Abstract
Genomic information on tumors from 50 cancer types catalogued by The International Cancer Genome Consortium (ICGC) shows that only few well-studied driver genes are frequently mutated, in contrast to many infrequently mutated genes that may also contribute to tumor biology. Hence there has been large interest in developing pathway and network analysis methods that group genes and illuminate the processes involved. We provide an overview of these analysis techniques and show where they guide mechanistic and translational investigations.
Introduction
As sequencing costs continue to decrease, it is becoming common to assay genomic information from a cohort of cancer patients at the level of single nucleotide variants (SNVs) and copy number alterations (CNAs). Other alterations including structural changes, fusion transcripts and epigenetic reprogramming are also studied routinely. These genomic data are associated with rich clinical annotation, and some groups have begun to incorporate sequencing into standard clinical practice1. Recent studies have painted a portrait of the mutation landscape for multiple cancers2 including pancreatic3, lung4, breast5, brain6, and ovarian7. In each case, the distribution of somatic single nucleotide variants (SNVs) across the samples typically includes a few altered genes at frequencies higher than 10% and a “long tail” of many genes mutated at frequencies of 5% or lower2,8. Interestingly, some tumor types, including prostate cancer and some pediatric cancers, have relatively few SNVs or CNAs9; their biology is presumably driven by other types of somatic variation like DNA methylation10. Driver genes are mostly detected using signals of positive selection in the mutation patterns of individual genes across tumors11. However, this approach will miss less frequently mutated but functionally important genes that a typical cohort with hundreds of tumor samples is not statistically powered to detect12. Recent pan-cancer analyses have detected cancer genes using several thousand samples of different tumor types, however these studies still remain limited in power due to tissue-specific drivers such as APC in colorectal and ovarian cancers, VHL in renal cell carcinoma, and ERG fusion genes in prostate cancers. Alternatively, grouping of genetic alterations using prior knowledge about cellular mechanisms allows investigation of the full complement of mutations in a tumor and the determination of affected oncogenic pathways.
In this Perspective, the term “pathway and network analysis” denotes any analytic technique that benefits from biological pathway or molecular network information to gain insight into a biological system. The fundamental aim is to reduce data involving thousands of altered genes and proteins to a smaller and more interpretable set of altered processes (see recent reviews13,14). This process-oriented view helps generate testable hypotheses, identify drug targets, find tumor subtypes with clinically distinct outcomes, and identify both cancer-specific and cross-cancer pathways.
Pathways and networks are similar concepts with certain distinctions. Both comprise systems of interacting genes, proteins, and other biomolecules that carry out biological functions. Pathways are small-scale systems of well-studied processes where interactions comprise biochemical reactions and events of regulation and signaling. Pathways represent consensus systems based on decades of research and can be visualized in detailed linear diagrams. In contrast, networks comprise genome- or proteome-wide interactions derived from large-scale screens or integrative analyses of multiple datasets. Network interactions are simplified abstractions of complex cellular logic. For instance, physical protein-protein interactions may be represented as directionless edges and directed edges may stand for inhibitory or activating gene regulation. Networks are noisy and challenging to visualize and interpret, however they likely contain novel information not covered in well-defined pathways. A related concept to pathways and networks is a functionally annotated gene set that comprises all genes involved in a particular process or pathway without their interactions. Annotated gene sets of the Gene Ontology and other resources are based on multiple types of evidence and are broader in scope than pathways.
Pathway and network analysis has a number of benefits relative to analyzing genomics data at the level of individual genes. First, these techniques aggregate molecular events across multiple genes in the same pathway or network neighborhood, thus increasing the likelihood that the events will pass a statistical detection threshold and reducing the number of hypotheses tested15. Second, results are often easier to interpret as genomic alterations are related to familiar concepts such as cell cycle or apoptosis. Third, potential causal mechanisms can be identified, for instance by predicting a particular microRNA or transcription factor that explains expression differences between tumor samples and controls. Fourth, results obtained from related datasets may become more comparable since pathway information allows interpretation in a common feature space. Finally, the techniques facilitate integration of diverse inputs such as genomic, transcriptomic and proteomic data into a unified view of tumor biology, improving statistical and interpretative power.
Pathway and network analyses have been applied to cancer data sets to find driver genes and pathways16,17, to identify hidden tumor subtypes distinguished by common patterns of network alteration18, to propose cancer mechanisms and biomarkers17,18, and to identify key regulators of cancer-related gene networks19,20.
