Abstract
Background:
The diagnosis of myelodysplastic syndrome (MDS), especially hypoplastic MDS, and MDS with low blast counts or normal karyotype may be problematic. This study characterized ID4 gene methylation in patients with MDS and aplastic anemia (AA).
Methods:
The methylation status of ID4 was analyzed by bisulfite sequencing polymerase chain reaction (PCR) and quantitative real-time methylation-specific PCR (MethyLight PCR) in 100 patients with MDS and 31 patients with AA.
Results:
The MDS group had a higher ID4 gene methylation positivity rate (22.22%) and higher methylation levels (0.21 [0–3.79]) than the AA group (P < 0.05). Furthermore, there were significant differences between the hypoplastic MDS and AA groups, the MDS with low blast count and the AA groups, and the MDS with normal karyotype and the AA groups. The combination of genetic and epigenetic markers was used in much more patients with MDS (62.5% [35/56]) than the use of genetic markers only (51.79% [29/56]).
Conclusions:
These results showed that the detection of ID4 methylation positivity rates and levels could be a useful biomarker for MDS diagnosis.
Keywords: Diagnosis, ID4 Gene, Myelodysplastic Syndrome, Methylation, Quantitative
INTRODUCTION
Myelodysplastic syndrome (MDS) is a heterogenous group of hematopoietic stem cell disorders that are multifactorial in their etiology and often difficult to distinguish from hematological benign diseases with unilineage or multilineage cytopenia such as aplastic anemia (AA) both in clinic and laboratory. MDS is a heterogenous group of bone marrow (BM) clonal hematopoiesis with unilineage or multilineage dysplasia and cytogenetic abnormalities. However, all of these characterized manifestations are insufficient for an MDS diagnosis.[1]
In a proportion of patients, hypoplastic MDS and MDS with a low blast count or a normal karyotype are difficult to distinguish from AA. In approximately 50% of patients with MDS, the BM shows a normal karyotype.[2] In particular, only 12.5% of patients with hypoplastic MDS or MDS with a low blast count have chromosomal abnormalities.[3] BM dyserythropoiesis and BM clone chromosomal abnormalities involving chromosomes 5, 7, and 8 have been seen in patients with MDS or AA.[4] All these factors make it more difficult to distinguish MDS from AA.
Multiparameter flow cytometry (MFC) and molecular genetics were recently shown to be very helpful in MDS diagnostics. For MFC, a panel of frequently described phenotypic abnormalities in myeloid lineages in MDS has been performed in the clinical setting. Although no abnormal marker profile is specific for MDS, such phenotypic changes may help with the distinguishing of a normal or reactive BM from a clonal myeloid malignancy. However, MFC testing involves very strict requirements for BM specimens.[5] The molecular abnormalities of MDS are less well-analyzed. Only some studies analyzed the occurrence of molecular markers that are typical for acute myeloid leukemia in MDS such as FLT3, RAS, and mixed lineage leukemia (MLL) and so on. It may be problematic to widely use applied molecular techniques for sample collection and preservation since they have a low coverage rate.[6]
The initiation of MDS is believed to be a multistep process requiring gene expression abnormalities to accumulate malignant clone proliferation and differentiation inhibition. Gene expression abnormalities are believed to require the accumulation of both genetic and epigenetic alterations. The aberration of epigenetic regulation especial hypermethylation of tumor suppressor gene (TSG) promoter region plays a functionally equivalent role to genetic alterations in the gene silencing mechanism.[7,8,9] In the past few years, the transcriptional inactivation of TSG by promoter CpG island hypermethylation has been a subject of intense interest as a causal factor in hematological malignancies.
