex mixtures through the identification and quantification of as many metabolites as possible are referred to as metabolomics. The primary goal of metabolomics is to gain a bird’s-eye view of cell/tissue/organ composition at a specific time point to understand the molecular signature of macroscopic biological influences (1–4). Three types of analytic techniques are used in metabolomics: vibrational spectroscopy such as infrared and Raman spectroscopy, NMR spectroscopy, and MS (5–8). Taken individually, none of these techniques meets the requirements for the measurement of all metabolites present in a sample. Comprehensive metabolomic analysis can only be obtained by merging data from these different platforms. Both MS and NMR are recognized as the most important complementary techniques that cover a wide range of metabolites at different concentrations (6–8). The former determines m/z of molecular ions, their fragmentation patterns, and the relative ionic intensities of the metabolites, generally after LC or GC. NMR measures the resonance frequency of the metabolites’ nuclei, such as 1H and 31P, under the influence of a magnetic field. Particularly, NMR-based metabolomics favors the measurement of protons (1H), owing to its relatively high sensitivity, natural abundance, and nearly ubiquitous presence in organic compounds (9–11). The analysis of 1-dimensional (1D) 1H-NMR spectra acquired under quantitative conditions (qHNMR) enables the structural identification and subsequent quantification of several metabolites present in a sample (12–15). From a nomenclature point of view, the term metabolite fingerprinting was preferred when spectroscopic techniques, such as NMR, were used, whereas metabolite profiling was mostly associated with LC-based methods. Now, the border has become blurred, and both terms are used interchangeably in the literature, together with the terms metabolomics and metabonomics (1, 2, 5, 10).
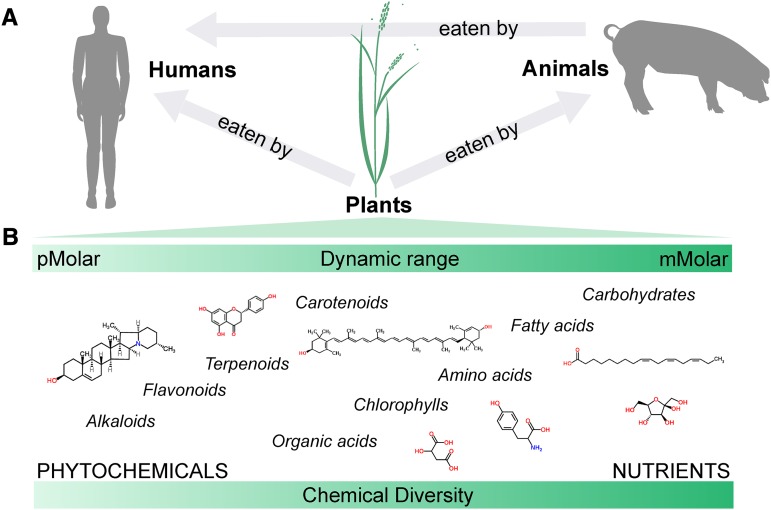
In nutrition research, metabolomics aims to understand the effects of diet on human health. Because the diet clearly affects the human metabolome, nutritional investigations usually require the analysis of metabolomes of both the food and the consuming animal/human (Figure 1A). A comprehensive study of the food metabolome is necessary to generate relevant hypotheses that relate the effects of the diet on humans (16, 17). In line with this consideration, the recently introduced concept of foodomics integrates multiple omics techniques to describe the food composition with the same high standards generally reserved for the analyses of the human organism (18–20). Moreover, knowledge of the plant metabolome is fundamental for an accurate understanding of the human diet, considering that edible fruits, grains, and vegetables represent most of the consumed food (21). The holistic study of the plant metabolome faces several challenges linked to the chemical diversity (nutrients and phytochemicals), varying physicochemical properties of plant-derived metabolites, and the accommodation of their large dynamic ranges in terms of concentration (pM to mM; Figure 1B) (3).
FIGURE 1.
Dietary dependence (A) and analytic challenges (B) of the plant metabolome.
Food analysis and nutritional research have used metabolomic tools for ∼15 y. The implementation of metabolomics has increased since 2010, supported by the improvement of analytic instruments, statistical software, and accessible databases. Review articles already cover the experimental designs and methodologies for 1H-NMR–based (9, 10, 22) and MS-based (23) metabolomics. The complex and large data sets obtained from the various analytic platforms require data mining resources, including bioinformatics and multivariate data analysis (MVDA), to extract meaningful information. To date, the most common MVDA approaches for untargeted analysis are principal component analysis and hierarchical clustering analysis. Details on data preparation and processing with the use of chemometrics and bioinformatics were also thoroughly reviewed (24–27).
In recent literature, MS is more abundantly applied to metabolomics than NMR. Therefore, the present review focuses on the role and relevance of NMR-based untargeted metabolomics in food plant analysis. A metabolomic workflow is proposed on the basis of NMR’s complementarity with MS-based techniques. The surveyed literature covers diverse applications of 1H-NMR–based metabolomics, from the assessment of nutritional properties, to quality controls (QCs), and the determination of organoleptic quality of edible plants before human metabolism.
Use of 1D 1H-NMR as a First-Pass Screen for Metabolomics
NMR-based metabolomics in plant (5, 28) and food analyses has matured considerably (29, 30). This section addresses the main characteristics of NMR-based metabolomics from sample preparation to metabolite identification and quantification, while highlighting its complementarity with MS-based techniques (Table 1) (26, 31–33).
TABLE 1.
Comparison of NMR and hyphenated MS systems in metabolomics1
| 1H NMR | LC-MS | GC-MS | |
| Sample preparation | Extraction required for solid samples | Preparation and/or extraction required for both liquid and solid samples | |
| Derivatization required for polar metabolites | |||
| Direct analysis | Liquid samples and HR-MAS for tissues and cells | DART or DESI-MS for tissues and cells | |
| Recovery | Sample recovery | Sample destruction | |
| Detected metabolites | Detection of 1H frequencies under a magnetic field | Detection based on structure polarity and ionizability | |
| Ionization: ESI/APCI | Ionization: EI, CI | ||
| All structural types and polarity | Polar to medium polar | Volatile, thermostable | |
| Identification | Chemical shifts, coupling constants, multiplicity | Retention time, m/z, fragmentation pattern | Kovats Index, m/z, EI, fragmentation pattern |
| Two-dimensional data | Need for authentic standards | ||
| Few databases | Various databases | Rich databases | |
| Identification limitations | Variability of NMR data because of solvent effects | Variability of non-EI data | Derivatization often required |
| Potential adduct formation | Potential formation of artifacts | ||
| MW range (<1000 Da) | |||
| Lack of database information for rare phytochemicals | Lack of standards for rare phytochemicals | ||
| Sensitivity | Low μM | pM, low nM | |
| Favorable identification of most abundant metabolites | Favorable identification of trace metabolites | ||
| Detection of numerous nonidentifiable compounds | |||
| Specificity | Less than LC/GC-based techniques because of signal overlap | Greater than NMR because of chromatographic separation, ion selection, and fragmentation patterns | |
| Reproducibility robustness | Higher cross-laboratory and cross-platform reproducibility | Lower because of the inherent variations of the LC/GC systems associated with each part of the hyphenation | |
| Quantification | Direct proportionality | Semiquantitative, in certain cases, based on molecular ion relative intensity | |
| Various calibrations possible | |||
| No need for structurally identical reference standard | Need for structurally identical reference standards for calibration and accurate quantification | ||
| Metabolomic approach | High-throughput first-pass screen | Lower throughput: sample preparation and calibration | |
| Fingerprinting | Profiling | ||
| Untargeted/unbiased overview | Targeted analysis, hypothesis driven | ||
APCI, atmospheric pressure chemical ionization; CI, chemical ionization; DART, direct analysis in real time; DESI, desorption electrospray ionization; EI, electron impact; ESI, electrospray ionization; HR-MAS, high-resolution magic angle spinning; MW, molecular weight.