The Mutation Consequences and Pathway Analysis (MUCOPA) working group of the International Cancer Genome Consortium (ICGC)21 has developed standard operating procedures for the analysis of cancer genome data generated by the ICGC. In a recent review11 we outlined our recommendations for prioritizing somatic mutations using gene-level statistics, including criteria for the functional impact of mutations and positive selection for mutations in genes within the patient population. Here we describe diverse analytic techniques to prioritize altered gene sets, pathways, and networks consisting of multiple interacting genes. While we focus on somatic SNVs and altered RNA expression, the concepts are generally applicable to other oncogenic alterations such as CNAs, epigenetic changes, and genomic rearrangements, though the details of analysis, including data processing and confounding factor control, can be different for other data types.
Major Types of Pathway and Network Analysis Techniques
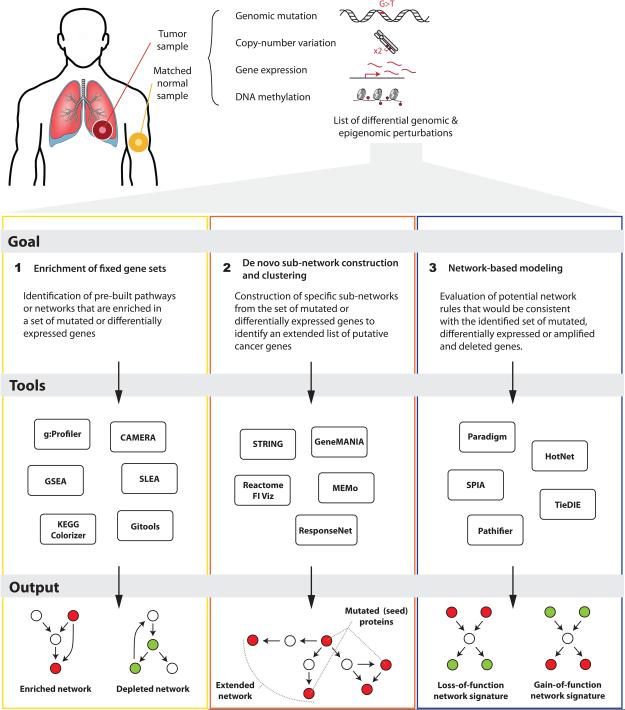
We consider three major approaches to network and pathway analyses to interpret somatic cancer mutations, listed from the simplest to the more complex (Fig. 1). The simplest analysis provides a high-level summary of pathways affected in the tumor, whereas more complex methods provide detailed hypotheses about affected cellular mechanisms. We recommend that approaches from each of these classes be applied to cancer genome sequencing projects wherever feasible.
Figure 1. Major approaches to pathway and network analysis of cancer data.
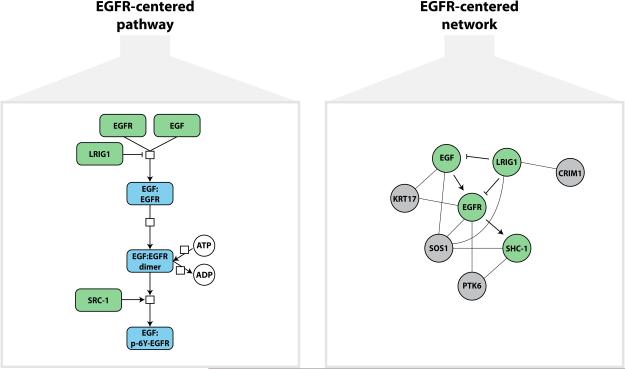
All three approaches require two general resources. The first is a list of oncogenic alterations that affect protein-coding genes. The second is a database of gene sets, pathways, or network interactions22. Pathway databases represent biological processes as series of biochemical reactions and other physical events (e.g. complex formation, phosphorylation events, sub-cellular localization, conformational changes), while network databases use a simpler data model that treats biological processes as sets of bimolecular interactions. A simplified version of the Epidermal Growth Factor pathway illustrates the essential difference between pathway and network interaction databases (Fig. 2). The first approach, fixed gene set enrichment analysis, analyses functionally annotated gene sets that can be extracted from either type of database. Network interaction databases provide inputs to the second approach, de novo network construction and clustering, while both types of databases are used in the most sophisticated approach, network-based modeling.
Figure 2. Pathway and network representation of EGF signaling.
(A) In the simplified pathway representation, heterogeneous nodes and edges correspond to genes, proteins, small molecules, and their regulatory and catalytic relationships. Nodes do not interact directly but participate in reaction events designated by white squares. (B) In the network representation, all nodes correspond to the same type of biological entity (gene products). Edges derived from curated pathways are shown as bold arrows. Additional gene-gene interactions derived from gene co-expression and physical protein interactions are shown as light lines.