One of the most frequently and best studied epigenetic events in MDS is the silencing of the cyclin-dependent kinase inhibitor gene p15INKB, which controls the progression of cells from the G1 to S phase.[10] Hypermethylation of the p15INKB promoter region occurs in approximately 50% of MDS cases.[11] It has been reported to be acquired during disease progression[12,13] and associated with leukemic transformation[14] and poor prognosis. Yu et al.[15] found that aberrant methylation of CpG islands of the ID4 gene promoter region occurred in a T/NK acute lymphoblastic leukemia mouse model by restriction landmark genomic scanning. ID4 gene aberrant hypermethylation has a relationship with the initial and development of many kinds of hematological malignant diseases. In addition, ID4 is also involved in regulation of cell cycle, cell proliferation, and differentiation by several pathways such as aberrant hypermethylation of the promoter region.[16,17,18,19,20,21,22,23,24,25] A recent research showed a notable relationship of ID4 gene methylation status with the initial and development of MDS.[26]
To investigate the role of ID4 gene aberrant methylation in the diagnosis of MDS, the methylation status of ID4 was analyzed in patients with newly diagnosed MDS and AA by bisulfite sequencing polymerase chain reaction (PCR) (BSP) and MethyLight PCR. We studied the differences in ID4 gene methylation statuses between patients with hypoplastic MDS and those with MDS and low blast counts or a normal karyotype with AA. We not only detected the occurrence of ID4 methylation in patients with MDS or AA, but the methylation levels were also tested to make the initial accurate diagnosis. ID4 gene methylation testing may be a new biomarker for the diagnosis of MDS.
METHODS
Samples
One hundred and thirty-one BM samples were obtained from adult patients diagnosed with different types of MDS (n = 100) or AA (n = 31). More detailed clinical information about the patients is presented in Table 1. Consent for sample collection was obtained from every patient who was enrolled in this study following institutional guidelines. Leukemia cell line NB4 and renal cell line 293 played positive and negative roles in our study, respectively.
Table 1.
Patients’ clinical characteristics by group
| Groups | Number of patients |
|---|---|
| AA | 31 |
| Karyotype abnormal | 0/9 |
| MDS | 100 |
| WHO subtype | |
| RA | 41 |
| RARS | 7 |
| RCMD | 22 |
| RAEB1 | 21 |
| RAEB2 | 9 |
| Karyotype | |
| Abnormal | 18 |
| Normal | 19 |
| Low BM blast counts | 47 |
| Hypoplastic BM cellularity | 16 |
WHO: World Health Organization; AA: Aplastic anemia; MDS: Myelodysplastic syndrome; RA: Refractory anemia; RARS: Refractory anemia with ringed sideroblasts; RCMD: Refractory cytopenia with multilineage dysplasia; RCMD-RS: Refractory cytopenia with multilineage dysplasia and ringed sideroblasts; RAEB: Refractory anemia with excess of blasts; BM: Bone marrow, Karyotype abnormal, −5/5q−, −7/7q−, +8, 20q− and complex (≥3 abnormalities).
Genomic DNA extraction and bisulfite modification
Genomic DNA was extracted following the genomic DNA extraction kit (Promega, USA) guidelines. Next, 1 μg DNA was modified using an EpiTect Bisulfite kit (Qiagen, USA) according to the manufacturer's instructions. All of the unmethylated CpG sites of the DNA were converted to TG, while the methylated CpG sites remained CG. All of the bisulfite-treated DNA samples were stored at −20°C.
Detection of CpG site methylation frequency (bisulfite sequencing polymerase chain reaction)
A bisulfite-treated ID4 DNA sequence was used to design the primers. ID4 primers [Table 2] were designed in promoter regions near the known transcription start sites. The forward primer sequence had no CpG site, while the reverse primer sequence had one CpG site. As such, the reverse primers were designed with a mixture of CG and CA to equally amplify the methylated and unmethylated sites. A final volume of 25 μl of the reaction mixture contained 0.5 μl of HotStarTaq (Qiagen), 20 pmol of each primer, 2.5 μl of × 10 Herman's PCR buffer, 1.25 μl of 25 mmol/L deoxynucleotide, and 2 μl of bisulfite-treated DNA. This PCR reaction consisted of 15 min of heating at 95°C, 40 PCR cycles of 50 s at 94°C, 45 s at 53°C, and 60 s at 72°C with a final 10 min at 72°C for elongation. The PCR product was a plasmid incorporated with Escherichia coli cells by the T4 ligase (Takara, China) and pGEM®-T Easy vector (Promega). Ten colonies were chosen for sequencing (Invitrogen).
Table 2.