Sample preparation and representativeness
Producing samples that are compatible with the analytic platform with minimal loss and without adulteration is essential in metabolomics (34). The first potential bias in the metabolomic workflow is linked to the extraction process of solid samples. The conditions of extraction affect the sample composition, and, thus, the range of metabolites that can be detected (30, 34). To avoid these effects, high-resolution magic angle spinning-NMR can be used to analyze freeze-dried tissues or cells dissolved in buffered deuterium oxide before acquisition. The spectra are acquired with the use of the 1D nuclear Overhauser effect with presaturation to remove the signals of residual water and proteins, thereby leading to high-quality, solution-like 1H spectra (35–38). Developments in MS systems, such as desorption electrospray ionization (DESI) and direct analysis in real time, also allow the evaluation of biological samples without pretreatment and chromatographic separation (32). These direct measurements shorten the acquisition and preparation time, but their applications are limited because of the overlap of small signals by larger ones during detection and the occurrence of ion suppression (2).
For liquid samples such as biological fluids (30), beverages, or edible oils, NMR is highly suited. These samples are directly prepared in NMR tubes, buffered, and diluted with deuterated solvents before analysis (9, 30). In contrast, most of the MS-based methods require sample cleanup procedures before analysis (Table 1). Any sample preparation step involves manipulation of the number, identity, and abundance of detected metabolites, potentially biasing the final results. Hence, one of the important advantages of NMR is its minimal sample manipulation, thereby providing more authenticity and less opportunity for human error (4).
Reproducibility and robustness
The fundamental variables that modulate an NMR spectrum are the solvent, the magnetic field strength, and the pulse sequence. When these parameters are kept constant, 1H-NMR results obtained under quantitative conditions are highly reproducible (12, 15, 30, 39). Because of minimal sample preparation, a high-sample throughput with little instrumental drift is achievable, making 1H-NMR suitable for rapid first-pass screening. Markus et al. (40) recently reported the robustness of 1H-NMR analysis for Vaccinium extracts acquired at different sites; although field strength, console, probe, sample diameter, and software differed, the results were found to cluster tightly together in principal component analysis score plots. As stated by the investigators, “material validation … is possible using NMR on spectrometers of various configurations and architecture” (40). Comparatively, chromatographic separations cause the most critical source of variation and experimental errors in LC/GC-MS (1, 11, 23). Altogether, the robustness of NMR and the reproducibility of the obtained results promote the comparability and exchange of data.
Metabolite identification and databases
The identification of metabolites related to certain biological effects is fundamental in metabolomics. Measuring the variation of certain proton signal intensities between samples and connecting them with defined structural characteristics can be challenging. Powerful MVDA is required to interrogate the data and to identify key MS or NMR signals associated with the analysis (24–27).
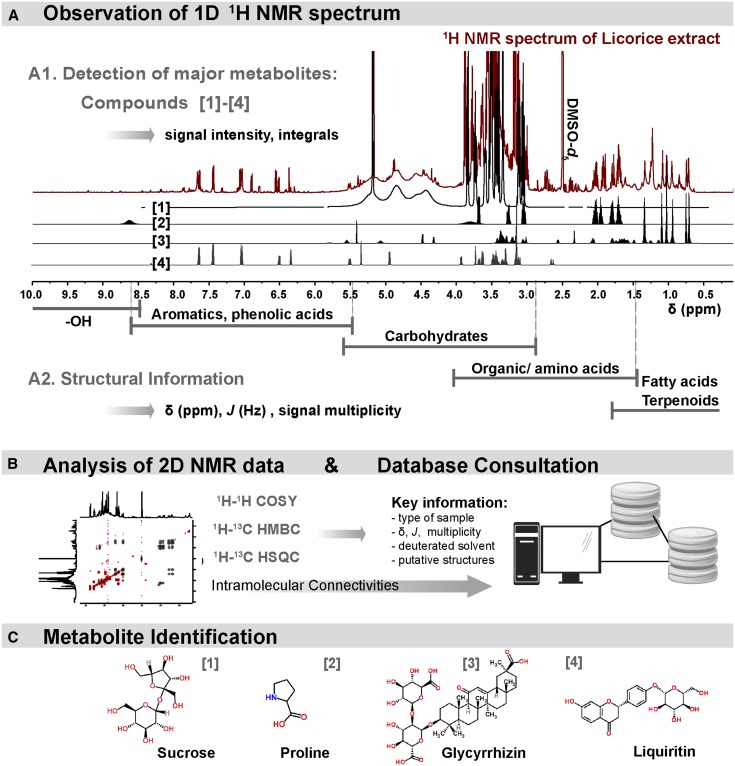
The 1H-NMR spectrum of a complex mixture represents the superposition of spectra from all individual metabolites present in the sample at their relative abundance (Figure 2A). Hence, the spectrum of a mixture accounts for the full set of metabolites, but only the most abundant (in μM to mM) can be easily identified and quantified (4, 11). The metabolic complexity of NP mixtures generates a large number of 1H signals from diverse structures, all located in a narrow chemical shift window (0–15 ppm), hindering metabolite identification and quantification because of signal overlap. Identification requires careful analysis of proton resonances throughout the 1H-NMR spectrum, acquisition of various 2-dimensional spectra (42), and consultation of databases (26, 31) (Figure 2). Merging all of these data sets leads to the documentation of molecular/structural connections, thereby constituting evidence for the characterization of abundant metabolites.
FIGURE 2.
Key steps for the NMR identification of metabolites in complex mixtures. (A) The interpretation of the 1D 1H resonances in terms of chemical shifts (δ in ppm), multiplicity, coupling constants (J in Hz), and intensities leads to the determination of the relative molar concentrations (A1) and structural classes (A2) of the most abundant metabolites. (B) The acquisition of 2D NMR data enables the determination of structural connections. (C) The consultation of databases guides the analysts in the identification of known metabolites. The entire process is illustrated with metabolomic fingerprinting of licorice extract (41), leading to the simultaneous identification/quantification of major nutrients (sucrose [1], proline [2]) and phytochemicals (glycyrrhizin [3] and liquiritin [4]). COSY, correlation spectroscopy; HMBC, heteronuclear multiple-bond correlation spectroscopy; HSQC, heteronuclear single quantum coherence spectroscopy; 1D, 1-dimensional; 2D, 2-dimensional.