Approach 1: Fixed gene set enrichment analysis
The first approach treats pathways, biological processes, and networks simply as gene sets and discards information about their interactions. Fixed gene set enrichment analysis identifies genes in pathways (or any other functionally-related grouping) that are present in a gene list more frequently than expected by chance. The gene sets are usually collected from curated community databases or the gene annotation tables of the Gene Ontology23, but may also be experimentally derived (e.g. genes up-regulated in a cell line exposed to low oxygen levels). Several recommended software tools are available (Supplementary Table 1). The simplest input to such analysis is a list of genes that is most differentially expressed or frequently mutated in a dataset. A typical analysis workflow consists of two steps: (1) a gene list is defined by filtering experimental data for genes with significant gene-level statistics, and (2) enrichment analysis is performed to determine processes and pathways over-represented in the gene list.
A hypergeometric distribution (Fisher’s exact test) is commonly used to calculate the statistical significance of this over-representation, followed by a correction for multiple testing to estimate the proportion of enriched gene sets that would occur by chance given the number of tested gene sets. The basic form of this test is applied in many tools (Supplementary Table 1) including the widely used but no longer updated web service DAVID24. However, the key drawback of this approach is that an arbitrary threshold is used to select the input genes and potentially informative genes below the threshold are excluded. An alternative approach enables interpretation of a ranked list of genes in the experiment (e.g. by strength of differential expression) with the assumption that top-ranking genes are more important in terms of biological function. One recommended web service g:Profiler25 applies a modified hypergeometric test to analyze increasingly complete ranked lists of input genes and determines a sub-list with the strongest level of enrichment. The GSEA method26 is designed to work with continuous data and searches for gene sets that are enriched at the top (over-expressed vs. control) or bottom (under-expressed) of a ranked list of all genes. Both methods score each gene set separately and compute additional statistics to estimate p-values and make multiple-testing corrections with false discovery rate (FDR).
Enhancements of these approaches allow enrichment analysis for each tumor sample, thereby enabling the discovery of distinct cancer subtypes from different enrichment patterns. Examples of methods that allow comparisons among samples include sample level enrichment analysis (SLEA)27, single-sample GSEA (ssGSEA)26, and gene set variation analysis (GSVA)28.
Rank-based enrichment methods do best when genes are easily ranked but may be suboptimal in scenarios such as cancer mutation analysis in which most genes are difficult to rank due to low mutation counts. A pathway association analysis may be helpful in case of a two-class experimental design (e.g. cases vs. controls). This resembles a genome-wide association analysis and uses pathways and other gene sets instead of genetic markers. For each experimental class and gene set, one counts all samples containing a mutation that may affect that gene set. A series of Fisher’s exact tests identify gene sets significantly mutated in cases versus controls, followed by multiple-testing correction.
Fixed gene set enrichment analysis generates a list of processes and pathways and provides a bird’s-eye view of affected biological systems. However, sometimes many related gene sets are enriched. The key functional themes in these large pathway lists can be identified using tools like Enrichment Map29 app of the Cytoscape network visualization software30. Another useful approach is to overlay the original genomics data on a detailed pathway diagram or high-level molecular interaction network. For example, the databases KEGG31, Reactome32, and HumanCyc33 enable diagrams of enriched pathways with colors highlighting the genes of interest. This may help researchers to move beyond asking what pathways are enriched among alterations towards understanding the functional consequences of the altered gene set.
This family of techniques is still evolving. For instance, most enrichment statistics assume that genes in the list occur independently, an assumption that does not hold true for co-regulated genes in gene expression data, overlapping or shared exons in point mutation data, or co-located paralogous genes with similar functions. The quality and coverage of gene sets can also affect interpretation of fixed gene set enrichment analysis, as databases report genes and their functions with variable levels of detail and confidence. Combined use of multiple databases, filtering, and visualization help overcome these problems. Another issue is that many annotated pathways represent normal physiology that may be altered in disease. New methods are developed to address these issues; for example CAMERA34 corrects gene set enrichment statistics for inter-gene correlations. A more fundamental limitation of this class of algorithms is their ignorance of interactions between genes and proteins, as neither network topology nor dynamics is taken into account. These limitations are addressed by the next two approaches.