Primer and probe sequences
| Genes | Forward | Reverse |
|---|---|---|
| ID4 BSP primer | GTTTGATTGGTTGGTTATTTTAGAT | ACCGAAAAAAAAATAACCCAC |
| CACCAAAAAAAAAATAACCCAC | ||
| ID4 methylight | ||
| Primer | TCGGAGTTTTCGTTTTCGTT | CGATACTACTCACAACCGCG |
| Probe | AGCGGGTTTCGTTCGGTTCG | |
| MYOD1 methylight | ||
| Primer | CCAACTCCAAATCCCCTCTCTAT | TGATTAATTTAGATTGGGTTTAGAGAAGGA |
| Probe | TCCCTTCCTATTCCTAAATCCAACCTAAATACCTCC |
BSP: Bisulfite sequencing polymerase chain reaction.
Quantitative detection of DNA methylation patterns by MethyLight
In our previous study, a complete MethyLight system for ID4 methylation quantitative detection was established. This quantitative detection system had both high sensitivity and good specificity.[27] We used MethyLight PCR to detect the DNA methylation status and determine the ID4 gene methylation positivity rate and methylation level. The pairs of primers and the responding probe with 10 total CpG sites were designed according to the bisulfite-treated DNA sequence [Table 2]. The MYOD1 gene, whose primers and probe described as prescribed, was chosen as the internal control gene to normalize for suitable amounts of bisulfite-treated DNA from every sample.[28] All of the primers and probes that were labeled with FAM at the 5′ end and the quencher TAMRA at the 3′ end were synthesized by Invitrogen. The final reaction mixture consisted of 2.5 μmol of each primer, 1.25 μmol of probe, 2 μl of bisulfite-converted DNA, and TaqMan Universal PCR Master mix (Qiagen) according to the manufacturer's protocol. PCR was run under the following conditions: 95°C for 10 min, followed by 45 cycles at 95°C for 30 s, and 57°C for 1 min. MethyLight PCR was performed at M × 3000p (Stratagene) for the amplification and analyses.
CpG methylation was quantitated as follows: Fraction of methylated molecules = 100 × aim gene values/MYOD1 gene values.
Statistical analyses
The Mann-Whitney nonparametric test was performed to compare the scale measures. The Chi-square or Fisher's exact test was used to compare the categorical variables. All of the statistical analyses were performed using the Statistical Package for the Social Sciences version 18.0 for Windows (SPSS, Chicago, IL, USA). P < 0.05 was considered significant, and all were two-tailed.
RESULTS
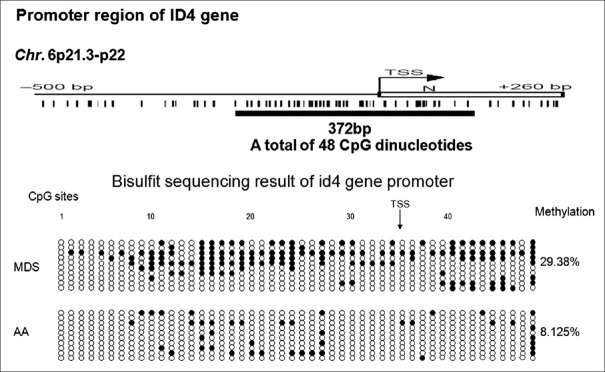
ID4 gene CpG site methylation frequency comparison between one patient with aplastic anemia and one patient with myelodysplastic syndrome using bisulfite sequencing polymerase chain reaction
Bisulfite sequencing polymerase chain reaction was used to directly detect the methylation status of every CpG site of the given region. We analyzed the methylation status of BM samples of one patient with MDS and one patient with AA using Bisulfite sequencing polymerase chain reaction. For the ID4 gene, every sample included 10 clones, each of which had 48 CpG sites. The ID4 gene methylation positivity site frequency of the patients with MDS (29.38% [141/480]) was significantly higher than that of the patients with AA (8.125% [39/480]) (P = 0.000, Figure 1).
Figure 1.
ID4 genes CpG sites methylation frequencies between patients with aplastic anemia and those with myelodysplastic syndrome. ●: One positive CpG site (CG); ○: One negative CpG site (TG). TSS: Transcriptional start site; NBM: Normal bone marrow. The ID4 gene methylation positivity site frequency in patients with myelodysplastic syndrome (29.38% [141/480]) was significantly higher than that of the patients with AA (8.125% [39/480]) (P = 0.000).