To promote metabolite identification, a recently developed fast NMR method involves the acquisition of three 2-dimensional NMR spectra (43). To improve the selectivity of 1H-NMR measurements specific pulse sequences (e.g., J-resolved experiments, diffusion-ordered spectroscopy) (10, 20, 42) can be implemented. Computer-assisted deconvolution algorithms of processed 1H-NMR spectra can also facilitate the identification of metabolites (44, 45). The use of cryogenic and microcoil probes and the implementation of polarization methods such as Dynamic Nuclear Polarization leads to an increase in the sensitivity of NMR measurements (39, 46). However, Dynamic Nuclear Polarization has not been fully implemented in metabolomic analysis (15, 20–22, 34).
The availability of databases, such as the Human Metabolome Database or the Biological Magnetic Resonance Data Bank, has fostered the identification of metabolites by NMR. Lists and descriptions of NMR/MS databases for structural identification/dereplication are available in reviews (26, 31). Most of these collections enable the identification of nutrients, often designated as primary metabolites (1°Ms), and ubiquitous phytochemicals such as quercetin or β-sitosterol. The NMR spectra in these databases were mainly acquired in deuterated water, methanol, or chloroform. Because complex plant samples usually contain a wide range of structurally diverse chemicals, these solvents cannot cover the broad polarity range of a true metabolomic window. Hence, the use of a more universal solvent, covering a wide range of polarity, such as deuterated dimethyl sulfoxide, could improve the utility of NMR databases. Moreover, the identification of less common metabolites, particularly rare phytochemicals, remains an intricate task, because their spectra are usually not freely available (Table 1) (28, 34).
In plants, 1°Ms are the most abundant metabolites and, thus, are easily identified by NMR. Accordingly, most food plant NMR-based metabolomic studies (see next section) focus on the 1°M composition. Generally, phytochemicals are less concentrated than the designated 1°Ms, explaining the popularity and use of MS in metabolomics.
Hyphenated MS systems are capable of identifying, but not necessarily quantifying, several thousand metabolites in biological samples. The limit for detecting metabolites is reliant on their concentrations and nonoptimal performances in chromatographic systems and MS detection (e.g., poor retention, insufficient ionization). Depending on the metabolite structures, polarities, and type of desired analysis (quantitative or not), different MS systems can be used. Thanks to their sensitivity, triple-quadrupole mass spectrometers are good platforms for targeted quantitative analysis. With their high resolution and ability to detect multiple ions simultaneously, Time-of-Flight, Orbitrap, and Fourier transform ion cyclotron resonance mass spectrometers are more suitable for broader metabolomic screening (7, 16, 26). In these systems, soft ionization methods such as electrospray ionization (ESI) and atmospheric pressure chemical ionization are commonly used. For the identification of nonpolar or derivatized volatile thermostable metabolites, GC-MS systems with hard ionization methods, such as electron impact or chemical ionization, are preferred. To increase the ionization of diverse metabolites and to enhance the metabolite coverage, dual-source MS instruments, combining atmospheric pressure chemical ionization and ESI for LC systems (47) and soft and hard ionization for GC systems (48), can be used.
In MS systems, metabolite identification relies on the retention time, abundance of molecular ions, and fragmentation patterns, which altogether will be searched in available databases (6, 26). Particularly, GC-MS spectral libraries are highly suitable for metabolite identification because results (e.g., Kovats indexes and fragmentation patterns) obtained from different GC-MS instruments are highly consistent (1, 2, 31). However, LC-MS and MS/MS results are subject to variation, because of the component nature and properties of LC systems and the lack of consistent fragmentation and ionization across different MS platforms. Despite the existence of various MS databases, robust compound identification and quantification still relies on direct comparison with identical reference compounds.
Unique characteristics of qHNMR
NMR has unique quantitative capabilities, provided that appropriate acquisition and processing conditions are used (12, 39, 49). The fundamental characteristic of qHNMR is that the 1H signal intensity is directly proportional to the number of nuclei responsible for that particular resonance (I = kN). In a mixture, the normalized areas of 1H signals are directly proportional to the molar concentrations of the components. NMR can use different calibration methods, which were shown to be equally suitable for determining the constant k. Calibration can use internal or external reference compounds (synonym: calibrants), implement an electronic reference, or under certain circumstances simply normalize the full spectrum, as reviewed elsewhere (12, 39, 50, 51).
NMR quantification requires no measurement of specific response factors for each metabolite, which is compulsory for LC/GC quantification. Hence, another important distinction from LC-based calibration concepts is that the calibrant does not need to be structurally identical to the metabolite of interest (Table 1) and can be any suitable compound of known purity (50, 51). Another advantage of qHNMR is its ability to quantify simultaneously, in a single experiment, several metabolites, including rare and not commercially available phytochemicals. Performing quantification by MS usually limits the number of measurable compounds because of the requirement for individual calibration curves.
The inherent signal overlap in metabolomic qHNMR spectra generally requires the implementation of computer-assisted deconvolution for the accurate measurement of integrals. There are mainly two types of deconvolution methods: 1) linear deconvolution, including those that use Bayesian models (27, 45, 52), and 2) quantum-mechanical deconvolution that use the proton NMR variables or spin systems, referred to as 1H iterative full spin analysis and/or quantitative quantum mechanical spectral analysis (10, 13, 44).
Place of 1H NMR in the metabolomic workflow
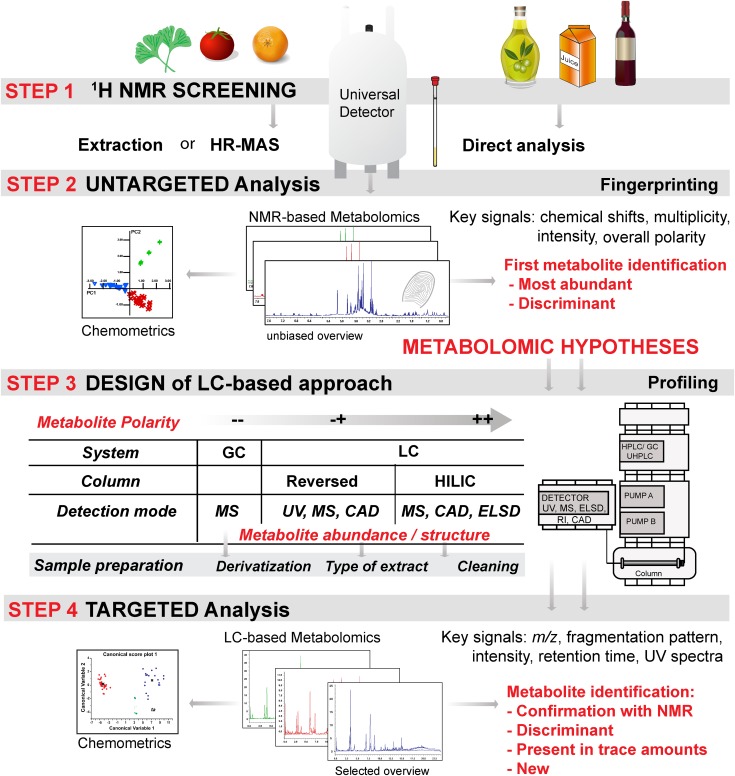
As stated by McGhie et al. (16), when designing a metabolomic experiment, “analytical options are often (but not ideally) selected based on underlying assumptions about the types of metabolites that are of interest to the experimenter.” NMR, however, does not require underlying assumptions of the sample composition. NMR offers a comprehensive unbiased overview of all metabolites in the sample without destruction, making it highly suitable for first-pass untargeted metabolomic screening (Figure 3). The information acquired through the interpretation of 1H-NMR spectra (e.g., putative identity and relative concentrations of key metabolites) can help the analyst in developing or refining chemical hypotheses and in designing appropriate MS-based targeted analyses (1).