Approach 2: de novo Network Construction and Clustering
Methods in this section construct cancer gene networks de novo by analyzing networks of molecular or functional interactions. These methods begin with a list of mutated or otherwise altered genes, and one or more databases of gene or protein interactions, such as those compiled by iRefIndex35, BioGRID36, IntAct37, STRING38, or GeneMANIA39 (Supplementary Table 2). The altered genes and a subset of their neighbors are then extracted from the databases and reconstructed as an interaction network. The resulting network reveals interactions of input genes and helps discover additional related genes by “guilt by association”, highlighting non-mutated genes that likely participate in tumor biology due to their interactions. By clustering and annotating the discovered networks with the enrichment and colorization approaches described above, one may reveal similarities and differences among distinct tumors that would not be apparent at the gene level.
Examples of recommended network construction algorithms include GeneMANIA39, ReactomeFIViz40, STRING38, ResponseNet41, NetBox42, MEMo43, and EnrichNet44 (Supplementary Table 2). GeneMANIA is an interactive web service and a Cytoscape app that uses a diverse set of interaction databases. It suggests genes that are related to those in the experimental data set using network analysis. ReactomeFIViz (previously called Reactome FI Plugin) runs in Cytoscape and features a number of algorithms for clustering and annotating sets of interacting genes, and relating these clusters to tumor phenotype and patient clinical characteristics. For example, ReactomeFIViz identified prognostic biomarkers in breast and ovarian cancer45. NetBox is conceptually similar to ReactomeFIViz and reports functional network modules by identifying clusters of altered genes on a background network derived from databases. MEMo studies mutual exclusivity of cancer alterations in groups of genes across tumor samples to discover sub-networks of synthetic lethality and other functional groupings. It nominates sets of oncogenic alterations that have a particularly strong selective effect, potentially pointing to therapeutic combinations where mutual exclusivity reflects synthetic lethality.
A key use of networks is to search for alteration patterns in interacting genes that correlate with clinical information46. The HyperModules method47 identifies subnetworks with cancer mutations that are maximally correlated with clinical characteristics such as patient survival, tumor stage, or relapse. This tool can also be used to study tumor subtypes by extracting subnetworks whose mutations are significantly enriched in a particular subtype. HyperModules was applied to the kinase-signaling network in ovarian cancer and revealed network modules with mutations in phosphorylation sites and kinase domains that significantly correlated with patient survival48.
A drawback of de novo network construction and clustering techniques is their use of a simplified data model that discards much information known about biological networks. For example, an alteration may act at the DNA level by deleting a portion of a gene, at the transcriptional level by disrupting a promoter, or at the protein level by altering a catalytic site. The activating effect of a mutation in a transmembrane receptor can be masked by inactivation of a downstream effector of the same signaling pathway. These subtleties are not easily captured in a binary interaction network. In addition, the molecular interactions in databases are derived from specific experiments such as yeast-two-hybrid assays that may or may not matter for cancer biology. Thus it is advisable to consult the literature underlying the network interactions when forming hypotheses based on patterns observed in interaction networks; several text-mining tools are available to automate this task49.
Approach 3: Network-Based Modeling
The approaches discussed in this section infer how network states are disrupted in cancer. Network-based modeling approaches use qualitative and quantitative measurements to infer the activities and interactions of various genetic components in pathway or networks. These methods relate the activities of some components with their influences and consequences on other components. Such modeling approaches have been applied to infer the mechanisms of NRAS signaling in melanoma50 to map transcriptional regulatory networks in physiologically normal and diseased states19,20,51,52,53, to build maps of phosphorylation networks54, and to identify cancer drivers16. Below, we briefly describe several network modeling algorithms that are available as user installable software packages and have been applied to cancer (Supplementary Table 3).
The HotNet55 tool treats the gene network as a metallic lattice, and then uses the physics of heat diffusion to model the effects of gene alterations. Each gene in the query “heats up” its local region of the network, and the effect is then metaphorically propagated along metallic wires defined by gene-gene linkages, leading to “hot” (highly relevant) network neighborhoods. This approach mitigates some of the ascertainment biases in curated gene interaction networks. For example, because TP53 is exceedingly well studied, it is an artificially inflated “hub” of known linkages to other genes; but because of TP53’s high degree of connectedness, heat diffuses away from it rapidly, reducing its overall influence. The related method TieDIE56 extends the network diffusion concept to integrated analysis of multiple types of genomic alterations.
The Pathifier method57 transforms gene-level information to network-level information by quantifying molecular activities on a continuous sample-by-sample curve in the multidimensional space of gene expression values. It ranks cancer samples along a gradient of clinical or biological attributes such as tumor aggressiveness or patient survival. The method generates hypotheses and identifies testable markers to predict clinical outcomes.