ID4 gene promoter methylation status (by MethyLight) correlated with clinical characters in myelodysplastic syndrome and aplastic anemia patients
As summarized in Table 3, the MDS group had a higher ID4 gene methylation positivity rate (P = 0.001) and higher methylation levels (P = 0.001) than the AA group. There were no significant differences in clinical features, such as gender, initial white blood cell counts, platelet counts, or the incidence of molecular genetic abnormalities between the MDS and AA groups (P > 0.05). The proportion of initial hemoglobin level, age, and the incidence of chromosomal abnormalities were significantly different between the MDS and AA groups (P < 0.05). However, these three factors did not differ between patients with ID4 methylation and those with unimethylation (P > 0.05). There was a significant difference in BM blast levels between patients with MDS and those with AA (P = 0.000) as well as between patients with MDS with or without hypermethylation (P = 0.001). In this study, 51.79% (29/56) of the patients with MDS harbored identical cytogenetic and molecular genetic markers (such as −5/5q−, −7/7q−, +8, 20q−, complex karyotype, N-RAS mutation, AML1 mutation, p53 mutation, EVI-1 overexpression, TEL fusions, MLL fusions) for distinguishing MDS. In the other 27 patients, the methylation positivity rate of the ID4 gene was 22.22% (6/27). Therefore, 62.5% (35/56) of patients with MDS could be distinguished using a combination of genetic and epigenetic markers.
Table 3.
Comparison of clinical characteristics and methylation status between patients with aplastic anemia and those with MDS
| Indices | AA | MDS | P |
|---|---|---|---|
| Patients (n) | 31 | 100 | |
| Age (years) | 34.36 (12–71) | 46.61 (13–86) | 0.001 |
| Male/female (n) | 20/11 | 55/45 | 0.349 |
| WBC (×109/L) | 3.88 (0.49–8.27) | 4.12 (0.35–25.9) | 0.130 |
| Hemoglobin (g/L) | 100.5 (53–166) | 86.17 (44–140) | 0.036 |
| Platelet (×109/L) | 87.83 (1–307) | 84.19 (1–459) | 0.680 |
| BM blast (%) | 0.5 (0–1.6) | 2.99 (0–17.2) | 0.000 |
| Karyotype abnormal | 0/9 | 18/37 | 0.021 |
| Gene abnormal | 0/9 | 13/43 | 0.138 |
| ID4 methylation | |||
| Methylation positive rate | 0/31 | 27/100 | 0.001 |
| Methylation level | 0 (0–0) | 0.21 (0–3.79) | 0.001 |
MDS: Myelodysplastic syndrome; RA: Refractory anemia; RARS: Refractory anemia with ringed sideroblasts; RCMD: Refractory cytopenia with multilineage dysplasia; RCMD-RS: Refractory cytopenia with multilineage dysplasia and ringed sideroblasts; RAEB: Refractory anemia with excess of blasts; BM: Bone marrow; AA: Aplastic anemia; WBC: White blood cell counts; gene abnormal, N-RAS, FLT3, AML1, EVI-1, TEL, MLL, p53, IRF-1. Karyotype abnormal, −5/5q−, −7/7q−, +8, 20q− and complex (≥3 abnormalities).
ID4 gene promoter methylation status (by MethyLight) in patients with hypoplastic myelodysplastic syndrome or myelodysplastic syndrome with low blast counts or normal karyotype
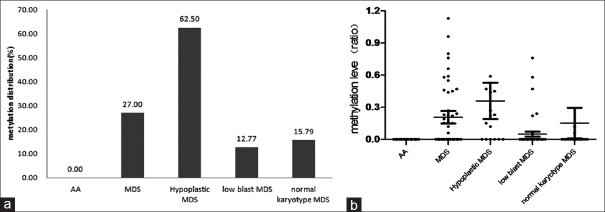
The methylation positivity rates (P = 0.000) and methylation levels (P = 0.000) of the ID4 gene were significantly different between patients with hypoplastic MDS and those with AA. The same results were found in patients with MDS and a normal karyotype and those with AA (P = 0.049, P = 0.024, Table 4 and Figure 2). Between patients with MDS and a low BM blast count and patients with AA, only methylation levels (P = 0.040, Table 4 and Figure 2) of the ID4 gene had apparent differences, whereas ID4 gene methylation positivity rates did not (P = 0.076, Table 4 and Figure 2). Both patients with hypoplastic MDS and those with low BM blast counts showed differences in the abnormal karyotype from those of patients with AA (P = 0.014, P = 0.038, Table 4 and Figure 2).
Table 4.