FIGURE 3.
1H NMR as a first-pass metabolomic screening. The 4 steps of the metabolomic workflow show the analytic progression, starting with a sample of unknown composition and leading to its metabolomic description. In step 2, results obtained from NMR-based metabolomics assist in the generation of chemical hypotheses, enabling the design of LC-based analyses. In step 3, the use of specific detection methods, hyphenated with chromatographic systems, enhances the detection specificity for certain types of metabolites according to their physical properties (11). CAD, charged aerosol detector; ELSD, evaporating light-scattering detection; HR-MAS, high-resolution magic angle spinning; RI, refractive index; UHPLC, ultra-HPLC.
Subsequently, the choice of orthogonal analytic methods will depend on the nature of the samples, the structural characteristics (e.g., polarity and volatility) of the metabolites, and the choice of targeted or untargeted analysis (Figure 3) (53). In LC or GC systems, the metabolic signature depends on the type of separation and detection, making unbiased metabolomic analysis problematic.
LC- or GC-MS are well suited for targeted metabolomic studies that require the measurement of selected metabolites with a high level of precision and accuracy (26). The combination of analytic techniques is finally determined by the overall objectives related to either the accurate quantification of selected metabolites, the detection of unusual/discriminant metabolites, or the comprehensive overview of the metabolite composition. The use of both NMR and MS-based metabolomics, in a strategic manner, can increase the number of identified metabolites and provide a more comprehensive, accurate biochemical understanding of the samples under investigation (4, 10).
NMR-Based Metabolomics Can Assess the Quality and Nutritional Value of Edible Plants
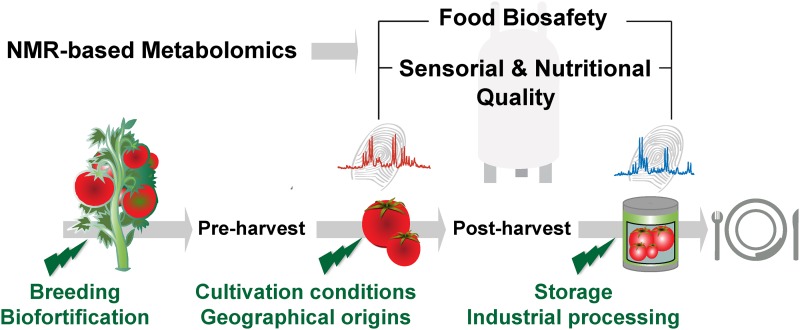
The quality of dietary plant products is greatly affected by variables before (genetic and botanical origins, cultivation conditions, type of soil, climate) and after (milling, storage conditions, industrial processing) harvest (Figure 4). Various metabolomic techniques can be applied to understand the plant’s biochemical responses to these diverse conditions. High-field (400–600 MHz), liquid state 1H-NMR–based metabolomics has been used increasingly in food analyses to assess the overall nutritional quality and authenticity of a large number of fruits, vegetables, and plant-derived food products. This includes tomatoes, potatoes, various fruits and their juices, lettuce, tea, wines and beers, honeys, and edible oils (9, 29). Complementing the 2 reviews published in 2012 (9) and 2013 (29), the following sections address the main applications of NMR-based metabolomics for the evaluation of cultivation conditions of various crops, the assessment of their nutritional qualities, the determination of authenticity and QC, and the evaluation of organoleptic properties of food plants.
FIGURE 4.
Applications of NMR-based metabolomics for food plant analysis. 1H NMR-based metabolomics can evaluate the effects of crop breeding, cultivation, geographical origins, and industrial processing on nutritional composition and organoleptic properties of food plants. By offering an unbiased metabolomic overview NMR has a fundamental place in food biosafety and for the determination of the metabolomic equivalence.
Agricultural optimization and nutritionally improved food
The nutritional quality and safety of crops is a direct function of metabolite content. Biofortification and breeding methods may alter the nutritional content of a crop and affect its taste, texture, aroma, shelf-life, and even its safety (31). Therefore, the identification of molecular markers can be useful to guide the development of crop plants and crop-derived foods (54, 55). Metabolomics establishes a link between genotype and phenotype to assist plant breeders in producing crops of high quality and high yield (3). In this regard, 1H-NMR–based metabolomic analysis was applied to map the carrot metabolome and to identify metabolomic differences between varieties mainly based on their 1°M composition (carbohydrates, lipids, and amino acids) and β-carotene content (54). Collectively, the difference in lipid and amino acid composition was suggested to be responsible for the sensory properties of the carrot varieties. 1H-NMR–based metabolomics favors the understanding of the effects of cultivation conditions on the metabolite (mainly 1°Ms/nutrients) profiles of oilseed and turnip rape (55) and of sea buckthorn berries (56) and mandarin oranges (57). 1H-NMR fingerprinting was used to characterize the metabolite composition associated with resistance to fruit fly attacks in 2 varieties of peaches (58) and revealed that the more resistant variety had a lower concentration of the flavor precursors valine and isoleucine, both considered highly attractive to the fruit fly. The 1°M composition of tomatoes is regarded as most responsible for the final taste and, thus, was monitored during fruit development, breeding, and as a function of cultivation conditions (59–61). For example, differences in the lipophilic fractions (e.g., tocopherol, unsaturated lipids, phospholipids, and chlorophylls) were observed between 2 varieties of cherry tomatoes as a function of harvest time (59), and γ-aminobutyric acid was identified as a marker of ripening transition (60). Efforts toward a better annotation of the rice metabolome combined MS and NMR analyses to identify 36 specialized metabolites in the leaves, including new flavonoids and flavonolignan isomers (62).
The development of genetically modified (GM) plants with improved nutritional and sensorial/organoleptic characteristics and better agronomic properties has raised safety concerns about the effects of such transgenic crops on both human and animal health. Challenges in food biosafety rely on the adequate monitoring of possible unintended biochemical changes to assess the equivalence between GM crops and their wild-type parents (9, 63). Maize, various cereals such as wheat and rice (62), soybean (64), tomatoes, and potatoes (31, 65) are the most-studied GM plants. In food biosafety, untargeted and unbiased metabolomic analyses that combine MS and NMR are preferred to determine unexpected metabolic effects of genetic transformation. It was found repeatedly that the most important sources of metabolite variation were environmental and cultivation conditions rather than genetic modifications (3, 63–65). Thorough determination of substantial equivalence requires the integration of metabolomic information with data generated by other -omics platforms, such as proteomics and transcriptomics, as proposed in the foodomics concept (18, 19, 66).