Signaling Pathway Impact Analysis (SPIA)58 applies a recursive algorithm similar to that used by Google to rank search results. SPIA scores a gene product as highly impactful if it points to other impactful gene products in the network diagram. By ranking the effects hierarchically, SPIA distinguishes primary changes in gene activity and secondary effects of the regulatory network.
Several methods use information theoretical principles to reconstruct regulatory networks from gene expression data. Application of these methods to cancer genomics has led to insights into tumor biology and identification of actionable drug treatments. ARACNe applies mutual information to discover regulatory networks of transcription factors and target genes59, while MARINa interrogates these networks to identify master regulators19,20. For example, application of these tools to the reconstruction of the gene regulatory network in glioblastoma and follow-up experimental validations revealed C/EBPβ and STAT3 to be master regulators of mesenchymal transformation20.
Other methods integrate gene expression and CNA data to identify cancer driver genes and downstream regulatory networks. For example, CONEXIC assumes that copy number gains and losses alter gene expression16. It employs a Bayesian Network algorithm to find significantly altered genes regulating modules of differentially expressed genes. The approach was applied to predict and experimentally validate multiple cancer driver genes in melanoma and glioblastoma16,60.
Several approaches have been developed to “fit” gene interactions to the data rather than taking the interactions as prior knowledge. Thus interactions are not interpreted as direct physical interactions but rather as measures of influence between network nodes. Functions of discrete logic were used to connect gene products through “gates”61 and to infer functions best capturing the observed dynamics in the data. This was extended to fuzzy logic62 that relaxes the rules of gene interactions and allows for biological noise and uncertainty. Similar approaches were developed for partial least squares regression (PLSR) models63 in which parameters are fit to dependent variables typically reflecting a cellular phenotype. These approaches were applied to interpret drug response in triple negative breast cancer and to suggest effective therapeutic treatments64. The DataRail package65 allows users to experiment with multiple similar model-fitting methods for gene networks.
Probabilistic graphical models (PGMs) have been applied to cancer network analysis. PGMs are widely used in machine learning and statistics for modeling complex dependencies among multiple variables. PathOlogist66 analyses pathways from curated databases to derive a set of network interactions. It then uses the inhibitory and excitatory regulatory connections in each pathway-derived network model to determine if 1) a given cancer gene expression dataset is consistent with the model and 2) if the pathway-derived network’s components are activated. Thus a collection of known gene interactions with details of co-regulation helps interpret gene expression data. This family of algorithms was applied to develop predictors of drug sensitivity in cancer cell lines67.
PARADIGM68 extends the PGM framework of PathOlogist by formally modeling the “central dogma” of gene expression to represent pathway and network effects of alterations at the DNA, RNA and protein levels. This method uses factor graphs to assign weights to each molecular interaction and to integrate the effects of multiple simultaneous alterations (e.g. copy number changes, simple somatic mutations, expression changes). The tool provides predicted pathway activity scores by integrating all observed variations to assess whether the activities of each pathway are increased, decreased, or unaffected. The algorithm was used to identify new tumor subtypes based on shared pathway activation patterns18. An extension called PARADIGM-SHIFT infers whether somatic mutations are neutral, loss-of-function, or gain-of-function69. This method has detected several well-known examples of pathway alterations, including loss of function events in the tumor suppressor TP53 in breast cancer, and gain of function events in oncogene NFE2L2 in lung squamous cell tumors. More recent PGM approaches include the application of Dynamic Bayesian Networks to consider tumorigenesis as a temporally evolving system. The inferred network of breast cancer cell lines contributes an important proof-of-concept in this area70.
Higher-resolution modeling of cellular wiring in cancer requires quantitative data that is not yet readily available from patient tissue samples. Established cell lines, organoids, and xenograft models will enable collection of more data for integrative analysis. Time courses and perturbation experiments on such cancer models will contribute key data points that will help parameterize more realistic models such as systems of differential equations. Large interacting systems of differential equations such as full cell models71 also show promise but are in their infancy in their application to cancer.
Challenges and future perspectives
Pathway and network analysis can effectively uncover biological systems perturbed in tumor cells. However, our knowledge of pathways and networks both in normal cells and more acutely in cancer cells is far from complete. Many approaches, particularly the modeling techniques of the third section, require accurate, detailed, and comprehensive pathway descriptions with regulatory relationships, orthogonal data (DNA, RNA, protein), and deep quantitative data. Even among protein-coding genes, high-resolution data are only available for well-studied biological processes and are scarce for pathways involving many non-coding genomic elements. This argues for an expanded effort in the development of pathway databases and the systematic reconstruction of regulatory and signaling networks.