Comparison of clinical characteristics and methylation status among patients with aplastic anemia and those with hypoplastic MDS, those with MDS and a low BM blast count, and those with MDS and a normal karyotype
| Indices | AA | Hypoplastic MDS | MDS with low BM blast counts | MDS with normal karyotype | ||||
|---|---|---|---|---|---|---|---|---|
| Values | P | Values | P | Values | P | |||
| Patients (n) | 31 | 16 | 47 | 19 | ||||
| BM blast (%) | 0.5 (0–1.6) | 4.9 (0–17.2) | 0.000 | 0.6 (0–1.8) | 0.202 | 3.8 (0–17.2) | 0.002 | |
| Karyotype | ||||||||
| Abnormal | 0/9 | 3/4 | 0.014 | 9/19 | 0.038 | – | – | |
| Gene | ||||||||
| Abnormal | 0/9 | 2/6 | 0.143 | 6/23 | 0.15 | 6/15 | 0.052 | |
| ID4 methylation | ||||||||
| Methylation positive rate | 0/31 | 10/16 | 0.000 | 6/47 | 0.076 | 3/19 | 0.049 | |
| Methylation level | 0 (0–0) | 0.36 (0–2.8) | 0.000 | 0.39 (0–0.76) | 0.040 | 0.15 (0–2.72) | 0.024 | |
AA: Aplastic anemia; MDS: Myelodysplastic syndrome; BM: Bone marrow; gene abnormal, N-RAS, FLT3, AML1, EVI-1, TEL, MLL, p53, and IRF-1. Karyotype abnormal, −5/5q−, −7/7q−, +8, 20q− and complex (≥3 abnormalities).
Figure 2.
The ID4 gene methylation distribution and levels of patients with aplastic anemia (AA) and myelodysplastic syndrome. (a) The ID4 gene methylation distribution in patients with AA and those with myelodysplastic syndrome (MDS). In the patients with AA, no methylation abnormalities in the ID4 gene were found; (b) ID4 gene methylation level of patients with AA and MDS. The methylation positivity rates (P = 0.000) and methylation levels (P = 0.000) of the ID4 gene were significantly different between patients with MDS and those with AA (0/31, 0 [0–0]).
DISCUSSION
Recently, epigenetic alterations, such as the aberrant methylation of CpG islands in the promoter regions of genes have been reported to inactivate TSG, and many genes were found to be silenced by methylation in solid tumors and hematological malignancies[29] to contribute to tumor genesis,[30] including MDS. Based on difficulty distinguishing MDS and hematological benign diseases such as AA that are only dependent on morphology and genetics, we turned to epigenetics, especially abnormal methylation of the TSG promoter region. Several studies have reported that the P15 gene, which encodes a cyclin-dependent kinase inhibitor, is frequently methylated in patients with MDS but not in those with AA, which provides a key clue in the elucidation of ID4 gene methylation status and levels to distinguish patients with hypoplastic MDS from those with MDS and low blast counts or a normal karyotype from those with AA. Because of the limited reports of the methylation status of ID4 genes in MDS,[26] especially regarding distinguishing MDS from AA, we were prompted to examine the genetics of patients with MDS and AA.
In our study, BSP revealed that the CpG sites of the ID4 gene were more methylated in patients with MDS and refractory anemia than in those with AA. As the most direct technology for CpG site methylation status detection, BSP provides the most investible theoretic basis to further identify the different methylated status of the ID4 gene.
This study demonstrated that both methylation positivity rates and methylation levels of the ID4 gene were significantly higher in patients with MDS than in those with AA on MethyLight PCR, which was consistent with the results using BSP. A total of 27% of the patients with MDS had the higher methylation status of the ID4 gene, whereas none of the patients with AA had the higher status. There was a strong correlation between BM blast level and ID4 gene methylation (P = 0.001), and BM blast levels showed an obvious difference between patients with MDS and those with AA. Thus, to some extent, ID4 gene methylation levels give clues to aid in the estimation of BM blast levels. The ID4 methylation status may be independent of many clinical factors that are the genetic features of diseases. There were no significant differences in the incidence of molecular genetic abnormalities between patients with MDS and those with AA due to the lack of sufficient samples, so future studies with additional samples are necessary to confirm our findings. We estimated that there were no significant differences in the BM blasts in patients with MDS and low BM blast counts and those of AA patients; however, a significant difference in methylation levels was seen. Therefore, even in patients with low BM blasts, ID4 gene methylation could help with the distinguishing of some patients with MDS from those with AA. Unfortunately, there was no significant difference in the ID4 gene methylation positivity rate between patients with MDS and low BM blast counts and those with AA, which may also be due to the insufficient number of samples. However, our findings demonstrate the significance of the difference in ID4 gene methylation levels between the two groups.