Quality assessment and adulteration
The geographical origins of edible plants give an indication of both quality and phytochemical composition, which may collectively denote unique nutritional and organoleptic properties (16, 67). Recent metabolomic studies have focused on the evaluation of food plant composition as a function of their regional provenance, comprising the influences of climate, soil, and cultivation conditions. Through the application of various labels [e.g., protected geographical indications (PGIs)], the European Union valorizes regional provenances to protect unique farming systems. With the benefit from their protective labels, the resulting products have higher prices, triggering fraudulent activities through adulteration (68). General market globalization has also led to the occurrence of food safety issues. Hence, for obvious economic and safety concerns, the ability to trace and authenticate the origin of food products is of major interest in the industry (67, 68). Similarly, in the area of herbal medicines, metabolomics can assist with botanical characterization, chemical standardization, and traceability of geographical origin, collectively constituting the quality and safety aspects of herbal products (5, 69, 70). In addition, fraudulent products can be spiked with individual metabolites to appear authentic in a targeted analysis (5). 1H-NMR metabolite fingerprinting offers an untargeted overview of the sample composition and, thus, may be more suitable to identify subtle differences between authentic and fraudulent products.
Publications on NMR-based metabolomics for QC and authentication of geographical origins were found mainly for the following dietary products: olive oil (71, 72), wines/grapes (73), spices such as saffron (74), tea (75), honey (76), and nuts such as hazelnuts (77) and pistachios (78, 79). Relating to the importance of European protective labels, 1H-NMR–based metabolomics has proved useful in the determination of the 1°M signature to evaluate the traceability of PGI compared with non-PGI chicory (35) and to promote the authentication of Italian sweet peppers (37) from different cultivars and geographical origins.
According to general public opinion, organic foods are regarded as healthier than conventional ones, although little scientific evidence can currently support this consideration (19, 36, 72, 80). Recently, 1H-NMR fingerprinting combined with MVDA was used to evaluate major metabolomic differences between conventionally and organically grown vegetables such as tomatoes (80) and potatoes (36). In both cases, organic and conventionally farmed vegetables could be distinguished by the unique pattern of their 1°M fingerprints, which could potentially be used to facilitate the authentication and traceability of farming systems.
Industrial processing
Untargeted NMR-based metabolomics is useful for monitoring changes in food composition during processing such as fermentation, thermal and mechanical processing, and ripening (81–83). Maintaining the balance between nutritional and sensorial quality of food is one of the main challenges in the food industry. For these purposes, changes in the composition of 1°Ms are usually investigated by 1H-NMR metabolomics, mainly because of the quantitative abundance of 1°Ms and available database information. As highlighted in recent articles (73, 81–83), 1H-NMR–based metabolomics can assist food industries in optimizing and designing various processing steps for the preservation of the product qualities. This approach was applied to monitor and understand metabolic changes during the production of black garlic in which a thermal process leads to the formation of new constituents (e.g., pyroglutamic acids and cycloalliin) and modifications in the amino acid, sugar, and organic acid content (81). 1H-NMR fingerprinting was applied to assess the effect of industrial processing on the polar metabolite composition of vegetable purees (82) and wines (73). The lipid and flavonoid composition of olive paste and pomace obtained during the production of extra virgin olive oil were also monitored by 1H-NMR, revealing that both paste and pomace had a higher content in phenolic compounds than the oil (83).
Magnetic tongue and sensory analyses
The organoleptic properties of edible plants and derived products were subject to new types of 1H-NMR investigations through the concept of the magnetic tongue. This idea was introduced by Malmendal et al. (84) who analyzed 18 canned tomato products and reported that 1H-NMR fingerprinting could be used to determine the specific 1°M composition linked with characteristic sensory descriptors, including saltiness, tomato, metallic taste, and density. The concept was subsequently adapted to evaluate the aromatic and flavor qualities of other plant products such as olive oil (85), roasted coffee beans (86), and fresh melons (87).
Conclusions
NMR spectroscopy, particularly 1H-NMR, offers an unbiased view of sample composition without chromatographic separation, allowing the simultaneous identification and quantification of diverse metabolites, in one experiment, while avoiding sample destruction. Both liquid and HR-MAS 1H-NMR require minimal sample preparation while giving reproducible results, which is of paramount importance for the application of MVDA and the exchange of information between platforms. Although NMR-based metabolomics has matured considerably in the past decade, it continues to face challenges related to its lower sensitivity and selectivity/specificity, compared with MS. These limitations are overcome by improved NMR technologies and through the desirable use of orthogonal MS techniques. Approaching a holistic analysis of complex NPs requires the strategic combination of complementary techniques and efficient data management to reach a comprehensive understanding of biological systems. To this end, NMR is an important, unbiased technique in the metabolomic toolset.
The objective of metabolomics in food/diet analyses is to gain additional insight into food composition in both quality and nutritional contexts to better understand the interactions between metabolites found in the diet and humans (3, 16). Food plant metabolomics constitutes a building block of dietetic investigations. NMR and MS-based metabolomics are fundamental tools to assess the agricultural influences on the plant metabolite composition, thus guiding efforts toward the improvement of nutritional and flavor qualities of edible plants (3, 88). N MR-based metabolomic studies were found to assist researchers in the comprehensive determination of food quality, authenticity, and biosafety. However, such methods are not yet validated by the regulatory agencies.
The standardization of metabolomic methods is essential for the development of databases that foster a rapid identification of key metabolites (6, 26, 30, 68). Attempts to harmonize metabolomic studies supported by the Metabolomics Standards Initiative (89) have enhanced the transparency of results and facilitated the exchange of information. The use of NMR for metabolomics will greatly benefit from the development of freely available databases that contain raw data and full NMR spectra of a wide variety of metabolites found in the diet, including 1°Ms, as well as rare and ubiquitous phytochemicals (10).
The holistic analysis of dietary NPs involves the application of various skills and knowledge, from analytic chemistry to statistics and biology. Mastering the multiple challenges of food analysis in nutritional investigations requires multidisciplinary collaboration with NP chemists, plant biologists, food analysts, nutritionists, and statisticians. To gain a deeper biochemical understanding, the resulting metabolomic data sets can be combined with information from different -omics platforms (24, 90).
Acknowledgments
All authors read and approved the final manuscript.
Footnotes
Abbreviations used: DESI, desorption electrospray ionization; GM, genetically modified; MVDA, multivariate data analysis; NP, natural product; PGI, protected geographical indication; QC, quality control; qHNMR, quantitative 1H NMR; 1D, 1-dimensional; 1°M, primary metabolite.