A second challenge is the computationally expensive modeling of biological networks that can consume weeks of CPU time, particularly for permutation-based estimates of statistical significance. This problem will only grow as reference pathways and networks and experimental datasets increase in size. As cancer genomics data become available for progressively larger patient cohorts, fundamental computer science research is needed to optimize these algorithms to scale to thousands of samples72.
A third challenge arises from the abundant interdependencies in complex biological systems. It is well established that the role of a mutation, such as its functional impact or its role in suppressing or enabling a tumor, is not static. Instead it depends on cell state and the presence of other mutations73 and could have effects on multiple cellular processes. The establishment of annotation standards that can encapsulate such dependencies also represents a major challenge for the field.
A final challenge is the evaluation of pathway and network methods in patient care. With a sufficient battery of pathway-specific therapeutics, one can envision the selection of therapies based on networks constructed from the molecular alterations present in individual tumors. It will be a major statistical challenge to devise adaptive clinical trials that leverage such information74. The difficulties of communicating genomic information to clinicians and patients will certainly be exacerbated by the complexity of network-level alterations75.
Our understanding of cancer biology through the lens of pathway and network analyses is nascent, but holds the potential to transform our thinking on disease etiology and treatment.
Supplementary Material
Acknowledgements
We gratefully acknowledge the assistance of J. Jennings, Ontario Institute for Cancer Research, during preparation of this manuscript. J.M.S. acknowledges support from National Cancer Institute (R01-CA180778 and U24-CA143858), Stand Up To Cancer, Prostate Cancer Foundation, and the Movember Foundation. P.C. is currently funded by a Ludwig Fund Postdoctoral Fellowship. P.C.B. and L.D.S. were supported by the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. P.C.B. was also supported by a Terry Fox Research Institute New Investigator Award and a Canadian Institutes of Health Research New Investigator Award. L.D.S. and G.W. acknowledge support from the US National Institutes of Health and National Human Genome Research Institute (P41 HG003751). G.D.B is supported by NRNB (US National Institutes of Health, National Institute of General Medical Sciences grant number P41 GM103504).
Footnotes
Competing Financial Interests
The authors declare they have no competing financial interests as defined by Nature Publishing Group, or other interests that might be perceived to influence the results and/or discussion reported in this article.
References
- 1.Newman WG, Black GC. Delivery of a clinical genomics service. Genes. 2014;6:1001–17. doi: 10.3390/genes5041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawrence MS, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biankin AV, et al. Pancreatic cancer genomes reveal aberrations in axon guidance pathway genes. Nature. 2012;491:399–405. doi: 10.1038/nature11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Imielinski M, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cancer Genome Atlas Research Network Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garraway LA, Lander ES. Lessons from the Cancer Genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. This review discusses the advances and findings in cancer genome sequencing as well as current challenges of the field, including the “long tail” of infrequently mutated genes, and the need for functional validation of cancer mutations. [DOI] [PubMed] [Google Scholar]
- 9.Zack T, et al. Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mack SC, et al. Epigenomic alterations define lethal CIMP-positive ependymomas of infancy. Nature. 2014;506:445–50. doi: 10.1038/nature13108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez-Perez A, et al. Computational approaches to identify functional genetic variants in cancer genomes. Nat Methods. 2013;10:723–729. doi: 10.1038/nmeth.2562. This review from the ICGC-MUCOPA working group discusses methods and recommendations to distinguish functional cancer mutations and to predict cancer driver genes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leiserson MDM, et al. Simultaneous Identification of Multiple Driver Pathways in Cancer. PLoS Computational Biology. 2013;9 doi: 10.1371/journal.pcbi.1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pe'er D, Hacohen N. Principles and strategies for developing network models in cancer. Cell. 2011;144(6):864–73. doi: 10.1016/j.cell.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Califano A, et al. Leveraging models of cell regulation and GWAS data in integrative network-based association studies. Nat Genet. 2012;44:841–847. doi: 10.1038/ng.