Approximately 50% of patients with MDS have no distinctive genetic markers (detected by cytogenetics or molecular genetics) for distinguishing them from patients with AA;[2] thus, epigenetics, especially gene CpG site methylation, has become increasingly more popular and useful. Many studies have shown that the methylation positivity rates and the levels of many genes differed between patients with malignant diseases and those with normal bone marrow.[8,31,32,33,34,35] Few genes can currently be used for methylation detection to distinguish between MDS and AA. In this study, patients with MDS and a low BM blast count and those with MDS and a normal karyotype had significantly different abnormal karyotypes from those with AA. We found that patients with MDS had higher ID4 gene methylation positivity rates and methylation levels than those with AA, and none of the 31 patients with AA had a positive methylation status. As such, detecting the ID4 gene methylation status can distinguish a proportion of patients with MDS from those with AA in the absence of distinctive genetic markers, and many more patients with MDS could be correctly diagnosed using a combination of genetic and epigenetic markers.
Here, we reported the initial results of MDS diagnosis. Many more samples must be examined in a future study to confirm the role of this new marker and promote its application in the clinical setting. Our findings show that the combination use of genetic and epigenetic methods to improve the discriminability between MDS and AA.
In conclusion, this is the first study to examine the methylation status and levels of the ID4 gene in patients with MDS and AA. A total of 27% of patients with MDS harbor the higher methylation status of the ID4 gene compared to patients with AA, which provided the theoretical basis of the use of demethylating agents in patients with MDS, suggesting hypermethylation as a major causative agent in hematopoietic clonal disorders that can be used to distinguish MDS from AA.
ACKNOWLEDGMENTS
We thank Xu-Feng Luo, Jing-Xin Li, Xiao-Ning Gao, Li-Ping Dou, Yuan-Yuan Xu, and Yi Ding for discussion and technical assistance.
Footnotes
Edited by: Xiu-Yuan Hao
Source of Support: This work was supported by grants from the National Basic Research Program of China (2005CB522400), National Natural Science Foundation of China (90919044, 30971297, and 81000221), and National Key Scientific Instrument and Equipment Development Projects (2012YQ03026107).
Conflict of Interest: None declared.
REFERENCES
- 1.Scott BL, Deeg HJ. Myelodysplastic syndromes. Annu Rev Med. 2010;61:345–58. doi: 10.1146/annurev.med.051308.132852. [DOI] [PubMed] [Google Scholar]
- 2.Ohyashiki K, Kodama A, Ohyashiki JH. Cytogenetics in myelodysplastic syndromes. Methods Mol Biol. 2011;730:79–88. doi: 10.1007/978-1-61779-074-4_6. [DOI] [PubMed] [Google Scholar]
- 3.Marisavljevic D, Cemerikic V, Rolovic Z, Boskovic D, Colovic M. Hypocellular myelodysplastic syndromes: Clinical and biological significance. Med Oncol. 2005;22:169–75. doi: 10.1385/MO:22:2:169. [DOI] [PubMed] [Google Scholar]
- 4.Kim SY, Lee JW, Lee SE, Cho BS, Kim M, Eom KS, et al. The characteristics and clinical outcome of adult patients with aplastic anemia and abnormal cytogenetics at diagnosis. Genes Chromosomes Cancer. 2010;49:844–50. doi: 10.1002/gcc.20793. [DOI] [PubMed] [Google Scholar]