References
- 1.Holmes E, Tang H, Wang Y, Seger C. The assessment of plant metabolite profiles by NMR-based methodologies. Planta Med 2006;72:771–85. [DOI] [PubMed] [Google Scholar]
- 2.Hegeman AD. Plant metabolomics--meeting the analytical challenges of comprehensive metabolite analysis. Brief Funct Genomics 2010;9:139–48. [DOI] [PubMed] [Google Scholar]
- 3.Hall RD, Brouwer ID, Fitzgerald MA. Plant metabolomics and its potential application for human nutrition. Physiol Plant 2008;132:162–75. [DOI] [PubMed] [Google Scholar]
- 4.Zulyniak MA, Mutch DM. Harnessing metabolomics for nutrition research. Curr Pharm Biotechnol 2011;12:1005–15. [DOI] [PubMed] [Google Scholar]
- 5.Bilia AR. Science meets regulation. J Ethnopharmacol 2014;158:487–94. [DOI] [PubMed] [Google Scholar]
- 6.Scalbert A, Brennan L, Manach C, Andres-Lacueva C, Dragsted LO, Draper J, Rappaport SM, Van Der Hooft JJ, Wishart DS. The food metabolome: a window over dietary exposure. Am J Clin Nutr 2014;99:1286–308. [DOI] [PubMed] [Google Scholar]
- 7.Zhang A, Sun H, Wang P, Han Y, Wang X. Modern analytical techniques in metabolomics analysis. Analyst 2012;137:293–300. [DOI] [PubMed] [Google Scholar]
- 8.Brennan L. Metabolomics in nutrition research: current status and perspectives. Biochem Soc Trans 2013;41:670–3. [DOI] [PubMed] [Google Scholar]
- 9.Mannina L, Sobolev AP, Viel S. Liquid state 1H high field NMR in food analysis. Prog Nucl Magn Reson Spectrosc 2012;66:1–39. [DOI] [PubMed] [Google Scholar]
- 10.Larive CK, Barding GA, Dinges MM. NMR spectroscopy for metabolomics and metabolic profiling. Anal Chem 2015;87:133–46. [DOI] [PubMed] [Google Scholar]
- 11.Simmler C, Napolitano JG, McAlpine JB, Chen S-N, Pauli GF. Universal quantitative NMR analysis of complex natural samples. Curr Opin Biotechnol 2014;25:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pauli GF, Gödecke T, Jaki BU, Lankin DC. Quantitative 1H NMR. Development and potential of an analytical method: an update. J Nat Prod 2012;75:834–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Napolitano JG, Lankin DC, Chen S-N, Pauli GF. Complete 1H NMR spectral analysis of ten chemical markers of Ginkgo biloba. Magn Reson Chem 2012;50:569–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohmann M, Koospal V, Bauer-Christoph C, Christoph N, Wachter H, Diehl B, Holzgrabe U. Quantitative 1H NMR analysis of egg yolk, alcohol, and total sugar content in egg liqueurs. J Agric Food Chem 2015;63:4112–9. [DOI] [PubMed] [Google Scholar]
- 15.Barding GA, Salditos R, Larive CK. Quantitative NMR for bioanalysis and metabolomics. Anal Bioanal Chem 2012;404:1165–79. [DOI] [PubMed] [Google Scholar]
- 16.McGhie TK, Rowan DD. Metabolomics for measuring phytochemicals, and assessing human and animal responses to phytochemicals, in food science. Mol Nutr Food Res 2012;56:147–58. [DOI] [PubMed] [Google Scholar]
- 17.Wang L, Chen C. Emerging applications of metabolomics in studying chemopreventive phytochemicals. AAPS J 2013;15:941–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garcia-Cañas V, Simó C, Herrero M, Ibáñez E, Cifuentes A. Present and future challenges in food analysis: foodomics. Anal Chem 2012;84:10150–9. [DOI] [PubMed] [Google Scholar]
- 19.Bordoni A, Capozzi F. Foodomics for healthy nutrition. Curr Opin Clin Nutr Metab Care 2014;17:418–24. [DOI] [PubMed] [Google Scholar]
- 20.Laghi L, Picone G, Capozzi F. Nuclear magnetic resonance for foodomics beyond food analysis. TrAC Trends Anal Chem. 2014;59:93–102. [Google Scholar]
- 21.Slavin JL, Lloyd B. Health benefits of fruits and vegetables. Adv Nutr 2012;3:506–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smolinska A, Blanchet L, Buydens LMC, Wijmenga SS. NMR and pattern recognition methods in metabolomics: From data acquisition to biomarker discovery: a review. Anal Chim Acta 2012;750:82–97. [DOI] [PubMed] [Google Scholar]
- 23.Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics 2009;5:435–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skov T, Honoré AH, Jensen HM, Næs T, Engelsen SB. Chemometrics in foodomics: handling data structures from multiple analytical platforms. TrAC Trends Anal Chem. 2014;60:71–9. [Google Scholar]
- 25.Jansen JJ, Smit S, Hoefsloot HCJ, Smilde AK. The photographer and the greenhouse: how to analyse plant metabolomics data. Phytochem Anal 2010;21:48–60. [DOI] [PubMed] [Google Scholar]
- 26.Alonso A, Marsal S, Juliá A. Analytical methods in untargeted metabolomics: state of the art in 2015. Front Bioeng Biotechnol 2015;3:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson CH, Ivanisevic J, Benton HP, Siuzdak G. Bioinformatics : the next frontier of metabolomics. Anal Chem 2015;87:147–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim HK, Choi YH, Verpoorte R. NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol 2011;29:267–75. [DOI] [PubMed] [Google Scholar]
- 29.Marcone MF, Wang S, Albabish W, Nie S, Somnarain D, Hill A. Diverse food-based applications of nuclear magnetic resonance (NMR) technology. Food Res Int 2013;51:729–47. [Google Scholar]
- 30.Brennan L. NMR-based metabolomics: from sample preparation to applications in nutrition research. Prog Nucl Magn Reson Spectrosc 2014;83:42–9. [DOI] [PubMed] [Google Scholar]
- 31.Shepherd LV, Fraser P, Stewart D. Metabolomics: a second-generation platform for crop and food analysis. Bioanalysis 2011;3:1143–59. [DOI] [PubMed] [Google Scholar]
- 32.Pan Z, Raftery D. Comparing and combining NMR spectroscopy and mass spectrometry in metabolomics. Anal Bioanal Chem 2007;387:525–7. [DOI] [PubMed] [Google Scholar]
- 33.Beltran A, Suarez M, Rodríguez MA, Vinaixa M, Samino S, Arola L, Correig X, Yanes O. Assessment of compatibility between extraction methods for NMR- and LC/MS-based metabolomics. Anal Chem 2012;84:5838–44. [DOI] [PubMed] [Google Scholar]
- 34.Kim HK, Choi YH, Verpoorte R. NMR-based metabolomic analysis of plants. Nat Protoc 2010;5:536–49. [DOI] [PubMed] [Google Scholar]
- 35.Ritota M, Casciani L, Valentini M. PGI chicory (Cichorium intybus L.) traceability by means of HRMAS-NMR spectroscopy: a preliminary study. J Sci Food Agric 2013;93:1665–72. [DOI] [PubMed] [Google Scholar]