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi YY, et al. Power calculation for overall hypothesis testing with high-dimensional commensurate outcomes. Stat Med. 2013;33:812–27. doi: 10.1002/sim.5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Akavia UD, et al. An integrated approach to uncover drivers of cancer. Cell. 2010;143:1005–1017. doi: 10.1016/j.cell.2010.11.013. One of the first studies to integrate molecular data at different network levels to pinpoint tumor dependencies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Danussi C, et al. RHPN2 drives mesenchymal transformation in malignant glioma by triggering RhoA activation. Cancer Res. 2013;73:5140–5150. doi: 10.1158/0008-5472.CAN-13-1168-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158:929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonabend AM, et al. The transcriptional regulatory network of proneural glioma determines the genetic alterations selected during tumor progression. Cancer Res. 2014;74(5):1440–51. doi: 10.1158/0008-5472.CAN-13-2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carro MS, et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.International Cancer Genome Consortium International Network of Cancer Genome Projects. Nature. 2010;465:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–6. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.The Gene Ontology Consortium The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–5. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang DW, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler--a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–200. doi: 10.1093/nar/gkm226. g:Profiler is a frequently updated web server for conducting fixed gene set enrichment analysis of plain and ranked gene lists. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:5545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gundem G, Lopez-Bigas N. Sample-level enrichment analysis unravels shared stress phenotypes among multiple cancer types. Genome Med. 2012;4:28. doi: 10.1186/gm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010;5:e13984–e13984. doi: 10.1371/journal.pone.0013984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nature Protocols. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanehisa M, et al. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Croft D, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caspi R, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2014;42:D459–D471. doi: 10.1093/nar/gkt1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu D, Smyth GK. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012;40:e133. doi: 10.1093/nar/gks461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:2105–9. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–23. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kerrien S, et al. The IntAct molecular interaction database in 2012. Nucleic Acids Res. 2012;40:D841–6. doi: 10.1093/nar/gkr1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Franceschini A, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warde-Farley D, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–20. doi: 10.1093/nar/gkq537. GeneMANIA is a web server for integrative analysis of gene lists in the context of molecular interaction networks. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu G, Dawson E, Duong A, Haw R, Stein L. ReactomeFIViz: a Cytoscape app for pathway and network-based data analysis. F1000Res. 2014;3:146. doi: 10.12688/f1000research.4431.1. ReactomeFIViz is a Cytoscape app with multiple algorithms for network-based clustering and analysis of the Reactome functional interaction network. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lan A, et al. ResponseNet: revealing signaling and regulatory networks linking genetic and transcriptomic screening data. Nucleic Acids Res. 2011;39:W424–9. doi: 10.1093/nar/gkr359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cerami E, Demir E, Schultz N, Taylor BS, Sander C. Automated network analysis identifies core pathways in glioblastoma. PLoS One. 2010;5:e8918. doi: 10.1371/journal.pone.0008918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ciriello G, Cera E, Sander C, Schultz N. Mutual exclusivity analysis identifies oncogenic network modules. Genome Res. 2012;22:398–406. doi: 10.1101/gr.125567.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glaab E, Baudot A, Krasnogor N, Schneider R, Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012;28:i451–i457. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu G, Stein L. A network module-based method for identifying cancer prognostic signatures. Genome Biol. 2012;13:R112. doi: 10.1186/gb-2012-13-12-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:140–140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Leung A, Bader GD, Reimand J. HyperModules: identifying clinically and phenotypically significant network modules with disease mutations for biomarker discovery. Bioinformatics. 2014;30:2230–2232. doi: 10.1093/bioinformatics/btu172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimand J, Bader GD. Systematic analysis of somatic mutations in phosphorylation signaling predicts novel cancer drivers. Mol Syst Biol. 2013;9:637–637. doi: 10.1038/msb.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krallinger M, et al. The Protein-Protein Interaction tasks of BioCreative III: classification/ranking of articles and linking bio-ontology concepts to full text. BMC Bioinformatics. 2011;12:S3. doi: 10.1186/1471-2105-12-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwong LN, et al. Oncogenic NRAS signaling differentially regulates survival and proliferation in melanoma. Nat Med. 2012;18:1503–1510. doi: 10.1038/nm.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schadt EE, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Aytes A, et al. Cross-species analysis of genome-wide regulatory networks identifies a synergistic interaction between FOXM1 and CENPF that drives prostate cancer malignancy. Cancer Cell. 2014;25:638–651. doi: 10.1016/j.ccr.