- 5.Valent P, Horny HP, Bennett JM, Fonatsch C, Germing U, Greenberg P, et al. Definitions and standards in the diagnosis and treatment of the myelodysplastic syndromes: Consensus statements and report from a working conference. Leuk Res. 2007;31:727–36. doi: 10.1016/j.leukres.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Huh HJ, Chae SL, Lee M, Hong KS, Mun YC, Seong CM, et al. CD34, RAB20, PU.1 and GFI1 mRNA expression in myelodysplastic syndrome. Int J Lab Hematol. 2009;31:344–51. doi: 10.1111/j.1751-553X.2008.01056.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhou L, Opalinska J, Sohal D, Yu Y, Mo Y, Bhagat T, et al. Aberrant epigenetic and genetic marks are seen in myelodysplastic leukocytes and reveal Dock4 as a candidate pathogenic gene on chromosome 7q. J Biol Chem. 2011;286:25211–23. doi: 10.1074/jbc.M111.235028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benetatos L, Dasoula A, Hatzimichael E, Syed N, Voukelatou M, Dranitsaris G, et al. Polo-like kinase 2 (SNK/PLK2) is a novel epigenetically regulated gene in acute myeloid leukemia and myelodysplastic syndromes: Genetic and epigenetic interactions. Ann Hematol. 2011;90:1037–45. doi: 10.1007/s00277-011-1193-4. [DOI] [PubMed] [Google Scholar]
- 9.Xu F, Li X, Wu L, Zhang Q, Yang R, Yang Y, et al. Overexpression of the EZH2, RING1 and BMI1 genes is common in myelodysplastic syndromes: Relation to adverse epigenetic alteration and poor prognostic scoring. Ann Hematol. 2011;90:643–53. doi: 10.1007/s00277-010-1128-5. [DOI] [PubMed] [Google Scholar]
- 10.Aggerholm A, Holm MS, Guldberg P, Olesen LH, Hokland P. Promoter hypermethylation of p15INK4B, HIC1, CDH1, and ER is frequent in myelodysplastic syndrome and predicts poor prognosis in early-stage patients. Eur J Haematol. 2006;76:23–32. doi: 10.1111/j.1600-0609.2005.00559.x. [DOI] [PubMed] [Google Scholar]
- 11.Lübbert M. Gene silencing of the p15/INK4B cell-cycle inhibitor by hypermethylation: An early or later epigenetic alteration in myelodysplastic syndromes? Leukemia. 2003;17:1762–4. doi: 10.1038/sj.leu.2403045. [DOI] [PubMed] [Google Scholar]
- 12.Uchida T, Kinoshita T, Nagai H, Nakahara Y, Saito H, Hotta T, et al. Hypermethylation of the p15INK4B gene in myelodysplastic syndromes. Blood. 1997;90:1403–9. [PubMed] [Google Scholar]
- 13.Quesnel B, Guillerm G, Vereecque R, Wattel E, Preudhomme C, Bauters F, et al. Methylation of the p15(INK4b) gene in myelodysplastic syndromes is frequent and acquired during disease progression. Blood. 1998;91:2985–90. [PubMed] [Google Scholar]
- 14.Tien HF, Tang JH, Tsay W, Liu MC, Lee FY, Wang CH, et al. Methylation of the p15(INK4B) gene in myelodysplastic syndrome: It can be detected early at diagnosis or during disease progression and is highly associated with leukaemic transformation. Br J Haematol. 2001;112:148–54. doi: 10.1046/j.1365-2141.2001.02496.x. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, Liu C, Vandeusen J, Becknell B, Dai Z, Wu YZ, et al. Global assessment of promoter methylation in a mouse model of cancer identifies ID4 as a putative tumor-suppressor gene in human leukemia. Nat Genet. 2005;37:265–74. doi: 10.1038/ng1521. [DOI] [PubMed] [Google Scholar]
- 16.Dell’Orso S, Ganci F, Strano S, Blandino G, Fontemaggi G. ID4: A new player in the cancer arena. Oncotarget. 2010;1:48–58. doi: 10.18632/oncotarget.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen SS, Claus R, Lucas DM, Yu L, Qian J, Ruppert AS, et al. Silencing of the inhibitor of DNA binding protein 4 (ID4) contributes to the pathogenesis of mouse and human CLL. Blood. 2011;117:862–71. doi: 10.1182/blood-2010-05-284638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carey JP, Asirvatham AJ, Galm O, Ghogomu TA, Chaudhary J. Inhibitor of differentiation 4 (Id4) is a potential tumor suppressor in prostate cancer. BMC Cancer. 2009;9:173. doi: 10.1186/1471-2407-9-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeon HM, Jin X, Lee JS, Oh SY, Sohn YW, Park HJ, et al. Inhibitor of differentiation 4 drives brain tumor-initiating cell genesis through cyclin E and notch signaling. Genes Dev. 2008;22:2028–33. doi: 10.1101/gad.1668708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. doi: 10.1186/1479-5876-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noetzel E, Veeck J, Horn F, Hartmann A, Knüchel R, Dahl E. Promoter methylation of ID4. A marker for recurrence-free survival in human breast cancer. Pathologe. 2008;29(Suppl 2):319–27. doi: 10.1007/s00292-008-1038-7. [DOI] [PubMed] [Google Scholar]