- 36.Pacifico D, Casciani L, Ritota M, Mandolino G, Onofri C, Moschella A, Parisi B, Cafiero C, Valentini M. NMR-based metabolomics for organic farming traceability of early potatoes. J Agric Food Chem 2013;61:11201–11. [DOI] [PubMed] [Google Scholar]
- 37.Ritota M, Marini F, Sequi P, Valentini M. Metabolomic characterization of Italian sweet pepper (Capsicum annum L.) by means of HRMAS-NMR spectroscopy and multivariate analysis. J Agric Food Chem 2010;58:9675–84. [DOI] [PubMed] [Google Scholar]
- 38.Beckonert O, Coen M, Keun HC, Wang Y, Ebbels TMD, Holmes E, Lindon JC, Nicholson JK. High-resolution magic-angle-spinning NMR spectroscopy for metabolic profiling of intact tissues. Nat Protoc 2010;5:1019–32. [DOI] [PubMed] [Google Scholar]
- 39.Bharti SK, Roy R. Quantitative 1H NMR spectroscopy. TrAC Trends Anal Chem. 2012;35:5–26. [Google Scholar]
- 40.Markus MA, Ferrier J, Luchsinger SM, Yuk J, Cuerrier A, Balick MJ, Hicks JM, Killday KB, Kirby CW, Berrue F, et al. . Distinguishing Vaccinium species by chemical fingerprinting based on NMR spectra, validated with spectra collected in different laboratories. Planta Med 2014;80:732–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simmler C, Nikolić D, Lankin DC, Yu Y, Friesen JB, van Breemen RB, Lecomte A, Le Quémener C, Audo G, Pauli GF. Orthogonal analysis underscores the relevance of primary and secondary metabolites in licorice. J Nat Prod 2014;77:1806–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahrous EA, Farag MA. Two dimensional NMR spectroscopic approaches for exploring plant metabolome: a review. J Adv Res 2015;6:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pudakalakatti SM, Dubey A, Jaipuria G, Shubhashree U, Adiga SK, Moskau D, Atreya HS. A fast NMR method for resonance assignments: application to metabolomics. J Biomol NMR 2014;58:165–73. [DOI] [PubMed] [Google Scholar]
- 44.Tiainen M, Soininen P, Laatikainen R. Quantitative Quantum Mechanical Spectral Analysis (qQMSA) of 1H NMR spectra of complex mixtures and biofluids. J Magn Reson 2014;242:67–78. [DOI] [PubMed] [Google Scholar]
- 45.Astle W, De Iorio M, Richardson S, Stephens D, Ebbels T. A Bayesian model of NMR spectra for the deconvolution and quantification of metabolites in complex biological mixtures. J Am Stat Assoc 2011;107:500:1259–71. [Google Scholar]
- 46.Sze K-H, Wu Q, Tse H-S, Zhu G. Dynamic nuclear polarization: new methodology and applications. Top Curr Chem 2012;326:215–42. [DOI] [PubMed] [Google Scholar]
- 47.Cheng S-C, Jhang S-S, Huang M-Z, Shiea J. Simultaneous detection of polar and nonpolar compounds by ambient mass spectrometry with a dual electrospray and atmospheric pressure chemical ionization source. Anal Chem 2015;87:1743–8. [DOI] [PubMed] [Google Scholar]
- 48.Hejazi L, Guilhaus M, Hibbert DB, Ebrahimi D. Gas chromatography with parallel hard and soft ionization mass spectrometry. Rapid Commun Mass Spectrom 2015;29:91–9. [DOI] [PubMed] [Google Scholar]
- 49.Pauli GF, Chen S, Simmler C, Lankin DC, Gödecke T, Jaki BU, Friesen JB, Mcalpine JB, Napolitano JG. Importance of purity evaluation and the potential of quantitative 1H NMR as a purity assay. J Med Chem 2014;57:9220–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giraudeau P, Tea I, Remaud GS, Akoka S. Reference and normalization methods: essential tools for the intercomparison of NMR spectra. J Pharm Biomed Anal 2014;93:3–16. [DOI] [PubMed] [Google Scholar]
- 51.Cullen CH, Ray GJ, Szabo CM. A comparison of quantitative nuclear magnetic resonance methods: internal, external, and electronic referencing. Magn Reson Chem 2013;51:705–13. [DOI] [PubMed] [Google Scholar]
- 52.Hao J, Astle W, De Iorio M, Ebbels TM. BATMAN--an R package for the automated quantification of metabolites from nuclear magnetic resonance spectra using a Bayesian model. Bioinformatics 2012;28:2088–90. [DOI] [PubMed] [Google Scholar]
- 53.Manach C, Hubert J, Llorach R, Scalbert A. The complex links between dietary phytochemicals and human health deciphered by metabolomics. Mol Nutr Food Res 2009;53:1303–15. [DOI] [PubMed] [Google Scholar]
- 54.Clausen MR, Edelenbos M, Bertram HC. Mapping the variation of the carrot metabolome using 1H NMR spectroscopy and consensus PCA. J Agric Food Chem 2014;62:4392–8. [DOI] [PubMed] [Google Scholar]
- 55.Kortesniemi M, Vuorinen AL, Sinkkonen J, Yang B, Rajala A, Kallio H. NMR metabolomics of ripened and developing oilseed rape (Brassica napus) and turnip rape (Brassica rapa). Food Chem 2015;172:63–70. [DOI] [PubMed] [Google Scholar]
- 56.Kortesniemi M, Sinkkonen J, Yang B, Kallio H. 1H NMR spectroscopy reveals the effect of genotype and growth conditions on composition of sea buckthorn (Hippophae rhamnoides L.) berries. Food Chem 2014;147:138–46. [DOI] [PubMed] [Google Scholar]
- 57.Zhang X, Breksa AP, Mishchuk DO, Slupsky CM. Elevation, rootstock, and soil depth affect the nutritional quality of mandarin oranges. J Agric Food Chem 2011;59:2672–9. [DOI] [PubMed] [Google Scholar]
- 58.Capitani D, Sobolev AP, Tomassini A, Sciubba F, De Salvador FR, Mannina L, Delfini M. Peach fruit: metabolic comparative analysis of two varieties with different resistances to insect attacks by NMR spectroscopy. J Agric Food Chem 2013;61:1718–26. [DOI] [PubMed] [Google Scholar]
- 59.Masetti O, Ciampa A, Nisini L, Valentini M, Sequi P, Dell’Abate MT. Cherry tomatoes metabolic profile determined by 1H-high resolution-NMR spectroscopy as influenced by growing season. Food Chem 2014;162:215–22. [DOI] [PubMed] [Google Scholar]
- 60.Iglesias MJ, García-López J, Collados-Luján JF, López-Ortiz F, Díaz M, Toresano F, Camacho F. Differential response to environmental and nutritional factors of high-quality tomato varieties. Food Chem 2015;176:278–87. [DOI] [PubMed] [Google Scholar]
- 61.Perez-Fons L, Wells T, Corol DI, Ward JL, Gerrish C, Beale MH, Seymour GB, Bramley PM, Fraser PD. A genome-wide metabolomic resource for tomato fruit from Solanum pennellii. Sci Rep 2014;4:3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Z, Nakabayashi R, Okazaki Y, Mori T, Takamatsu S, Kitanaka S, Kikuchi J, Saito K. Toward better annotation in plant metabolomics: Isolation and structure elucidation of 36 specialized metabolites from Oryza sativa (rice) by using MS/MS and NMR analyses. Metabolomics 2014;10:543–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mehrotra S, Goyal V. Evaluation of designer crops for biosafety--a scientist’s perspective. Gene 2013;515:241–8. [DOI] [PubMed] [Google Scholar]