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Piovan E, et al. Direct Reversal of Glucocorticoid Resistance by AKT Inhibition in Acute Lymphoblastic Leukemia. Cancer Cell. 2013;24:766–776. doi: 10.1016/j.ccr.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bandyopadhyay S, et al. A human MAP kinase interactome. Nat Methods. 2010;7:801–805. doi: 10.1038/nmeth.1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vandin F, Upfal E, Raphael BJ. Algorithms for detecting significantly mutated pathways in cancer. J Comput Biol. 2011;18:507–22. doi: 10.1089/cmb.2010.0265. The HotNet algorithm uses a heat diffusion model to analyze molecular interaction networks and detect significantly mutated modules in cancer. [DOI] [PubMed] [Google Scholar]
- 56.Paull EO, et al. Discovering causal pathways linking genomic events to transcriptional states using Tied Diffusion Through Interacting Events (TieDIE) Bioinformatics. 2013;29:2757–64. doi: 10.1093/bioinformatics/btt471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drier Y, Sheffer M, Domany E. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci U S A. 2013;110:6388–93. doi: 10.1073/pnas.1219651110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tarca AL, et al. A novel signaling pathway impact analysis. Bioinformatics. 2009;25:75–82. doi: 10.1093/bioinformatics/btn577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Margolin AA, et al. ARACNE: an algorithm for the reconstruction of gene regulatory networks in a mammalian cellular context. BMC Bioinformatics. 2006;7(Suppl 1):S7–S7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Danussi C, et al. RHPN2 drives mesenchymal transformation in malignant glioma by triggering RhoA activation. Cancer Res. 2013;73:5140–5150. doi: 10.1158/0008-5472.CAN-13-1168-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris MK, Saez-Rodriguez J, Sorger PK, Lauffenburger DA. Logic-based models for the analysis of cell signaling networks. Biochemistry. 2010;49:3216–3224. doi: 10.1021/bi902202q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morris MK, Saez-Rodriguez J, Clarke DC, Sorger PK, Lauffenburger DA. Training signaling pathway maps to biochemical data with constrained fuzzy logic: quantitative analysis of liver cell responses to inflammatory stimuli. PLoS Comput Biol. 2011;7:e1001099–e1001099. doi: 10.1371/journal.pcbi.1001099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–53. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 64.Lee MJ, et al. Sequential application of anticancer drugs enhances cell death by rewiring apoptotic signaling networks. Cell. 2012;149:780–794. doi: 10.1016/j.cell.2012.03.031. This article utilized integrative network analysis to identify key rewiring cellular events that informed a combination-based therapeutic strategy for specific tumors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saez-Rodriguez J, et al. Flexible informatics for linking experimental data to mathematical models via DataRail. Bioinformatics. 2008;24:840–847. doi: 10.1093/bioinformatics/btn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Greenblum SI, Efroni S, Schaefer CF, Buetow KH. The PathOlogist: an automated tool for pathway-centric analysis. BMC Bioinformatics. 2011;12:133. doi: 10.1186/1471-2105-12-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Brubaker D, et al. Drug Intervention Response Predictions with PARADIGM (DIRPP) identifies drug resistant cancer cell lines and pathway mechanisms of resistance. Pac Symp Biocomput. 2014:125–135. [PMC free article] [PubMed] [Google Scholar]
- 68.Vaske CJ, et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–245. doi: 10.1093/bioinformatics/btq182. The PARADIGM algorithm predicts the impact of oncogenic alterations on downstream pathway and network activity by modeling the central dogma of gene expression. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ng S, et al. PARADIGM-SHIFT predicts the function of mutations in multiple cancers using pathway impact analysis. Bioinformatics. 2012;28:i640–i646. doi: 10.1093/bioinformatics/bts402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hill SM, et al. Bayesian inference of signaling network topology in a cancer cell line. Bioinformatics. 2012;28:2804–10. doi: 10.1093/bioinformatics/bts514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanghvi JC, et al. Accelerated discovery via a whole-cell model. Nat Methods. 2013;10:1192–5. doi: 10.1038/nmeth.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dittrich MT, Klau GW, Rosenwald A, Dandekar T, Muller T. Identifying functional modules in protein-protein interaction networks: an integrated exact approach. Bioinformatics. 2008;24:i223–i231. doi: 10.1093/bioinformatics/btn161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu M, Pastor-Pareja JC, Xu T. Interaction between Ras(V12) and scribbled clones induces tumour growth and invasion. Nature. 2010;463:545–8. doi: 10.1038/nature08702. This paper demonstrated the importance of cooperation between mutations in cancer in the RAS signaling pathway. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Berry DA. Adaptive clinical trials: the promise and the caution. J Clin Oncol. 2011;29:606–9. doi: 10.1200/JCO.2010.32.2685. [DOI] [PubMed] [Google Scholar]
- 75.Green RC, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med. 2013;15:565–74. doi: 10.1038/gim.2013.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.