- 22.Noetzel E, Veeck J, Niederacher D, Galm O, Horn F, Hartmann A, et al. Promoter methylation-associated loss of ID4 expression is a marker of tumour recurrence in human breast cancer. BMC Cancer. 2008;8:154. doi: 10.1186/1471-2407-8-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uhm KO, Lee ES, Lee YM, Park JS, Kim SJ, Kim BS, et al. Differential methylation pattern of ID4, SFRP1, and SHP1 between acute myeloid leukemia and chronic myeloid leukemia. J Korean Med Sci. 2009;24:493–7. doi: 10.3346/jkms.2009.24.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Borinstein SC, Conerly M, Dzieciatkowski S, Biswas S, Washington MK, Trobridge P, et al. Aberrant DNA methylation occurs in colon neoplasms arising in the azoxymethane colon cancer model. Mol Carcinog. 2010;49:94–103. doi: 10.1002/mc.20581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara K, Nagai H, Li Y, Ohashi H, Hotta T, Saito H. Frequent DNA methylation but not mutation of the ID4 gene in malignant lymphoma. J Clin Exp Hematop. 2007;47:15–8. doi: 10.3960/jslrt.47.15. [DOI] [PubMed] [Google Scholar]
- 26.Wang H, Wang XQ, Xu XP, Lin GW. ID4 methylation predicts high risk of leukemic transformation in patients with myelodysplastic syndrome. Leuk Res. 2010;34:598–604. doi: 10.1016/j.leukres.2009.09.031. [DOI] [PubMed] [Google Scholar]
- 27.Liu Y, Kang HY, Wang LL, Lu XC, Zhu HL, Yu L. Establishment of methylation-specific quantitative PCR system for ID4 gene in acute leukemia cells and its specificity and sensitivity (in Chinese) J Experimental Hematol. 2014;22:269–74. doi: 10.7534/j.issn.1009-2137.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 28.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: A high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–54. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 30.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet. 1999;21:163–7. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 31.Voso MT, Scardocci A, Guidi F, Zini G, Di Mario A, Pagano L, et al. Aberrant methylation of DAP-kinase in therapy-related acute myeloid leukemia and myelodysplastic syndromes. Blood. 2004;103:698–700. doi: 10.1182/blood-2003-07-2249. [DOI] [PubMed] [Google Scholar]
- 32.Valencia A, Cervera J, Such E, Ibañez M, Gómez I, Luna I, et al. Aberrant methylation of tumor suppressor genes in patients with refractory anemia with ring sideroblasts. Leuk Res. 2011;35:479–83. doi: 10.1016/j.leukres.2010.08.012. [DOI] [PubMed] [Google Scholar]
- 33.Mori N, Yoshinaga K, Tomita K, Ohwashi M, Kondoh T, Shimura H, et al. Aberrant methylation of the RIZ1 gene in myelodysplastic syndrome and acute myeloid leukemia. Leuk Res. 2011;35:516–21. doi: 10.1016/j.leukres.2010.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Rodrigues EF, Santos-Rebouças CB, Gonçalves Pimentel MM, Mencalha AL, Dobbin J, Da Costa ES, et al. Epigenetic alterations of p15(INK4B) and p16(INK4A) genes in pediatric primary myelodysplastic syndrome. Leuk Lymphoma. 2010;51:1887–94. doi: 10.3109/10428194.2010.505820. [DOI] [PubMed] [Google Scholar]
- 35.Greco M, D’Alò F, Scardocci A, Criscuolo M, Fabiani E, Guidi F, et al. Promoter methylation of DAPK1, E-cadherin and thrombospondin-1 in de novo and therapy-related myeloid neoplasms. Blood Cells Mol Dis. 2010;45:181–5. doi: 10.1016/j.bcmd.2010.05.008. [DOI] [PubMed] [Google Scholar]