- 64.Harrigan GG, Skogerson K, MacIsaac S, Bickel A, Perez T, Li X. Application of 1H NMR profiling to assess seed metabolomic diversity. A case study on a soybean era population. J Agric Food Chem 2015;63:4690–7. [DOI] [PubMed] [Google Scholar]
- 65.Simó C, Ibáez C, Valdés A, Cifuentes A, García-Cañas V. Metabolomics of genetically modified crops. Int J Mol Sci 2014;15:18941–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Valdés A, Simó C, Ibáñez C, García-Cañas V. Foodomics strategies for the analysis of transgenic foods. TrAC Trends Anal Chem. 2013;52:2–15. [Google Scholar]
- 67.Oms-Oliu G, Odriozola-Serrano I, Martín-Belloso O. Metabolomics for assessing safety and quality of plant-derived food. Food Res Int 2013;54:1172–83. [Google Scholar]
- 68.Esslinger S, Riedl J, Fauhl-Hassek C. Potential and limitations of non-targeted fingerprinting for authentication of food in official control. Food Res Int 2014;60:189–204. [Google Scholar]
- 69.Kwon YK, Bong YS, Lee KS, Hwang GS. An integrated analysis for determining the geographical origin of medicinal herbs using ICP-AES/ICP-MS and 1H NMR analysis. Food Chem 2014;161:168–75. [DOI] [PubMed] [Google Scholar]
- 70.Walch SG, Lachenmeier DW, Kuballa T, Stühlinger W, Monakhova YB. Holistic control of herbal teas and tinctures based on sage (Salvia officinalis L.) for compounds with beneficial and adverse effects using NMR spectroscopy. Anal Chem Insights 2012;7:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dais P, Hatzakis E. Quality assessment and authentication of virgin olive oil by NMR spectroscopy: a critical review. Anal Chim Acta 2013;765:1–27. [DOI] [PubMed] [Google Scholar]
- 72.Cubero-Leon E, Peñalver R, Maquet A. Review on metabolomics for food authentication. Food Res Int 2014;60:95–107. [Google Scholar]
- 73.Hong YS. NMR-based metabolomics in wine science. Magn Reson Chem 2011;49:S13–21. [DOI] [PubMed] [Google Scholar]
- 74.Petrakis EA, Cagliani LR, Polissiou MG, Consonni R. Evaluation of saffron (Crocus sativus L.) adulteration with plant adulterants by 1H NMR metabolite fingerprinting. Food Chem 2015;173:890–6. [DOI] [PubMed] [Google Scholar]
- 75.Lee J-E, Lee B-J, Chung J-O, Kim H-N, Kim E-H, Jung S, Lee H, Lee S-J, Hong Y-S. Metabolomic unveiling of a diverse range of green tea (Camellia sinensis) metabolites dependent on geography. Food Chem 2015;174:452–9. [DOI] [PubMed] [Google Scholar]
- 76.Simova S, Atanassov A, Shishiniova M, Bankova V. A rapid differentiation between oak honeydew honey and nectar and other honeydew honeys by NMR spectroscopy. Food Chem 2012;134:1706–10. [DOI] [PubMed] [Google Scholar]
- 77.Sciubba F, Di Cocco ME, Gianferri R, Impellizzeri D, Mannina L, De Salvador FR, Venditti A, Delfini M. Metabolic profile of different Italian cultivars of hazelnut (Corylus avellana) by nuclear magnetic resonance spectroscopy. Nat Prod Res 2014;28:1075–81. [DOI] [PubMed] [Google Scholar]
- 78.Sciubba F, Capuani G, Di Cocco ME, Avanzato D, Delfini M. Nuclear magnetic resonance analysis of water soluble metabolites allows the geographic discrimination of pistachios (Pistacia vera). Food Res Int 2014;62:66–73. [Google Scholar]
- 79.Rychlik M. Challenges in food chemistry. Front Nutr. 2015;2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hohmann M, Christoph N, Wachter H, Holzgrabe U. 1H NMR profiling as an approach to differentiate conventionally and organically grown tomatoes. J Agric Food Chem 2014;62:8530–40. [DOI] [PubMed] [Google Scholar]
- 81.Liang T, Wei F, Lu Y, Kodani Y, Nakada M, Miyakawa T, Tanokura M. Comprehensive NMR analysis of compositional changes of black garlic during thermal processing. J Agric Food Chem 2015;63:683–91. [DOI] [PubMed] [Google Scholar]
- 82.Lopez-Sanchez P, de Vos RC, Jonker HH, Mumm R, Hall RD, Bialek L, Leenman R, Strassburg K, Vreeken R, Hankemeier T, et al. . Comprehensive metabolomics to evaluate the impact of industrial processing on the phytochemical composition of vegetable purees. Food Chem 2015;168:348–55. [DOI] [PubMed] [Google Scholar]
- 83.Del Coco L, De Pascali SA, Iacovelli V, Cesari G, Schena FP, Fanizzi FP. Following the olive oil production chain: 1D and 2D NMR study of olive paste, pomace, and oil. Eur J Lipid Sci Technol 2014;116:1513–21. [Google Scholar]
- 84.Malmendal A, Amoresano C, Trotta R, Lauri I, De Tito S, Novellino E, Randazzo A. NMR spectrometers as “magnetic tongues”: prediction of sensory descriptors in canned tomatoes. J Agric Food Chem 2011;59:10831–8. [DOI] [PubMed] [Google Scholar]
- 85.Lauri I, Pagano B, Malmendal A, Sacchi R, Novellino E, Randazzo A. Application of “magnetic tongue” to the sensory evaluation of extra virgin olive oil. Food Chem 2013;140:692–9. [DOI] [PubMed] [Google Scholar]
- 86.Wei F, Furihata K, Miyakawa T, Tanokura M. A pilot study of NMR-based sensory prediction of roasted coffee bean extracts. Food Chem 2014;152:363–9. [DOI] [PubMed] [Google Scholar]
- 87.Allwood JW, Cheung W, Xu Y, Mumm R, De Vos RC, Deborde C, Biais B, Maucourt M, Berger Y, Schaffer AA, et al. . Metabolomics in melon: a new opportunity for aroma analysis. Phytochemistry 2014;99:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hall RD. Plant metabolomics in a nutshell: potential and future challenges. Annual Plant Reviews. Hokoken (NJ): Wiley; 2011. p. 1–24. [Google Scholar]
- 89.Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, Koddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, et al. . The metabolomics standards initiative. Nat Biotechnol 2007;25:846–8. [DOI] [PubMed] [Google Scholar]
- 90.Vernocchi P, Vannini L, Gottardi DI, Del Chierico F, Serrazanetti D, Ndagijimana M, Guerzoni ME. Integration of datasets from different analytical techniques to assess the impact of nutrition on human metabolome. Front Cell Infect Microbiol 2012;2:156. [DOI] [PMC free article] [PubMed] [Google Scholar]